Abstract
Lactococci inoculated into cheese grow as colonies producing lactic acid. The pH microgradients were investigated around colonies in a complex food such as cheese. The results, obtained using a nondestructive technique, demonstrated that pH microgradients did not occur regardless of the acidification kinetics and the size of the colony.
TEXT
Lactic acid bacteria are used as starters in cheese manufacture. They are immobilized within the cheese curd during the coagulation step regardless of the cheese-making process. Immobilized cells then grow as colonies (1), producing lactic acid within the cheese matrix. The hypothesis of an accumulation of lactic acid around bacterial colonies has been formulated as a consequence of mass transfer limitations from the colony (2, 3), lowering the pH on the edge of the colony. Since low pH is known to modify the metabolic activity of lactic acid bacteria (4, 5) and to have even more influence when cells are immobilized (5), it is important to determine whether or not, in solid and complex foods such as cheese, the local pH around colonies differed from the “global” pH usually measured. Microgradients of pH occur in gelatin media around large colonies of Salmonella enterica serovar Typhimurium (diameters from 450 μm to 2.5 mm) (3, 6) and Lactobacillus curvatus (diameters from 380 μm to 430 μm) but do not occur around L. curvatus colonies with a diameter of 155 μm (7). These results, obtained in gelatin medium, cannot be applied to cheese since the diffusion coefficients have been shown to depend on the microstructure (8).
Different techniques have been used to monitor pH in and around bacterial colonies. Microelectrodes were first used to measure pH in and around submerged colonies of S. Typhimurium (3). Later, a nondestructive technique based on fluorescence microscopy was developed to describe the pH microgradient around colonies of L. curvatus (7). More recently, the pH-sensitive fluoroprobe, C-SNARF-4F, has been applied to measure the pH within biofilms using a ratiometric correlation to pH (9). Our final aim is to explore the pH microenvironment around bacterial colonies in a complex and solid food such as cheese. Our strategy, in this first study, was to use a nonfat model cheese which was a reproducible and homogeneous cheese matrix directly molded in imaging chambers for nondestructive microscopic examination. As we previously demonstrated that the size of colonies depends on the inoculation level in cheese (10), the model cheeses were inoculated at three different levels with Lactococcus lactis, generating three different sizes of colonies. Bigger colonies (lower inoculation levels) than in traditional cheeses were generated in order to enhance the phenomenon of lactic acid accumulation. We monitored the pH both at the macroscopic scale, so called here macro-pH, and at the microscopic scale, so called here micro-pH, using a nondestructive microscopic examination.
Monitoring macro-pH and micro-pH in model cheeses.
Model cheeses were made from ultrafiltrated (UF) milk retentate both in 15-ml flasks and 1-ml cassettes as described by Floury et al. (11), except that we used CoverWell imaging chambers (Sigma-Aldrich, Saint-Quentin Fallavier, France). The model cheese produced is a soft-type cheese, similar to a Pavé d'Affinois cheese without fat. The dry matter of the final model cheese was 21%. The strain L. lactis subsp. lactis bv. diacetylactis LD61 (Soredab, La Boissière Ecole, France), previously studied in similar model cheeses (12, 13), was used because it grows up to 5 × 109 CFU/g and acidifies down to pH 4.8 in 24 h (13), which is the lowest pH observed in UF cheeses because of a high buffer capacity. It was inoculated at 1.3 × 103, 4 × 104 and 1.6 × 105 CFU/g in the UF retentate in both 15-ml flasks and 1-ml imaging chambers. The addition of 0.03% of a coagulant agent (MAXIREN 180; DSM Food, Delft, Netherlands) and incubation for 1 h at 30°C allowed complete coagulation. Then, flasks and imaging chambers were incubated for 72 h at 19°C. Macro-pH was measured in the 15-ml flasks using a pH meter (Inlab pH level 1; WTW, Germany) and pH electrode (Sebtix 41; WTW, Germany). Micro-pH around a colony was monitored in the imaging chambers using the pH-sensitive probe C-SNARF-4F (Molecular Probes, Invitrogen, Villebon-sur-Yvette, France) and an inverted confocal laser-scanning microscope (CLSM) (Eclipse-TE2000-C1; Nikon, Champigny-sur-Marne, France). SYTO9 (Molecular Probes, Invitrogen, Villebon-sur-Yvette, France), used as a dye to detect bacterial colonies, was excited at 488 nm and detected at 515 ± 15 nm. C-SNARF-4F was excited at 543 nm and detected at 590 ± 50 nm and over 650 nm. The ratio (R) between the two emission peaks of C-SNARF-4F was calculated along 100 μm from the edge of the colony, at 10-μm steps, at opposite sides of the colony. R was calculated using the plugin “ratio plus” of the open source software FIJI (14), under our conditions, according to the following equation: R = E590/E650, where Eλ is the mean fluorescence of a region of interest (ROI), diameter of 10 μm, at an emission wavelength λ (R was calculated for 10 ROI on each side of the colony). The background of images has been measured as negligible at both wavelengths.
