Abstract
The uncultured miscellaneous crenarchaeotic group (MCG) archaea comprise one of the most abundant microbial groups in the Earth's subsurface environment. However, very little information is available regarding the lifestyle, physiology, and factors controlling the distribution of members of this group. We established a novel method using both cultivation and molecular techniques, including a pre-PCR propidium monoazide treatment, to investigate viable members of the MCG in vitro. Enrichment cultures prepared from estuarine sediment were provided with one of a variety of carbon substrates or cultivation conditions and incubated for 3 weeks. Compared with the samples from time zero, there was an order-of-magnitude increase in the number of MCG 16S rRNA genes in almost all cultures, indicating that MCG archaea are amenable to in vitro cultivation. None of the tested substrates or conditions significantly stimulated growth of MCG archaea more than the basal medium alone; however, glycerol (0.02%) had a significantly inhibitory effect (P < 0.05). Diversity analysis of populations resulting from four culture treatments (basal medium, addition of amino acids, H2-CO2 as the gas phase, or initial aerobic conditions) revealed that the majority of viable MCG archaea were affiliated with the MCG-8 and MCG-4 clusters. There were no significant differences in MCG diversity between these treatments, also indicating that some members of MCG-4 and MCG-8 are tolerant of initially oxic conditions. The methods outlined here will be useful for further investigation of MCG archaea and comparison of substrates and cultivation conditions that influence their growth in vitro.
INTRODUCTION
With the application of gene-based technologies in microbial ecology, it has become increasingly evident that the diversity of microbial life in natural ecosystems far exceeds that which has been revealed by cultivation-based studies (1, 2). Subsurface environments are quite typical in this regard, and almost every phylogenetic analysis of a sedimentary ecosystem has revealed an abundance of microorganisms from presently uncultivated and often deeply branching phylogenetic lineages of both Bacteria and Archaea (3). While rapid advances in sequencing technologies are affording deeper insight into the phylogenetic composition of microbial communities, the metabolic function of most members of these communities remains speculative or is completely unknown. Metagenomics, proteomics, and transcriptomic approaches have helped to obtain insights into metabolic capabilities of communities in general or specific members thereof (4–7). However, cultivation, i.e., growth on specific substrates, remains the final proof of metabolic activity and is required for detailed physiologic study. Although the majority of microorganisms are not yet cultivable in artificial media as pure cultures, the combination of enrichment cultivation and gene-based analyses can provide valuable insight into the function of microorganisms, often not possible using gene-based techniques alone (2).
In the present study, we sought to use a combination of molecular and cultivation-based techniques to investigate the possible phenotype of members of the miscellaneous crenarchaeotic group (MCG) archaea. MCG archaea are regularly detected in subsurface ecosystems (marine and estuarine sediments), and they have also been found in a variety of other habitats, including hydrothermal vents, water columns, aquifers, and soils (e.g., see reference 8). The MCG is a phylogenetically diverse group, with 16S rRNA gene sequence identities between the most distant members being as low as 76% (8). As a comparison within the domain Bacteria, this level of divergence would represent at least order-level diversity, and as one example, the most distant members of the very heterogeneic order Clostridiales share approximately 77% 16S rRNA gene identity. The wide distribution of MCG archaea in sediments as well as evidence from the carbon isotopic composition of archaeal cells in MCG-dominated sediments (9) have led to the hypothesis that MCG archaea are anaerobic heterotrophs (3). Webster et al. (10) found evidence for MCG involvement in acetate cycling, and recent genomic and metagenomic information suggested that members of the MCG are involved in protein degradation (11) and that others may be involved in protocatechuate degradation (12). However, beyond that, there are no clear indications about the function of this widespread microbial group in sediments and their potentially significant role in elemental cycling in Earth's biosphere.
The aim of the present study was to establish a method for quantifying and comparing the growth of MCG archaea in enrichment cultures, in order to examine the response of MCG archaea to various substrates and cultivation conditions. We sought to employ the membrane-impermeant dye propidium monoazide (PMA) (13) in our analyses, in order to exclude DNA from nonviable cells and therefore examine viable MCG archaea in enrichment cultures.
MATERIALS AND METHODS
Sample collection.
Sediment cores were collected from a 1.5-m water depth in the White Oak River estuary, NC (34°44.141′N, 77°07.298′W), in January 2012, a sedimentary system known as a “natural enrichment” of a dominant and highly diverse assemblage of MCG archaea (8). Sediment from various horizon depths (14 to 42 cm) was transferred into sterilized glass Schott bottles (500 ml, precombusted at 450°C for 6 h), using utensils sterilized with 70% (vol/vol) ethanol. Bottles were then sealed with autoclaved (121°C for 20 min) butyl rubber stoppers that had been prewashed in 1 N potassium hydroxide. The headspace gas was replaced with nitrogen, and samples were stored at 4°C for subsequent cultivation experiments over the following 9 months.
Media and cultivation conditions.
