Abstract
Communities of arbuscular mycorrhizal fungi (AMF) are crucial for promoting plant productivity in most terrestrial systems, including anthropogenically managed ecosystems. Application of AMF inocula has therefore become a widespread practice. It is, however, pertinent to understand the mechanisms that govern AMF community composition and their performance in order to design successful manipulations. Here we assess whether the composition and plant growth-promotional effects of a synthetic AMF community can be altered by inoculum additions of the isolates forming the community. This was determined by following the effects of three AMF isolates, each inoculated in two propagule densities into a preestablished AMF community. Fungal abundance in roots and plant growth were evaluated in three sequential harvests. We found a transient positive response in AMF abundance to the intraspecific inoculation only in the competitively weakest isolate. The other two isolates responded negatively to intra- and interspecific inoculations, and in some cases plant growth was also reduced. Our results suggest that increasing the AMF density may lead to increased competition among fungi and a trade-off with their ability to promote plant productivity. This is a key ecological aspect to consider when introducing AMF into soils.
INTRODUCTION
It is an ongoing objective in ecology to understand the mechanisms that shape the community structure and productivity of ecosystems, in order to ultimately maintain the services ecosystems provide. Thereby, soil communities belowground are known to be a key element in maintaining the productivity and diversity of communities aboveground (1, 2). Arbuscular mycorrhizal fungi (AMF) are a guild of soil organisms that are dependent upon plant hosts to acquire carbon and that provide in exchange many services for the plants, such as improving their nutrient acquisition, productivity, coexistence, and pathogen protection (3). Considering the large potential of these symbiotic fungi to contribute to the ecological sustainability of managed ecosystems, efforts are being made to improve the resource use efficiency of arable and degraded soils by introducing AMF inocula. However, there are many questions remaining regarding the conditions under which introduction of AMF into soils is successful at improving the plant growth-promotional effects of AMF communities (4). The current challenge for improving soil productivity by AMF community manipulations is in understanding the ecological constraints, such as competitive and complementary interactions with AMF genotypes present in the soil (5).
AMF differ in their life traits and nutrient-foraging strategies (6, 7). These differences can be the basis for the complementary effect of greater AMF richness (8) or may favor the more-beneficial partners under the given conditions (9, 10). On the other hand, AMF colonizing a root system compete for space and the plant-derived carbohydrates (11), with potential trade-offs with the beneficial effects of the symbiosis with the host plants (12–14). Understanding the interactions among coexisting AMF is therefore important not only from the point of view of basic community ecology, but also for predicting the effects of AMF community manipulations, such as when AMF are inoculated into soils containing AMF communities. Inoculation may decrease AMF diversity in roots, resulting in both positive and negative plant growth responses (15–17). Shifts in propagule densities of AMF in soil can affect the quantitative composition of the communities, with potential effects on plant growth (18–20). Yet, little is currently known about the interactions among different AMF colonizing one root system and the factors responsible for the presence and abundance of particular AMF taxa among roots from the pool of propagules available in the soil.
Here we assess the responses in both the mycorrhizal fungal partners and the plant host when additional AMF propagules are introduced into a preestablished AMF community of three Glomaceae isolates. We thus mimicked the approach of inoculation with native AMF (17) and changed the relative infectivity of these isolates in the soil. We expected that additional AMF propagules would increase the abundance of the inoculated fungus and suppress the development of the other two fungi. Second, we assessed whether the AMF inoculation could improve plant growth-promotional effects of the AMF community. In order to test this, we grew medic (Medicago sativa L.) in soil with a previously established AMF community or in the same soil after it had been sterilized. The three fungal isolates were introduced at different propagule amounts into both background treatments, and the systems were sampled at 6, 12, and 24 weeks to assess both fungal development and plant growth.
MATERIALS AND METHODS
Experimental system.
The experiment was based on a ruderal soil-fungi system of a freshly leveled spoil bank of a surface mine near Chomutov, North Bohemia, Czech Republic (see reference 21 for a site description). The cultivation substrate was prepared by mixing homogenized gray miocene clay collected on the spoil bank surface with sand, in the ratio of 1:2. The mixture had the following main parameters: pH (H2O), 7.5; Corg, 1.15%; N, 0.06%; Olsen-P (0.5 M NaHCO3 extractable), 2.93 mg kg−1.
Two of the three fungal isolates used originated from the same spoil bank as the clay substrate: Glomus claroideum Schenck & Smith Chomutov (isolate referred to herein as G. claroideum) and G. intraradices sensu lato Chomutov (isolate referred to herein as G. intraradices; for closer specification, see reference 22). The third isolate used, G. mosseae (Nicol. & Gerd.) Gerd. & Trappe BEG95 (isolate referred to herein as G. mosseae), originated from another spoil bank near Most, in the same geographic region (labeled G. mosseae ALB in reference 23).
Several cultures were established for each isolate in a mixture of zeolite and sand (1:1) with medic (Medicago sativa L. cv. Vlasta) as the host plant, using 200 spores collected from a pooled soil sample from three multispore cultures. The established cultures were used for the characterization of genetic diversity and as the source of inoculum.
Design and optimization of quantiative PCR (qPCR) assays.
The genetic variations of the studied isolates in the large subunit of the nuclear ribosomal DNA (nrDNA LSU) were characterized in detail as described for the isolate G. intraradices in reference 22. The obtained sequences were aligned together with sequences deposited in GenBank under the respective species names by using Clustal X (24), and the alignments were corrected manually in BioEdit (25). Initially, alignments for the individual species were done separately and subsequently combined into a final alignment. Maximum parsimony analyses were performed based on both the individual as well as the complete sequence alignments by using PAUP* (26) (see Fig. S1 in the supplemental material).
Primers discriminating between the studied isolates were designed for G. claroideum and G. mosseae (see Table S1 in the supplemental material) based on the complete sequence alignment obtained using Primer 3 Plus (27). Variations of nrDNA LSU sequences in the G. mosseae isolate prevented the design of a single primer pair that would amplify all the sequence variants of this isolate but not cross-amplify the other fungal species. Two specific primer pairs were therefore developed, each amplifying only a part of the G. mosseae ribotypes, and applied as a duplex assay. For the G. intraradices isolate, previously designed primers (22) were used, as they were found suitable for the present study. The specificity of these qPCR assays was tested in silico using the Fast PCR software (28) as well as experimentally by cross-amplification experiments using different templates (plasmid standards, medic DNA extracted from roots and leaves, fungal DNA extracted from spores, and DNA extracted from medic roots colonized by each fungal isolate). The preparation of plasmid standards, qPCR, and the estimate of amplification efficiencies followed in general the procedures described in reference 22. The details on annealing temperatures, primer concentrations, amplicon lengths, and the estimated amplification efficiencies for each qPCR assay are summarized in Table S1 of the supplemental material.
