Abstract
Clostridium difficile is the leading cause of antibiotic-associated diarrheal disease in health care settings across the world. Despite its pathogenic capacity, it can be carried asymptomatically and has been found in terrestrial and marine ecosystems outside hospital environments. Little is known about these environmental strains, and few studies have been conducted on estuarine systems. Although prophage abundance and diversity are known to occur within clinical strains, prophage carriage within environmental strains of C. difficile has not previously been explored. In this study, we isolated C. difficile from sites sampled in two consecutive years in an English estuarine system. Isolates were characterized by PCR ribotype, antibiotic resistance, and motility. The prevalence and diversity of prophages were detected by transmission electron microscopy (TEM) and a phage-specific PCR assay. We show that a dynamic and diverse population of C. difficile exists within these sediments and that it includes isolates of ribotypes which are associated with severe clinical infections and those which are more frequently isolated from outside the hospital environment. Prophage carriage was found to be high (75%), demonstrating that phages play a role in the biology of these strains.
INTRODUCTION
Clostridium difficile is an endospore-forming, anaerobic, low-G+C-content, Gram-positive bacterium belonging to the phylum Firmicutes. It is of major health care importance, as it causes C. difficile infection (CDI), typically a nosocomial disease that has a high incidence of relapse and treatment failure (1). It has been estimated that CDI costs Europe €3 billion and the United States $800 million annually, making it a significant health and economic problem (2).
The disease is mediated by the production of up to three toxins, TcdA, TcdB, and the Clostridium difficile binary toxin (CDT) (3). Usually, CDI is associated with previous antibiotic treatment, including with clindamycin and fluoroquinolones, such as ciprofloxacin (4). Three drugs, vancomycin, metronidazole, and fidaxomicin, are used to treat infection, but they are not always effective (5, 6). Genes conferring antibiotic resistance to other drugs, such as erythromycin and tetracycline, have been identified on potentially mobile elements and on the chromosome (7, 8). C. difficile also encodes additional virulence factors, including adhesins, flagella, and sporulation factors, which may aid the bacterium during host colonization, infection, and persistence in the environment to facilitate transmission (9).
The species is genetically diverse, and PCR ribotyping is the most commonly used method to type strains via band profiles produced by amplifying the variable intergenic region between the 16S rRNA and 23S rRNA genes (10–12). Monitoring of ribotypes has revealed C. difficile strain epidemiology changes between countries and over time (2). The emergence of new ribotypes in outbreaks, such as R027 and R078, highlights the importance of the ongoing strain evolution of this pathogen (13, 14). Whole-genome sequence analysis has also examined the global spread of R027 strains and documented the continued evolution of this ribotype in clinics and its transmission between humans and animals (15).
C. difficile can also be isolated from environmental samples, such as soils, river water, and sand (16–23). Despite a crossover between ribotypes of clinical, livestock, and environmental strains, some ribotypes appear to be more associated with particular sources (24, 25). R078, which is associated with both livestock and clinical isolates, has been suggested to have evolved from an atoxigenic strain that may have occurred within reservoirs outside clinical settings (26, 27). One factor in the evolution of the C. difficile genome is acquisition of mobile genetic elements (27). Carriage of prophages is high, as detected by induction of prophages (see, e.g., reference 28), determination of molecular markers to probe their diversity (29), and identification of prophages in sequenced strains (30) and temperate phages that can infect in a lytic manner other C. difficile isolates (31). Whether environmental reservoirs are sources of novel strains is not known, as only a few studies have examined the genetic diversity or physiological traits of these strains in detail (see, e.g., references 20 and 22), and none have examined prophage carriage.
In order to investigate environmental C. difficile strains, the prevalence and distribution of strains within an estuarine system in the south of England were determined. The phenotypic and genetic diversity of the isolates was assessed using ribotyping analysis, detection of toxin genes, motility assays, and antibiotic susceptibility testing. Prophage carriage was examined using transmission electron microscopy (TEM) and screening for specific C. difficile phages by PCR.
MATERIALS AND METHODS
Sampling and site selection.
In October 2009, 22 samples were obtained from 19 sites in the estuarine system of Langstone and Chichester Harbors, Hampshire, United Kingdom. These included 17 sediment samples from a depth of ∼2 cm, 2 sediment samples from a depth of ∼5 cm, and 3 water samples. During 2010, 2 extra sediment samples from one location and a sample of foam were obtained. All samples were stored in the dark at 4°C until processed (within 72 h). Locations were chosen for the sampling of C. difficile around the south coast estuarine system in a nationally designated site of special scientific interest (SSSI) by Natural England (unit 13, unit identifier 1016965). Sampling sites included those thought likely to be polluted with sewage or other anthropomorphic input and those less likely to be contaminated from these sources. The overall pollution levels were informally assessed for our purposes by reviewing the relative development of intertidal communities. Intertidal phanergogamic communities, such as Spartina spp., reduce polluted areas, as do the grasses of the salt marsh communities. Development and expression of the algal species Blidingia, Ulva, and Fucus are well known effective indicators of trophic status (32). The relative expression of populations of the macrofaunas Hydrobia, Corophium, and species of Arenicola were used as indicators of oxic status. Finally, physical features of sedimentation rate and nature were used to determine the level of the anoxic boundary layer. To determine whether sediment depth and oxygen status impacted C. difficile recovery, surface and anoxic layer sediments were tested at two sites (Thorney Island [site 9] and Kench Beach [site 14]). Status was assigned based on multiple features, including the presence and absence of trophic indicator species, and ranged from 1 (worst) to 5 (least polluted).
Method for isolation of C. difficile from environmental sources and their routine laboratory culture.
Approximately 2 cm3 of sample was transferred into either 10 ml fastidious anaerobic broth (FA; Oxoid, United Kingdom) or brain heart infusion (BHI; Oxoid, United Kingdom) broth. This was supplemented with 250 μg ml−1 cycloserine and 8 μg ml−1 cefoxitin as selective agents and 0.1% sodium taurocholate to enhance spore germination (as Wilson et al. [33] used in solid media). Cultures were incubated under anaerobic conditions (10% hydrogen, 10% carbon dioxide, and 80% nitrogen gases) at 37°C for 10 days in a MiniMACS anaerobic chamber (Don Whitley Scientific, United Kingdom). Following enrichment, cultures were centrifuged for 10 min at 3,398 × g. The pellet was resuspended by vortexing it in an equal volume of industrialized methylated spirit (IMS) and incubated for 30 min at room temperature. Five to 15 μl of sample was subcultured on Brazier's cycloserine, cefoxitin, and egg yolk (CCEY) agar plates (BioConnections, United Kingdom) and incubated for 24 to 48 h. C. difficile colonies were purified through two further rounds of subculturing on BHI–7% defibrinated horse blood (DHB; Oxoid, United Kingdom)–1% bacteriological agar (BA; Oxoid, United Kingdom) plates. Glycerol cryogenic stocks were produced by growing single colonies from the third round of purification in 5 ml FA broth. Cultures were centrifuged at 10,000 × g for 10 min, and the pellet was resuspended in glycerol. Strain stocks were stored at −80°C. Routine culturing was performed in the MiniMACS anaerobic chamber on BHI–7% DHB–1% BA plates using single colonies for liquid broth cultures.