A standard curve was drawn from the macro-pH measurements and R values which were fitted to a third-degree polynomial equation with a determination coefficient, r2 = 0.99 (Fig. 1). CLSM images (Fig. 1) show the change of fluorescence intensity of the two emission peaks during curd acidification in imaging chambers. Fluorescence is shown in red when the intensity of fluorescence of the first peak is high and in blue when the intensity of fluorescence of the second peak is high.
Fig 1.
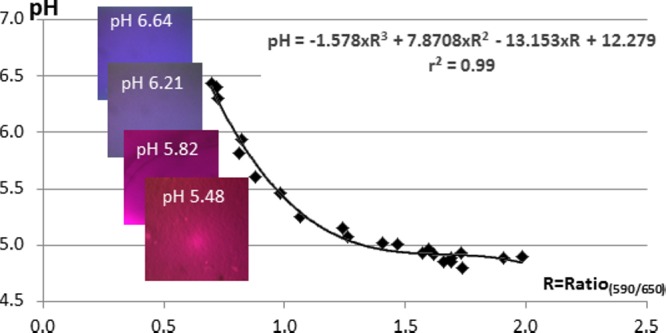
Standard curve is pH = f(R590/650), where pH is the macro-pH values measured in the model cheeses inoculated at the 3 different inoculation levels (1.6 × 105, 4 × 104, and 1.3 × 103 CFU/g) through time, and ratio (R590/650) is calculated from the corresponding imaging chambers (CLSL images, 318 by 318 μm) mixed with C-SNARF-4F of which intensity of fluorescence of the two emission peaks was measured at 590 ± 50 and >650 nm.
No pH microgradients occurred around colonies in the model cheese.
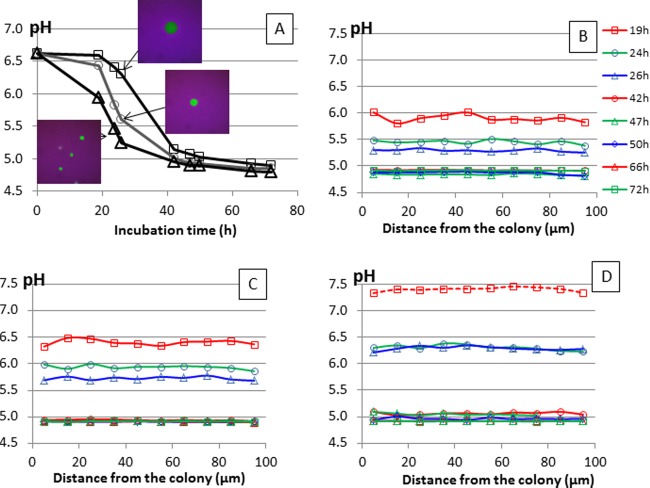
Macro-pH decreased in the model cheese (Fig. 2A) from pH 6.62 to pH 4.9 to 4.8, as previously observed using the same strain (13). The lower the inoculation level, the slower the acidification kinetics in cheeses inoculated from 1.3 × 103 CFU/g to 1.6 × 105 CFU/g, as expected. CLSM images (Fig. 2A) confirmed that the lower the inoculation level, the bigger the colonies at 24 to 26 h of growth. The maximum diameters of colonies were 35 ± 4 μm (n = 10), 65 ± 4 μm (n = 3), and 111 ± 1 μm (n = 3) in cheeses inoculated at 1.6 × 105, 4 × 104, and 1.3 × 103 CFU/g, respectively. Micro-pH was measured around colonies by calculating R. Whatever the size of colonies, R was constant, and its value was the same along a distance of 100 μm from the colony (Fig. 2B, C, and D), on both sides. These results clearly demonstrate that no pH microgradients occurred under these conditions regardless of the colony size. Furthermore, we showed that C-SNARF-4F was pH sensitive enough to discriminate the local acidification kinetics generated by lactococci inoculated at different levels.