Precultivation of the synthetic AMF community.
The three AMF isolates were cultivated together to establish a background AMF community. In order to give them the same chance to develop within the community, the mycorrhizal inoculation potential of the initial inoculum was standardized to equal numbers of infective propagules (IP) of each isolate. To achieve this, the substrates from five 6-month-old cultures of each isolate were checked microscopically for the presence of spores and absence of contamination, homogenized, and air dried. Five serial dilutions of the inocula with the γ-irradiated experimental substrate (1:10 to 1:105, vol:vol) were planted with medic in five replicates. When the roots of the plants had grown through the whole soil volume of 85 ml (after 5 weeks), the presence and absence of root colonization in the root systems was scored and used to calculate the number of infective propagules by the most probable number (MPN) method (29). The G. claroideum and G. intraradices inocula had an identical inoculation potential of 150 IP ml−1 of substrate, while the inoculation potential of G. mosseae was distinctly lower (4 IP ml−1).
The soil-sand cultivation substrate described above was sterilized by γ-irradiation and used to fill 33 pots, each 10 liters in volume. Every pot was inoculated with 230 IP of each fungus, which corresponded to 1.6 g of G. claroideum and G. intraradices and 50 g of the G. mosseae inoculum. The mixed inoculum was prepared for each pot separately by weighting and thoroughly mixing the air-dried culture substrates of known inoculation potentials. The inoculum was placed into the center of the pot, about 3 cm below the surface, and the pot was planted with eight pregerminated medic plantlets. The pots were cultivated in a glasshouse with a light supplement (12 h, metalhalide lamps, 400 W) and fertilized once in 2 weeks with 200 ml of the P2N3 nutrient solution (30).
After 6 months of cultivation, the shoot biomass was cut. From the center of each pot, a root sample was extracted using a soil corer and wet sieving. Half of the obtained root samples were immediately frozen in liquid N, stored at −80°C, and later used for the determination of copy numbers (CN) of each fungus (as described below), and another portion was used for a check of root colonization after trypan blue staining (31). The substrates from the pots were air dried and homogenized, including most of the fine roots; thick roots were removed in order to decrease the amount of root biomass in the homogenized substrate. The resulting substrate was subsequently termed the AMF substrate and contained a synthetic AMF background community. Its infectivity was determined by the MPN method as described above, together with determination of the infectivity of the inocula used later in the experiment. The inoculation potential of the established AMF community was 22 IP ml−1. The frequency of root colonization as obtained from 10 randomly selected root samples was 84% (±2% standard error of the mean [SEM]). The G. intraradices isolate was present in the roots with the highest nrDNA copy numbers (on average, 120 × 103 CN ng−1 isolated DNA, ±28 × 103), followed by G. claroideum (3 × 103 CN ng−1, ±0.57 × 103), while G. mosseae was almost absent. It was detected in very low copy numbers (50 and 276 CN ng−1 isolated DNA) in only two out of seven analyzed root samples.
Half of the substrate was then sterilized by γ-irradiation to be used as control substrate without AMF.
Establishment and cultivation of the experiment.
Seven inoculation treatments were established, one each in the AMF substrate and the control substrate (14 treatments in total). The treatments were replicated 24 times, to be harvested at three consecutive harvests (eight replicates per harvest). This resulted in 336 pots in total, each 0.7 liters in volume and planted with one medic plant. The inoculation treatments were as follows: (1) noninoculated; (2 and 3) inoculated with G. claroideum at two inoculum levels; (4 and 5) inoculated with G. intraradices at two levels; (6 and 7) inoculated with G. mosseae at two levels (see Fig. 1 for an overview).
Fig 1.
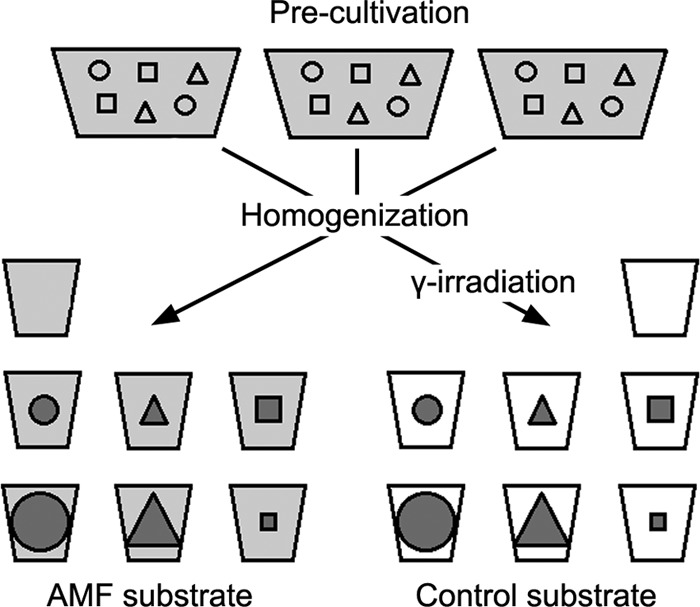
Schematic of the experimental design. The precultivation stage rendered the substrate colonized by a synthetic community of three arbuscular mycorrhizal fungal isolates (AMF substrate) and sterilized substrate without AMF background (control substrate). Each treatment of the experiment is represented by one small pot in the lower part of the diagram. Besides noninoculated treatments (empty pots), each of the three isolates of the community was inoculated with 150 IP (middle-sized geometric symbols). Additionally, the G. claroideum and G. intraradices pots were inoculated with 400 IP (large circles and triangles) and the G. mosseae isolate was inoculated with 20 IP (small squares). For specification of the isolates, see Materials and Methods.
The inocula were prepared and standardized as described above based on the MPN method. Again, the inocula of G. claroideum and G. intraradices had higher infectivities (81 and 57 IP ml−1, respectively) than the G. mosseae inoculum (4 IP ml−1). The inoculation level of 150 IP per pot, applied for all three isolates, was selected as on the order of magnitude corresponding with the recommendations of inoculum producers for inoculation. A higher inoculation level of 400 IP per pot was applied with G. claroideum and G. intraradices, but this level could not be applied with G. mosseae because of the low infectivity of the G. mosseae inoculum. Instead, the remaining G. mosseae inoculum was used to establish at least a low inoculation level of 20 IP per pot.
