Abstract
Escherichia coli K-12 utilizes 3-(3-hydroxyphenyl)propionate (3HPP) as a sole carbon and energy source. Among the genes in its catabolic cluster in the genome, mhpT was proposed to encode a hypothetical transporter. Since no transporter for 3HPP uptake has been identified, we investigated whether MhpT is responsible for 3HPP uptake. MhpT fused with green fluorescent protein was found to be located at the periphery of cells by confocal microscopy, consistent with localization to the cytoplasmic membrane. Gene knockout and complementation studies clearly indicated that mhpT is essential for 3HPP catabolism in E. coli K-12 W3110 at pH 8.2. Uptake assays with 14C-labeled substrates demonstrated that strain W3110 and strain W3110ΔmhpT containing recombinant MhpT specifically transported 3HPP but not benzoate, 3-hydroxybenzoate, or gentisate into cells. Energy dependence assays suggested that MhpT-mediated 3HPP transport was driven by the proton motive force. The change of Ala-272 of MhpT to a histidine, surprisingly, resulted in enhanced transport activity, and strain W3110ΔmhpT containing the MhpT A272H mutation had a slightly higher growth rate than the wild-type strain at pH 8.2. Hence, we demonstrated that MhpT is a specific 3HPP transporter and vital for E. coli K-12 W3110 growth on this substrate under basic conditions.
INTRODUCTION
Microbes play a major role in the degradation of phenylpropanoid compounds, which result largely from the degradation of lignin and other aromatic constituents of plants (1, 2). Among these phenylpropanoid compounds, phenylpropionate and its hydroxylated derivatives have been reported to be degraded by various bacterial strains (3–7). In particular, a number of Escherichia coli strains (strains B, C, W, K-12, and NCTC 5928) were shown to be able to utilize 3-(3-hydroxyphenyl)propionate (3HPP), a major member of the phenylpropanoids, as their sole carbon source (7). A 9.8-kb mhp cluster conferred the ability to grow on 3HPP to E. coli MC4100, Salmonella enterica serovar Typhimurium LT-2, Pseudomonas putida KT2442, and Rhizobium meliloti Rm1021Rif, suggesting that this cluster contained all of the catabolic genes necessary for the catabolism of this aromatic acid into Krebs cycle intermediates and also the regulatory element of the pathway (8). The expression of mhp catabolic genes was shown to be induced by 3HPP (9). In addition to the genes mhpRABCDFE encoding the regulatory function and 3HPP catabolism (8, 9), mhpT (formerly orfT) was tentatively proposed to encode a potential transporter (8). Nevertheless, the function of MhpT has so far not been identified genetically or biochemically. As a matter of fact, no report on the involvement of any transporter in the microbial degradation of phenylpropionate and its hydroxylated derivatives can be found in the literature.
The robust ability of microorganisms to grow on a variety of aromatic compounds partially relies on multiple transporters for their uptake into cells (10, 11). Among the various categories of transporters classified in the Transporter Classification Database (TCDB; http://www.tcdb.org/) (12), the functionally identified aromatic acid transporters (13–18) belong to the aromatic acid/H+ symporter (AAHS) family (cluster 2.A.1.15 in TCDB), except for the 4-hydroxyphenylacetate transporter HpaX in E. coli W (18). In this study, we report that MhpT, which shares 27 to 37% identities with other AAHS transporters, is specifically involved in 3HPP catabolism of E. coli K-12 and transports 14C-labeled 3HPP but not 14C-labeled benzoate, 3-hydroxybenzoate, or gentisate into cells.
MATERIALS AND METHODS
Strains, plasmids, media, growth conditions, and chemicals.
The bacterial strains and plasmids used in this study are listed in Table 1. The substrate 3HPP obtained was the highest purity grade (≥98%) commercially available. The tracers [carboxyl-14C]3HPP (55 mCi/mmol), [carboxyl-14C]gentisate (55 mCi/mmol), [carboxyl-14C]3-hydroxybenzoate (55 mCi/mmol), and [ring-U-14C]benzoate (70 mCi/mmol) were purchased from American Radiolabeled Chemicals, Inc. (St. Louis, MO). Carbonyl cyanide m-chlorophenylhydrazone (CCCP) was purchased from Sigma-Aldrich Co. (St. Louis, MO). Restriction enzymes were purchased from TaKaRa Biotechnology Co. Ltd. (Dalian, China). Plasmid DNA extraction and DNA gel extraction kits were purchased from Omega Bio-Tek Inc. (Doraville, GA). E. coli strains were grown in lysogeny broth (LB) (19, 20) or mineral salts medium (MM; at pH 8.2 with Na2HPO4·12H2O at 35.6987 g and KH2PO4 at 0.0362 g, at pH 7.2 with Na2HPO4·12H2O at 25.5646 g and KH2PO4 at 4.2694 g, and at pH 6.2 with Na2HPO4·12H2O at 7.6000 g and KH2PO4 at 10.7194 g, along with the addition of 0.28 mg MnSO4·H2O, 0.3 mg FeSO4·7H2O, 0.06 mg MgSO4·7H2O, 1 mg CaCl2, 0.05 mg CuSO4, 0.05 mg ZnSO4, 0.05 mg H3BO3, and finally, H2O to a total volume of 1 liter) with 2 mM 3HPP and 6 mM (NH4)2SO4 on a rotary shaker (200 rpm) at 37°C. When necessary, ampicillin, kanamycin, and tetracycline were used at final concentrations of 100, 50, and 25 μg/ml, respectively. Isopropyl β-d-1-thiogalactopyranoside (IPTG) and 3HPP were used as inducers at final concentrations of 0.1 and 0.5 mM, respectively.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Strains | ||
| E. coli DH5α | supE44 lacU169 (ϕ80dlacZΔM15) hsdR17 recA1 endA1 gyrA96 Δthi relA1 | 38 |
| E. coli BL21(DE3) | F− ompT hsdSB(rB− mB−) gal dcm lacY1 (DE3) | Novagen |
| E. coli K-12 W3110 | 3HPP utilizer, Kms Tcs | 26 |
| E. coli K-12 W3110ΔmhpT | E. coli K-12 W3110 with the DNA fragment encoding MhpT deleted | This study |
| Plasmids | ||
| pGFPe | Kmr, ColE1 replicon, derivative of pWaldo digested with XbaI and EcoRI, yielding a 14-amino-acid linker sequence (SVPGSENLYFQGQF) followed by GFP | 23 |
| pGFPe-mhpT | PCR-amplified fragment containing mhpT without the stop codon inserted into pGFPe at the XhoI/BamHI restriction sites | This study |
| pGFPe-mhpTE27A | PCR-amplified fragment containing mhpT (mhpTE27A) without the stop codon inserted into pGFPe at the XhoI/BamHI restriction sites | This study |