Molecular confirmation of C. difficile isolates, PCR ribotyping, and toxin gene carriage.
C. difficile was identified according to its colony morphology, distinctive C. difficile odor of horse dung/urine, and characteristic chartreuse fluorescence under long-wave UV light (365 nm). C. difficile genomic DNA was extracted using 5% Chelex resin (Bio-Rad, United Kingdom). Samples were heated to 100°C for 12 min, cooled for 5 min, and then centrifuged at 16,000 × g for 10 min. The supernatant was then stored at 4°C until use. PCR confirmation for C. difficile was performed by amplification of the 16S rRNA gene using C. difficile-specific primers described by Rinttila et al. (34). PCRs were performed in a total volume of 50 μl, containing template DNA, 4 μM forward and reverse primers, 0.25 mM deoxynucleoside triphosphates (dNTPs), 4 mM MgCl2, 1× PCR buffer, and 0.5 U of BioTaq polymerase (Bioline, United Kingdom). Amplification conditions were 95°C for 5 min and 40 cycles of 95°C for 30 s, 44°C for 30 s, and 72°C for 45 s, with a final extension of 5 min at 72°C. Products were separated (alongside a 1-kbp ladder [GeneRuler, Fermentas, United Kingdom]) on a 1% Helena agarose gel, which was prepared in 1× Tris-acetate-EDTA (TAE) buffer, stained with GelRed, and run at 90 V for 1 h in TAE buffer, and DNA was visualized. PCR ribotyping was performed on Chelex-extracted DNA as previously reported (35). The PCR products were analyzed using capillary gel electrophoresis performed by the Protein Nucleic Acid Chemistry Laboratory (PNACL) at the University of Leicester in an ABI 3730 genetic analyzer (Applied Biosystems, United Kingdom) and, where possible, assigned to known ribotype profiles consistent with C. difficile Ribotyping Network (CDRN) profiles.
Toxin gene carriage was determined using primers NK2, NK3, NK9, NK11, NK104, and NK105, which amplify the partial tcdA tcdA repeat region and partial tcdB gene (36, 37). Primers NK2, NK3, NK9, and NK11 were used in a multiplex reaction mixture in a 50-μl volume containing template DNA, 1 μM forward and reverse primers, 0.25 mM dNTPs, 1.5 mM MgCl2, 1× PCR buffer, and 0.5 U of BioTaq DNA polymerase. Primers NK104 and NK105 were used in PCRs in 50-μl volumes with template DNA, 2 μM forward and reverse primers, 0.25 mM dNTPs, 1.5 mM MgCl2, 1× PCR buffer, and 0.5 U of BioTaq DNA polymerase. Amplification conditions for all NK primers were 95°C for 5 min and 32 cycles of 95°C for 20 s, 62°C for 120 s, and 72°C for 2 min, with a final extension step at 72°C for 5 min. Also, carriage of the binary toxin genes cdtA and cdtB was determined by using primers cdtApos, cdtArev, cdtBpos, and cdtBrev (38). These primers were used in a 50-μl multiplex reaction mixture containing template DNA, 1 μM forward and reverse primers, 0.25 mM dNTPs, mM MgCl2, 1× PCR buffer, and 0.5 U of BioTaq DNA polymerase. PCR conditions were 95°C for 5 min and 30 cycles of 94°C for 45 s, 52°C for 60 s, and 72°C for 2 min, with a final extension step at 72°C for 5 min. Products were visualized alongside 10 μl of the 1-kbp GeneRuler DNA ladder. Samples were loaded on a 1% Helena agarose gel prepared in 1× TAE buffer and stained with GelRed. It was run at 90 V for 1 h in TAE buffer. Gels were imaged using Syngene software.
Phenotypic characterization of environmental C. difficile isolates by their motility and antibiotic susceptibility.
In order to characterize key physiological traits of the environmental strains, the motility of and antibiotic MICs for the 27 isolates were assessed. Motility was assessed following overnight growth in a low-percent-agar plate assay adapted from a method of Be'er et al. (39). Overnight liquid cultures were stabbed into 7 ml BHI–0.3% BA in 60-mm-diameter petri dishes and incubated overnight. Cultures were scored for the absence of or their levels of motile growth. Hazy growth was indicative of motility in soft-agar plate assays, and the absence of this hazy growth indicated that the organism was nonmotile. Assays were performed in triplicate from the same culture and repeated with fresh culture. Antibiotic MICs of vancomycin, metronidazole, clindamycin, and ciprofloxacin were measured using Etest strips (bioMérieux, United Kingdom) by following the manufacturer's guidelines for interpretation of results. A MIC breakpoint of 8 μg ml−1 was used to indicate resistance, and 4 μg ml−1 was used to indicate borderline resistance.
Prophage carriage as determined through TEM in induced and noninduced culture lysates of environmental isolates.
Liquid cultures were induced with either mitomycin (MC) at a concentration of 3 μg ml−1 or norfloxacin (NFX) at a concentration of 6 μg ml−1 or were exposed to no antibiotic treatment. Following overnight incubation, lysates were centrifuged at 3,400 × g for 10 min and the supernatant was filtered through 0.22-μm-pore-size Millipore filters. The lysates were further purified and concentrated using polyethylene glycol 6000 (PEG 6000). NaCl was added to the lysates to a final concentration of 0.5 M, and the lysates were mixed thoroughly and left on ice for 1 h. They were then centrifuged at 3,400 × g for 40 min at 4°C, and the supernatant was transferred to a sterile tube. Ten percent (wt/vol) added PEG 6000 was dissolved by rocking the tubes at room temperature for 1 h, followed by overnight incubation at 4°C. Phages were then precipitated at 3,400 × g for 40 min at 4°C and resuspended in SM buffer (0.1 M NaCl, 1 mM MgSO4, 0.2 M Tris-HCl, pH 7.5). Samples were treated with 1.4-μg μl−1 DNase and 3-μg μl−1 RNase and incubated for 1 h at 37°C. An equal volume of chloroform was added to each sample, and the tubes were centrifuged for 3,398 × g for 10 min at 4°C. The supernatant was stored at 4°C in the dark. Samples were viewed by transmission electron microscopy (TEM) on 0.25% Pioloform-coated copper electron microscopic grids (Athene type, 3 mm; Agar Scientific, United Kingdom) that had, before use, been carbon coated and subjected to high voltage to produce a glow discharge. Samples were negatively stained with 0.1% uranyl acetate and examined with a JEOL 1220 TEM (JEOL, United Kingdom).
Detection of myovirus diversity in environmental C. difficile lysogens.