The inocula were weighted for each replicate separately and mixed with sterilized zeolite-sand mixture to constant volumes corresponding to the highest volume of inoculum added (38 ml in the G. mosseae 150-IP treatment). Seedlings of medic were germinated in autoclaved sand and precultivated for 3 weeks in the sterilized experimental substrate. They were inoculated at planting into the experiment by an inoculum layer inserted about 3 cm below the surface. In order to equalize the microbial community compositions with the different treatments, all pots were irrigated with bacterial filtrate from the AMF substrate and the inoculum. The filtrates were obtained by shaking a sample of the AMF substrate (about 500 g) and samples of the inocula (about 200 g each) overnight in distilled water, passing the resulting soil suspensions twice through a filter paper (Whatman no. 1), and adjusting the volume to that required. The filtrate from the AMF substrate was added into every pot with control substrate (10 ml each). The filtrates from the inocula were mixed together and added into every experimental pot (10 ml each). The experimental pots were cultivated in a glasshouse with a light supplement (12 h, metalhalide lamps, 400 W) and fertilized once in 2 weeks with 50 ml per pot of P2N3 nutrient solution (30).
Harvest and data collection.
After 6, 12, and 24 weeks, eight pots per treatment and harvest were destructively harvested. At each harvest, root systems were carefully washed from the substrate, weighed, and cut into 1-cm segments. A subsample of 100 mg (fresh weight) was immediately frozen in liquid N and stored at −80°C. Another part was used for microscopic determination of root colonization after staining with 0.05% trypan blue in lactoglycerol (31). The remaining roots and the shoots were dried at 80°C for 24 h.
Shoot and root dry weights were determined for the experimental plants. Root colonization was evaluated by microscopy (Olympus BX60, 100× magnification) according to the methods described in reference 32. The intensity of colonization of the root system was determined using the program Mycocalc (http://www.dijon.inra.fr/mychintec/Mycocalc-prg/download.html).
Genomic DNA was extracted from the root samples by using a DNeasy plant minikit (Qiagen) according to the manufacturer's instructions. DNA extracts from root samples were quantified spectrophotometrically, and 10 ng of total genomic DNA was used as the template for qPCR, which was performed as described in reference 22. Six replicate root systems per treatment and harvest were analyzed. All target sequences were quantified in all root samples.
Analyses of fungal root colonization.
Root colonization parameters were assessed by an analysis of variance (ANOVA) for variations among harvest period, the identity of the AMF inoculated, and the IP level, and by a three-way ANOVA separately for plants with and without a background AMF community present. The nrDNA copy numbers of each fungus were used to assess the response in the abundance of each AMF to the various AMF inoculation treatments. This was done using the relative interaction intensity (RII; see reference 33) to calculate an index reflecting the competitive ability of AMF to colonize roots in the presence of a background AMF community relative to the intensity when it is the only AMF present. This response to a background AMF community was calculated as follows: RIIcommunity = (O − M)/(O + M), where O was the observed abundance of an AMF in roots when a background AMF community was present and M was the mean of the same AMF detected in roots in the absence of a background community of the same IP level and harvest period combination. We tested whether the RIIcommunity of each AMF was influenced by the harvest period and the IP level at which the fungi were inoculated by using a two-way ANOVA.
The RII metric was also used to assess the response of AMF to the addition of inter- and intraspecific inocula (RIIinoculation). In this case, O was the observed abundance of an AMF when the AMF inoculum was added and M was the mean abundance of a fungus detected in roots in the background AMF community with no additional AMF inoculum added. This RII measure was assessed for variations among AMF identity, the IP level at which the fungus was inoculated, and the harvest period, by using a three-way ANOVA. All model means were compared to an RII of 0 to test for a significant response in the AMF abundance within the host plant roots.
Analyses of plant performance.
All biomass data were square-root transformed before analyses to improve homoscedasticity. Separate ANOVAs for each harvest and AMF community background level were used to test for differences in shoot and root biomass between the noninoculated control treatments and each of the AMF-inoculated treatments. In order to determine whether the IP level (150 IP or an alternative level) and the presence of a background AMF community influenced the growth-promotional effect of an AMF inoculum, separate two-way ANOVAs for each AMF species, with the IP level and the background as sources of variation, were used.
Additionally, the RII was used to calculate the response of total plant biomass to AMF inoculum additions into the AMF background. Here, M was the mean total biomass of plants grown with a background AMF community, and O was the observed biomass produced when additional AMF inoculum was added. This plant response index was assessed for variations among identities of the fungal species inoculated, the IP level at which the fungus was inoculated, and the harvest period, by using a three-way ANOVA.
For all ANOVAs, nonsignificant interaction terms were removed to capture the full amount of variation explained by the main effects. However, full ANOVA results are presented in Tables S3 and S4 in the supplemental material to show the magnitudes of these interaction effects. All data manipulations and statistical analyses were performed in R for Mac OS X version 2.15.1 (R Foundation for Statistical Computing).
Nucleotide sequence accession numbers.
Representative sequences of AMF nrDNA LSU were submitted to GenBank under the accession numbers KC522414 to KC522421.
RESULTS
Root colonization.
Root colonization by AMF was not observed in any replicates at any harvest where there was no background AMF community present and no addition of AMF inoculum. When AMF were inoculated into the control background (no previous AMF present), colonization intensity varied among the AMF species inoculated, depending on the time of harvest (F4,89 = 28.0; P < 0.0001) (Table 1). At 6 weeks, root colonization was lower for G. mosseae than for G. intraradices or G. claroideum. At 12 weeks, both G. mosseae and G. claroideum resulted in significantly lower colonization intensities than G. intraradices, while at 24 weeks all three fungi differed from each other (G. claroideum < G. mosseae < G. intraradices) (Table 1). Additionally, for the control background the colonization intensity also differed overall between plants receiving 150 IP versus alternative IP levels, depending on the identity of the fungus inoculated (F2,95 = 4.61; P = 0.01). Inoculation of G. mosseae at 150 IP resulted in a greater colonization intensity than at 20 IP, while neither G. claroideum nor G. intraradices results differed significantly overall between IP levels (Table 1). In the presence of a background AMF community, the intensity of colonization only varied among harvests depending on the AMF species inoculated (F2,102 = 3.04; P = 0.02), as colonization intensity was greater at 24 than at 12 weeks for G. mosseae-inoculated plants and greater at 24 weeks than at 6 weeks for G. intraradices-inoculated plants (Table 1).
Table 1.