| pGFPe-mhpTE27D | PCR-amplified fragment containing mhpT (mhpTE27D) without the stop codon inserted into pGFPe at the XhoI/BamHI restriction sites | This study |
| pGFPe-mhpTD75A | PCR-amplified fragment containing mhpT (mhpTD75A) without the stop codon inserted into pGFPe at the XhoI/BamHI restriction sites | This study |
| pGFPe-mhpTD75E | PCR-amplified fragment containing mhpT (mhpTD75E) without the stop codon inserted into pGFPe at the XhoI/BamHI restriction sites | This study |
| pGFPe-mhpTA272H | PCR-amplified fragment containing mhpT (mhpTA272H) without the stop codon inserted into pGFPe at the XhoI/BamHI restriction sites | This study |
| pGFPe-mhpTK276D | PCR-amplified fragment containing mhpT (mhpTK276D) without the stop codon inserted into pGFPe at the XhoI/BamHI restriction sites | This study |
| pVLT31 | Broad-host-range expression vector (IncQ, RSF1010 replicon), Tcr Ptac lacIq | 39 |
| pVLT31-mhpT | PCR fragment containing mhpT inserted into pVLT31 at the EcoRI/HindIII restriction sites | This study |
| pVLT31-mhpTE27A | PCR fragment containing mutant mhpT (mhpTE27A) inserted into pVLT31 at the EcoRI/HindIII restriction sites | This study |
| pVLT31-mhpTE27D | PCR fragment containing mutant mhpT (mhpTE27D) inserted into pVLT31 at the EcoRI/HindIII restriction sites | This study |
| pVLT31-mhpTD75A | PCR fragment containing mutant mhpT (mhpTD75A) inserted into pVLT31 at the EcoRI/HindIII restriction sites | This study |
| pVLT31-mhpTD75E | PCR fragment containing mutant mhpT (mhpTD75E) inserted into pVLT31 at the EcoRI/HindIII restriction sites | This study |
| pVLT31-mhpTA272H | PCR fragment containing mutant mhpT (mhpTA272H) inserted into pVLT31 at the EcoRI/HindIII restriction sites | This study |
| pVLT31-mhpTK276D | PCR fragment containing mutant mhpT (mhpTK276D) inserted into pVLT31 at the EcoRI/HindIII restriction sites | This study |
General molecular biology methods.
Plasmid DNA was isolated using the alkaline lysis method (21) for all plasmids except plasmid pVLT31 and its derivatives, which were isolated by the boiling lysis method (22). Restriction endonuclease digestion and ligation with T4 DNA ligase were conducted in accordance with the manufacturer's instructions. E. coli BL21(DE3) and DH5α were both transformed by standard procedures (21). All inserts were sequenced by Invitrogen Biotechnology Co. Ltd. (Shanghai, China). Analyses and comparison of amino acid sequences were performed with BLAST programs on the National Center for Biotechnology Information website.
Cellular localization of MhpT-green fluorescent protein (GFP) fusion protein.
Cultures of E. coli BL21(DE3) carrying pGFPe (23) and its derivatives containing mhpT and mutant mhpT genes (all without a stop codon) (Table 1) were harvested and washed twice with 50 mM phosphate buffer (pH 7.4) after being induced with IPTG at 37°C for 4 h. Then, the cells were resuspended in the same buffer mixed with agarose (0.3%) to immobilize cells for imaging under a confocal microscope with a 490-nm excitation filter and a 520-nm emission filter (24). The imaging experiments were performed using a TCS SP2 Leica laser scanning spectral confocal microscope equipped with a cooled charge-coupled-device camera (Leica TCS SP2; Leica Microsystems, Mannheim, Germany). The background cell fluorescence was subtracted.
mhpT disruption.
Temperature-sensitive vector pKD46 (25) was introduced into E. coli K-12 W3110 (26) for preparation of competent cells. Primers mhpT1F and mhpT1R (Table 2) into which 51-bp 5′ and 3′ homologous arms of mhpT were incorporated, respectively, were used to amplify the kanamycin resistance gene by PCR using pKD4 (25) as the template. The resulting PCR fragment was electroporated into E. coli K-12 W3110(pKD46) for deletion of the mhpT gene as described previously (25), with minor modifications. Shocked cells were incubated in 1 ml LB at 37°C for 1 h, and cells were then spread onto agar to select kanamycin-resistant transformants. Verified transformants were grown in liquid LB at 42°C for 2 h for loss of pKD46.
Table 2.
Primers in this study
| Primer | Sequencea | Purpose |
|---|---|---|
| mhpT1 | 5′-CCGGAATTCATGTCGACTCGTACCCCTT-3′ | Forward primer for mhpT |
| mhpT2 | 5′-CGGAAGCTTTCAGGCATCGGCGCACGGC-3′ | Reverse primer for mhpT |
| mhpT1D | 5′-CCGCTCGAGATGTCGACTCGTACCCCTTC-3′ | Forward primer for mhpT without a stop codon |
| mhpT2D | 5′-CGCGGATCCGGCATCGGCGCACGGCTGTATTC-3′ | Reverse primer for mhpT without a stop codon |
| mhpT1F | 5′-GATGTCGACTCGTACCCCTTCATCATCTTCATCCCGCCTGATGCTGACCATATATGAATATCCTCCTTAG-3′ | Forward primer for kanamycin resistance gene with the homologous arm of mhpT |
| mhpT2R | 5′-TCAGGCATCGGCGCACGGCTGTATTCGTGATCTCCGGCTCATCAAAATAATGTAGGCTGGAGCTGCTTCG-3′ | Reverse primer for kanamycin resistance gene with the homologous arm of mhpT |
The introduced restriction sites are boldface, and the homologous arms of mhpT are underlined.
Site-directed mutagenesis.
The desired mutants of MhpT were obtained by overlap extension PCR (27). The outer primers for amplification were mhpT1 and mhpT2 (Table 2). The inner primers were designed to incorporate one codon change. The overlap extension PCR products were digested before ligating into the similarly digested pVLT31, resulting in the expression constructs (Table 1). All mutant mhpT genes were verified by DNA sequencing to ensure that only the desired mutations had occurred.
Measurement of bacterial growth on 3HPP.
The bacterial growth of E. coli K-12 W3110 and its variants with 2 mM 3HPP was measured at an optical density at 600 nm (OD600). Such experiments were repeated three times, and similar results were obtained. The data acquired were transformed into the corresponding number of cells. Then, the growth curves were fitted by the modified Gompertz equation (28) with OriginPro (version 8) software, and all points represent the mean value of triplicate trials. Their maximum specific growth rates (μmax h−1) were calculated and are expressed as the mean values of triplicate trials.
