Abstract
The circulation of human parechoviruses (HPeVs) in the population was studied by environmental surveillance comprising of molecular analyses of sewage samples (n = 89) that were collected from 15 different locations in the Netherlands. Samples were taken from sewage originating from schools (n = 9) or from parts of municipalities (n = 6) during the Dutch school year 2010-2011. At 13/15 locations HPeV1, HPeV3, or HPeV6 RNA was detected at least once; however, sequence diversity did not reflect associations in time or place. A higher percentage of positives was observed in the samples originating from the municipalities. It was demonstrated that HPeV circulated in the studied population to a higher extent than would be expected from the current knowledge on infections predominating in young children.
INTRODUCTION
Human parechoviruses (HPeVs) are small, nonenveloped, single-stranded RNA viruses belonging to the family Picornaviridae. Although infections with these viruses in humans are often asymptomatic (1), they can also cause mild symptoms such as diarrhea and flu-like disease (2). Occasionally, more severe disease outcomes such as meningitis, sepsis, and paralysis occur (3–6). Parechovirus infections are predominantly seen in children. Especially neonates are at increased risk for more severe disease due to their immature immune systems. HPeVs cause infections in the central nervous system, and particularly HPeV3 but occasionally also HPeV1 may lead to severe sequelae (3, 7, 8). The burden of disease caused by HPeV infection is probably still underestimated.
Although HPeV infections are globally endemic throughout the year, a different periodical occurrence of infections is observed for the different types of HPeV. In temperate climates, HPeV3 infections are more particularly noted in summer and fall, whereas the other types of HPeV circulate more in fall and winter (8–10). Human parechoviruses, like other enteric viruses, are assumed to be transmitted by the fecal-oral route. Upon infection, these viruses can multiply in the gastrointestinal but also in the respiratory tract. Infected individuals, both symptomatic and asymptomatic, can shed these viruses in large amounts in their feces (11) that finally might end up in the environment. Furthermore, viruses present in sewage, directly or after treatment, are discharged onto surface waters and may spread through the water. Subsequently, humans may be exposed to those viruses because surface water is used as source water for the production of drinking water, for recreational purposes, for irrigation of crops, and for shellfish cultivation for human consumption.
Human parechovirus types 1 and 2, the former enteroviruses echoviruses 22 and 23, were previously reassigned to a new picornavirus genus, Parechovirus (12, 13). Since then, 14 other types (HPeV3 to -16) have been discovered (www.picornaviridae.com/parechovirus/parechovirus.htm). The genome organization of HPeV consists of two uncoding regions (5′ end and 3′ end) and a single open reading frame that encodes a polyprotein, cleaved to give the individual structural (N-terminal part of the polyprotein) and nonstructural (C-terminal part) proteins. Parechoviruses, like most RNA viruses, have a high mutation rate due to the lack of proofreading activity during genome replication, and also recombination events occur frequently. As a result, HPeVs show rapid sequence changes over time (14).
Classical HPeV detection methods are based on virus isolation by cell culture, followed by virus identification by antigenic typing methods and molecular typing. More recently, direct molecular virus detection methods have increasingly been applied in clinical and environmental virology studies, because of their sensitivity, specificity, and ability to relatively rapidly detect a large group of viruses. Many reverse transcription-PCR (RT-PCR) assays have been described for HPeV detection that target the highly conserved 5′ untranslated region (5′ UTR) (15–18). Although useful for detection, the 5′ UTR sequence does not provide enough sequence information to type the detected virus. The part of the HPeV genome encoding the structural proteins, such as VP1 or the VP3/VP1 junction, is more appropriate for typing purposes (10, 17, 18). Because of the high sequence variability in these structural regions, it is difficult to design primers that can detect all the HPeV types with sufficient sensitivity, and highly degenerated primers are needed to obtain sufficient specificity. Especially in environmental samples, where the virus concentrations are generally low and many PCR-inhibitory substances are present, sensitivity of the PCR may be a complicating factor. Identification of circulating HPeVs in the human population may help in understanding the epidemiology of HPeV infections, which may contribute in obtaining a better understanding of the burden of disease caused by parechoviruses. To be able to monitor the circulation of HPeV, it is important to detect viruses from both asymptomatic and symptomatic infected individuals. This can be achieved by environmental surveillance for the presence of enteric viruses in sewage. Environmental surveillance programs are frequently used to monitor circulation of wild-type poliovirus (19, 20) and of vaccine-derived polioviruses (21–23). Early detection of poliovirus circulation is important, especially in a susceptible population, to prevent further spread and possible new poliomyelitis cases. In the Netherlands, environmental poliovirus surveillance is done in an area where many unvaccinated individuals, due to religious beliefs, live close together in the so-called Bible belt (24). The presence of poliovirus and other enteric pathogens, such as human parechoviruses, detected in the sewage by environmental surveillance is representative of the circulation of these viruses in the sampled population (25).