Intensity of root colonization by AMF by week, AMF species, and IP
| Week of harvest | AMF inoculated | No. of IP | Mean (SE) intensity of root colonizationa |
|
|---|---|---|---|---|
| Control background | AMF background | |||
| 6 | None | 0 | 62.0 (9.12) abc | |
| 6 | G. mosseae | 150 | 28.5 (13.8) ad, C | 60.0 (16.0) abc |
| 6 | G. mosseae | 20 | 7.30 (7.03) ad, B | 67.5 (15.4) abc |
| 6 | G. intraradices | 150 | 48.0 (26.4) e, A | 52.3 (15.4) ab |
| 6 | G. intraradices | 400 | 50.5 (12.9) e, A | 55.7 (8.33) ab |
| 6 | G. claroideum | 150 | 37.5 (10.5) bce, BC | 56.2 (12.0) ab |
| 6 | G. claroideum | 400 | 34.0 (4.60) bce, C | 52.3 (11.5) ab |
| 12 | None | 0 | 62.2 (12.4) abc | |
| 12 | G. mosseae | 150 | 34.5 (8.41) abd | 59.5 (9.48) a |
| 12 | G. mosseae | 20 | 13.0 (9.12) abd | 40.7 (7.00) a |
| 12 | G. intraradices | 150 | 89.3 (3.88) f | 68.3 (4.84) abc |
| 12 | G. intraradices | 400 | 88.2 (3.19) f | 58.8 (9.52) abc |
| 12 | G. claroideum | 150 | 33.7 (13.7) abc | 59.7 (13.8) ab |
| 12 | G. claroideum | 400 | 24.8 (4.17) abc | 54.8 (7.41) ab |
| 24 | None | 0 | 68.5 (21.1) abc | |
| 24 | G. mosseae | 150 | 44.0 (11.2) ce | 71.8 (9.43) bc |
| 24 | G. mosseae | 20 | 42.0 (17.3) ce | 68.0 (8.22) bc |
| 24 | G. intraradices | 150 | 85.3 (10.8) f | 80.2 (3.77) c |
| 24 | G. intraradices | 400 | 87.3 (8.48) f | 72.0 (14.1) c |
| 24 | G. claroideum | 150 | 12.8 (7.44) d | 67.3 (10.6) bc |
| 24 | G. claroideum | 400 | 14.5 (11.8) d | 69.2 (18.6) bc |
Calculated according to the methods described by Trouvelot et al. (32). Means of the same background community not sharing a common letter differed significantly (based on Tukey's honestly significant difference comparisons, P < 0.05). Lowercase letters (a to f) indicate differences among a harvest by interaction with the AMF species inoculated. Capital letters (A to C) indicate differences among an IP level by interaction with the AMF species inoculated (only indicated at harvest 1).
AMF response to background community.
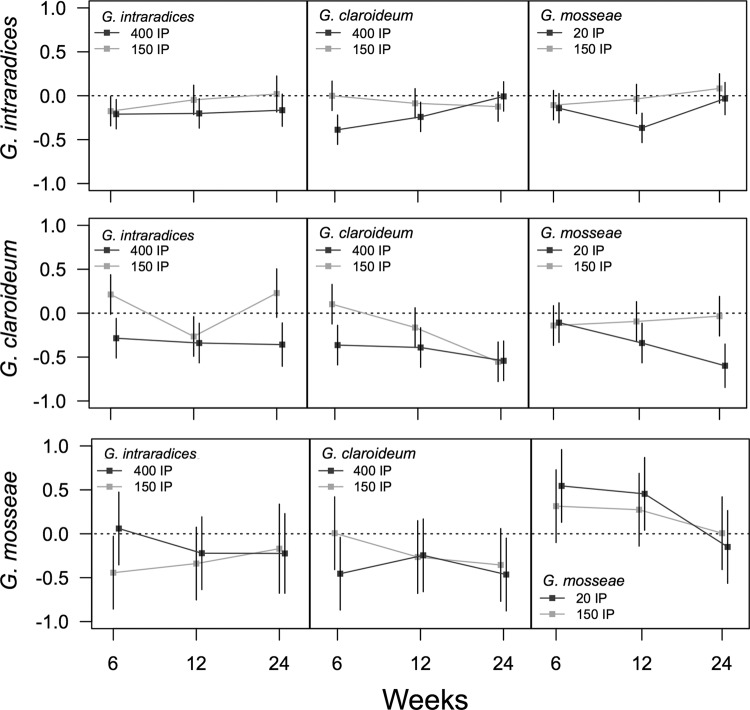
In general, the introduced AMF were less abundant in roots in the presence of a background AMF community than when inoculated at the same levels in the control background (Fig. 2; see also Table S2 in the supplemental material for raw means and standard errors of AMF nrDNA detection). The suppression was most pronounced for G. mosseae. Additionally, the performance of G. mosseae only differed among harvests (F2,32 = 9.16; P = 0.0007; see Table S3 in the supplemental material for full ANOVA results), where it was least detected at 24 weeks relative to earlier harvests despite IP level (Fig. 2). For both G. intraradices and G. claroideum, the abundance response was generally smaller and depended upon the time of harvest and level of IP added (F2,27 = 3.76, P = 0.04 and F2,30 = 9.85, P = 0.0005 for G. intraradices and G. claroideum, respectively). Only at 24 weeks did G. intraradices differ between IP levels, with 150 IP leading to a similar abundance as when no AMF background was present (Fig. 1). At 24 weeks, G. claroideum was the most suppressed in abundance despite IP level, while at 6 and 12 weeks, it reached similar abundance as without AMF background either at 150 IP (6 weeks) or at 400 IP (12 weeks) (Fig. 2).
Fig 2.
Abundance of AMF in roots when a preestablished AMF background community is present relative to when AMF are inoculated in the absence of a background AMF community (RIIcommunity). This reflects the ability of AMF to achieve colonization levels in the presence of an AMF community relative to when no interspecific competition is present. The dotted black line indicates the colonization abundance when no background AMF community was present. Error bars indicate the 95% confidence intervals. Lines connecting means highlight the trend between harvests.
AMF responses to additional inoculation.