Uptake assays.
The assays for the uptake of aromatic acids were performed with 14C-labeled substrates as described previously (16), with minor modifications. Cells of E. coli K-12 W3110 and its variants grown in LB with 3HPP and/or with IPTG induction at 37°C were washed twice and resuspended in 100 mM Na2HPO4-KH2PO4 buffer (pH 6.2, 7.2, or 8.2) to an OD600 of 1 to 2. Before the uptake assay, cells were incubated at 37°C for 3 min with 10 mM glucose for energy generation (14). The 14C-labeled substrate concentration for use in the uptake assay system with E. coli was 10 μM. The amount of substrate accumulated in the cells on the filters was determined in a scintillation counter (1450 MicroBeta TriLux; PerkinElmer Life Sciences, Boston, MA). All assays were performed at least in triplicate. The transport activity was expressed as the nanomoles of substrate taken up per milligram of protein. Cell protein contents were determined as described previously (14).
Cellular starvation and inhibitor treatments.
The assays for uptake by starved or reenergized cells of strain W3110ΔmhpT(pVLT31-mhpT) (grown in LB with IPTG induction) were carried out as described previously (14), with a slight modification. Cellular energy stores were depleted by washing and resuspending the cells in an original volume of medium using Na2HPO4-KH2PO4 buffer (100 mM, pH 8.2), and the cells were incubated with shaking at 37°C for 18 h. Then, starved cells were washed and divided into three aliquots in the same buffer, and one of them was treated with CCCP (50 μM) at 37°C for 1 min. After inhibitor treatment, the treated aliquot together with another aliquot was reenergized with 10 mM glucose. Then, all three aliquots were incubated at 37°C for an additional 10 min before the uptake assay.
RESULTS
Bioinformatics analysis and cellular localization of MhpT.
MhpT was indicated to contain 12 predicted α-helix transmembrane spanners (TM1 to TM12), a typical feature of the AAHS family within the major facilitator superfamily (11), through each of the four following topology prediction methods: TMHMM (http://www.cbs.dtu.dk/services/TMHMM/), MEMSAT (http://saier-144-21.ucsd.edu/barmemsat.html), PSIPRED (http://bioinf.cs.ucl.ac.uk/psipred/), and HMMTOP (http://www.enzim.hu/hmmtop/). By amino acid sequence comparison, MhpT exhibited 27 to 37% identities with other AAHS members, including the 3-hydroxybenzoate transporter MhbT from Klebsiella pneumoniae M5a1 (16), 4-hydroxybenzoate and protocatechuate transporter PcaK from P. putida (14), benzoate transporter BenK from Acinetobacter sp. strain ADP1 (13), 2,4-dichlorophenoxyacetate (2,4-D) transporter TfdK from Ralstonia eutropha JMP134 (15), and gentisate transporter GenK from Corynebacterium glutamicum (17). In order to confirm the prediction that MhpT was a membrane protein, mhpT was tagged with the GFP gene in plasmid pGFPe-mhpT (Table 1) to facilitate the detection of its localization in bacterial cells. Cells examined using confocal microscopy clearly showed that the MhpT-GFP fusion protein in E. coli BL21(DE3)(pGFPe-mhpT) was located at the periphery of cells (Fig. 1A), consistent with the cytoplasmic membrane location of MhpT predicted by bioinformatics. In contrast, GFP was distributed in the cytoplasm in the negative control of E. coli BL21(DE3)(pGFPe) (Fig. 1H).
Fig 1.
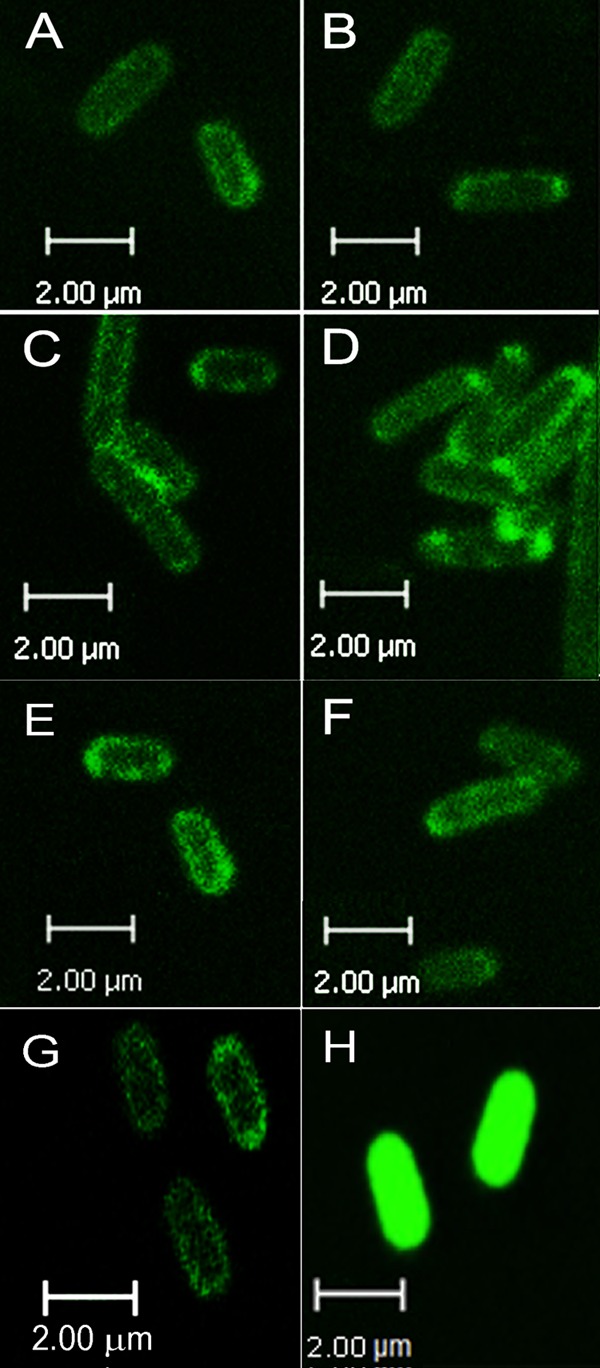
Localization of GFPs under a confocal microscope. (A) MhpT-GFP expressed in E. coli BL21(DE3)(pGFPe-mhpT); (B) MhpTE27A-GFP expressed in E. coli BL21(DE3)(pGFPe-mhpTE27A); (C) MhpTE27D-GFP expressed in E. coli BL21(DE3)(pGFPe-mhpTE27D); (D) MhpTD75A-GFP expressed in E. coli BL21(DE3)(pGFPe-mhpTD75A); (E) MhpTD75E-GFP expressed in E. coli BL21(DE3)(pGFPe-mhpTD75E); (F) MhpTA272H-GFP expressed in E. coli BL21(DE3)(pGFPe-mhpTA272H); (G) MhpTK276D-GFP expressed in E. coli BL21(DE3)(pGFPe-mhpTK276D). Panels A to G show that the fusion proteins are located at the periphery of the cells. (H) GFP expressed in E. coli BL21(DE3)(pGFPe) as a negative control, showing that GFP is distributed throughout the cytoplasm.