Fig 2.
(A) Acidification of the model cheeses inoculated at levels of 1.6 × 105 CFU/g (△), 4 × 104 CFU/g (○), and 1.3 × 103 CFU/g (□): macro-pH measured using a pH meter electrode and CLSM images (318 by 318 μm) of colonies at 24 to 26 h of growth using C-SNARF-4F; micro-pH calculated around a colony using the standard curve equation at 19, 24, 26, 42, 47, 50, 66, and 72 h for model cheeses inoculated at levels of 1.6 × 105 CFU/g (B), 4 × 104 CFU/g (C), and 1.3 × 103 CFU/g (D). The key shown applies to panels B, C, and D.
No pH microgradients occurred around Lactococcus colonies in a model cheese regardless of the size of colonies, from diameters of 35 to 111 μm. Previous studies have been performed only in gelatin or agar medium. The ratiometric method using C-SNARF-4F is a relevant tool because it allows stable R values to be obtained. It is independent of the experimental parameters, such as intensity of fluorescence, detection, and probe concentration. Moreover, C-SNARF-4F was shown to have no interactions with bacterial components (proteins, polysaccharides) at pH 7 (9). This study aimed to determine whether pH microgradients occur or not around colonies. Even if microgradients occur in gelatin and agar medium only around bacterial colonies bigger than 400 μm (3, 6), it was important to demonstrate that they did not occur in a model cheese around smaller colonies for several reasons. First, an adaptation to acid stress has previously been shown at the gene level for strain LD61 in the same model cheese (12). Second, the few data on lactic acid diffusion suggest that its diffusion coefficient is lower in cheese than in gelatin or agar medium. It is known to be 2.81 × 10−10 ± 0.21 × 10−10 m2/s in a buffered gelatin medium at 20°C (15) and 4.63 × 10−10 m2/s in an agar medium at 12°C (16), while it is around 1 × 10−10 m2/s in a semihard cheese, Pategras cheese (17). Consequently, the lower diffusion coefficient in cheese could have led to an accumulation of lactic acid around colonies. However, our results show that the lactic acid did not accumulate at the edge of colonies, suggesting that it migrates faster than it is produced. This is finally in agreement with the results observed in gelatin in which no pH microgradients were observed around colonies smaller than 150 μm (7). In these model cheeses, we decreased the inoculation level to 1.3 × 103 CFU/g, leading to a maximum colony size of 111 μm, because colonies formed at lower inoculation levels are difficult to observe in microscopy within the opaque cheese matrix. Practically, the inoculation levels used in this study are below the inoculation levels of lactic acid bacteria used in cheese manufacture (about 106 to 107 CFU/ml) in order to enhance the potential accumulation of lactic acid around colonies.
In conclusion, the macro-pH is the pH that cells effectively perceive in a model cheese during acidification. These results are innovative because they have been performed on a solid-food matrix. Even if this model cheese is different from a traditional cheese, we think that the results are likely to be the same in a traditional cheese. In traditional cheese, bacterial colonies are smaller and less dense than in this model cheese (18, 19), thus decreasing the probability of generating pH microgradients compared to the conditions in the present study. It would also be interesting to investigate the influence of heterogeneity on local pH microgradients in cheeses exhibiting different compositions and structures and for other lactic acid bacterial species.
ACKNOWLEDGMENTS
This study was part of the CheesOmic project cofunded by the regions Bretagne and Pays-de-la-Loire and supported by the Bretagne Biotechnologie Alimentaire (BBA) association.