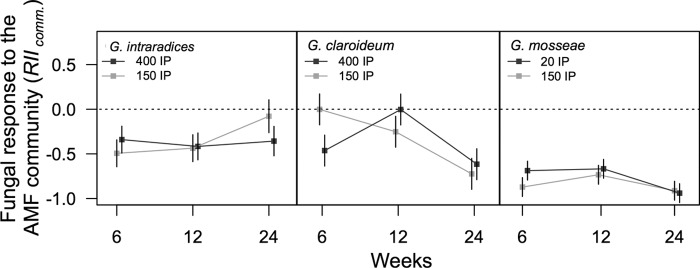
Both G. intraradices and G. claroideum varied in their responses to inoculum additions (RIIinoc.), depending on the time of harvest, the identity of the fungus inoculated, and the IP level at which the fungus was inoculated (F4,86 = 2.55, P = 0.04 and F4,86 = 3.64, P = 0.009, respectively; see Table S3 in the supplemental material for full ANOVA results). Generally, the effects of inoculum addition were frequently negative (Fig. 3).
Fig 3.
The response for AMF abundance in roots to an additional AMF inoculum in the presence of a preestablished AMF background community (RIIinoc.). The dotted black line indicates the AMF abundance in roots achieved in the background AMF community when no additional AMF propagules were added. This reflects the influences of intra- and interspecific AMF densities on their abundance within roots. The dotted black line indicates the abundance of AMF when no AMF inoculum was added. Error bars indicate the 95% confidence intervals. Lines connecting means highlight the trends between harvests.
The IP level of G. intraradices added did not influence its overall relative abundance; however, at 400 IP, G. intraradices was reduced in abundance relative to its abundance without additional inocula at 6 and 12 weeks. G. intraradices was also suppressed by 400 IP of G. claroideum at 6 and 12 weeks, and by 20 IP of G. mosseae at 12 weeks, relative to its abundance without additional inocula. At 24 weeks, G. intraradices was not affected by any inoculum additions.
G. claroideum was consistently reduced in abundance with the addition of 400 IP of intraspecific inoculum at all three harvests. Conversely, 150 IP of G. claroideum inoculum had no effect on G. claroideum abundance after 6 weeks, but it had an increasingly suppressive effect at the later harvests. Inoculation of G. intraradices at 150 IP marginally increased G. claroideum abundance at 6 and 24 weeks but reduced its abundance at 12 weeks. However, with 400 IP of G. intraradices inoculated, G. claroideum was consistently suppressed in abundance at all three harvests. Inoculation with 150 IP of G. mosseae had no effect, but 20 IP of G. mosseae increasingly suppressed G. claroideum in abundance at the later harvests.
The abundance of G. mosseae was only influenced by the identity of the AMF inoculated (F2,99 = 12.1, P < 0.0001; see Table S3 in the supplemental material). In general, intraspecific inoculum additions of G. mosseae increased its overall abundance within roots relative to its background levels, whereas inoculation with G. intraradices or G. claroideum significantly suppressed the abundance of G. mosseae in comparison with the noninoculated treatment (Fig. 3).
Plant performance.
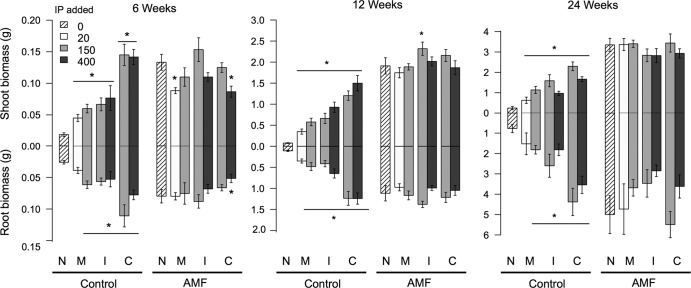
Overall plant biomass, both above- and belowground, increased with time and was generally greater in the presence of a background AMF community (Fig. 4). Plant shoot and root biomass were improved by the addition of AMF at all three harvests when no background AMF community was present. Only inoculation with G. mosseae at 20 IP did not significantly improve root biomass at 6 and 24 weeks (P = 0.12 and P = 0.11, respectively) relative to when no AMF were present in the control background. Conversely, comparing individual inoculated treatments to the uninoculated plants in the presence of the background AMF community, only G. intraradices inoculated at 150 IP after 12 weeks resulted in improved shoot biomass (Fig. 3). Inoculation with G. mosseae at 20 IP reduced shoot productivity, and G. claroideum at 400 IP reduced both root and shoot productivity at 6 weeks (Fig. 4).
Fig 4.
Means (with SE) for the biomass of roots and shoots grown in each inoculation treatment with and without an arbuscular mycorrhizal fungal background community (N, no inoculum added; M, G. mosseae isolate; I, G. intraradices isolate; C, G. claroideum isolate; AMF, with preestablished background community; control, without AMF background community). Shading indicates the different amounts of IP added of each isolate. Asterisks above/below means indicate differences between the individual inoculated treatments means and the corresponding noninoculated treatment (N). When not indicated, the effect of inoculation was nonsignificant.
Plant performance differed in some cases, depending on the IP level of inoculation (see Table S4 in the supplemental material for full ANOVA results). At both 6 and 12 weeks, G. mosseae-inoculated plants had a higher shoot biomass when inoculated with 150 IP than with 20 IP, despite the presence of an AMF background community (overall IP effects: F1,21 = 4.65, P = 0.04 and F1,21 = 4.93, P = 0.04, respectively). Additionally, at 24 weeks after inoculation with G. claroideum at the higher 400-IP level, shoot biomass was depressed in comparison to inoculation with G. claroideum at 150 IP in both control and AMF background treatments (overall IP effect of G. claroideum: F1,21 = 4.90; P = 0.04). The IP levels at which G. intraradices was inoculated did not significantly alter shoot production in any case (see Table S4). Furthermore, inoculation with 400 IP of G. claroideum reduced root biomass at 6 and 24 weeks in comparison to the 150-IP inoculation (F1,21 = 6.79, P = 0.02 and F1,21 = 5.15, P = 0.03, respectively), despite the background treatment. At 12 weeks, inoculation with G. intraradices at the higher level of 400 IP improved root biomass in the absence of an AMF community, but it reduced root production in the presence of an AMF community, resulting in an IP level by background interaction (F1,20 = 10.7; P = 0.003) (Fig. 3). The IP levels at which G. mosseae was inoculated did not significantly alter root production in any case (see Table S4).
The response in total plant biomass to AMF inoculation (RIIinoc.) into the AMF background differed between the two IP levels overall (F2,95 = 14.4; P = 0.0003). The 150-IP level generally resulted in greater plant performance than the alternative IP levels (Fig. 5). The response in total plant biomass also varied among a harvest by AMF identity interaction (F4,93 = 3.38; P = 0.01). Inoculation with G. intraradices at 24 weeks suppressed total biomass despite inoculation level (P = 0.002), while the other two AMF did not vary greatly overall in response to AMF inoculum additions.