MhpT is essential for 3HPP catabolism under basic conditions.
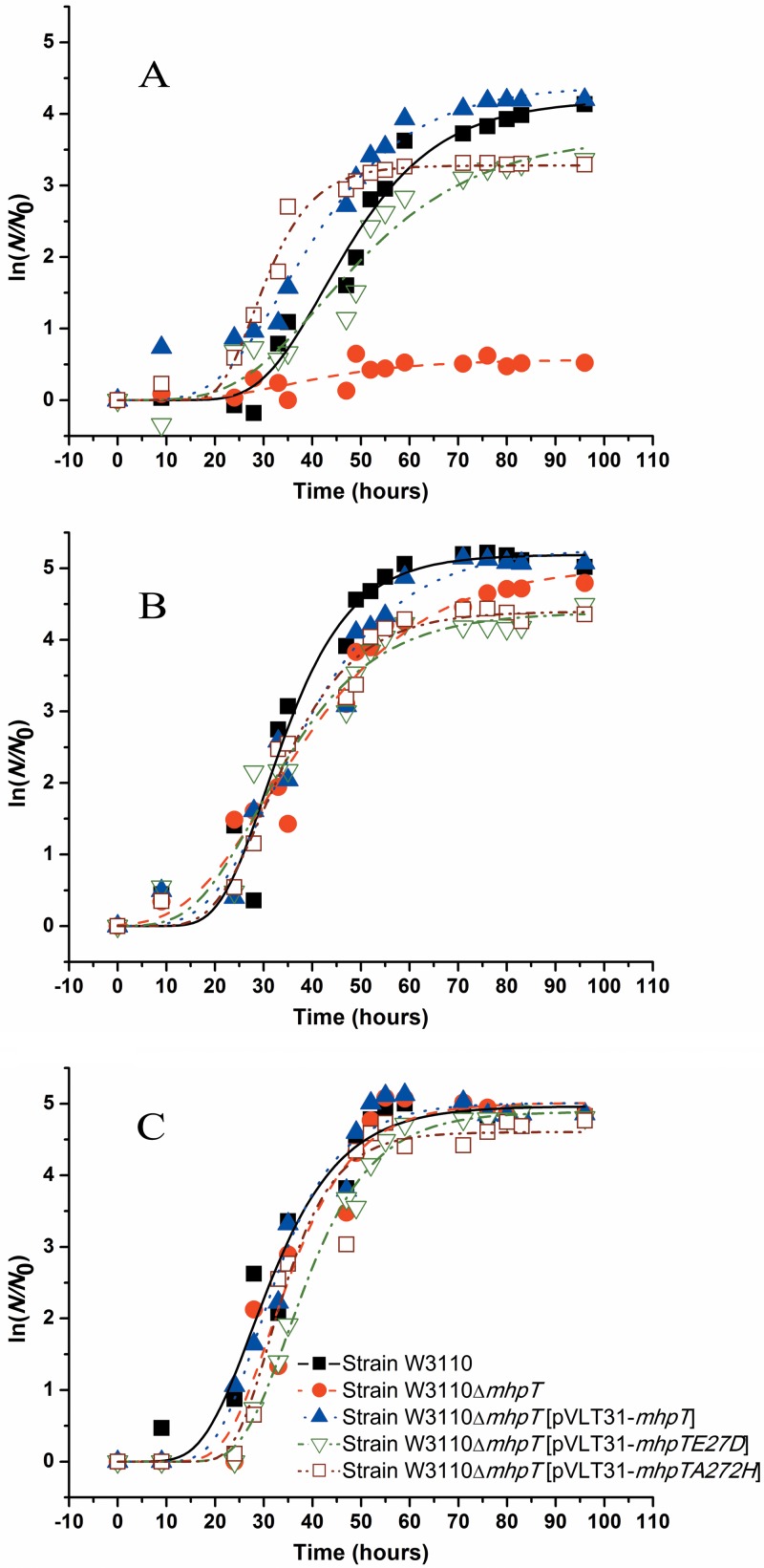
To identify the possible involvement of mhpT in 3HPP catabolism, this gene was deleted to create a mutant strain, W3110ΔmhpT. Strain W3110, mutant strain W3110ΔmhpT, and complemented strain W3110ΔmhpT(pVLT31-mhpT) were grown on 3HPP at three pH values. Growth study data are presented in semilog plots (Fig. 2), and the maximum specific growth rates of these strains are indicated in Table 3. As shown in Fig. 2A, strain W3110ΔmhpT virtually lost the ability to utilize 3HPP at pH 8.2, but its ability to grow on 3HPP was restored after pVLT31-mhpT was introduced, and it had a growth rate similar to that of the wild-type strain. However, at pH 6.2 (Fig. 2C), all three strains described above were able to grow on 3HPP with similar growth rates, regardless of the presence of mhpT. Even though the three E. coli strains had the ability to utilize 3HPP at pH 7.2 (Fig. 2B), the growth rate of the mutant strain was slightly lower than the growth rates of the wild type and the complemented strains. This clearly indicated that mhpT is essential for 3HPP catabolism in E. coli K-12 W3110 under basic conditions.
Fig 2.
Growth curves of cells of E. coli K-12 and its variants on 3HPP. Strains were grown with 2 mM 3HPP in MM at 37°C. (A) pH 8.2; (B) pH 7.2; (C) pH 6.2. N, number of cells; N0, initial number of cells. The growth curves were fitted by the modified Gompertz equation (28) with OriginPro (version 8) software, and all points represent the mean values of triplicate trials. The symbol definitions in panel C apply to the curves in both panels A and B.
Table 3.