Strictly anaerobically prepared (according to standard techniques [14]) 1/2SMEbc medium was the basal medium used for almost all cultures in this study. 1/2SMEbc medium was a half-strength modification of the synthetic seawater SME medium described previously by Stetter et al. (15) and was chosen as a suitable medium for the present study after preliminary investigations testing various basal media (dilutions of SME medium with and without added bicarbonate) showed that MCG archaea were most readily detectable in successive transfers on this medium (data not shown), suggesting that it was suitable for their growth. 1/2SMEbc medium contained (per liter) 13.85 g NaCl, 3.5 g MgSO4 · 7H2O, 3 g NaHCO3, 2.75 g MgCl2 · 6H2O, 0.5 g KH2PO4, 0.5 g NH4Cl, 0.38 g CaCl2 · 2H2O, 0.33 g KCl, 0.05 g NaBr, 0.015 g H3BO3, 7.5 mg SrCl2 · 6H2O, 1 mg resazurin, 25 μg KI, 1 ml of 10× Wolfe's minerals, and 1 ml of 10× Wolfe's vitamin stock [solutions described previously by Wolin et al. (16) except prepared as 10× stock solutions, and the mineral stock solution was prepared at pH 1.0 without a chelating agent and also including (per liter) 2.8 g (NH4)2Ni(SO4)2 · 6H2O and 0.1 g each of Na2WO4 · 2H2O and Na2SeO4]. The final pH of the medium was adjusted to 7.0, to correspond with the reported in situ pH for the White Oak River basin (17). Medium was usually dispensed by placing 50 ml into 120-ml serum bottles in an anaerobic chamber (atmosphere of 95% [vol/vol] N2 and 5% [vol/vol] H2; Coy Laboratory Products Inc., Ann Arbor, MI, USA). The bottles were sealed with butyl rubber stoppers and aluminum crimp seals, evacuated three times, and provided with 200 kPa of the desired gas mixture before sterilization.
Slurry preparation and sediment incubations for testing of specific substrates.
Sediment slurries were prepared freshly before each experiment. Approximately 22.5 g of sediment from a depth of 25 to 38 cm below the sea floor was added to 70 ml of 1/2SMEbc medium by using sterile spoons inside an anaerobic chamber, as described above. Aliquots (1.5 ml) of this sediment slurry were introduced into 50 ml standard basal medium (1/2SMEbc medium with 200 kPa H2-N2-CO2 [15:65:20, vol/vol/vol] in the headspace, unless otherwise specified) via a syringe. Prereduced sterile stock solutions of additives were introduced into the medium in a proportion of 0.5 ml per 50 ml immediately before inoculation. The stock solution of the additive “sediment dissolved organic matter” (SDOM) that was used in some treatments (as 1 ml in 50 ml) was previously prepared by Soxhlet extraction of 72 g of White Oak River estuary sediment (at a depth of 0 to 50 cm) with 400 ml water over 24 h.
Each culture treatment was performed in triplicate. Exact details of each treatment are listed in Table 2. Briefly, tested additives or cultivation conditions included (separately) acetate, glucose, a mixture of 20 amino acids, methanol, protocatechuic acid, pyruvate, SDOM, SDOM with acetate, glycerol, a mixture of complex undefined organics, selected citric acid cycle intermediates, headspace containing H2-CO2, headspace containing N2-CO2, an aerobic variation of 1/2SMEbc medium which did not contain the reducing agent sodium sulfide or the resazurin indicator and which had an air headspace, and a variation of 1/2SMEbc medium that contained 1 g liter−1 cysteine hydrochloride as a reducing agent instead of sodium sulfide. Triplicate control cultures consisting of basal medium were incubated along with tests. Triplicate cultures on basal medium were also harvested at the beginning of each experiment for determinations at time zero. Triplicate killed controls on basal medium were prepared by autoclaving at 121°C for 40 min after inoculation and before incubation. Cultures were incubated horizontally, without shaking, in the dark at 20°C for 3 weeks and were inverted once after 10 days of incubation. The incubation temperature was chosen as a midpoint between measured summer and winter temperatures at the White Oak River basin (18), where MCG populations have been found to be stable throughout the year.
Table 2.