In this study, we determined which HPeV types circulated in sewage to obtain a high coverage of a large part of the population with a relative easy applicable and sensitive method. The detected viruses were typed by sequence and phylogenetic analysis. The number of positive samples obtained from the different sampling locations, schools and municipalities, and the detected sequence diversity were compared to each other and to the known sequences in GenBank. These data may contribute to a better understanding of the HPeV circulation in the Dutch population and may also aid in developing strategies to prevent these HPeV infections.
MATERIALS AND METHODS
Sewage samples.
As part of the environmental poliovirus surveillance in the Netherlands, 1-liter grab sewage samples were taken in the Bible belt area, where many people that have not been vaccinated against poliomyelitis on religious grounds live close together. In April 2010 (pilot samples) and during the Dutch 2010-2011 school year, sewage samples were collected from 15 locations, either from sewer drains with sewage directly originating from schools where a high percentage of children with a Reformed upbringing attend or from sewer drains in a municipality with a high percentage of Reformed individuals (Fig. 1). The sampling sites were divided into two sampling routes with an interval scheme, resulting in six complete sampling sets (April 2010 and from September 2010 until June 2011) (Table 1).
Fig 1.
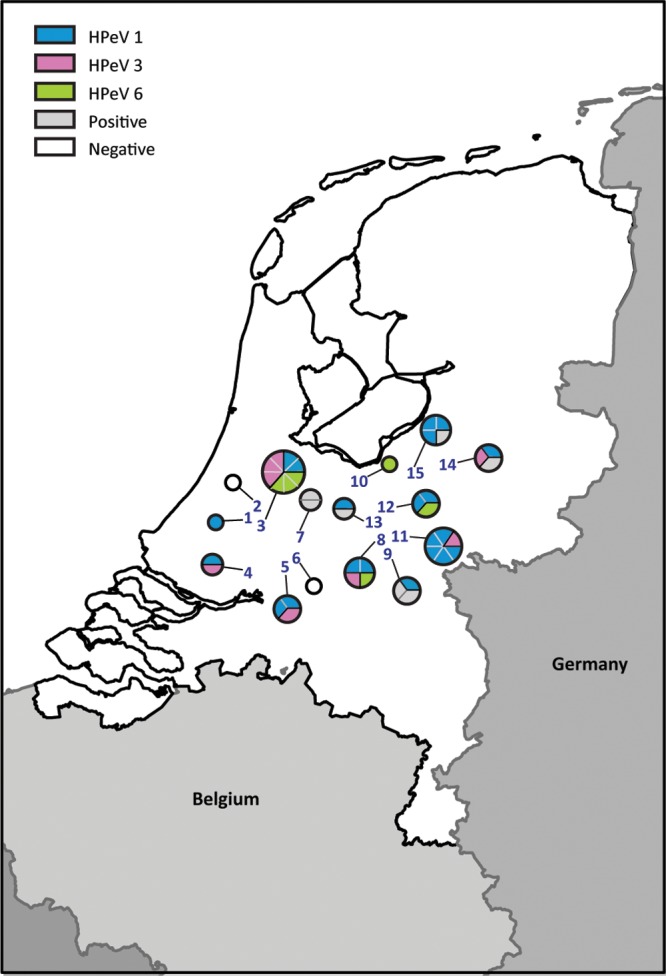
Sample locations (numbers) correspond to the numbers in Table 1. Each circle represents the types of HPeV found (color) at each location. The number of pie parts of one color represents the number of samples in which this HPeV type was detected over time.
Table 1.