Effects of pH values on the maximum specific growth rates and transport activities of wild-type strain W3110 and its variants on 3HPP
| Strain | Maximum specific growth rate (μmax h−1) |
Transport activity at 1st min (nmol/mg protein) |
||||
|---|---|---|---|---|---|---|
| pH 6.2 | pH 7.2 | pH 8.2 | pH 6.2 | pH 7.2 | pH 8.2 | |
| W3110 | 0.18 | 0.19 | 0.12 | 107.37 ± 0.98 | 46.86 ± 0.71 | 43.91 ± 0.59 |
| W3110ΔmhpT | 0.20 | 0.11 | 0.01 | 25.29 ± 0.18 | 15.07 ± 0.87 | 5.45 ± 0.43 |
| W3110ΔmhpT(pVLT31-mhpT) | 0.21 | 0.14 | 0.11 | 178.33 ± 0.98 | 75.59 ± 0.71 | 60.07 ± 0.59 |
| W3110ΔmhpT(pVLT31-mhpTE27D) | 0.19 | 0.12 | 0.08 | NAa | NA | NA |
| W3110ΔmhpT(pVLT31-mhpTA272H) | 0.22 | 0.17 | 0.18 | NA | NA | NA |
NA, not applicable (no detection carried out).
MhpT specifically transports 3HPP.
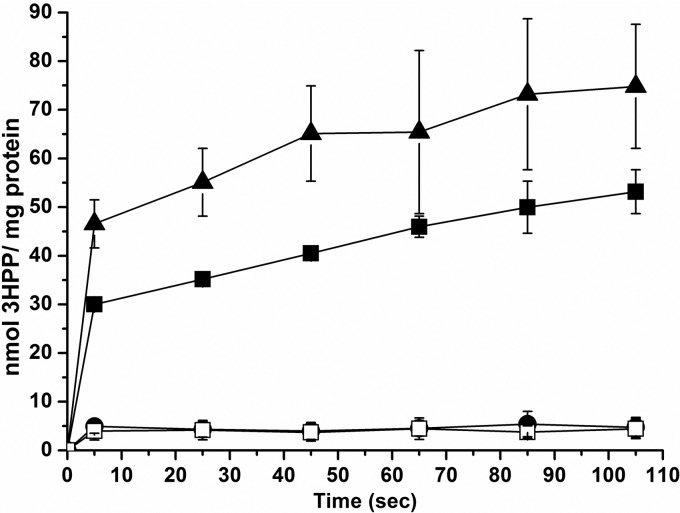
To detect the ability to transport aromatic acids by MhpT, intracellular radiolabel accumulation by resting cells of E. coli K-12 W3110 and its variants was measured at pH 8.2. As shown in Fig. 3, it was evident that, when grown in LB with 3HPP and IPTG induction, strain W3110 was able to accumulate 14C-labeled 3HPP but strain W3110ΔmhpT virtually lost this ability. However, this ability was restored in strain W3110ΔmhpT(pVLT31-mhpT). On the other hand, 3HPP-induced strain W3110 cells also had 3HPP accumulation activity similar to that of 3HPP- and IPTG-induced ones, but IPTG-induced cells had no such uptake activity (data not shown). These findings indicate that the expression of the transporter gene mhpT in strain W3110 is also induced by 3HPP, together with other mhp catabolic genes, as described previously (9). On the other hand, with IPTG induction only, LB-grown strain W3110ΔmhpT(pVLT31-mhpT) was able to accumulate 14C-labeled 3HPP, but strain W3110ΔmhpT(pVLT31)was not (Fig. 4). Notably, the 3HPP- and IPTG-induced cells, in which all the 3HPP catabolic genes were expressed, had a much higher transport activity than IPTG-induced cells. This observation was similar to observations in previous studies on the 3-hydroxybenzoate transporter (16) and the 4-hydroxybenzoate transporter (14). Detection of accumulation was also performed with the available 14C-labeled 3HPP analogues benzoate, 3-hydroxybenzoate, and gentisate in the above-described systems, but no uptake of these three aromatic acids by 3HPP-induced strain W3110 or IPTG-induced strain W3110ΔmhpT(pVLT31-mhpT) was detected (data not shown).
Fig 3.
MhpT-mediated 14C-labeled 3HPP accumulation in E. coli K-12. Assays of 3HPP uptake by 3HPP- and IPTG-induced cells of E. coli K-12 W3110 and its variants were performed at pH 8.2. ■, E. coli K-12 W3110; •, E. coli K-12 W3110ΔmhpT; □, E. coli K-12 W3110ΔmhpT(pVLT31); ▲, E. coli K-12 W3110ΔmhpT(pVLT31-mhpT). All points represent the mean values of triplicate trials, with error bars denoting the standard deviations.
Fig 4.
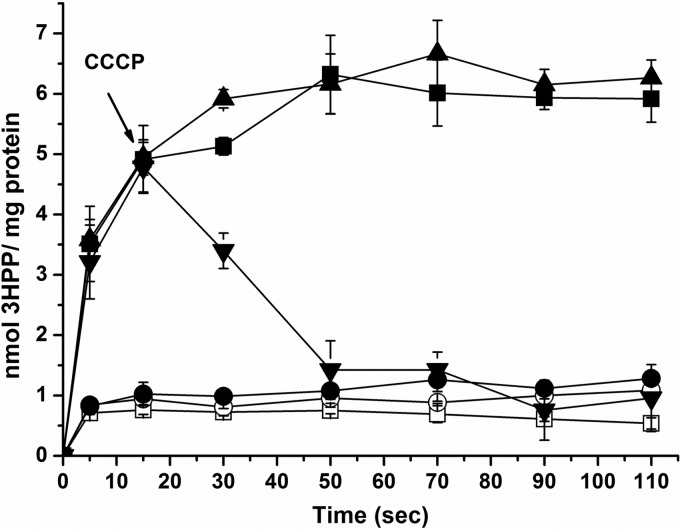
Energy requirements in MhpT-mediated 3HPP transport. Assays of 3HPP uptake by IPTG-induced strains W3110ΔmhpT(pVLT31-mhpT) and W3110ΔmhpT(pVLT31) were performed at pH 8.2. ▲, strain W3110ΔmhpT(pVLT31-mhpT) before starvation; ○, starved strain W3110ΔmhpT(pVLT31-mhpT) cells; ■, starved strain W3110ΔmhpT(pVLT31-mhpT) cells reenergized with glucose; ▼, starved strain W3110ΔmhpT(pVLT31-mhpT) cells reenergized with glucose (the addition of CCCP at the 15th second is indicated by an arrow); •, starved CCCP-treated strain W3110ΔmhpT(pVLT31-mhpT) cells reenergized with glucose; □, strain W3110ΔmhpT(pVLT31) before starvation. All points represent the mean values of triplicate trials, with error bars denoting the standard deviation.
Environmental pH values affect transport activity.