Fig 5.
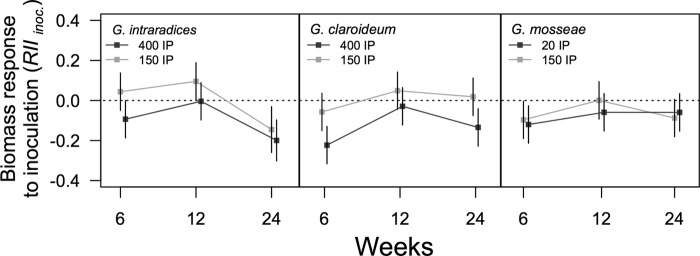
The response in total plant biomass to an additional AMF inoculum when a preestablished AMF background community is present (RIIinoc.). The dotted black line indicates the biomass production when no additional AMF propagules were added. Error bars indicate the 95% confidence intervals. Lines connecting means highlight the trends between harvests.
DISCUSSION
Our results show that addition of AMF propagules results in changes in the abundance of AMF away from the equilibrium state achieved in a previously established AMF community. Importantly, high levels of propagules of the inoculated fungus resulted in lower abundance of the fungi in the roots. In some cases, this corresponded with a decrease in plant growth.
A number of studies have demonstrated that the additional host benefits following inoculation with multiple AMF are likely context dependent (10, 34–36). Competition among AMF for host resources may result in a trade-off with the host growth promotion abilities of the AMF community (12, 13). Our results support these previous findings in that increased AMF competition, here by increasing fungal density and/or shifts in the established propagule balance, can affect the root colonization process and decrease the promotion of host growth by the AMF symbiosis. The performance of both the host plant and the AMF was dependent not only on the identity of the AMF species inoculated, but also on the density of infective propagules within the inoculum. Such results suggest that the addition of AMF propagules into established AMF communities can result in a trade-off with the ability of the AMF community to support the productivity of the plant host.
Inoculation effects on fungal development.
The pool of nrDNA copies of the AMF community was dominated by G. intraradices, in accordance with observations of highly skewed natural communities being usually dominated by a single taxon (37). However, nrDNA copy numbers per nucleus may vary among AMF isolates (38), and the overall high levels of G. intraradices copy numbers, compared to the other two isolates, cannot be unambiguously related to high levels of biologically relevant units such as nuclei or intraradical fungal structures. It is therefore preferable to avoid direct comparisons of nrDNA copy numbers among AMF taxa and perform comparisons among experimental treatments, such as by using the RII index.
The RIIcommunity index clearly showed that G. mosseae was competitively the weakest isolate of the synthetic community, while G. intraradices and G. claroideum were more successful competitors, maintaining their abundance when challenged by the presence of the other two AMF. This competitive weakness of G. mosseae corresponded with the low infectivity of its inocula as determined in the MPN tests. Both features may be related to the different origin and cultivation history of this isolate (23) in comparison with G. intraradices and G. claroideum and may reflect its lower compatibility with the experimental conditions.
Quick development of root colonization by the AMF background affirms the generally high inoculation potential of the precultivated AMF community. Under such conditions, inoculation may not be expected to increase root colonization further but instead lead to shifts in the abundances of the community members (18, 39). However, the expectation that inoculum additions increase the abundance of the inoculated fungus was only confirmed for the G. mosseae isolate. It is known that the root colonization of monoinoculated Glomus isolates typically reach a maximum level over time, depending on plant and fungal isolate identity, as well as environmental conditions. For instance, higher infectivity in soil accelerates the establishment of this plateau level but does not increase its height (19, 35, 40, 41). Our results demonstrated that the G. mosseae isolate was suppressed below this potential maximum by interspecific competition and therefore could profit in its competitive advantage for host roots from additional propagules. In contrast, the other two isolates may have achieved their potential maximum abundance within the AMF community and thus did not respond to intraspecific inoculation by a higher abundance.
The simple concept of a colonization plateau level, however, fails to explain the observed abundance decreases of G. claroideum and G. intraradices following intraspecific inoculation, especially with 400 IP, and these findings point to more complex dynamics of the root colonization process. The plateau level is a feature of microscopically determined root colonization dynamics after nonvital staining of all fungal structures. In contrast, vital staining and quantification of fungal nuclei based on nrDNA copy numbers often reveal a peak of fungal vitality after a few weeks of cultivation, followed by a decline (19, 22, 35, 42). Assuming acceleration of the root colonization process by higher propagule levels, the negative abundance responses to intraspecific inoculation with G. intraradices and G. claroideum may in fact reflect shifts in colonization dynamics, with an earlier vitality peak and onset of the vitality decline, both occurring before the first harvest after 6 weeks of cultivation. Indeed, the G. claroideum isolate displayed this dynamic with its progressive decline in nrDNA copy numbers as well as root colonization by the second and third harvests. Consistent with the assumption of accelerated vitality peak and decline, the response of G. claroideum to intraspecific inoculation was progressively negative with time. In contrast to the negative abundance response to intraspecific inoculation, the abundance decreases following additions of interspecific propagules could be more parsimoniously explained as consequences of more intensive competition for space and carbohydrates (11–14).
The observed inoculation effects thus highlight the dynamic character of both the root colonization process and interactions of coexisting AMF observed in earlier studies (41, 43). Nevertheless, an AMF community in responding to inoculation tends to stabilize in most treatments at the end of the cultivation period of 24 weeks, which indicates its resilience to shifts in propagule densities.
Inoculation effects on plant performance.
The overall positive plant growth response to inoculation into the control background showed that all three isolates behaved as mutualists, as would be expected from the vast literature previously demonstrating this (3). The dependence of G. mosseae on inoculum additions to improve the plant performance relates to the slower establishment of symbiosis with the 20-IP treatment (44, 45). The high 400-IP level of G. intraradices and G. claroideum, in contrast, did not further improve plant performance compared to the 150-IP level, indicating that 150 IP was sufficient to induce maximum benefit to the plant. The overall magnitude of the response in the control soil, however, and especially the differences between plant growth in the control and in the AMF background soil should not be overinterpreted in view of other potential microbial factors. Soil microbial community and especially rhizobia associated with legumes such as Medicago may significantly influence plant performance (46). Despite the additions of microbial filtrates, rhizobia (and other microbes) were certainly less abundant in the control soil than in the AMF soil in the beginning of the cultivation, which may explain the generally better performance of plants growing in AMF soil compared to control soil.