Sample dates and locations and different types of human parechovirus detected in Dutch sewage samplesa
| Location no. | Location | Virus type(s) (no. of clones) detected on sampling date |
|||||
|---|---|---|---|---|---|---|---|
| 28 April 2010 | 2010-2011 school year |
||||||
| 28 September 2010 | 2 November 2010 | 15 March 2011 | 12 April 2011 | 7 June 2011 | |||
| 1 | Gouda (school A) | − | − | HPeV1 (5) | − | − | − |
| 2 | Gouda (school B) | − | − | − | − | − | − |
| 3 | Gouda (school C) | HPeV3 (4) HPeV6 (7) | HPeV3 (13) | HPeV1 (6) | HPeV6 (10) | − | HPeV1 (3) HPeV3 (4) HPeV6 (1) |
| 4 | Dordrecht (school) | HPeV1 (4) HPeV3 (11) | − | − | − | − | |
| 5 | Gorinchem | HPeV3 (11) | HPeV1 (12) | HPeV1 (9) | − | − | − |
| 6 | Gorinchem (school) | − | − | − | − | − | − |
| 7 | Leerbroek | + | + | − | − | − | − |
| Sampling date | 13 April 2010 | 6 October 2010 | 16 November 2010 | 1 March 2011 | 29 March 2011 | 17 May 2011 | |
| 8 | Kesteren | HPeV6 (7) | HPeV3 (5) | HPeV1 (8) | − | − | HPeV1 (5) |
| 9 | Kesteren (school) | + | − | + | HPeV1 (24) | − | − |
| 10 | Amersfoort (school) | − | − | HPeV6 (8) | − | − | − |
| 11 | Opheusden | + | HPeV1 (6) HPeV3 (6) | HPeV1 (7) | HPeV1 (10) | HPeV1 (4) | HPeV1 (10) |
| 12 | Veenendaal | HPeV3 (10) | HPeV1 (3) HPeV6 (1) | − | − | − | − |
| 13 | Veenendaal (school) | + | − | − | − | − | HPeV1 (10) |
| 14 | Apeldoorn (school) | + | − | HPeV1 (2) HPeV3 (7) | − | − | ND |
| 15 | Kootwijk | − | HPeV1 (6) | HPeV1 (7) | − | + | HPeV1 (4) |
−, negative; +, 5′ UTR PCR positive but negative with the VP3-VP1 PCR; ND, not done.
RNA extraction.
Genomic material was isolated from 5 ml of raw sewage using the NucliSens miniMAG (bioMérieux, Zaltbommel, the Netherlands) nucleic acid isolation kit as described before (26). Viral genomes were eluted from the silica in 50 μl of elution buffer with RNase inhibitor (Promega, Leiden, the Netherlands), and the eluate either was used directly in the reverse transcription reaction or was stored at −70°C until use. On samples from which no VP3/VP1 product could be obtained, an additional purification and concentration step with the extracted RNA was done with the RNeasy MinElute cleanup kit (Qiagen, Hilden, Germany) to reduce the volume and to further remove inhibitory substances from the samples.
Detection by qPCR.
Human parechovirus RNA was detected in these sewage samples by a real-time RT-PCR method, targeting the 5′ UTR of the parechovirus genome as previously described (16). Briefly, per sample, 5 μl of undiluted, 10-fold diluted, and 100-fold diluted RNA was added to 1.5 μg of random hexamers, and the mixture was heated at 70°C for 5 min and chilled on ice for 5 min. Subsequently, 1× first-strand buffer (Invitrogen, Leek, the Netherlands), 2.5 mmol/liter of each deoxynucleoside triphosphate (dNTP; Roche, Almere, the Netherlands), 2.5 mmol/liter of dithiothreitol (DTT; Roche), 4 U of RNase inhibitor (Promega), and 100 U of Superscript II (Invitrogen) were added at room temperature to a final volume of 20 μl. The RT reaction mixture was incubated in a thermal incubator at 42°C for 60 min; the synthesized cDNA was either used directly in a PCR or stored at −70°C until use.
A Lightcycler 480 Probes Master kit (Roche) was used according to the manufacturer's instructions. In brief, an aliquot of 5 μl of the synthesized cDNA was added to 15 μl of the quantitative PCR (qPCR) mixture containing 1× Lightcycler 480 Probes Master (Roche), an 18 μM concentration of each primer (primers F31 and K30), and a 4 μM concentration of the probe. The qPCR protocol was as follows: a preincubation step at 95°C for 10 min and 45 cycles of 95°C for 15 s and 50°C for 60 s (at a temperature transition rate of 2.2°C s−1), with a cooling step at 40°C for 10 s. The qRT-PCR assays were performed in a LightCycler 480 system (Roche), and the LightCycler software automatically determined the cycle threshold (CT) point of each qPCR. All samples were tested in triplicate, and each run included a negative- and positive-control reaction.
Typing by nested PCR.
To type the detected viruses, cDNA samples that tested positive by the HPeV 5′ UTR PCR were subsequently amplified by a nested PCR using primers targeting the VP3/VP1 junction region, as described previously (18). The second-round PCR products were separated on 2% agarose gels and visualized under UV illumination after staining with SYBR gold nucleic acid gel stain (Molecular Probes, Leiden, the Netherlands). HPeV DNA fragments of 300 bp were purified from agarose gels using a QIAquick PCR purification kit (Qiagen) according to the manufacturer's instructions. All purified PCR products were stored at −20°C until further use.