Average MCG 16S rRNA gene copy numbers from triplicate culture treatments incubated for 3 weeks with various additives or cultivation conditions
| Treatment | Additive/culture conditiona | Avg no. of MCG 16S rRNA gene copies per ml of culturef | No. of MCG 16S rRNA gene copies relative to incubated control |
|---|---|---|---|
| Time zero | None | (2.34 ± 2.04) × 104 | |
| Incubated control | None | (3.18 ± 0.56) × 105 A* | |
| Acetate | Acetate (2 mM) | (3.02 ± 0.10) × 105 | 0.95 |
| Glucose | Glucose (2 mM) | (2.33 ± 0.39) × 105 A* | 0.73 |
| Amino acid mix | Equal concn of 20 amino acidsb to final total amino acid concn of 2 mM | (4.84 ± 0.20) × 105 A*** | 1.52 |
| Methanol | Methanol (2 mM) | (3.46 ± 0.20) × 105 A* | 1.09 |
| Protocatechuic acid | Protocatechuic acid (2 mM) | (3.55 ± 0.07) × 105 A** | 1.12 |
| Pyruvate | Pyruvate (2 mM) | (2.07 ± 0.59) × 105 | 0.65 |
| SDOM | Sediment dissolved organic matterc (1 ml) | (1.95 ± 0.17) × 105 A** | 0.61 |
| SDOM + acetate | Sediment dissolved organic matterc (1 ml) + acetate (2 mM) | (1.54 ± 0.33) × 105 A* | 0.48 |
| Glycerol | Glycerol (0.02%, vol/vol) | (7.37 ± 2.64) × 104 B* | 0.23 |
| Complex undefined | Yeast extract (0.01%, wt/vol) and peptone and Casamino Acids (each at 0.005%, wt/vol) | (2.65 ± 0.45) × 105 A* | 0.83 |
| Citric acid cycle intermediates | Malate, succinate, citrate, fumarate, and oxaloacetate (1 mM each) | (1.94 ± 0.41) × 105 A* | 0.61 |
| H2-CO2 | H2-CO2 headspaced | (3.72 ± 0.38) × 105 A** | 1.17 |
| N2-CO2 | N2-CO2 headspaced | (5.22 ± 3.02) × 105 | 1.64 |
| Aerobic medium | Medium prepared aerobically and reducing agent and resazurin indicator omitted; air headspacee | (6.22 ± 1.62) × 105 | 1.96 |
| Cysteine | Cysteine hydrochloride monohydrate (1 g liter−1) instead of Na2S · 3H2O as reducing agent | (3.53 ± 0.15) × 105 | 1.11 |
| Autoclaved control | None | (1.30 ± 0.58) × 102 B* | 0.00 |
Unless otherwise indicated, basal medium was anaerobically prepared in 1/2SMEbc, and initial headspaces were 200 kPa H2-N2-CO2 (15:65:20 [vol/vol]).
Amino acids used were the l-forms of arginine, histidine, lysine, aspartic acid, glutamic acid, serine, threonine, asparagine, glutamine, cysteine, glycine, proline, alanine, valine, isoleucine, leucine, methionine, phenylalanine, tyrosine, and tryptophan, each at a final concentration of 0.1 mM.
Total organic carbon and total nitrogen concentrations of the SDOM stock were determined to be 7 mM and 0.5 mM, respectively, by using a Shimadzu TOC/TN 5000 analyzer, calibrated with consensus reference materials for dissolved organic carbon provided by the NSF-CRM program (http://www.rsmas.miami.edu/groups/biogeochem/CRM.html).
N2-CO2 was 80:20 (vol/vol), and H2-CO2 was 80:20 (vol/vol) at 200 kPa.
Air headspace was not provided at overpressure.
Error is represented as the standard error of the mean (n = 3). Significant differences in treatments compared to cultures at time zero or to the incubated control are indicated by the letters A and B, respectively, and asterisks indicate the level of significance, where * indicates a P value of <0.05, ** indicates a P value of <0.01, and *** indicates a P value of <0.001.
DNA extraction, PMA treatment, and qPCR.
DNA was extracted from 20 ml of harvested (13,000 × g for 10 min at 4°C) sediment slurries or cultures by using part of a cetyltrimethylammonium bromide (CTAB) bead-beating-based method outlined previously (19) in combination with column-based purification of nucleic acids. Rather than isopropanol precipitation of nucleic acids (19), the aqueous phase after phenol-chloroform-isoamyl alcohol treatment was processed by using the final column-based steps of the UltraClean soil DNA isolation kit (Mo Bio, Carlsbad, CA). Pre-PCR propidium monoazide (PMA) treatment to intercalate DNA from nonviable cells was performed as outlined by Nocker et al. (13), with minor modifications, as follows: pellets from harvested sediment slurries were immediately resuspended in 1.5 ml of cold phosphate-buffered saline (PBS) (pH 7.2), and PMA (in water) (Biotium Inc., Hayward, CA) was added to a final concentration of 50 μM, before incubation in the dark at room temperature for 10 min, with continuous gentle inversion throughout. Sample tubes were then placed on ice, with the lids opened, in a biohazard cabinet and exposed to light from two halogen lamps (500 W and 400 W) held side by side at a distance of 20 cm for 5 min; samples were on a shaker during this process to ensure adequate exposure of all parts of each sample to the light. For comparison, samples that were not PMA treated were subjected to the same incubation in the dark and exposure to light as the PMA-treated samples.