However, this factor does not preclude comparisons among the inoculation treatments that were dependent upon the AMF background soil, as the microbial community developed in the precultivation step was preserved. The negative growth response to inoculation after 6 weeks is interesting and may imply increased competition among AMF following inoculum additions. Bennett and Bever (12) raised a question regarding resource allocation in AMF between competition and providing benefits to the host plant. The increased competition following inoculation due to higher propagule number and disturbance of the preestablished propagule balance possibly redirects resources to competitive interactions. For example, the competing AMF may have invested more into the formation of structures related to carbon consumption and space occupation than in structures involved in nutrient uptake and transfer to the host. This also supports the hypothesis described by Gange and Ayres (47), who proposed a model in which the relation between plant benefit and mycorrhizal density was curvilinear, or rather hump-shaped, where there is an optimal range of AMF density for maximum plant benefit and a negative plant response when AMF density approaches its maximum.
Conclusion.
Our results suggest potentially undesirable effects of AMF inoculation in systems where an AMF community is established. The observed changes in the AMF community and plant performance indicate there may be an optimal level of AMF propagule density and composition in soil. Changes in these levels may then lead to increased competition among the root-colonizing AMF and decrease the AMF community potential to promote plant growth. This ecological aspect has been rather neglected in inoculation studies and should be further explored.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Grant Agency of the Czech Republic, number 526/09/0838, and the institutional projects AV0Z60050516 and Z5038910 of the Grant Agency of the Academy of Sciences of the Czech Republic. C. Wagg was supported by a grant from the Swiss National Science Foundation (number 406840_143097).
We are grateful to Jana Maršíčková for excellent technical assistance.
Footnotes
Published ahead of print 16 August 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02135-13.
REFERENCES
- 1.van der Heijden MGA, Klironomos JN, Ursic M, Moutoglis P, Streitwolf-Engel R, Boller T, Wiemken A, Sanders IR. 1998. Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity. Nature 396:69–72 [Google Scholar]
- 2.Wardle DA, Bardgett RD, Klironomos JN, Setala H, van der Putten WH, Wall DH. 2004. Ecological linkages between aboveground and belowground biota. Science 304:1629–1633 [DOI] [PubMed] [Google Scholar]
- 3.Smith SE, Read DJ. 2008. Mycorrhizal symbiosis, 3rd ed. Academic Press, Amsterdam, Netherlands [Google Scholar]
- 4.Vosátka M, Látr A, Gianinazzi S, Albrechtová J. 2012. Development of arbuscular mycorrhizal biotechnology and industry: current achievements and bottlenecks. Symbiosis 58:29–37 [Google Scholar]
- 5.Verbruggen E, van der Heijden MGA, Rillig MC, Kiers ET. 2013. Mycorrhizal fungal establishment in agricultural soils: factors determining inoculation success. New Phytol. 197:1104–1109 [DOI] [PubMed] [Google Scholar]
- 6.Hart MM, Reader RJ. 2002. Taxonomic basis for variation in the colonization strategy of arbuscular mycorrhizal fungi. New Phytol. 153:335–344 [Google Scholar]
- 7.Powell JR, Parrent JL, Hart MM, Klironomos JN, Rillig MC, Maherali H. 2009. Phylogenetic trait conservatism and the evolution of functional trade-offs in arbuscular mycorrhizal fungi. Proc. R. Soc. B Biol. Sci. 276:4237–4245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koide RT. 2000. Functional complementarity in the arbuscular mycorrhizal symbiosis. New Phytol. 147:233–235 [Google Scholar]
- 9.Kiers ET, Duhamel M, Beesetty Y, Mensah JA, Franken O, Verbruggen E, Fellbaum CR, Kowalchuk GA, Hart MM, Bago A, Palmer TM, West SA, Vandenkoornhuyse P, Jansa J, Bücking H. 2011. Reciprocal rewards stabilize cooperation in the mycorrhizal symbiosis. Science 333:880–882 [DOI] [PubMed] [Google Scholar]
- 10.Wagg C, Jansa J, Schmid B, van der Heijden MGA. 2011. Belowground biodiversity effects of plant symbionts support aboveground productivity. Ecol. Lett. 14:1001–1009 [DOI] [PubMed] [Google Scholar]
- 11.Pearson JN, Abbott LK, Jasper DA. 1994. Phosphorus, soluble carbohydreates and the competition between 2 arbuscular mycorrhizal fungi clonizing subterranean clover. New Phytol. 127:101–106 [DOI] [PubMed] [Google Scholar]
- 12.Bennett AE, Bever JD. 2009. Trade-offs between arbuscular mycorrhizal fungal competitive ability and host growth promotion in Plantago lanceolata. Oecologia 160:807–816 [DOI] [PubMed] [Google Scholar]
- 13.Bever JD, Richardson SC, Lawrence BM, Holmes J, Watson M. 2009. Preferential allocation to beneficial symbiont with spatial structure maintains mycorrhizal mutualism. Ecol. Lett. 12:13–21 [DOI] [PubMed] [Google Scholar]
- 14.Graham JH, Abbott LK. 2000. Wheat responses to aggressive and non-aggressive arbuscular mycorrhizal fungi. Plant Soil 220:207–218 [Google Scholar]
- 15.Mummey DL, Antunes PM, Rillig MC. 2009. Arbuscular mycorrhizal fungi pre-inoculant identity determines community composition in roots. Soil Biol. Biochem. 41:1173–1179 [Google Scholar]
- 16.Koch AM, Antunes PM, Barto EK, Cipollini D, Mummey DL, Klironomos JN. 2011. The effects of arbuscular mycorrhizal (AM) fungal and garlic mustard introductions on native AM fungal diversity. Biol. Invasions 13:1627–1639 [Google Scholar]
- 17.Pellegrino E, Bedini S, Avio L, Bonari E, Giovannetti M. 2011. Field inoculation effectiveness of native and exotic arbuscular mycorrhizal fungi in a Mediterranean agricultural soil. Soil Biol. Biochem. 43:367–376 [Google Scholar]