Cloning and sequencing.
The purified, nested PCR products were cloned into a pCRII-Topo vector (Invitrogen) according to the manufacturer's instructions, and the construct was subsequently transformed into JM109 competent cells. Approximately 9 clones (minimum of 1 and maximum of 24 per sample) (Table 1) were randomly selected per purified PCR product and were checked using M13 primers supplied by the manufacturer (Invitrogen). Both strands were sequenced using a BigDye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems, CA).
Phylogenetic analysis.
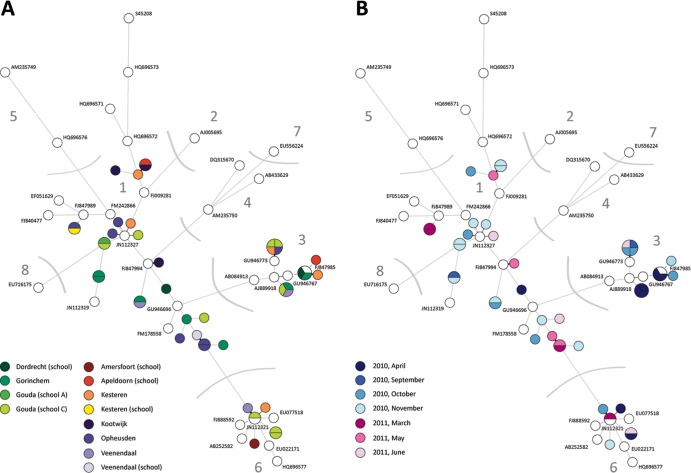
The obtained HPeV sequences were aligned and clustered using BioNumerics software, version 6.6 (Applied Maths, Kortrijk, Belgium), and compared to available sequences present in the NCBI/GenBank database to subsequently type the detected parechoviruses. Phylogenetic trees were constructed using the 256-nucleotide (nt) sequences of the VP3-VP1 region derived from the sewage samples and all the available sequences in the NCBI/GenBank database, using the neighbor-joining (NJ) method with 1,000 bootstrap replications, as implemented in the BioNumerics software. A minimum spanning tree (MST) was estimated from the sequence data, using Prim's algorithm, to graphical illustrate the genetic diversity using the BioNumerics software. The accession numbers of the reference strains used in the MST are shown in Fig. 2.
Fig 2.
Minimum spanning tree, based on the alignment of 256 nucleotides of the VP3-VP1 region of the HPeV genome, showing genetic distances between sequence variants isolated from Dutch sewage samples and prototype strains (GenBank). The circles represent the various types and variants. Lines linking two circles in the tree denote sequence differences between those types and variants. Pie parts indicate how many times this variant was detected. Numbers indicate HPeV type. (A) Locations from which HPeV sequences were obtained are indicated (colored circles), as are the prototype strains of HPeV1 to -8, obtained from GenBank (white circles), and their accession numbers. (B) Months of sampling are indicated (April 2010 and September 2010 to June 2011) (colored circles), as are the prototype strains of HPeV1 to -8, obtained from GenBank (white circles), and their accession numbers.
Nucleotide sequence accession numbers.
The nucleotide sequences of the VP3-VP1 junction region are deposited in GenBank with accession numbers KF434777 to KF435026.
RESULTS
In April 2010 and between 28 September 2010 and 7 June 2011, sewage samples from the 15 different sampling points in 10 different municipalities were collected on six occasions (89 samples in total) and analyzed for the presence of HPeV RNA. In the respective screenings, in 67%, 47%, 60%, 20%, 13%, and 36% of the 15 samples, HPeV RNA was detected by qPCR targeting the 5′ UTR of the virus genome (Table 1). All samples were tested in triplicate by qPCR, and the percentage of positives varied per sample location. In general, when all three replicates were positive, the CT value was lower than for the samples for which only one of the three replicates was positive.
On average, 9 clones were typed with conclusive typing results, with a minimum of 1 clone and a maximum of 24 clones per sample (Table 1), sometimes originating from several PCR products. Based on sequence analysis of the VP3-VP1 junction region, HPeV types were assigned by comparing the sequence isolated from the sewage to strains in the GenBank database (with the highest identity score ranging from 89 to 98%) (Fig. 1 and 2). Three different HPeV types were observed, with HPeV1 (in 20/89 samples) being the most prevalent (Table 1). Furthermore, HPeV3 (8/89) and HPeV6 (6/89) were detected. Nevertheless, in 8 out of the 35 5′ UTR-positive samples typing failed, since no VP3/VP1 PCR product could be obtained by nested PCR.