Four sets of primers were designed to detect genes specific for different myoviruses in C. difficile isolates; they were based on (i) the capsid gene sequences from phiC2 (GenBank accession no. NC_009231) and phiCD119 (accession no. NC_007917) to differentiate two types of medium myoviruses (MMs), (ii) a candidate capsid gene in the genomes of phiCD27 (accession no. NC_011398) and phiMMP02 (accession no. NC_019421) to differentiate long-tailed myoviruses (LTMs), and (iii) the capsid gene of phiMMP04 (accession no. NC_019422) to differentiate small myoviruses (SMVs). The primers were designed using Primer3 on the basis of alignments of capsid nucleotide sequences determined with CLUSTALW (67). DNA was extracted using Chelex. We used two sets of target MM capsid sequences, MM1F (5′-AACTTCGGGGATTTGTATGC-3′) and MM1R (5′-CAACAAATTGTATTGCATCAGC-3′), which produce an expected product of 814 bp, and MM2F (5′-TGGTTGGATGGATTCTAATGCT-3′) and MM2R (5′-GACCAAGCATTTGCTGTTTG-3′), which produce an expected product of 771 bp. The CD27 group oligonucleotides LTMF (5′-GAGGGCAGGAATAAGAAAAGC-3′) and LTMR (5′-GATTCCCTATCCTCAACTACGC-3′) produce an expected product of 711 bp, and the SMV group oligonucleotides SMVF (5′-GAGCGGAAGTTCAAACAAGC-3′) and SMVR (5′-AGCAAGAATCTCGCCATCTG-3′) produce an expected product of 707 bp.
PCRs were performed for each primer set, individually, in a total volume of 25 μl containing template DNA, 4 μM forward and reverse primers, 0.25 mM dNTPs, 3 mM MgCl2, 1× PCR buffer, and 0.5 U of BioTaq DNA polymerase. Amplification conditions were 94°C for 5 min and 30 cycles of 94°C for 45 s, 48°C for 45 s, and 72°C for 60 s, with a final extension of 10 min at 72°C. Products were visualized on a 1% Helena agarose gel prepared in 1× TAE buffer and stained with ethidium bromide. Samples were loaded with 10 μl of the 1-kbp GeneRuler DNA ladder, and the gel was run at 90 V for 1 h in TAE buffer. Gels were imaged using Syngene software.
RESULTS
Isolation and ribotyping of C. difficile isolates from Hampshire samples from 2009 and 2010.
C. difficile was isolated from 54% (12/22) of the samples tested from 2009 and 60% (15/25) of the samples from 2010 (Table 1). All isolates were confirmed to be C. difficile using 16S rRNA gene-targeted PCR when each produced the expected ∼157-bp amplicon. Of the 19 sites sampled in both years, C. difficile was isolated from 11 in 2009 and 12 in 2010. There were three sites where C. difficile was not isolated from samples of either year, eight where C. difficile was isolated in samples from both years, and four where C. difficile was isolated only once. C. difficile was not isolated from any of the six water samples tested, which may be due either to sample type or to the absence of C. difficile at these sites. The bacterium was isolated from sites throughout the estuarine system regardless of assessed trophic status. The isolation of C. difficile from this environment indicates that its distribution is heterogeneous within sediments, as rates of recovery varied between proximal sites, as well as between years at the same site.
Table 1.
Sampling site locations and C. difficile isolate ribotypes, motility, and toxin gene carriagea
| Site | Source | Sample type | Statusb | 2009 |
2010 |
||||
|---|---|---|---|---|---|---|---|---|---|
| Ribotype | Motilityc | Toxin gene carriage | Ribotype | Motility | Toxin gene carriage | ||||
| 1 | Langstone Bridge | Sediment | 3 | -ve | 031 | +/− | A−B−CDT− | ||
| Foam | ND | 010 | −/− | A−B−CDT− | |||||
| 2 | Langstone Bridge | Sediment | 3 | -ve | -ve | ||||
| 3 | Langstone Bridge Bay | Sediment | 3 | 220 | +/+ | A+B+CDT− | 010 | +/− | A−B−CDT− |
| 4 | Ship Inn beach | Sediment | 3 | -ve | 001 | +/+ | A+B+CDT− | ||
| 5 | Sewage pipe | Sediment | 1 | 220 | ++/− | A+B+CDT− | 005 | +/− | A−B+CDT− |
| Sediment | ND | 078 | −/− | A+B+CDT+ | |||||
| Sediment | ND | 010 | −/− | A−B−CDT− | |||||
| Water | -ve | -ve | |||||||
| 6 | Bedhampton Creek | Sediment | 1 | 002 | ++/− | A+B+CDT− | -ve | ||
| 7 | Bosham Channel (west) | Sediment | 3 | -ve | 001 | ++/− | A−B+CDT− | ||
| 8 | Bosham Channel (east) | Sediment | 2 | -ve | N/A | −/− | A−B−CDT− | ||
| 9 | Thorney Island (coastal) | Sediment | 3 | 046 | −/− | A−B+CDT− | -ve | ||
| Anoxic layer of sediment | 014 | ++/− | A−B+CDT− | 031 | ++/++ | A−B−CDT− | |||
| 10 | Thorney Island (inland) | Sediment | 4 | 005 | ++/+ | A−B+CDT− | 220 | +/+ | A−B+CDT− |
| 11 | Emsworth Beach | Sediment | 3 | 012 | +/− | A−B+CDT− | -ve | ||
| 12 | Stoke boatyard | Sediment | 1 | 021 | +/+ | A−B+CDT− | 002 | ++/++ | A+B+CDT− |
| 13 | Stoke oyster beds | Sediment | 3 | NA | +/− | A−B−CDT− | -ve | ||
| 14 | Kench Beach (west) | Sediment | 3 | 027 | +/++ | A+B+CDT+ | 078 | ++/++ | A+B+CDT+ |
| Anoxic layer of sediment | -ve | 001 | ++/+ | A+B+CDT− | |||||
| 15 | Kench Beach (east) | Sediment | 3 | 001 | +/+ | A+B+CDT− | 106 | ++/− | A−B+CDT− |
| 16 | Kench boat homes | Sediment | 4 | 010 | −/− | A−B−CDT− | -ve | ||
| 17 | Kench boat homes (site 2) | Sediment | 3 | -ve | -ve | ||||
| 18 | Ferry point | Water | -ve | -ve | |||||
| 19 | Eastern Road bridge | Water | -ve | -ve | |||||
Abbreviations: ND, not done; NA, not assignable; -ve, negative for C. difficile; A, tcdA; B, tcdB; CDT, cdtA and cdtB.
Status was assigned based on multiple features, including the presence and absence of trophic indicator species, and ranged from 1 (worst) to 5 (least polluted).
++, highly motile growth; +, motile growth; −, no motile growth. The slash separates results of different assays.
To examine the genetic diversity of the isolates, their PCR ribotype profiles were generated. The distribution of C. difficile ribotypes varied both with location and year. In total, 15 different ribotypes were detected; 2 of these were not able to be assigned to a known profile. The ribotypes that we detected include those generally associated with environmental sources, such as R010, and those of clinical importance, including R027 and R078. The most frequently isolated ribotypes were R010 and R001, both of which were present at four sites. In terms of their temporal distribution, five ribotypes were isolated in samples from both years (R010, R220, R002, R001, and R005), five distinct ribotypes were isolated in samples from 2009 (R046, R014, R012, R021, and R027), and a further three were isolated in samples from 2010 (R078, R106, and R031). Different ribotypes were observed at each site between years, with one exception (at the sewage pipe [site 5]). In summary, these findings show that C. difficile in marine sediments is genetically diverse and that its presence varies with time and by location.