For quantitative real-time PCR (qPCR) of MCG 16S rRNA genes, the MCG-specific primer pair 528f and 732r (Table 1) was used. qPCR mixtures (10-μl reaction mixtures) contained final concentrations of 1× QuantiTect SYBR green PCR mix (Qiagen, Hilden, Germany), 0.3 μM each forward and reverse primer, and 1 μl of template DNA. qPCRs were performed by using a Rotor Gene 6000 instrument and analyzed by using Rotor Gene 6000 series software 1.7 (Qiagen). qPCR cycling was performed as follows: 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 15 s, annealing at 60°C for 25 s, and extension at 72°C for 20 s, with fluorescence acquisition at the end of each cycle, followed by dissociation curve analysis from 65°C to 95°C increasing at a rate of 0.2°C s−1, with continuous fluorescence measurement. qPCRs were performed in triplicate. Negative controls were included in each assay by omitting the DNA template. Results were expressed as the number of 16S rRNA gene copies per μl DNA. A standard curve of MCG 16S rRNA genes was generated from a 10-fold dilution series of 108 gene copies to 103 gene copies per μl using PCR amplicons obtained from White Oak River estuary sediment with primers 493f and 1069r (Table 1) (used at an annealing temperature of 50°C) and then cloned as previously described (19) and purified.
Table 1.
Primers used in this study
| Primer | Sequence (5′–3′) | Reference | Target group |
|---|---|---|---|
| 493f | GGAATAAGGAGAGGGCAAG | 8a | MCG archaea |
| 915r | GTGCTCCCCCGCCAATTCCT | 36 | General archaea |
| 528f | CGGTAATACCAGCTCTCCGAG | 8 | MCG archaea |
| 732r | CGCGTTCTAGCCGACAGC | 8 | MCG archaea |
| 1069r | ACCTCACGGCACGAA | 37 | Group C2 archaea |
Modified from the 493 fluorescence in situ hybridization (FISH) probe described by Kubo et al. (8).
Statistical analyses.
16S rRNA gene copies per μl DNA were converted to 16S rRNA gene copies per ml of culture by assuming 100% DNA recovery and equal DNA extraction efficiency for all samples. Results are presented as means ± standard errors. Student's t test was used to compare differences in means, and differences were considered significant at a P value of <0.05.
Clone libraries and diversity analyses.
MCG diversity analyses were performed on DNA from four culture treatments: (i) incubated control (CON), (ii) H2-CO2 headspace (HY), (iii) amino acid mix (AA), and (iv) initial aerobic conditions (OX). PCR mixtures (25 or 50 μl) contained final concentrations of 1× PCR buffer (10× buffer stock [200 mM Tris-HCl {pH 8.4} and 500 mM KCl]), 3 mM MgCl2, 0.2 mM deoxyribonucleotide triphosphates, 0.14 μM each primer, 1 U of Platinum Taq DNA polymerase (Invitrogen, Carlsbad, CA), and 1 μl of template DNA. PCR conditions were as follows: initial denaturation step at 95°C for 5 min followed by 30 cycles of denaturation at 95°C for 30 s, annealing at 58°C for 30 s, and extension at 72°C for 30 s, followed by a final extension step at 72°C for 7 min. MCG 16S rRNA genes were amplified from triplicate cultures of each treatment separately. Amplicons were then purified, pooled according to treatment, and cloned and sequenced as previously described (19). Sequences were aligned by using MOTHUR (20) and the Greengenes reference alignment (21). Fine alignments were performed manually in ARB (22). Bootstrapped neighbor-joining trees of 16S rRNA gene sequences were constructed in ARB (22) with 100 resamplings. Sequences were clustered into operational taxonomic units (OTUs) at a distance of ≤0.03 by using MOTHUR (20). LIBSHUFF in MOTHUR (20) was used to compare the structures of 16S rRNA gene libraries.
Nucleotide sequence accession numbers.
Sequences of the clones have been submitted to the GenBank database under accession numbers KF308184 to KF308270 (n = 18 for CON, n = 24 for HY, n = 22 for AA, and n = 23 for OX). Sequences of the clones used for qPCR of MCG 16S rRNA have been submitted to GenBank under accession numbers KF308271 and KF308272.
RESULTS AND DISCUSSION
A diverse, abundant, and active population of MCG archaea exists within the top meter of White Oak River estuary sediments throughout the year (8), making it an ideal sampling site for in vitro studies of the MCG archaea. Initial studies (data not shown) using sediment collected from this site indicated that MCG archaea were amenable to in vitro cultivation and subculture (2 to 4% inoculum) up to six times. However, while growth, as indicated by an increase in MCG 16S rRNA gene copy numbers in cultures, was evident in subsequent subcultures, it was poorer with each subculture, and MCG archaea were never detectable beyond the sixth transfer despite the use of a wide range of tested carbon substrates in the culture medium and culture conditions.
Therefore, the present study was designed to establish a method for detailed quantitative comparison of MCG archaea in cultures grown on various substrates and/or under different culture conditions, which may be useful in shedding light on the factors that influence the growth of MCG archaea in vitro. A range of substances from various broad groups of compound classes (e.g., gases, sugars, short-chain fatty acids, common microbial metabolism intermediates, and complex undefined organics) or specific tests based on the literature (e.g., acetate, amino acids, protocatechuate, and citric acid cycle intermediates [10–12]) were tested in cultures as possible growth stimulants for MCG archaea.