At 9 of the 15 sampling locations, sewage samples were taken directly from schools. For the six remaining locations, sewage originated from a part of a municipality. At three locations, sewage from both schools and parts of the same municipality were sampled. No correlations between the HPeV positivity on the different sampling dates and HPeV types were found for these three locations.
In sewage sampled at seven of the nine schools (78%) and at all six municipalities (100%), HPeV RNA could be detected at least once. As shown in Table 1, HPeV was detected generally less frequently in the samples taken from the schools than in the samples taken from the municipalities, with one exception (Gouda school C), where on five of the six sampling dates HPeV was detected. Interestingly, this school was the only elementary school (average age of the children is 4 to 12 years); the other schools were high schools (average age of the children is 13 to 18 years). The age distributions of the sampled municipalities (total population) were overall the same: approximately 5% 0 to 3 years, 12% 4 to 12 years, 8% 13 to 18 years, 62% 19 to 65 years, and 13% 65+ years. Although the exact age distribution of people who discharged on the sampled sewerage in the municipalities is not known, it can be expected that the age distribution is more diverse than that of the schools.
In five samples, two different HPeV types could be detected, and in one sample taken at Gouda school C (7 June 2011), three HPeV types were detected (Table 1). In addition, Fig. 2 demonstrates the variation in HPeV sequences per location (Fig. 2A) as well as the variation per sampling month (Fig. 2B), showing that different HPeV1 variants and, to a lesser extent, HPeV3 variants were present in a single sample. The detected sequence diversity was high, but also several identical HPeVs were detected. However, no obvious correlation in time and place was seen.
Although no obvious seasonal pattern was seen in our limited data set, the samples taken in March (5/23) and April (0/7) 2011 clearly showed the lowest overall percentage of HPeV positives. However, in 10 of the 15 samples taken in April 2010, HPeV RNA could be detected (Table 1). HPeV3 was detected mainly in samples taken in 2010 but also once in 2011. Furthermore, the samples taken in November 2010 showed the highest diversity of detected HPeV sequences (Fig. 2B).
DISCUSSION
Sewage samples were molecularly analyzed for the presence of HPeVs during the 2010-2011 school year from 15 different locations in the Bible belt in the Netherlands. The PCR-based methods for virus detection and typing provide a useful tool to assess the circulation of HPeV variants in a population. By environmental surveillance, it is clearly demonstrated that several HPeV types and variants circulated in the studied population.
Several studies reported a biennial cycle of HPeV3 infections, with a much higher frequency of HPeV3 cases in even-numbered years between 2000 and 2010 and the virtual absence in intervening years in Western Europe (5, 9, 17, 27). We also found HPeV3 mainly in April, September, and November 2010 (6 sampling times) but also once in a sample taken from a school in Gouda in 2011, indicating circulation also in the odd years. Although parechovirus infections are globally endemic throughout the year, periodical occurrence of cases is described. For instance, in temperate climates HPeV1 cases are seen particularly more in fall and winter (9). Furthermore, a higher frequency of HPeV3 was reported in summer and fall in Western Europe (8–10). We did not observe a clear seasonal pattern in the samples taken in our study, although in the samples taken in March and April 2011, we found the smallest amount of positives compared to the other sampling dates, which is to be expected according to published literature.
Several studies where age was taken into account describe HPeV infection to be mostly restricted to children under 2 years of age (28–31). Other studies more specifically indicate that HPeV infections were found in young infants (<3 months), whereas older children and adults were not infected (27, 30). Furthermore, seroprevalence studies have shown that over 90% of children have been infected with at least one HPeV type by the age of 2 years (29, 32). The incidence of HPeV infections is primarily based on data obtained from symptomatic individuals (mainly young children), but testing both symptomatic and asymptomatic infected individuals will give a more reliable indication of the prevalence of HPeV and the circulating types. In our study, we found several HPeV types and variants over time in sewage originating from eight high schools (with children between approximately 12 and 19 years of age). For sewage originating from an elementary school (with children between approximately 4 and 12 years of age), more samples were positive and also more HPeV types were detected, suggesting a more prevalent human parechovirus circulation in this younger age group. In the samples taken from the municipalities, with a more diverse age distribution, more samples were positive for the presence of HPeV RNA and more different types of HPeV were found. When samples were taken from a school as well as from the municipality, generally more samples tested positive for the municipalities than for the school (Table 1), possibly explained by the shedding of HPeV by the younger children in the population. Nevertheless, more research has to be done to obtain more clarity on the age distribution of the people shedding HPeVs.