Isolates include atoxigenic strains that were negative for the targeted regions tcdA, tcdB, cdtA, and cdtB (A−B−CDT−), e.g., R010 isolates, tcdA- and tcdB-positive (A+B+CDT−) strains, e.g., R106 and R012 isolates, tcdB-only-positive (A−B+CDT−) strains, e.g., R005 isolates, and strains positive for all four genes (A+B+CDT+), e.g., R027 and R078 isolates. Two of the three isolates belonging to R220 had the same A+B+CDT− gene carriage, but the third was negative for tcdA and the repeat region.
Phenotypic characterization of individual C. difficile isolates reveals variation in motility and antibiotic susceptibility.
The environmental isolates showed considerable diversity in motility and exhibited no, low-motility, or high-motility growth (Table 1). Levels of motile growth differed for 9/27 isolates between repeats of the assay, but in total, 22 isolates showed motile growth in at least one assay.
To assess the levels of antibiotic resistance in our environmental isolates, the MICs of vancomycin, metronidazole, clindamycin, and ciprofloxacin were measured (Fig. 1) (also see Table S2 in the supplemental material). All 27 isolates were sensitive to metronidazole and vancomycin; 1 isolate, CD105HS26 (R078), had borderline resistance to vancomycin at a MIC of 4 μg ml−1, 5 isolates were resistant to clindamycin, with 1 being borderline resistant, and 12 isolates were ciprofloxacin resistant, with 7 being borderline resistant. The greatest ranges in MICs were observed for ciprofloxacin. Where multiple isolates representing a single ribotype were tested, for some ribotypes, both susceptible and resistant isolates were detected. For example, four isolates belonging to R001 include those which are resistant and sensitive to ciprofloxacin, whereas all the tested R010 isolates have borderline resistance to this antibiotic. The results show that there are both antibiotic-resistant and -susceptible strains present together in the environment.
Fig 1.
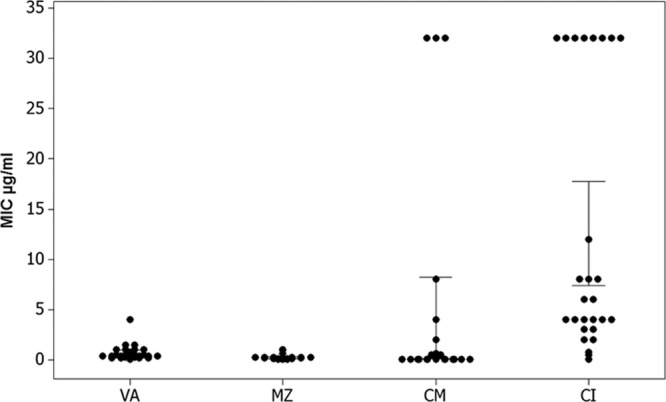
Individual value plot of MICs. MICs (on the y axis) for each antibiotic (on x axis) of the 27 isolates plotted in an individual value plot and analyzed in MiniTab. Abbreviations: VA, vancomycin; MZ, metronidazole; CM, clindamycin; CI, ciprofloxacin. MICS for isolates resistant to clindamycin and ciprofloxacin are >32 μg ml−1; in addition, there is greater variation in the MICs of these two antibiotics than in those of vancomycin and metronidazole.
High prevalence of lysogeny in environmental strains of C. difficile.
To determine if active prophages (those which can be released) are encoded in environmental strains of C. difficile, lysates from induced and noninduced cultures were examined for phage particles by TEM. The frequency of strains with phage particles observed in their lysates was 74% (20/27) (Table 2). Four main morphologies of C. difficile phage particles were observed across strains: medium myoviruses (MMs) (n = 9), long-tailed myoviruses (LTMs) (n = 3), small myoviruses (SMVs) (n = 3), and siphoviruses (SVs) (n = 7) (Fig. 2). Six isolates showed evidence of multiple-prophage carriage by the observation of phage particles belonging to different morphological groups. Additionally, 56% (15/27) of isolates produced particles resembling bacteriocins, termed phage-tail-like particles (PTLPs), often in lysates containing whole-phage particles (n = 7). There is considerable variation in prophage carriage within isolates of the same ribotype. For example, we observed R010 isolates (n = 4) with different phage particles of all four phage morphologies, LTMs and SMVs, SMVs and SVs, and MMs and LTMs. However, for isolates belonging to R001 (n = 4), only two morphologies were observed: SVs in one isolates and MMs in another. This suggests that prophage carriage may correlate only to specific ribotypes despite their genetic relatedness otherwise.
Table 2.
Lysogeny of strains according to the morphology of phage particles as observed by TEMa
| Ribotype | Isolate | Presence of: |
||||
|---|---|---|---|---|---|---|
| MMs | LTMs | SMVs | SVs | PTLPs | ||
| 010 | CD105HS14 | − | + | + | − | + |
| 010 | CD105HS15 | NAv | ||||
| 010 | CD105HS16 | − | − | + | + | − |
| 010 | CD105HS9 | + | + | − | − | + |
| 001 | CD105HS23 | − | − | − | + | − |
| 001 | CD105HS24 | NAv | ||||
| 001 | CD105HS25 | − | − | − | − | + |
| 001 | CD105HS12 | + | − | − | − | − |
| 220 | CD105HS22 | + | + | − | + | − |
| 220 | CD105HS2 | + | − | − | + | − |
| 220 | CD105HS6 | + | − | − | + | |
| 002 | CD105HS17 | − | − | − | + | + |
| 002 | CD105HS7 | − | − | − | + | − |
| 031 | CD105HS18 | − | − | − | − | + |
| 031 | CD105HS19 | − | − | − | − | + |
| 005 | CD105HS20 | − | − | − | − | + |
| 005 | CD105HS10 | + | − | − | − | + |
| 078 | CD105HS26 | NAv | ||||
| 078 | CD105HS27 | − | − | − | − | + |
| 046 | CD105HS3 | + | − | − | − | − |
| 014 | CD105HS4 | − | − | − | + | + |
| 021 | CD105HS5 | − | − | − | − | + |
| 027 | CD105HS8 | + | − | − | − | + |
| 012 | CD105HS1 | + | − | − | − | + |
| 106 | CD105HS21 | NAv | ||||
| NA | CD105HS28 | NAv | ||||
| NA | CD105HS11 | − | − | + | + | + |
All isolates were taken from sediment samples. Abbreviations: NA, not assignable, NAv, not available, MMs, medium myoviruses, LTMs, long-tailed myoviruses, SMVs, small myoviruses, SVs, siphoviruses, PTLPs, phage-tail-like particles.
Fig 2.
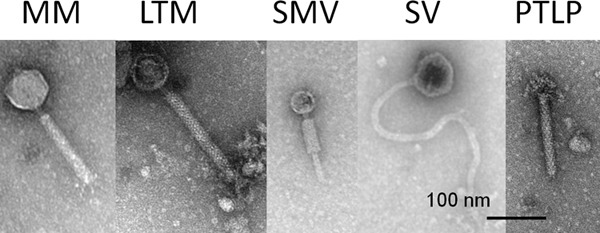
Morphological groups of temperate phages and phage-tail-like particles (PTLPs). Micrographs show representatives of the five main types of particle morphology observed: medium myoviruses (MMs), long-tailed myoviruses (LTMs), small myoviruses (SMVs), siphoviruses (SVs), and PTLPs. Assignation of the myovirus particles was made according to the capsid diameter and tail length; MMs had capsid diameters of 60 to 70 nm and contractile tail lengths of approximately ∼130 nm, LTMs had capsid diameters of 60 to 70 nm with tail lengths of 150 to 220 nm, and SMVs had capsid diameters of 40 to 50 nm and tail lengths of ∼105 nm. Bars represent 100 nm.