A recently developed pre-PCR PMA treatment (13) was utilized to exclude DNA from nonviable cells in cultures, as it seemed likely that there would be nonviable cells in our samples either from the sediment itself or from microorganisms adversely affected under the provided culture conditions. A comparison of cultures at time zero treated with and without PMA showed an order-of-magnitude decrease in the average number of MCG 16S rRNA genes detected in PMA-treated samples (2.02 × 104 ± 1.74 × 104 copies ml−1) compared with non-PMA-treated samples (2.11 × 105 ± 1.03 × 105 copies ml−1), confirming the importance of this type of treatment in culture-based studies. It is possible that not all DNA from nonviable cells was excluded from cultures in the present study, as the presence of particles in environmental samples has been found to decrease the efficiency with which PMA excludes DNA from nonviable cells (23). However, this was unavoidable using sediment as the inoculum. We considered the level of interference from sediment particles to be equal across all our treatments, which were amended with the same volume of sediment slurry.
After 3 weeks of incubation, MCG 16S rRNA gene copy numbers were approximately 1 order of magnitude higher than those in cultures that had been harvested at time zero, indicating growth of some members of the MCG archaea (Table 2). The variability in MCG 16S rRNA gene copy numbers within given treatments was quite large. Therefore, the increase in the gene copy numbers relative to the numbers at time zero was statistically significant for only some treatments (control incubation, glucose, amino acid mix, methanol, protocatechuic acid, SDOM, SDOM with acetate, complex organics, citric acid cycle intermediates, and H2-CO2 headspace) (Table 2). Surprisingly, growth of MCG archaea was not dependent on or even stimulated by the presence of any of the tested additives. None of the treatments resulted in a significant (P < 0.05) increase in MCG 16S rRNA gene copy numbers relative to the copy numbers of the incubated control on basal medium alone (Table 2). A mixture of 20 amino acids had the most significant effect on MCG 16S rRNA gene copy numbers in culture (i.e., it was the treatment demonstrating the lowest P value [P = 0.088] of the substrates that had higher average MCG 16S rRNA gene copy numbers than the control treatment), which is consistent with the findings of Lloyd et al. (11) that MCG archaea have a genomic potential for protein/amino acid metabolism. However, further investigation of this substrate in a subsequent subculture did not reveal any positive effect on MCG 16S rRNA genes relative to transferred controls. Also, a repeat of the amino acid treatment cultures and control cultures with more extensive sampling (nine replicates each) in order to further examine the statistical significance of this finding failed to show significant (P < 0.05) differences in MCG 16S rRNA gene copy numbers between the substrate and control.
Interestingly, glycerol at a final concentration of 0.02% had a significant (P < 0.05) inhibitory effect on the MCG in vitro. Whether this effect was a direct result of the glycerol itself or the end products from anaerobic glycerol fermentation by other members of the culture (e.g., ethanol [24]) is unknown, but this finding may be relevant for future cultivation-based investigations and for cryopreservation of samples containing MCG (25).
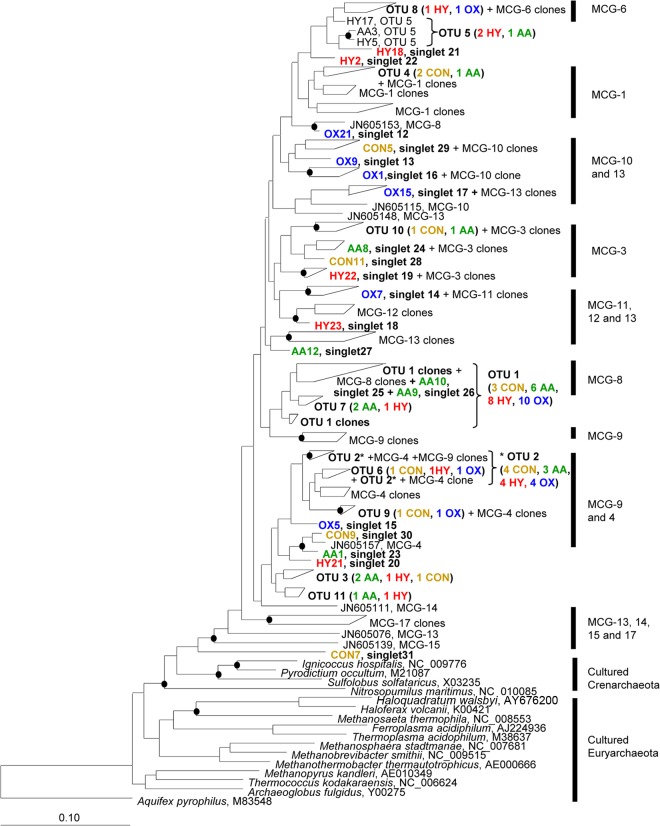
Given the large intragroup phylogenetic diversity within the MCG archaea (3, 8), we hypothesized that some substrates or culture conditions tested in the present study may have been stimulatory for particular subgroups of the MCG archaea and inhibitory for others, therefore not resulting in a significant net change in total MCG 16S rRNA gene copy numbers relative to those of control cultures. This possibility was investigated by MCG diversity analysis with four different culture treatments: the control treatment as well as the three cultures that had resulted in the most substantial increases in average MCG 16S rRNA gene copy numbers relative to those of the control incubation (i.e., amino acid mix, H2-CO2 headspace, and initial aerobic conditions). Surprisingly, quite a large diversity of MCG archaea was found for all four culture treatments (Fig. 1). Sequences clustered at ≤3% sequence divergence as an approximation for species-level clustering (26) formed 31 operational taxonomic units (OTUs). The two largest OTUs were affiliated with the MCG-8 and the MCG-4 subgroups, respectively, and contained sequences from all four of the culture treatments (Fig. 1). Members of MCG-8 and MCG-4 have been detected in a variety of sedimentary ecosystems (3, 8, 27, 28) (sometimes also called PM-2 and PM-7); however, to date, there are no indications about their potential metabolism or function. It is clear from the present study that members within these two MCG subgroups are amenable to in vitro