Footnotes
Published ahead of print 9 August 2013
REFERENCES
- 1.Parker ML, Gunning PA, Macedo AC, Malcata FX, Brocklehurst TF. 1998. The microstructure and distribution of micro-organisms within mature Serra cheese. J. Appl. Microbiol. 84:523–530 [DOI] [PubMed] [Google Scholar]
- 2.Malakar PK, Martens DE, van Breukelen W, Boom RM, Zwietering MH, van 't Riet K. 2002. Modeling the interactions of Lactobacillus curvatus colonies in solid medium: consequences for food quality and safety. Appl. Environ. Microbiol. 68:3432–3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wimpenny JWT, Leistner L, Thomas LV, Mitchell AJ, Katsaras K, Peetz P. 1995. Submerged bacterial colonies within food and model systems: their growth, distribution and interactions. Int. J. Food Microbiol. 28:299–315 [DOI] [PubMed] [Google Scholar]
- 4.Kajfasz JK, Quivey RG. 2011. Responses of lactic acid bacteria to acid stress, p 23–53 In Tsakalidou E, Papadimitriou K. (ed), Stress responses of lactic acid bacteria. Springer, New York, NY [Google Scholar]
- 5.Meldrum RJ, Brocklehurst TF, Wilson DR, Wilson PDG. 2003. The effects of cell immobilization, pH and sucrose on the growth of Listeria monocytogenes Scott A at 10°C. Food Microbiol. 20:97–103 [Google Scholar]
- 6.Walker SL, Brocklehurst TF, Wimpenny JWT. 1997. The effects of growth dynamics upon pH gradient formation within and around subsurface colonies of Salmonella typhimurium. J. Appl. Microbiol. 82:610–614 [PubMed] [Google Scholar]
- 7.Malakar PK, Brocklehurst TF, Mackie AR, Wilson PDG, Zwietering MH, van 't Riet K. 2000. Microgradients in bacterial colonies: use of fluorescence ratio imaging, a non-invasive technique. Int. J. Food Microbiol. 56:71–80 [DOI] [PubMed] [Google Scholar]
- 8.Aly S, Floury J, Famelart MH, Madec MN, Dupont D, Lortal S, Le Gouar Y, Jeanson S. 2011. Nisin quantification by ELISA allows the modeling of its apparent diffusion coefficient in model cheeses. J. Agric. Food Chem. 59:9484–9490 [DOI] [PubMed] [Google Scholar]
- 9.Hunter RC, Beveridge TJ. 2005. Application of a pH-sensitive fluoroprobe (C-SNARF-4) for pH microenvironment analysis in Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 71:2501–2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeanson S, Chadoeuf J, Madec M-N, Aly S, Floury J, Brocklehurst TF, Lortal S. 2011. Spatial distribution of bacterial colonies in a model cheese. Appl. Environ. Microbiol. 77:1493–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Floury J, Madec MN, Waharte F, Jeanson S, Lortal S. 2012. First assessment of diffusion coefficients in model cheese by fluorescence recovery after photobleaching (FRAP). Food Chem. 133:551–556 [DOI] [PubMed] [Google Scholar]
- 12.Cretenet M, Laroute V, Ulve V, Jeanson S, Nouaille S, Even S, Piot M, Girbal L, Le Loir Y, Loubiere P, Lortal S, Cocaign-Bousquet M. 2011. Dynamic analysis of the Lactococcus lactis transcriptome in cheeses made from milk concentrated by ultrafiltration reveals multiple strategies of adaptation to stresses. Appl. Environ. Microbiol. 77:247–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ulvé VM, Monnet C, Valence F, Fauquant J, Falentin H, Lortal S. 2008. RNA extraction from cheese for analysis of in situ gene expression of Lactococcus lactis. J. Appl. Microbiol. 105:1327–1333 [DOI] [PubMed] [Google Scholar]
- 14.Schindelin J, Arganda-Carreras I, Frise E, Kayning V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tivenez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9:676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malakar PK, Zwietering MH, Boom RM, Brocklehurst TF, Wilson PD, Mackie AR, van 't Riet K. 2002. Diffusion of lactic acid in a buffered gel system supporting growth of Lactobacillus curvatus. J. Sci. Food Agric. 82:1729–1734 [Google Scholar]
- 16.Aldarf M, Fourcade F, Amrane A, Prigent Y. 2004. Diffusion of lactate and ammonium in relation to growth of Geotrichum candidum at the surface of solid media. Biotechnol. Bioeng. 87:69–80 [DOI] [PubMed] [Google Scholar]
- 17.Floury J, Jeanson S, Aly S, Lortal S. 2010. Determination of the diffusion coefficients of small solutes in cheese: a review. Dairy Sci. Technol. 90:477–508 [Google Scholar]
- 18.Floury J, Jeanson S, Madec MN, Lortal S. 2013. Porosity of Lactococcus lactis subsp. lactis LD61 colonies immobilised in model cheese. Int. J. Food Microbiol. 163:64–70 [DOI] [PubMed] [Google Scholar]
- 19.Lopez C, Maillard MB, Briard-Bion V, Camier B, Hannon JA. 2006. Lipolysis during ripening of Emmental cheese considering organization of fat and preferential localization of bacteria. J. Agric. Food Chem. 54:5855–5867 [DOI] [PubMed] [Google Scholar]