Prophages could be released by different inducing agents, namely, MC and NFX, and also when no inducing agent was used (see Table S2 in the supplemental material). For five isolates, all methods were successful for phage particle production (CD105HS9, CD105HS22, CD105HS10, CD105HS8, and CD105HS1). For three isolates, phage particles (MMs) or PTLPs were observed only following antibiotic induction (CD105HS3, CD105HS6, and CD105HS5), and for two isolates, phage particles (SVs) were observed following treatment with MC and/or in uninduced cultures (CD105HS4 and CD105HS7).
The diversity of myovirus carriage was confirmed by using primers to amplify genes specific to the three different groups: the MMs, LTMs, and SMVs (Table 3). The PCR assay was largely consistent with results from the TEM analysis, with phage genes being detected at a high frequency (81%) and with MMs being the most commonly detected (24/27). However, the use of two different primers targeting this group revealed genetic diversity that TEM could not show. A comparison of the PCR assays versus TEM found that in 7 cases, both sets of results were consistent. However, in 8 cases, the PCR assays amplified markers where no phages of a corresponding morphology were observed in the TEM analysis. In contrast, in 2 cases, the TEM showed phage particles which did not have a corresponding marker detected in the PCR assay, and in 2 cases, conflicting results were obtained for each assay.
Table 3.
Myovirus capsid gene detection as a marker of prophage diversitya
| Ribotype | Isolate | Presence of capsid gene from: |
|||
|---|---|---|---|---|---|
| MM1 | MM2 | LTM | SMV | ||
| 010 | CD105HS14 | + | + | + | |
| 010 | CD105HS15 | + | + | ||
| 010 | CD105HS16 | + | |||
| 010 | CD105HS9 | + | + | + | |
| 001 | CD105HS23 | + | |||
| 001 | CD105HS24 | + | + | ||
| 001 | CD105HS25 | + | |||
| 001 | CD105HS12 | ||||
| 220 | CD105HS22 | + | |||
| 220 | CD105HS2 | + | |||
| 220 | CD105HS6 | + | |||
| 002 | CD105HS17 | NAv | |||
| 002 | CD105HS7 | + | + | ||
| 031 | CD105HS18 | ||||
| 031 | CD105HS19 | ||||
| 005 | CD105HS20 | + | |||
| 005 | CD105HS10 | + | + | ||
| 078 | CD105HS26 | + | |||
| 078 | CD105HS27 | ||||
| 046 | CD105HS3 | + | |||
| 014 | CD105HS4 | + | |||
| 021 | CD105HS5 | + | |||
| 027 | CD105HS8 | + | |||
| 012 | CD105HS1 | + | + | ||
| 106 | CD105HS21 | ||||
| NA | CD105HS28 | + | |||
| NA | CD105HS11 | + | |||
All isolates were from sediment samples. NA, not assignable; NAv, not available; MM1, medium myovirus group 1; MM2, medium myovirus group 2; LTM, long-tailed myovirus; SMV, small myovirus; +, positive.
DISCUSSION
In this study, C. difficile was isolated from sites located throughout the Langstone and Chichester Harbor estuarine system. Previously, C. difficile was found both free in marine water columns (16, 21) and in marine organisms, including plankton, shellfish, fish, otters, and a seal (21, 40–42). Detection of C. difficile in these organisms may arise following exposure to spores in contaminated water or by transmission through the food chain in marine ecosystems. Isolation of the bacterium from estuarine sediments has implications for fishery industries as well as tourism and for recreational use of these environments. The isolation of C. difficile from sediments highlights a new reservoir of this bacterium that may be involved in the bacterium's abundance, origin, transmission, and persistence in the marine ecosystem.
The presence of C. difficile in the environment may be due to contamination from infected hosts or sewage effluent (18, 23, 41). Although the sewage pipes in Langstone Harbor are currently nonoperational, they still serve as storm overflows and may be one source of C. difficile into the estuary system (43). Another source may be agricultural runoff, as the harbor areas sampled in this study have a known problem with agricultural runoff; fertilizer input has partly caused the area's ongoing problems with toxic algal overgrowth within the estuary (44). C. difficile has been previously detected in two different types of treated biosolids which are routinely used as agricultural fertilizers (45). Further evidence of input from agricultural runoff and the anthropomorphic effects of urbanization that affect bacterial presence comes from a Californian study of sea otters infected with C. difficile. It was observed that greater general bacterial loading, including that of C. difficile, was found in otter fecal samples at sewage inputs and interface areas of fresh and marine water (41). Similarly, some of the sites in Langstone Harbor where we isolated C. difficile also have excessive loading of coliform bacteria, particularly around the sewage pipe and Langstone Bridge sites sampled in this study (46). This strongly suggests that at least some of the C. difficile in this system may have originated from sewage or agricultural runoff and that it remains viable within the sediments.
From our ribotype analysis, we see a diverse set of strains present in the sediments that include strains of ribotypes that are associated with environmental or clinical sources, for example, R010 or R027, respectively (20, 47, 48). R010 and R001 were the most frequently isolated ribotypes in our study, and while R010 is not commonly isolated from United Kingdom clinical samples (48), it has been found in community samples, human wastewater, and other environmental sources (16, 25, 49).
The ribotype analysis does not indicate where C. difficile enters this estuarine system. While some clinical ribotypes, such as R078 and R027, were isolated from sites linked to anthropomorphic inputs, such as the sewage pipe (site 5) samples and the Kench boat homes (site 16) samples, R010 was isolated from the sewage pipe site in addition to sites not associated with potential sewage contamination. Furthermore, these sites also yielded ribotypes that are clinically prevalent, including R005, R046, R014, and R031. Additionally, we show that these isolates carry different toxin genes and that they include atoxigenic strains as well as those in which all four toxin genes were detected.
The prevalence of specific ribotypes may be due to different factors, such as their viability in the environment, or may reflect their relative abundances in inflow from contaminating inputs (e.g., sewage or agriculture runoff). We found that ribotypes can differ by site and year, which suggests that the C. difficile population in the estuarine sediments may be transient, similar to that of other environments, such as soil (22) and river water (20). It is unclear whether C. difficile environmental isolates exist as spores (perhaps under superdormancy) or are active in the sediments. The distribution of C. difficile isolates found in this study could also result from the movement of spores or cells throughout the estuarine system during the tidal cycles. Additionally, the turnover of strains may result from active growth in heterogeneous populations. The isolation of novel types from the environment could be new ecotypes or environmentally adapted strains. Studies to investigate this further are ongoing in our laboratory.