growth in synthetic medium (with residual sediment likely also providing additional essential growth factors). Additionally, members of MCG-8 and MCG-4 seem to tolerate oxygen to a certain extent, quite unlike some other anaerobes, which can be killed by oxygen after very short exposure times (e.g., <2 h in some cases [29]). Interestingly, MCG 16S rRNA gene sequences were recovered from a short-term aerobic culture previously (30), and MCG sequences from fermented seafood (a fermentation process which is not strictly anaerobic) have been reported to coexist with sequences from known aerobic archaea and bacteria (31), which also suggests that some MCG archaea could be tolerant of oxygen. Quite possibly, MCG archaea from the White Oak River basin exhibit oxygen tolerance mechanisms similar to those employed by some sulfate-reducing bacteria, which can remain viable after exposure to oxygen (e.g., for a review, see reference 32). To further test the oxygen tolerance or even oxygen metabolism of MCG archaea, completely oxic incubations (e.g., with shaking and vented headspaces) would be worthwhile in future studies. Attempts in our laboratory to cultivate MCG archaea from White Oak River sediment under the conditions appropriate for the aerobic, marine ammonia-oxidizing archaeon Nitrosopumilus maritimus (33), which is placed in a phylogenetic sister group to the uncultured MCG archaea, were unsuccessful in multiple incubations (our unpublished data).
Fig 1.
Neighbor-joining tree showing MCG sequences recovered from four different culture treatments, control (CON), amino acid mix (AA), H2-CO2 headspace (HY), or aerobic conditions (OX), in the present study. Sequences described previously by Kubo et al. (8) form the framework of the tree, and MCG subgroup labels were determined as indicated by Kubo et al. (see supplemental material in reference 8). GenBank accession numbers for reference species are shown after the species name. Branch nodes with ≥75% bootstrap support are marked with closed circles. The scale bar represents 10% sequence divergence.
The overall MCG community structure between the four investigated culture treatments in the present study (basal medium, addition of amino acids, H2-CO2 as the gas phase, or initial aerobic conditions) was not significantly different (significance of the ΔCXY score, by LIBSHUFF, was >0.05), suggesting that the basal medium itself or components from the sediment and/or interspecies metabolites were the more likely factors supporting and influencing the viable MCG archaea in the present study, rather than the tested additives or culture conditions. The low MCG copy numbers achieved in cultures as well as the repeated loss of MCG archaea in subsequent subcultures support the idea that components from the initial sediment itself, or perhaps other microbes in the sediment, were critical for growth and survival of MCG archaea.
While none of the tested substrates in this study positively influenced growth of MCG archaea in vitro, the medium used in this study was clearly suitable for their growth. The methods described here, particularly with a PMA step to remove background DNA from nonviable cells, will be useful for further in vitro studies on the MCG and will allow confirmation of the growth of these organisms in culture, something that has not been confirmed for MCG archaea in enrichment cultures previously (34). Additionally, the approach outlined here allows comparison of a range of substrates or conditions in parallel, therefore making it suitable for the testing of various hypotheses relatively rapidly without requiring expensive labeled compounds, which have been used to understand uncultured archaea in vitro previously (10). Immediate future studies using this approach to test a range of other possible substrates or substrate concentrations for the MCG would be worthwhile. For example, based on insights about the metabolism of MCG archaea revealed from single-cell genomics (11) and metagenomics (12) studies, cultivation attempts using genomically indicated potential substrates at various concentrations could provide powerful support for genomic evidence about the MCG. Once metabolic factors that select for the MCG have been determined and increased cell densities of MCG archaea have been achieved, other factors, such as temperature, salt, pH, and time of incubation, should be investigated by using a similar approach. Additionally, the relative abundance of MCG archaea in relation to the rest of the microbial population also could be monitored by using qPCR primers for total bacteria and total archaea (e.g., see reference 35), for example, to evaluate whether the presence or absence of other community members affects the MCG. This particular approach in combination with the testing of various microbial inhibitors (e.g., antibiotics and bromoethane sulfonate) is likely to add valuable information about the microbial interactions that affect the MCG archaea in vitro.