- 18.Alkan N, Gadkar V, Yarden O, Kapulnik Y. 2006. Analysis of quantitative interactions between two species of arbuscular mycorrhizal fungi, Glomus mosseae and G. intraradices, by real-time PCR. Appl. Environ. Microbiol. 72:4192–4199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thonar C. 2009. Synthetic mycorrhizal communities: establishment and functioning. Ph.D. dissertation. ETH Zürich, Zürich, Switzerland. 10.3929/ethz-a-005927506 [DOI] [Google Scholar]
- 20.Gustafson DJ, Casper BB. 2006. Differential host plant performance as a function of soil arbuscular mycorrhizal fungal communities: experimentally manipulating co-occurring Glomus species. Plant Ecol. 183:257–263 [Google Scholar]
- 21.Gryndler M, Sudová R, Püschel D, Rydlová J, Janoušková M, Vosátka M. 2008. Cultivation of high-biomass crops on coal mine spoil banks: can microbial inoculation compensate for high doses of organic matter? Bioresour. Technol. 99:6391–6399 [DOI] [PubMed] [Google Scholar]
- 22.Krak K, Janoušková M, Caklová P, Vosátka M, Scarontorchová H. 2012. Intraradical dynamics of two coexisting isolates of the arbuscular mycorrhizal fungus Glomus intraradices sensu lato as estimated by real-time PCR of mitochondrial DNA. Appl. Environ. Microbiol. 78:3630–3637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Enkhtuya B, Rydlová J, Vosatká M. 2000. Effectiveness of indigenous and non-indigenous isolates of arbuscular mycorrhizal fungi in soils from degraded ecosystems and man-made habitats. Appl. Soil Ecol. 14:201–211 [Google Scholar]
- 24.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis suite. Nucleic Acids Symp. Ser. 41:95–98 [Google Scholar]
- 26.Swofford DL. 2003. PAUP*: phylogenetic analysis using parsimony (*and other methods), 4th ed. Sinauer Associates, Sunderland, MA [Google Scholar]
- 27.Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JAM. 2007. Primer3Plus, an enhanced Web interface to Primer3. Nucleic Acids Res. 35:W71–W74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kalendar R, Lee D, Schulman AH. 2011. Java web tools for PCR, in silico PCR, and oligonucleotide assembly and analysis. Genomics 98:137–144 [DOI] [PubMed] [Google Scholar]
- 29.Alexander M. 1965. Most-probable-number method for microbial populations, p 1467–1472 In Black SA, Evans DD, Ensminger LE, White JL, Clark FE. (ed), Methods of soil analysis, part 2. Chemical and microbiological properties. American Society of Agronomy, Madison, WI [Google Scholar]
- 30.Gryndler M, Vejsadová H, Vančura V. 1992. The effect of magnesium ions on the vesicular arbuscular mycorrhizal infection of maize roots. New Phytol. 122:455–460 [DOI] [PubMed] [Google Scholar]
- 31.Koske RE, Gemma JN. 1989. A modified procedure for staining roots to detect VA mycorrhizas. Mycol. Res. 92:486–505 [Google Scholar]
- 32.Trouvelot A, Kough JL, Gianinazzi-Pearson V. 1986. Mesure du taux de mycorhization VA d'un systeme radiculaire. Recherche de methodes d'estimation ayant une signification fonctionnelle, p 217–221 In Gianinazzi-Pearson V, Gianinazzi S. (ed), Physiological and genetical aspects of mycorrhizae. INRA, Paris, France [Google Scholar]
- 33.Armas C, Ordiales R, Pugnaire FI. 2004. Measuring plant interactions: a new comparative index. Ecology 85:2682–2686 [Google Scholar]
- 34.Vogelsang KM, Reynolds HL, Bever JD. 2006. Mycorrhizal fungal identity and richness determine the diversity and productivity of a tallgrass prairie system. New Phytol. 172:554–562 [DOI] [PubMed] [Google Scholar]
- 35.Jansa J, Smith FA, Smith SE. 2008. Are there benefits of simultaneous root colonization by different arbuscular mycorrhizal fungi? New Phytol. 177:779–789 [DOI] [PubMed] [Google Scholar]
- 36.Hoeksema JD, Chaudhary VB, Gehring CA, Johnson NC, Karst J, Koide RT, Pringle A, Zabinski C, Bever JD, Moore JC, Wilson GWT, Klironomos JN, Umbanhowar J. 2010. A meta-analysis of context-dependency in plant response to inoculation with mycorrhizal fungi. Ecol. Lett. 13:394–407 [DOI] [PubMed] [Google Scholar]
- 37.Dumbrell AJ, Nelson M, Helgason T, Dytham C, Fitter AH. 2010. Idiosyncrasy and overdominance in the structure of natural communities of arbuscular mycorrhizal fungi: is there a role for stochastic processes? J. Ecol. 98:419–428 [Google Scholar]
- 38.Corradi N, Croll D, Colard A, Kuhn G, Ehinger M, Sanders IR. 2007. Gene copy number polymorphisms in an arbuscular mycorrhizal fungal population. Appl. Environ. Microbiol. 73:366–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gazey C, Abbott LK, Robson AD. 2004. Indigenous and introduced arbuscular mycorrhizal fungi contribute to plant growth in two agricultural soils from south-western Australia. Mycorrhiza 14:355–362 [DOI] [PubMed] [Google Scholar]
- 40.Wilson JM, Trinick MJ. 1983. Infection development and interactions between vesicular-arbuscular mycorrhizal fungi. New Phytol. 93:543–553 [Google Scholar]
- 41.Wilson GWT, Tommerup IC. 1992. Interactions between fungal symbionts. VA mycorrhizae, p 199–248 In Allen M. (ed), Mycorrhizal functioning: an integrative plant-fungal process. Springer, Berlin, Germany [Google Scholar]
- 42.Tisserant B, Gianinazzi S, GianinazziPearson V. 1996. Relationships between lateral root order, arbuscular mycorrhiza development, and the physiological state of the symbiotic fungus in Platanus acerifolia. Can. J. Bot. Rev. Can. Bot. 74:1947–1955 [Google Scholar]
- 43.Wilson JM. 1984. Competition for infection between vesicular arbuscular mycorrhizal fungi. New Phytol. 97:427–435 [Google Scholar]
- 44.Haas JH, Krikun J. 1985. Efficacy of endomycorrhizal-fungus isolates and inoculum quantities required for growth response. New Phytol. 100:613–621 [Google Scholar]
- 45.Al-Karaki GN, Clark RB. 1999. Varied rates of mycorrhizal inoculum on growth and nutrient acquisition by barley grown with drought stress. J. Plant Nutrit. 22:1775–1784 [Google Scholar]
- 46.Richardson AE, Barea JM, McNeill AM, Prigent-Combaret C. 2009. Acquisition of phosphorus and nitrogen in the rhizosphere and plant growth promotion by microorganisms. Plant Soil 321:305–339 [Google Scholar]
- 47.Gange AC, Ayres RL. 1999. On the relation between arbuscular mycorrhizal colonization and plant ‘benefit.' Oikos 87:615–621 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.