Motility may be involved in the bacterium's pathogenicity (50) and may aid the bacterium's adaptation to its environment (51). Our assays showed that some isolates produced nonmotile growth and others motile growth, which is consistent with previous observations (8, 52). Few strains of C. difficile have been designated nonmotile; for example, one study found that while 17 strains appeared to be nonflagellated, all expressed the flagellar gene fliC (53), but some strains, such as M120, lack the entire flagellin-encoding region F3 (8). Variability in the presence of the flagellin genes (fliC and fliD) indicates that they undergo recombination events and/or are subject to environmental selective pressures to evolve (54). We also showed that some isolates had a variable motility. This observed switch between motile and nonmotile growth between assays is consistent with work by Twine et al. (52), who suggest that motility may be phase variable. The variability in motile growth does demonstrate the diversity and plasticity of this trait in environmental isolates.
Populations of environmental bacteria are widely thought to serve as genetic reservoirs or “resistomes” for antibiotic resistance genes, which are then transferrable to pathogenic bacteria (see, e.g., reference 55). Previous work has shown that resistance to antibiotics used to treat C. difficile, i.e., vancomycin and metronidazole, is rare, but resistance to fluoroquinolones, such as ciprofloxacin, and to clindamycin, which was previously associated with CDI, is prevalent both in clinical isolates (56) and in nonclinical isolates (57). Our results show similar ranges of MICs, from sensitive to fully resistant, of clindamycin and ciprofloxacin and only one isolate for which the MIC is suggestive of moderate susceptibility to vancomycin. No definite correlation between MIC and ribotype was observed, but a greater sample size could determine if this is indeed the case. Although we did not observe resistance to metronidazole or vancomycin, it has been suggested that subinhibitory concentrations of antibiotics affect the ability of strains to colonize (15), and therefore, the presence of multiple fluoroquinolone-resistant isolates in this study may be of clinical relevance if there is a transmission route from the sediments back to human hosts.
An abundant and diverse prophage carriage is found within these environmental isolates, attesting to the facts that phage infection is a constituent part of their biology and that released phages have access to different strains in this environment. The frequency of detected prophage carriage in the examined C. difficile strains is comparable to previous rates determined in clinical and human isolates (29, 58, 59). We found that the most common morphology observed was that of myoviruses, which are frequently detected in other studies of this species (28, 29). Also consistent with previous work was the observation of bacteriocin-like particles (PTLPs) in several lysates.
The use of TEM to assess prophage carriage depends on the titers of phages present in a sample, and there is the chance that both prevalence and diversity may be underestimated for some strains. However, our results correspond to published accounts for a large screening of clinical strains (29), as does our finding that specific ribotypes encode different prophages (60, 61). The spontaneous release of C. difficile phages and their differential release by use of different antibiotics against clinical strains have been described previously (60, 62).
The PCR screen for known C. difficile myoviruses reveals that the isolates have genetically recognizable prophages. The disparate results from the TEM and PCR assays may be due to differences in the targeted gene sequences, which may explain our observation of a phage particle in the TEM analysis but no PCR product, or to the amplification of nonactive, cryptic, prophage elements, which may explain the cases observed in the TEM analysis where isolates produced an amplicon but no corresponding phage particle. Importantly, the known phages used as a basis for the PCR screen are all able to infect multiple strains of C. difficile (62–66). Detection of genes in the environmental isolates which are homologous to genes of infective phages suggests that these isolates may encode similar infective phages. Strains from the environment with the ability to spontaneously release phages may therefore have an impact on horizontal gene transfer within this system, depending on the activities of the strains in this environment and the infectivities of released phages.
To conclude, C. difficile strains are present in marine sediments, and they are genetically distinct and physiologically diverse in terms of antibiotic susceptibility, motility, and toxin gene carriage, with an abundant and morphologically diverse phage carriage.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by an MRC New Investigator Award (G0700855) given to M.R.J.C.
We thank Stefan Hyman and Natalie Allcock from the Advanced Microscopy Centre, University of Leicester, for their support and expertise and Julie Pratt for her useful comments on the manuscript.
Footnotes
Published ahead of print 2 August 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01849-13.
REFERENCES
- 1.Ananthakrishnan A. 2011. Clostridium difficile infection: epidemiology, risk factors and management. Nat. Rev. Gastroenterol. Hepatol. 8:17–26 [DOI] [PubMed] [Google Scholar]
- 2.Bouza E. 2012. Consequences of Clostridium difficile infection: understanding the healthcare burden. Clin. Microbiol. Infect. 18:5–12 [DOI] [PubMed] [Google Scholar]
- 3.Rupnik M, Wilcox MH, Gerding DN. 2009. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat. Rev. Microbiol. 7:526–536 [DOI] [PubMed] [Google Scholar]
- 4.Gerding DN. 2004. Clindamycin, cephalosporins, fluoroquinolones, and Clostridium difficile-associated diarrhea: this is an antimicrobial resistance problem. Clin. Infect. Dis. 38:646–648 [DOI] [PubMed] [Google Scholar]
- 5.Kuijper EJ, Wilcox MH. 2008. Decreased effectiveness of metronidazole for the treatment of Clostridium difficile infection? Clin. Infect. Dis. 47:63–65 [DOI] [PubMed] [Google Scholar]
- 6.Figueroa I, Johnson S, Sambol SP, Goldstein EJC, Citron DM, Gerding DN. 2012. Relapse versus reinfection: recurrent Clostridium difficile infection following treatment with fidaxomicin or vancomycin. Clin. Infect. Dis. 55:S104–S109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sebaihia M, Wren B, Mullany P, Fairweather N, Minton N, Stabler R, Thomson N, Roberts A, Cerdeno-Tarrraga A, Wang H, Holden MT, Wright A, Churcher C, Quail MA, Baker S, Bason N, Brooks K, Chillingworth T, Cronin A, Davis P, Dowd L, Fraser A, Feltwell T, Hance Z, Holroyd S, Jagels K, Moule S, Mungall K, Price C, Rabbinowitsch E, Sharp S, Simmonds M, Stevens K, Unwin L, Whithead S, Dupuy B, Dougan G, Barrell B, Parkhill J. 2006. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat. Genet. 38:779–786 [DOI] [PubMed] [Google Scholar]