Alternatively, more specific qPCR primers could be developed to target specific subgroups within the MCG, for example, MCG-4, MCG-8, or even OTUs within these groups, which have been shown to grow in vitro in the present study. This would provide a more focused approach to understanding the factors that affect specific uncultivable archaea in sedimentary ecosystems. Monitoring of these subgroups over time course incubations (e.g., using higher-volume cultures and subsampling periodically) will be particularly important in future enrichment and isolation attempts, particularly if a PMA step is included in analyses to further reveal which subgroups of the MCG are viable at which incubation time points. We anticipate that, ultimately, the combination of cultivation and molecular methods such as those outlined here will shed light on the factors that affect the MCG archaea in vitro and will eventually facilitate the enrichment and potential isolation of members of this group.
ACKNOWLEDGMENTS
We thank Cassandre Lazar and Andreas Teske for providing sediment samples from the White Oak River basin and for useful discussion about the research. We thank Christine Moissl-Eichinger and Alexander Probst for stimulating discussion, helpful advice, and critically reading the manuscript.
This work was primarily funded by the European Research Council (ERC) under the European Union's Seventh Framework Programme Ideas Specific Programme (ERC grant 247153 to K.-U.H.).
Footnotes
Published ahead of print 9 August 2013
REFERENCES
- 1.Amann R. 1995. Fluorescently labelled, rRNA-targeted oligonucleotide probes in the study of microbial ecology. Mol. Ecol. 4:543–554 [Google Scholar]
- 2.Stewart EJ. 2012. Growing unculturable bacteria. J. Bacteriol. 194:4151–4160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Teske A, Sørensen KB. 2008. Uncultured archaea in deep marine subsurface sediments: have we caught them all? ISME J. 2:3–18 [DOI] [PubMed] [Google Scholar]
- 4.Handelsman J. 2004. Metagenomics: application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 68:669–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riesenfeld CS, Schloss PD, Handelsman J. 2004. Metagenomics: genomic analysis of microbial communities. Annu. Rev. Genet. 38:525–552 [DOI] [PubMed] [Google Scholar]
- 6.Wilmes P, Bond PL. 2009. Microbial community proteomics; elucidating the catalysts and metabolic mechanisms that drive the Earth's biogeochemical cycles. Curr. Opin. Microbiol. 12:310–317 [DOI] [PubMed] [Google Scholar]
- 7.Xu J. 2006. Microbial ecology in the age of genomics and metagenomics: concepts, tools, and recent advances. Mol. Ecol. 15:1713–1731 [DOI] [PubMed] [Google Scholar]
- 8.Kubo K, Lloyd KG, Biddle JF, Amann R, Teske A, Knittel K. 2012. Archaea of the Miscellaneous Crenarchaeotal Group are abundant, diverse and widespread in marine sediments. ISME J. 6:1949–1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biddle JF, Lipp JS, Lever MA, Lloyd KG, Sørensen KB, Anderson R, Fredericks HF, Elvert M, Kelly TJ, Schrag DP, Sogin ML, Brenchley JE, Teske A, House CH, Hinrichs K-U. 2006. Heterotrophic Archaea dominate sedimentary subsurface ecosystems off Peru. Proc. Natl. Acad. Sci. U. S. A. 103:3846–3851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Webster G, Rinna J, Roussel EG, Fry JC, Weightman AJ, Parkes RJ. 2010. Prokaryotic functional diversity in different biogeochemical depth zones in tidal sediments of the Severn Estuary, UK, revealed by stable-isotope probing. FEMS Microbiol. Ecol. 72:179–197 [DOI] [PubMed] [Google Scholar]
- 11.Lloyd K, Schreiber GL, Petersen DG, Kjeldsen KU, Lever MA, Steen AD, Stepanauskas R, Richter M, Kleindienst S, Lenk S, Schramm A, Jørgensen BB. 2013. Predominant archaea in marine sediments degrade detrital proteins. Nature 496:215–220 [DOI] [PubMed] [Google Scholar]
- 12.Wang F, Meng J, Zheng Y, Qin D, Xu J, Xiao X. 2011. Genetic and functional properties of uncultivated Miscellaneous Crenarchaeotal Group (MCG): implications from the metagenome analysis. Miner. Mag. 75:2116 [Google Scholar]
- 13.Nocker A, Cheung C-Y, Camper AK. 2006. Comparison of propidium monoazide with ethidium monoazide for differentiation of live vs. dead bacteria by selective removal of DNA from dead cells. J. Microbiol. Methods 67:310–320 [DOI] [PubMed] [Google Scholar]
- 14.Balch WE, Fox GE, Magrum LJ, Woese CR, Wolfe RS. 1979. Methanogens: reevaluation of a unique biological group. Microbiol. Rev. 43:260–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stetter KO, König H, Stackebrandt E. 1983. Pyrodictium gen. nov., a new genus of submarine disc-shaped sulphur-reducing archaebacteria growing optimally at 105°C. Syst. Appl. Microbiol. 4:535–551 [DOI] [PubMed] [Google Scholar]