Given the fact that the growth of strain W3110ΔmhpT on 3HPP was affected by pH (Fig. 2 and Table 3), intracellular radiolabel accumulation was then also detected at three pH values to investigate the effect of environmental pH values on the transport activity by MhpT. Three equal portions of LB-grown cells for each strain were resuspended into buffers at pH 6.2, 7.2, and 8.2. After induction with 3HPP and IPTG for 4 h at 37°C, cells were resuspended in the same buffer before the uptake assays. The transport activities of all strains tested were decreased with the increase in the pH values, as shown in Table 3. Strain W3110 was capable of accumulating 14C-labeled 3HPP at three pH values, but the activities in mutant strain W3110ΔmhpT were decreased dramatically at each pH value, particularly at pH 8.2. On the other hand, this ability was restored in the complemented strain, W3110ΔmhpT(pVLT31-mhpT).
MhpT-mediated 3HPP transport is energized by the PMF.
To investigate the energy dependence of MhpT-mediated 3HPP transport, uptake assays were performed using unstarved, starved, and glucose-resupplied starved cells of IPTG-induced strain W3110ΔmhpT(pVLT31-mhpT) at pH 8.2. As shown in Fig. 4, the starved cells did not accumulate 3HPP, but its accumulation ability was restored when they were resupplied with glucose. This indicated that the MhpT-mediated 3HPP transport requires energy. CCCP was then introduced into starved cells to determine the type of energy requirement between the two main known sources of energy, ATP and the proton motive force (PMF). In contrast to the above-described experiment, the CCCP-treated starved cells with resupplied glucose were unable to accumulate 3HPP (Fig. 4). Meanwhile, addition of CCCP (at the 15th second) during the assays of uptake by glucose-resupplied starved cells resulted in a rapid loss of the accumulation capability and the efflux of 3HPP (Fig. 4). CCCP was thought to be able to dissipate the PMF by equilibrating protons across the membrane, but without a significant effect on intracellular ATP synthesis via glycolysis in glucose-reenergized cells (29). The evidence from this study presented above suggests that MhpT-mediated 3HPP transport is driven by the PMF and does not correlate with ATP, similar to PcaK-mediated 4-hydroxybenzoate transport (14).
Identification of critical residues in MhpT for 3HPP transport.
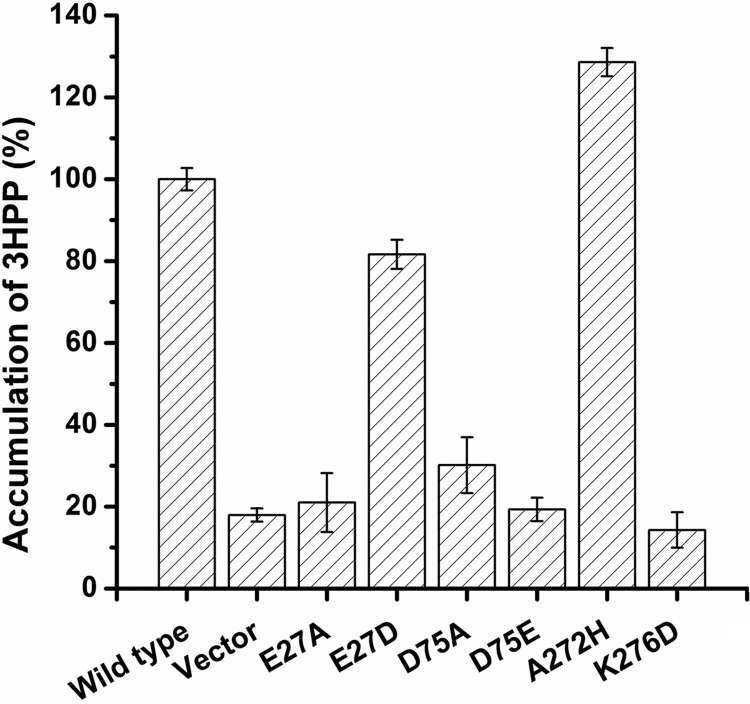
In AAHS family members, two conserved stretches with a motif of GXXXD(R/K)XGR(R/K) located in the cytoplasmic hydrophilic loops (the 2-3 and 8-9 loops) are required for substrate transport (30), and an essential motif (DGXD) containing aspartate residues located in the first transmembrane segment (TM1) is also conserved (31). Thus, four residues in TM1 and the 2-3 and 8-9 cytoplasmic loops were chosen for site-directed mutagenesis to examine the functional significance of these regions for 3HPP transport. Glu-27 in TM1, which was a conserved aspartate among the other AAHS members, was changed to an alanine (E27A) and an aspartate (E27D). Conserved Asp-75 in the 2-3 loop was replaced by an alanine (D75A) and a glutamate (D75E). In the 8-9 loop, less conserved Ala-272 and conserved Lys-276 were changed to a histidine (A272H) and an aspartate (K276D), respectively. By confocal microscopy, all these six mutant MhpT proteins were found to be fused with GFP and were also located at the periphery of cells (Fig. 1B to G), as in wild-type MhpT. Strains W3110ΔmhpT containing wild-type and mutant MhpT proteins were used for measuring their accumulation of 14C-labeled 3HPP at pH 8.2. All cells were grown in LB with IPTG induction, and the transport activity of strain W3110ΔmhpT(pVLT31-mhpT) was 5.37 ± 0.41 nmol/mg protein at the first minute. As shown in Fig. 5, the uptake assays showed that all mutants were devoid of 3HPP transport activity, except for MhpT mutants E27D and A272H, which retained activities approximately 0.8- and 1.3-fold, respectively, in comparison with the activity of wild-type MhpT. In an in vivo experiment, strain W3110ΔmhpT carrying E27D or A272H was able to grow on 3HPP at pH 6.2, 7.2, and 8.2 (as shown in Fig. 2; the maximum specific growth rates of these two strains are indicated in Table 3). Interestingly, strain W3110ΔmhpT containing A272H had a growth rate slightly higher than that of the wild-type strain at pH 8.2.
Fig 5.
Accumulation of 3HPP by E. coli K-12 W3110ΔmhpT containing mutant MhpT proteins. All strains were grown in LB with IPTG induction. The activity (5.37 ± 0.41 nmol/mg protein) of 3HPP uptake by strain W3110ΔmhpT(pVLT31-mhpT) at the first minute was set equal to 100%. Wild type, strain W3110ΔmhpT(pVLT31-mhpT); Vector, strain W3110ΔmhpT(pVLT31); E27A, strain W3110ΔmhpT(pVLT31-mhpTE27A); E27D, strain W3110ΔmhpT(pVLT31-mhpTE27D); D75A, strain W3110ΔmhpT(pVLT31-mhpTD75A); D75E, strain W3110ΔmhpT(pVLT31-mhpTD75E); A272H, strain W3110ΔmhpT(pVLT31-mhpTA272H); K276D, strain W3110ΔmhpT(pVLT31-mhpTK276D). All assays were performed in triplicate, and standard deviations are represented by error bars.