- 8.Stabler R, He M, Dawson L, Martin M, Valiente E, Corton C, Lawley T, Sebaihia M, Quail M, Rose G, Gerding DN, Gibert M, Popoff MR, Parkhill J, Dougan G, Wren BW. 2009. Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. Genome Biol. 10(9):R102. 10.1186/gb-2009-10-9-r102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vedantam G, Clark A, Chu M, McQuade R, Mallozzi M, Viswanathan VK. 2012. Clostridium difficile infection toxins and non-toxin virulence factors, and their contributions to disease establishment and host response. Gut Microbes 3:121–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cartwright CP, Stock F, Beekmann SE, Williams EC, Gill VJ. 1995. PCR amplification of ribosomal-RNA intergenic spacer regions as a method for epidemiologic typing of Clostridium difficile. J. Clin. Microbiol. 33:184–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.ONeill G, Ogunsola F, Brazier J, Duerden B. 1996. Modification of a PCR ribotyping method for application as a routine typing scheme for Clostridium difficile. Anaerobe 2:205–209 [Google Scholar]
- 12.Indra A, Huhulescu S, Schneeweis M, Hasenberger P, Kernbichler S, Fiedler A, Wewalka G, Allerberger F, Kuijper EJ. 2008. Characterization of Clostridium difficile isolates using capillary gel electrophoresis-based PCR ribotyping. J. Med. Microbiol. 57:1377–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goorhuis A, Bakker D, Corver J, Debast S, Harmanus C, Notermans D, Bergwerff A, Dekker F, Kuijper E. 2008. Emergence of Clostridium difficile infection due to a new hypervirulent strain, polymerase chain reaction ribotype 078. Clin. Infect. Dis. 47:1162–1170 [DOI] [PubMed] [Google Scholar]
- 14.McDonald L, Killgore G, Thompson A, Owens R, Kazakova S, Sambol S, Johnson S, Gerding D. 2005. An epidemic, toxin gene-variant strain of Clostridium difficile. N. Engl. J. Med. 353:2433–2441 [DOI] [PubMed] [Google Scholar]
- 15.He M, Miyajima F, Roberts P, Ellison L, Pickard DJ, Martin MJ, Connor TR, Harris SR, Fairley D, Bamford KB, D'Arc S, Brazier J, Brown D, Coia JE, Douce G, Gerding D, Kim HJ, Koh TH, Kato H, Senoh M, Louie T, Michell S, Butt E, Peacock SJ, Brown NM, Riley T, Songer G, Wilcox M, Pirmohamed M, Kuijper E, Hawkey P, Wren BW, Dougan G, Parkhill J, Lawley TJ. 2013. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat. Genet. 45:109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.al Saif N, Brazier JS. 1996. The distribution of Clostridium difficile in the environment of South Wales. J. Med. Microbiol. 45:133–137 [DOI] [PubMed] [Google Scholar]
- 17.Brazier JS, Borriello SP. 2000. Microbiology, epidemiology and diagnosis of Clostridium difficile infection. Curr. Top. Microbiol. Immunol. 250:1–33 [DOI] [PubMed] [Google Scholar]
- 18.Baverud V, Gustafsson A, Franklin A, Aspan A, Gunnarsson A. 2003. Clostridium difficile: prevalence in horses and environment, and antimicrobial susceptibility. Equine Vet. J. 35:465–471 [DOI] [PubMed] [Google Scholar]
- 19.del Mar Gamboa M, Rodriguez E, Vargas P. 2005. Diversity of mesophilic clostridia in Costa Rican soils. Anaerobe 11:322–326 [DOI] [PubMed] [Google Scholar]
- 20.Zidaric V, Beigot S, Lapajne S, Rupnik M. 2010. The occurrence and high diversity of Clostridium difficile genotypes in rivers. Anaerobe 16:371–375 [DOI] [PubMed] [Google Scholar]
- 21.Pasquale V, Romano VJ, Rupnik M, Dumontet S, Ciznar I, Aliberti F, Mauri F, Saggiomo V, Krovacek K. 2011. Isolation and characterization of Clostridium difficile from shellfish and marine environments. Folia Microbiol. (Praha) 56:431–437 [DOI] [PubMed] [Google Scholar]
- 22.Higazi T, Al-Saghir M, Burkett M, Pusok R. 2011. PCR detection of Clostridium difficile and its toxigenic strains in public places in southern Ohio. Int. J. Microbiol. Res. 2:105–111 [Google Scholar]
- 23.Simango C. 2006. Prevalence of Clostridium difficile in the environment in a rural community in Zimbabwe. Trans. R. Soc. Trop. Med. Hyg. 100:1146–1150 [DOI] [PubMed] [Google Scholar]
- 24.Janezic S, Ocepek M, Zidaric V, Rupnik M. 2012. Clostridium difficile genotypes other than ribotype 078 that are prevalent among human, animal and environmental isolates. BMC Microbiol. 12:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brazier JS. 2001. Typing of Clostridium difficile. Clin. Microbiol. Infect. 7:428–431 [DOI] [PubMed] [Google Scholar]
- 26.Dingle KE, Griffiths D, Didelot X, Evans J, Vaughan A, Kachrimanidou M, Stoesser N, Jolley KA, Golubchik T, Harding RM, Peto TE, Fawley W, Walker AS, Wilcox M, Crook DW. 2011. Clinical Clostridium difficile: clonality and pathogenicity locus diversity. PLoS One 6:e19993. 10.1371/journal.pone.0019993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stabler R, Dawson L, Valiente E, Cairns M, Martin M, Donahue E, Riley T, Songer J, Kuijper E, Dingle K, Wren BW. 2012. Macro and micro diversity of Clostridium difficile isolates from diverse sources and geographical locations. PLoS One 7:e31559. 10.1371/journal.pone.0031559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagy E, Foldes J. 1991. Electron microscope investigation of lysogeny of Clostridium difficile strains isolated from antibiotic-associated diarrhea cases and from healthy carriers. APMIS 99:321–326 [DOI] [PubMed] [Google Scholar]
- 29.Shan J, Patel K, Hickenbotham P, Nale J, Hargreaves K, Clokie M. 2012. Prophage carriage and diversity within clinically relevant strains of Clostridium difficile. Appl. Environ. Microbiol. 78:6027–6034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stabler R, Gerding D, Songer J, Drudy D, Brazier J, Trinh H, Witney A, Hinds J, Wren B. 2006. Comparative phylogenomics of Clostridium difficile reveals clade specificity and microevolution of hypervirulent strains. J. Bacteriol. 188:7297–7305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sell T, Schaberg O, Fekety F. 1983. Bacteriophage and bacteriocin typing scheme for Clostridium difficile. J. Clin. Microbiol. 17:1148–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clokie JJP, Boney AD. 1983. Marine algae and beach management in relation to nearby reclamation and construction work: the conservation of amenity. Biol. Conserv. 26:239–254 [Google Scholar]
- 33.Wilson KH, Kennedy MJ, Fekety FR. 1982. Use of sodium taurocholate to enhance spore recovery on a medium selective for Clostridium difficile. J. Clin. Microbiol. 15:443–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rinttila T, Kassinen A, Malinen E, Krogius L, Palva A. 2004. Development of an extensive set of 16S rDNA-targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR. J. Appl. Microbiol. 97:1166–1177 [DOI] [PubMed] [Google Scholar]
- 35.Griffiths D, Fawley W, Kachrimanidou M, Bowden R, Crook DW, Fung R, Golubchik T, Harding RM, Jeffery KJM, Jolley KA. 2010. Multilocus sequence typing of Clostridium difficile. J. Clin. Microbiol. 48:770–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kato N, Ou CY, Kato H, Bartley SL, Brown VK, Dowell VR, Ueno K. 1991. Identification of toxigenic Clostridium difficile by the polymerase chain reaction J. Clin. Microbiol. 29:33–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kato H, Kato N, Watanabe K, Iwai N, Nakamura H, Yamamoto T, Suzuki K, Kim SM, Chong Y, Wasito EB. 1998. Identification of toxin A-negative, toxin B-positive Clostridium difficile by PCR. J. Clin. Microbiol. 36:2178–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stubbs S, Rupnik M, Gibert M, Brazier J, Duerden B, Popoff M. 2000. Production of actin-specific ADP-ribosyltransferase (binary toxin) by strains of Clostridium difficile. FEMS Microbiol. Lett. 186:307–312 [DOI] [PubMed] [Google Scholar]