- 16.Wolin EA, Wolin MJ, Wolfe RS. 1963. Formation of methane by bacterial extracts. J. Biol. Chem. 238:2882–2886 [PubMed] [Google Scholar]
- 17.Martens CS, Goldhaber MB. 1978. Early diagenesis in transitional sedimentary environments of the White Oak River Estuary, North Carolina. Limnol. Oceanogr. 23:428–441 [Google Scholar]
- 18.Lloyd KG. 2009. Microbially-driven methane and sulfur cycling in a Gulf of Mexico methane seep and the White Oak River Estuary. Ph.D. dissertation. The University of North Carolina, Chapel Hill, NC [Google Scholar]
- 19.Gagen EJ, Denman SE, Padmanabha J, Zadbuke S, Al Jassim R, Morrison M, McSweeney CS. 2010. Functional gene analysis suggests different acetogen populations in the bovine rumen and tammar wallaby forestomach. Appl. Environ. Microbiol. 76:7785–7795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, Buchner A, Lai T, Steppi S, Jobb G, Forster W, Brettske I, Gerber S, Ginhart AW, Gross O, Grumann S, Hermann S, Jost R, Konig A, Liss T, Lussmann R, May M, Nonhoff B, Reichel B, Strehlow R, Stamatakis A, Stuckmann N, Vilbig A, Lenke M, Ludwig T, Bode A, Schleifer K-H. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bae S, Wuertz S. 2009. Discrimination of viable and dead fecal Bacteroidales bacteria by quantitative PCR with propidium monoazide. Appl. Environ. Microbiol. 75:2940–2944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gupta A, Murarka A, Campbell P, Gonzalez R. 2009. Anaerobic fermentation of glycerol in Paenibacillus macerans: metabolic pathways and environmental determinants. Appl. Environ. Microbiol. 75:5871–5883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hubálek Z. 2003. Protectants used in the cryopreservation of microorganisms. Cryobiology 46:205–229 [DOI] [PubMed] [Google Scholar]
- 26.Stackebrandt E, Goebel BM. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846–849 [Google Scholar]
- 27.Li Q, Wang F, Chen Z, Yin X, Xiao X. 2012. Stratified active archaeal communities in the sediments of Jiulong River estuary, China. Front. Microbiol. 3:1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Webster G, Parkes RJ, Cragg BA, Newberry CJ, Weightman AJ, Fry JC. 2006. Prokaryotic community composition and biogeochemical processes in deep subseafloor sediments from the Peru Margin. FEMS Microbiol. Ecol. 58:65–85 [DOI] [PubMed] [Google Scholar]
- 29.Brusa T, Canzi E, Pacini N, Zanchi R, Ferrari A. 1989. Oxygen tolerance of anaerobic bacteria isolated from human feces. Curr. Microbiol. 19:39–43 [Google Scholar]
- 30.Biddle JF, House CH, Brenchley JE. 2005. Enrichment and cultivation of microorganisms from sediment from the slope of the Peru trench (ODP site 1230), p 1–19 In Jørgensen BB, D'Hondt SL, Miller DJ. (ed), Proceedings of the Ocean Drilling Program, scientific results, vol 201 http://www-odp.tamu.edu/publications/201_SR/VOLUME/CHAPTERS/107.PDF
- 31.Roh SW, Kim K-H, Nam Y-D, Chang H-W, Park E-J, Bae J-W. 2010. Investigation of archaeal and bacterial diversity in fermented seafood using barcoded pyrosequencing. ISME J. 4:1–16 [DOI] [PubMed] [Google Scholar]
- 32.Sass H, Cypionka H. 2009. Response of sulphate reducing bacteria to oxygen, p 167–184 In Barton LL, Hamilton WA. (ed), Sulphate-reducing bacteria: environmental and engineered systems. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 33.Könneke M, Bernhard AE, de la Torre J, Walker CB, Waterbury JB, Stahl DA. 2005. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437:543–546 [DOI] [PubMed] [Google Scholar]
- 34.Plasencia A, Bañeras L, Llirós M, Casamayor EO, Borrego C. 2011. Maintenance of previously uncultured freshwater archaea from anoxic waters under laboratory conditions. Antonie Van Leeuwenhoek 99:403–408 [DOI] [PubMed] [Google Scholar]
- 35.Suzuki MT, Taylor LT, DeLong EF. 2000. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl. Environ. Microbiol. 66:4605–4616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stahl DA, Amann R. 1991. Development and application of nucleic acid probes, p 205–248 In Stackebrandt E, Goodfellow M.Nucleic acid techniques in bacterial systematics. Wiley, New York, NY [Google Scholar]
- 37.Sahl JW, Fairfield N, Harris JK, Wettergreen D, Stone WC, Spear JR. 2010. Novel microbial diversity retrieved by autonomous robotic exploration of the world's deepest vertical phreatic sinkhole. Astrobiology 10:201–213 [DOI] [PubMed] [Google Scholar]