DISCUSSION
Accumulating evidence indicates that the active transport of aromatic acids is widespread among bacteria (10, 13, 15, 16, 32–34). In addition to E. coli K-12 (7), Rhodococcus globerulus PWD1 (4), Comamonas testosteroni TA441 (5), and Cupriavidus necator JMP134 (6) were also reported to be able to grow on 3HPP. Several enzymes involved in 3HPP catabolism in various bacterial strains (4, 5, 35, 36) and its regulation in E. coli K-12 (9) have been thoroughly investigated. However, no biochemical or genetic analysis of 3HPP transport has been reported until this study, in which MhpT from E. coli K-12 was identified as an active 3HPP transporter involved in its catabolism. Among the other identified 3HPP utilizers, putative transporter-encoding genes hppK and mhpT were located on 3HPP catabolic clusters in strain PWD1 (4) and strain JMP134 (6), respectively. HppK (GenBank accession number AAB81315) from strain PWD1 and MhpT (GenBank accession number YP_299064) from strain JMP134 showed 26% and 49% identities, respectively, with MhpT in this study. Considering that both hppK and mhpT are located on the 3HPP catabolic clusters, it is likely that both of them encode 3HPP transporters. If this is the case, active transport driven by a specific transporter in the microbial catabolism of 3HPP probably occurs in phylogenetically divergent genera.
It is generally accepted that the undissociated forms of aromatic acids are able to diffuse across membranes through passive diffusion (16). In the current study, with the increase of pH values, more and more 3HPP (pKa = 4.68, calculated by ACD/Labs software) is changed to dissociated forms. This subsequently resulted in less 3HPP being able to enter into cells by passive diffusion. Thus, the significant decreases in transport activities (as shown in Table 3) with the increase in the pH value for the tested strains suggested that 3HPP diffusion is pH value dependent in strain W3110. These observations are somewhat different from those for 3-hydroxybenzoate transport in Pseudomonas putida (16) and gentisate transport in Corynebacterium glutamicum (17), in which the transport activities in transport-deficient strains were decreased with an increase in the pH values, but the activities were almost the same in the wild type and the complemented mutant strain at different pH values. On the other hand, the growth ability of strain W3110ΔmhpT at pH 6.2 and 7.2 and its inability to grow at pH 8.2 indicated that the absence of mhpT in the mutant strain resulted in a loss of active transport and a subsequent blockage of passive diffusion of 3HPP with increases in the pH value. This is because less than 0.1% 3HPP was present in the undissociated form at pH 8.2 and the 3HPP deficiency in the cells was unable to support the growth of the mutant strain. This is another example showing that the pH value of the growth medium is a major factor in determining whether microbes require certain transporters to grow on aromatic acids, as suggested previously (16).
Like other AAHS members, MhpT also contains 12 predicted α-helix transmembrane regions (TM1 to TM12). It was not surprising that the mutation of Glu-27 (in TM1) or Asp-75 (in the 2-3 loop) to an uncharged alanine resulted in the loss of 3HPP transport activity, similar to the findings of previous studies for PcaK (30, 31), BenK (37), and MhbT (16). On the other hand, the change from a positively charged lysine to a negatively charged aspartate at site 276 resulted in a complete loss of its transport activity. Although Lys-276 (in the 8-9 loop) is conserved at the corresponding residues of all AAHS members, its importance in substrate transport was not illustrated until this study. It can be tentatively proposed that this is also the case in other AAHS members. It is also worth noting that the less conserved Ala-272 in the 8-9 loop was different from the corresponding residues in PcaK (Trp-320), BenK (Trp-305), TfdK (Leu-314), MhbT (Val-311), and GenK (Trp-309). To our surprise, a 30% increase in 3HPP transport activity was observed after the simple and uncharged alanine was changed to a complicated and positively charged histidine. This suggests that although the characteristic features in the 8-9 loop are partially conserved in the AAHS family (30), the less conserved residue Ala-272 in MhpT may play an important role in transport efficiency during 3HPP transport. It would be intriguing to see if this is also the case when the corresponding residue in other AAHS members is mutated. Meanwhile, given the fact that strain W3110ΔmhpT(pVLT31-mhpTA272H) had improved 3HPP transport activity and a slightly higher growth rate, substrate transport is likely a limiting step for 3HPP utilization by E. coli K-12 W3110, and MhpT with His-272 is probably a better version of the 3HPP transporter for the growth of this strain on 3HPP.
ACKNOWLEDGMENTS
This work was supported by grants from the National Natural Science Foundation of China (grant 30900014) and the National Key Basic Research Program of China (973 Program, grant 2012CB725202).