- 39.Be'er A, Smith RS, Zhang HP, Florin EL, Payne SM, Swinney HL. 2009. Paenibacillus dendritiformis bacterial colony growth depends on surfactant but not on bacterial motion. J. Bacteriol. 191:5758–5764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Metcalf D, Avery BP, Janecko N, Matic N, Reid-Smith R, Weese JS. 2011. Clostridium difficile in seafood and fish. Anaerobe 17:85–86 [DOI] [PubMed] [Google Scholar]
- 41.Miller MA, Byrne BA, Jang SS, Dodd EM, Dorfmeier E, Harris MD, Ames J, Paradies D, Worcester K, Jessup DA, Miller WA. 2010. Enteric bacterial pathogen detection in southern sea otters (Enhydra lutris nereis) is associated with coastal urbanization and freshwater runoff. Vet. Res. 41:1. 10.1051/vetres/200904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McBee RH. 1960. Intestinal flora of some Antarctic birds and mammals. J. Bacteriol. 79:311–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Travis J. 4 October 2012. Investigation into water company after Langstone Harbour sewage spill. In The news. Johnston Press, Portsmouth, United Kingdom: http://www.portsmouth.co.uk/news/local/investigation-into-water-company-after-langstone-harbour-sewage-spill-1-4331144 [Google Scholar]
- 44.United Kingdom Environmental Agency (ed). 2010. Fact sheet: an environmental summary of Portsmouth. Environmental Agency, Rotherham, United Kingdom [Google Scholar]
- 45.Viau E, Peccia J. 2009. Survey of wastewater indicators and human pathogen genomes in biosolids produced by class A and class B stabilization treatments. Appl. Environ. Microbiol. 75:164–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Langstone Harbour Board 2008. Langstone Harbour management plan, p 1–96 Langstone Harbour Board, Langstone Harbour, United Kingdom [Google Scholar]
- 47.Al-Saif N, O'Neill G, Magee J, Brazier J, Duerden B. 1998. PCR-ribotyping and pyrolysis mass spectrometry fingerprinting of environmental and hospital isolates of Clostridium difficile. J. Med. Microbiol. 47:117–121 [DOI] [PubMed] [Google Scholar]
- 48.Wilcox MH, Shetty N, Fawley WN, Shemko M, Coen P, Birtles A, Cairns M, Curran MD, Dodgson KJ, Green SM, Hardy KJ, Hawkey PM, Magee JG, Sails AD, Wren MW. 2012. Changing epidemiology of Clostridium difficile infection following the introduction of a national ribotyping-based surveillance scheme in England. Clin. Infect. Dis. 55:1056–1063 [DOI] [PubMed] [Google Scholar]
- 49.Romano V, Pasquale V, Krovacek K, Mauri F, Demarta A, Dumontet S. 2012. Toxigenic Clostridium difficile PCR ribotypes from wastewater treatment plants in southern Switzerland. Appl. Environ. Microbiol. 78:6643–6646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aubry A, Hussack G, Chen WX, KuoLee R, Twine SM, Fulton KM, Foote S, Carrillo CD, Tanha J, Logan SM. 2012. Modulation of toxin production by the flagellar regulon in Clostridium difficile. Infect. Immun. 80:3521–3532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Borriello S. 1998. Pathogenesis of Clostridium difficile infection. J. Antimicrob. Chemother. 41:13–19 [DOI] [PubMed] [Google Scholar]
- 52.Twine SM, Reid CW, Aubry A, McMullin DR, Fulton KM, Austin J, Logan SM. 2009. Motility and flagellar glycosylation in Clostridium difficile. J. Bacteriol. 191:7050–7062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tasteyre A, Karjalainen T, Avesani V, Delmee M, Collignon A, Bourlioux P, Barc MC. 2000. Phenotypic and genotypic diversity of the flagellin gene (fliC) among Clostridium difficile isolates from different serogroups. J. Clin. Microbiol. 38:3179–3186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lemee L, Bourgeois I, Ruffin E, Collignon A, Lemeland JF, Pons JL. 2005. Multilocus sequence analysis and comparative evolution of virulence-associated genes and housekeeping genes of Clostridium difficile. Microbiology 151:3171–3180 [DOI] [PubMed] [Google Scholar]
- 55.Wright GD. 2010. Antibiotic resistance in the environment: a link to the clinic? Curr. Opin. Microbiol. 13:589–594 [DOI] [PubMed] [Google Scholar]
- 56.Wilcox MH, Fawley W, Freeman J, Brayson J. 2000. In vitro activity of new generation fluoroquinolones against genotypically distinct and indistinguishable Clostridium difficile isolates. J. Antimicrob. Chemother. 46:551–556 [DOI] [PubMed] [Google Scholar]
- 57.Norman KN, Harvey RB, Scott HM, Hume ME, Andrews K, Brawley AD. 2009. Varied prevalence of Clostridium difficile in an integrated swine operation. Anaerobe 15:256–260 [DOI] [PubMed] [Google Scholar]
- 58.Goh S, Riley T, Chang B. 2005. Isolation and characterization of temperate bacteriophages of Clostridium difficile. Appl. Environ. Microbiol. 71:1079–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fortier L, Moineau S. 2007. Morphological and genetic diversity of temperate phages in Clostridium difficile. Appl. Environ. Microbiol. 73:7358–7366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nale J, Shan J, Hickenbotham P, Fawley W, Wilcox M, Clokie M. 2012. Diverse temperate bacteriophage carriage in Clostridium difficile 027 strains. PLoS One 7:e37263. 10.1371/journal.pone.0037263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.He M, Sebaihia M, Lawley TD, Stabler RA, Dawson LF, Martin MJ, Holt KE, Seth-Smith HMB, Quail MA, Rance R, Brooks K, Churcher C, Harris D, Bentley SD, Burrows C, Clark L, Corton C, Murray V, Rose G, Thurston S, van Tonder A, Walker D, Wren BW, Dougan G, Parkhill J. 2010. Evolutionary dynamics of Clostridium difficile over short and long time scales. Proc. Natl. Acad. Sci. U. S. A. 107:7527–7532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meessen-Pinard M, Sekulovic O, Fortier L. 2012. Evidence of in vivo prophage induction during Clostridium difficile infection. Appl. Environ. Microbiol. 78:7662–7670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Govind R, Fralick J, Rolfe R. 2006. Genomic organization and molecular characterization of Clostridium difficile bacteriophage PhiCD119. J. Bacteriol. 188:2568–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Goh S, Ong P, Song K, Riley T, Chang B. 2007. The complete genome sequence of Clostridium difficile phage phiC2 and comparisons to phiCD119 and inducible prophages of CD630. Microbiology 153:676–685 [DOI] [PubMed] [Google Scholar]
- 65.Mayer M, Narbad A, Gasson M. 2008. Molecular characterization of a Clostridium difficile bacteriophage and its cloned biologically active endolysin. J. Bacteriol. 190:6734–6740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sekulovic O, Meessen-Pinard M, Fortier L. 2011. Prophage-stimulated toxin production in Clostridium difficile NAP1/027 lysogens. J. Bacteriol. 193:2726–2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. ClustalW and ClustalX version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