By environmental surveillance, the abundant presence of different types and variants of human parechoviruses in sewage was demonstrated, indicating that these viruses are circulating in the studied human population. Because the clinical manifestations of the circulating viruses were not investigated, it remains unknown to what extent clinical symptoms are related to HPeV infection. Interestingly, the HPeV types identified in sewage are those most frequently identified from patients, which implies that there are probably no additional HPeV types circulating to any great extent. Furthermore, there is probably no great difference between types in the level of virus shed in the feces, which could lead to masking of the presence of clinically important viruses by viruses which replicate more efficiently, or in particle stability, which could affect the recovery of virus sequences from sewage. This suggests that surveillance of sewage may be a useful way of analyzing HPeV circulation. In a previous study, it was shown that environmental poliovirus surveillance is able to identify 100 poliovirus-infected individuals, either symptomatic or asymptomatic, in a population of several tens of thousands of uninfected individuals if sampling is done regularly in strategic locations (25), and this assumption of detection might also be applicable for other picornaviruses such as HPeV.
The main route of transmission of human parechoviruses is by the fecal-oral route, but no other transmission routes have yet been elucidated, and the exact role of the aquatic environment has not yet been established. Epidemiological studies may aid in the better understanding of possible sources of HPeV infections, but also microbiological source tracking may give an impression of how the HPeVs are transmitted. Although the source of HPeV infection is hardly described, Eis-Hübinger et al. (33) have recently reported that two unlinked cases of HPeV infections with sepsis-like disease in 2 newborns were linked to preceding infections which caused only mild disease in their older siblings, indicating that these siblings were the source of infection.
As for other enteric viruses for which detection PCRs are generally more sensitive than typing PCRs, the sensitivity of the typing PCR for HPeV also remains a challenge. This is due to low but possibly clinically relevant virus loads in water, as well as the sequence variability in the structural gene region used for typing, such as VP1. Because of this high sequence variability, highly degenerated primers have to be used to be able to detect the different genotypes. This influences both the sensitivity and specificity of amplification. For HPeV, the detection RT-PCR (5′ UTR) is known to be more sensitive than the typing method based on the VP3-VP1 region. With 20% of the 5′ UTR-positive samples it was not possible to obtain a nested VP3-VP1 PCR product, and therefore, the viruses in these samples could not be typed. To increase the number of strains that can be typed, method optimization is needed. In the current study, 1 ml of sewage was tested directly in triplicate, but a higher volume with subsequent adjusted RNA extraction and a larger amount of replicates may result in more positive results.
Environmental surveillance of HPeV might aid in the understanding of human parechovirus-related disease. The detection of recently emerged virus strains and changes in the circulation of different HPeV types in often short periods provide the basis for more focused investigations on the role of community circulation and prevalence in the population. Our results, representing two relative small areas in the Netherlands, should not be considered representative for the entire Dutch population. Because three different types of HPeV (multiple virus lineages) were detected in sewage, it can be concluded that environmental surveillance in sewage samples using molecular methods is suitable to monitor the circulation of specific human-pathogenic picornaviruses in the population. No information on virus infectivity is obtained with these molecular detection methods; therefore, quantification of infectious HPeV using these methods is not possible. Because of this and the absence of a dose-response relation for human parechoviruses, a quantitative microbial risk assessment to assess the potential public health risks through environmental exposure to human parechovirus can be performed only using data obtained from other organisms and therefore is still complicated.
ACKNOWLEDGMENTS
The project was funded by the Strategic Research RIVM (SOR), project number S/330126/01/EP.
We gratefully acknowledge Harrie van der Avoort and Gokhan Uslu for providing important information on the Dutch environmental polio surveillance and Ron Altena and Edin Jusic for the sewage sampling.