Footnotes
Published ahead of print 9 August 2013
REFERENCES
- 1.Díaz E, Ferrández A, Prieto MA, García JL. 2001. Biodegradation of aromatic compounds by Escherichia coli. Microbiol. Mol. Biol. Rev. 65:523–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andreoni V, Bestetti G. 1986. Comparative analysis of different Pseudomonas strains that degrade cinnamic acid. Appl. Environ. Microbiol. 52:930–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dagley S, Chapman PJ, Gibson DT. 1965. The metabolism of β-phenylpropionic acid by an Achromobacter. Biochem. J. 97:643–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnes MR, Duetz WA, Williams PA. 1997. A 3-(3-hydroxyphenyl)propionic acid catabolic pathway in Rhodococcus globerulus PWD1: cloning and characterization of the hpp operon. J. Bacteriol. 179:6145–6153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arai H, Yamamoto T, Ohishi T, Shimizu T, Nakata T, Kudo T. 1999. Genetic organization and characteristics of the 3-(3-hydroxyphenyl)propionic acid degradation pathway of Comamonas testosteroni TA441. Microbiology 145(Pt 10):2813–2820 [DOI] [PubMed] [Google Scholar]
- 6.Pérez-Pantoja D, De la Iglesia R, Pieper DH, González B. 2008. Metabolic reconstruction of aromatic compounds degradation from the genome of the amazing pollutant-degrading bacterium Cupriavidus necator JMP134. FEMS Microbiol. Rev. 32:736–794 [DOI] [PubMed] [Google Scholar]
- 7.Burlingame R, Chapman PJ. 1983. Catabolism of phenylpropionic acid and its 3-hydroxy derivative by Escherichia coli. J. Bacteriol. 155:113–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferrández A, Garciá JL, Díaz E. 1997. Genetic characterization and expression in heterologous hosts of the 3-(3-hydroxyphenyl)propionate catabolic pathway of Escherichia coli K-12. J. Bacteriol. 179:2573–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Torres B, Porras G, García JL, Díaz E. 2003. Regulation of the mhp cluster responsible for 3-(3-hydroxyphenyl)propionic acid degradation in Escherichia coli. J. Biol. Chem. 278:27575–27585 [DOI] [PubMed] [Google Scholar]
- 10.Chaudhry MT, Huang Y, Shen XH, Poetsch A, Jiang CY, Liu SJ. 2007. Genome-wide investigation of aromatic acid transporters in Corynebacterium glutamicum. Microbiology 153:857–865 [DOI] [PubMed] [Google Scholar]
- 11.Pao SS, Paulsen IT, Saier MH., Jr 1998. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 62:1–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ren Q, Kang KH, Paulsen IT. 2004. TransportDB: a relational database of cellular membrane transport systems. Nucleic Acids Res. 32:D284–D288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collier LS, Nichols NN, Neidle EL. 1997. benK encodes a hydrophobic permease-like protein involved in benzoate degradation by Acinetobacter sp. strain ADP1. J. Bacteriol. 179:5943–5946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nichols NN, Harwood CS. 1997. PcaK, a high-affinity permease for the aromatic compounds 4-hydroxybenzoate and protocatechuate from Pseudomonas putida. J. Bacteriol. 179:5056–5061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leveau JH, Zehnder AJ, van der Meer JR. 1998. The tfdK gene product facilitates uptake of 2,4-dichlorophenoxyacetate by Ralstonia eutropha JMP134(pJP4). J. Bacteriol. 180:2237–2243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu Y, Gao XL, Wang SH, Liu H, Williams PA, Zhou NY. 2012. MhbT is a specific transporter for 3-hydroxybenzoate uptake by gram-negative bacteria. Appl. Environ. Microbiol. 78:6113–6120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu Y, Wang SH, Chao HJ, Liu SJ, Zhou NY. 2012. Biochemical and molecular characterization of the gentisate transporter GenK in Corynebacterium glutamicum. PLoS One 7:e38701. 10.1371/journal.pone.0038701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prieto MA, García JL. 1997. Identification of the 4-hydroxyphenylacetate transport gene of Escherichia coli W: construction of a highly sensitive cellular biosensor. FEBS Lett. 414:293–297 [DOI] [PubMed] [Google Scholar]
- 19.Bertani G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62:293–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bertani G. 2004. Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J. Bacteriol. 186:595–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 22.Walker AW, Keasling JD. 2002. Metabolic engineering of Pseudomonas putida for the utilization of parathion as a carbon and energy source. Biotechnol. Bioeng. 78:715–721 [DOI] [PubMed] [Google Scholar]
- 23.Rapp M, Drew D, Daley DO, Nilsson J, Carvalho T, Melén K, De Gier JW, Von Heijne G. 2004. Experimentally based topology models for E. coli inner membrane proteins. Protein Sci. 13:937–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Y, Yan DZ, Zhou NY. 2006. Heterologous expression and localization of gentisate transporter Ncg12922 from Corynebacterium glutamicum ATCC 13032. Biochem. Biophys. Res. Commun. 346:555–561 [DOI] [PubMed] [Google Scholar]
- 25.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bachmann BJ. 1972. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol. Rev. 36:525–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pogulis RJ, Vallejo AN, Pease LR. 1996. In vitro recombination and mutagenesis by overlap extension PCR. Methods Mol. Biol. 57:167–176 [DOI] [PubMed] [Google Scholar]
- 28.Zwietering MH, Jongenburger I, Rombouts FM, van't Riet K. 1990. Modeling of the bacterial growth curve. Appl. Environ. Microbiol. 56:1875–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joshi AK, Ahmed S, Ferro-Luzzi Ames G. 1989. Energy coupling in bacterial periplasmic transport systems. Studies in intact Escherichia coli cells. J. Biol. Chem. 264:2126–2133 [PubMed] [Google Scholar]
- 30.Ditty JL, Harwood CS. 1999. Conserved cytoplasmic loops are important for both the transport and chemotaxis functions of PcaK, a protein from Pseudomonas putida with 12 membrane-spanning regions. J. Bacteriol. 181:5068–5074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ditty JL, Harwood CS. 2002. Charged amino acids conserved in the aromatic acid/H+ symporter family of permeases are required for 4-hydroxybenzoate transport by PcaK from Pseudomonas putida. J. Bacteriol. 184:1444–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allende JL, Gibello A, Martin M, Garrido-Pertierra A. 1992. Transport of 4-hydroxyphenylacetic acid in Klebsiella pneumoniae. Arch. Biochem. Biophys. 292:583–588 [DOI] [PubMed] [Google Scholar]
- 33.Harwood CS, Nichols NN, Kim MK, Ditty JL, Parales RE. 1994. Identification of the pcaRKF gene cluster from Pseudomonas putida: involvement in chemotaxis, biodegradation, and transport of 4-hydroxybenzoate. J. Bacteriol. 176:6479–6488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miguez CB, Greer CW, Ingram JM, Macleod RA. 1995. Uptake of benzoic acid and chloro-substituted benzoic acids by Alcaligenes denitrificans BRI 3010 and BRI 6011. Appl. Environ. Microbiol. 61:4152–4159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bugg TD. 1993. Overproduction, purification and properties of 2,3-dihydroxyphenylpropionate 1,2-dioxygenase from Escherichia coli. Biochim. Biophys. Acta 1202:258–264 [DOI] [PubMed] [Google Scholar]
- 36.Spence EL, Kawamukai M, Sanvoisin J, Braven H, Bugg TD. 1996. Catechol dioxygenases from Escherichia coli (MhpB) and Alcaligenes eutrophus (MpcI): sequence analysis and biochemical properties of a third family of extradiol dioxygenases. J. Bacteriol. 178:5249–5256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang SH, Xu Y, Liu SJ, Zhou NY. 2011. Conserved residues in the aromatic acid/H+ symporter family are important for benzoate uptake by NCgl2325 in Corynebacterium glutamicum. Int. Biodeterior. Biodegrad. 65:527–532 [Google Scholar]
- 38.Woodcock DM, Crowther PJ, Doherty J, Jefferson S, Decruz E, Noyerweidner M, Smith SS, Michael MZ, Graham MW. 1989. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 17:3469–3478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Lorenzo V, Eltis L, Kessler B, Timmis KN. 1993. Analysis of Pseudomonas gene products using lacIq/Ptrp-lac plasmids and transposons that confer conditional phenotypes. Gene 123:17–24 [DOI] [PubMed] [Google Scholar]