Footnotes
Published ahead of print 9 August 2013
REFERENCES
- 1.Kolehmainen P, Oikarinen S, Koskiniemi M, Simell O, Ilonen J, Knip M, Hyoty H, Tauriainen S. 2012. Human parechoviruses are frequently detected in stool of healthy Finnish children. J. Clin. Virol. 54:156–161 [DOI] [PubMed] [Google Scholar]
- 2.Piralla A, Furione M, Rovida F, Marchi A, Stronati M, Gerna G, Baldanti F. 2012. Human parechovirus infections in patients admitted to hospital in Northern Italy, 2008–2010. J. Med. Virol. 84:686–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Verboon-Maciolek MA, Groenendaal F, Hahn CD, Hellmann J, van Loon AM, Boivin G, de Vries LS. 2008. Human parechovirus causes encephalitis with white matter injury in neonates. Ann. Neurol. 64:266–273 [DOI] [PubMed] [Google Scholar]
- 4.van Zwol AL, Lequin M, Aarts-Tesselaar C, van der Eijk AA, Driessen GA, de Hoog M, Govaert P. 2009. Fatal neonatal parechovirus encephalitis. BMJ Case Rep. 10.1136/bcr.05.2009.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harvala H, Simmonds P. 2009. Human parechoviruses: biology, epidemiology and clinical significance. J. Clin. Virol. 45:1–9 [DOI] [PubMed] [Google Scholar]
- 6.Schuffenecker I, Javouhey E, Gillet Y, Kugener B, Billaud G, Floret D, Lina B, Morfin F. 2012. Human parechovirus infections, Lyon, France, 2008–10: evidence for severe cases. J. Clin. Virol. 54:337–341 [DOI] [PubMed] [Google Scholar]
- 7.Wolthers KC, Benschop KS, Schinkel J, Molenkamp R, Bergevoet RM, Spijkerman IJ, Kraakman HC, Pajkrt D. 2008. Human parechoviruses as an important viral cause of sepsislike illness and meningitis in young children. Clin. Infect. Dis. 47:358–363 [DOI] [PubMed] [Google Scholar]
- 8.Harvala H, Robertson I, Chieochansin T, McWilliam Leitch EC, Templeton K, Simmonds P. 2009. Specific association of human parechovirus type 3 with sepsis and fever in young infants, as identified by direct typing of cerebrospinal fluid samples. J. Infect. Dis. 199:1753–1760 [DOI] [PubMed] [Google Scholar]
- 9.van der Sanden S, de Bruin E, Vennema H, Swanink C, Koopmans M, van der Avoort H. 2008. Prevalence of human parechovirus in the Netherlands in 2000 to 2007. J. Clin. Microbiol. 46:2884–2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benschop KS, Schinkel J, Minnaar RP, Pajkrt D, Spanjerberg L, Kraakman HC, Berkhout B, Zaaijer HL, Beld MG, Wolthers KC. 2006. Human parechovirus infections in Dutch children and the association between serotype and disease severity. Clin. Infect. Dis. 42:204–210 [DOI] [PubMed] [Google Scholar]
- 11.Kapusinszky B, Minor P, Delwart E. 2012. Nearly constant shedding of diverse enteric viruses by two healthy infants. J. Clin. Microbiol. 50:3427–3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stanway G, Kalkkinen N, Roivainen M, Ghazi F, Khan M, Smyth M, Meurman O, Hyypia T. 1994. Molecular and biological characteristics of echovirus 22, a representative of a new picornavirus group. J. Virol. 68:8232–8238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stanway G, Hyypia T. 1999. Parechoviruses. J. Virol. 73:5249–5254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calvert J, Chieochansin T, Benschop KS, McWilliam Leitch EC, Drexler JF, Grywna K, da Costa Ribeiro H, Jr, Drosten C, Harvala H, Poovorawan Y, Wolthers KC, Simmonds P. 2010. Recombination dynamics of human parechoviruses: investigation of type-specific differences in frequency and epidemiological correlates. J. Gen. Virol. 91:1229–1238 [DOI] [PubMed] [Google Scholar]
- 15.Nix WA, Maher K, Johansson ES, Niklasson B, Lindberg AM, Pallansch MA, Oberste MS. 2008. Detection of all known parechoviruses by real-time PCR. J. Clin. Microbiol. 46:2519–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benschop K, Molenkamp R, van der Ham A, Wolthers K, Beld M. 2008. Rapid detection of human parechoviruses in clinical samples by real-time PCR. J. Clin. Virol. 41:69–74 [DOI] [PubMed] [Google Scholar]
- 17.Benschop K, Thomas X, Serpenti C, Molenkamp R, Wolthers K. 2008. High prevalence of human parechovirus (HPeV) genotypes in the Amsterdam region and identification of specific HPeV variants by direct genotyping of stool samples. J. Clin. Microbiol. 46:3965–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harvala H, Robertson I, McWilliam Leitch EC, Benschop K, Wolthers KC, Templeton K, Simmonds P. 2008. Epidemiology and clinical associations of human parechovirus respiratory infections. J. Clin. Microbiol. 46:3446–3453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deshpande JM, Shetty SJ, Siddiqui ZA. 2003. Environmental surveillance system to track wild poliovirus transmission. Appl. Environ. Microbiol. 69:2919–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hovi T, Blomqvist S, Nasr E, Burns CC, Sarjakoski T, Ahmed N, Savolainen C, Roivainen M, Stenvik M, Laine P, Barakat I, Wahdan MH, Kamel FA, Asghar H, Pallansch MA, Kew OM, Gary HE, Jr, deGourville EM, El Bassioni L. 2005. Environmental surveillance of wild poliovirus circulation in Egypt—balancing between detection sensitivity and workload. J. Virol. Methods 126:127–134 [DOI] [PubMed] [Google Scholar]
- 21.Blomqvist S, Savolainen C, Laine P, Hirttio P, Lamminsalo E, Penttila E, Joks S, Roivainen M, Hovi T. 2004. Characterization of a highly evolved vaccine-derived poliovirus type 3 isolated from sewage in Estonia. J. Virol. 78:4876–4883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shulman LM, Manor Y, Handsher R, Delpeyroux F, McDonough MJ, Halmut T, Silberstein I, Alfandari J, Quay J, Fisher T, Robinov J, Kew OM, Crainic R, Mendelson E. 2000. Molecular and antigenic characterization of a highly evolved derivative of the type 2 oral poliovaccine strain isolated from sewage in Israel. J. Clin. Microbiol. 38:3729–3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshida H, Horie H, Matsuura K, Kitamura T, Hashizume S, Miyamura T. 2002. Prevalence of vaccine-derived polioviruses in the environment. J. Gen. Virol. 83:1107–1111 [DOI] [PubMed] [Google Scholar]
- 24.van der Avoort HG, Reimerink JH, Ras A, Mulders MN, van Loon AM. 1995. Isolation of epidemic poliovirus from sewage during the 1992–3 type 3 outbreak in The Netherlands. Epidemiol. Infect. 114:481–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lodder WJ, Buisman AM, Rutjes SA, Heijne JC, Teunis PF, de Roda Husman AM. 2012. Feasibility of quantitative environmental surveillance in poliovirus eradication strategies. Appl. Environ. Microbiol. 78:3800–3805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rutjes SA, Italiaander R, van den Berg HH, Lodder WJ, de Roda Husman AM. 2005. Isolation and detection of enterovirus RNA from large-volume water samples by using the NucliSens miniMAG system and real-time nucleic acid sequence-based amplification. Appl. Environ. Microbiol. 71:3734–3740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harvala H, McLeish N, Kondracka J, McIntyre CL, McWilliam Leitch EC, Templeton K, Simmonds P. 2011. Comparison of human parechovirus and enterovirus detection frequencies in cerebrospinal fluid samples collected over a 5-year period in Edinburgh: HPeV type 3 identified as the most common picornavirus type. J. Med. Virol. 83:889–896 [DOI] [PubMed] [Google Scholar]
- 28.Abed Y, Boivin G. 2006. Human parechovirus types 1, 2 and 3 infections in Canada. Emerg. Infect. Dis. 12:969–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tauriainen S, Martiskainen M, Oikarinen S, Lonnrot M, Viskari H, Ilonen J, Simell O, Knip M, Hyoty H. 2007. Human parechovirus 1 infections in young children—no association with type 1 diabetes. J. Med. Virol. 79:457–462 [DOI] [PubMed] [Google Scholar]
- 30.Harvala H, Wolthers KC, Simmonds P. 2010. Parechoviruses in children: understanding a new infection. Curr. Opin. Infect. Dis. 23:224–230 [DOI] [PubMed] [Google Scholar]
- 31.Pham NT, Takanashi S, Tran DN, Trinh QD, Abeysekera C, Abeygunawardene A, Khamrin P, Okitsu S, Shimizu H, Mizuguchi M, Ushijima H. 2011. Human parechovirus infection in children hospitalized with acute gastroenteritis in Sri Lanka. J. Clin. Microbiol. 49:364–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Joki-Korpela P, Hyypiä T. 1998. Diagnosis and epidemiology of echovirus 22 infections. Clin. Infect. Dis. 27:129–136 [DOI] [PubMed] [Google Scholar]
- 33.Eis-Hübinger AM, Eckerle I, Helmer A, Reber U, Dresbach T, Buderus S, Drosten C, Muller A. 2013. Two cases of sepsis-like illness in infants caused by human parechovirus traced back to elder siblings with mild gastroenteritis and respiratory symptoms. J. Clin. Microbiol. 51:715–718 [DOI] [PMC free article] [PubMed] [Google Scholar]