Abstract
SUMMARY
The negative impact of cytomegalovirus (CMV) infection on transplant outcomes warrants efforts toward improving its prevention, diagnosis, and treatment. During the last 2 decades, significant breakthroughs in diagnostic virology have facilitated remarkable improvements in CMV disease management. During this period, CMV nucleic acid amplification testing (NAT) evolved to become one of the most commonly performed tests in clinical virology laboratories. NAT provides a means for rapid and sensitive diagnosis of CMV infection in transplant recipients. Viral quantification also introduced several principles of CMV disease management. Specifically, viral load has been utilized (i) for prognostication of CMV disease, (ii) to guide preemptive therapy, (iii) to assess the efficacy of antiviral treatment, (iv) to guide the duration of treatment, and (v) to indicate the risk of clinical relapse or antiviral drug resistance. However, there remain important limitations that require further optimization, including the interassay variability in viral load reporting, which has limited the generation of standardized viral load thresholds for various clinical indications. The recent introduction of an international reference standard should advance the major goal of uniform viral load reporting and interpretation. However, it has also become apparent that other aspects of NAT should be standardized, including sample selection, nucleic acid extraction, amplification, detection, and calibration, among others. This review article synthesizes the vast amount of information on CMV NAT and provides a timely review of the clinical utility of viral load testing in the management of CMV in solid organ transplant recipients. Current limitations are highlighted, and avenues for further research are suggested to optimize the clinical application of NAT in the management of CMV after transplantation.
INTRODUCTION
Cytomegalovirus (CMV) is one of the most common pathogens that infect solid organ transplant (SOT) recipients (1). The impact of CMV on the outcome of SOT is enormous—the virus not only causes a highly morbid and potentially fatal illness but also indirectly influences other relevant outcomes, such as allograft rejection, risk of other opportunistic infections, and overall patient and allograft survival (1). Because of the magnitude of its direct and indirect impacts, there have been extraordinary efforts aimed at defining strategies for its prevention and treatment.
Critical to improving the management of CMV infection after SOT have been the remarkable advances in clinical virology. During the last 2 decades, significant breakthroughs in diagnostic virology have paralleled and facilitated improvements in CMV disease management (2). In particular, CMV diagnostics has been transformed from the use of laborious methods of cell culture, first to the more sensitive method of direct antigen testing and subsequently to the widespread use of viral load testing (reviewed in reference 2). During this period, nucleic acid amplification testing (NAT) moved from the realm of research laboratories to become one of the most commonly performed tests in clinical virology laboratories (2).
NAT provides a means for rapid and sensitive diagnosis of CMV infection in SOT recipients (2). The “real-time” nature of CMV NAT is of utmost importance in the management of the most immunocompromised patients by allowing rapid laboratory confirmation of clinically suspected CMV infections. Moreover, NAT allows for quantification of the amount of CMV present per volume of clinical specimen (known as the viral load) (2). Indeed, the advent of viral quantitation introduced several principles that have transformed the way that CMV disease is prevented and treated. Specifically, viral load has been utilized (i) for prognostication of CMV disease (i.e., viral load is directly correlated with risk or severity of disease), (ii) to guide the need to initiate preemptive therapy, (iii) to assess the virological efficacy of antiviral treatment, (iv) to guide the duration of treatment, and (v) to indicate the risk of clinical relapse or antiviral drug resistance (1–3).
However, there remain important limitations to such testing, including the lack (until recently) of widely adopted quantitative standards (4, 5). Several platforms and methodologies are used for CMV NAT (2, 6), and in the absence of calibration to a common international reference standard, their performance could not be compared directly and accurately. Accordingly, universal viral load thresholds for various clinical indications (i.e., for guiding preemptive therapy, treatment responses, and the risk of relapse) have not been defined accurately. Numerous studies have reported viral load values for various clinical indications, but these remain specific to the assay, institution, and patient population used in a given study. To address this limitation, the World Health Organization released the first international standard for CMV quantitation, in 2010 (7). The availability of this standard should aid in the generation of potential viral load thresholds that are comparable across centers and widely applicable for different clinical applications. In addition, standardization of other aspects of NAT may still be needed to further improve the portability of viral load reporting. Viral load thresholds may need to be generated for various patient groups, since thresholds may vary depending on risk profiles.
In this article, we review the various methodologies for CMV diagnosis, with particular emphasis on CMV NAT. We synthesize the vast amount of information published to date on CMV NAT and provide a timely review of the clinical utility of CMV load testing in the management of SOT recipients. We highlight its current limitations and suggest avenues for research to further optimize the clinical application of NAT in the management of CMV after transplantation.
VIRAL EPIDEMIOLOGY AND MECHANISMS OF INFECTION
CMV infection is widespread and occurs worldwide. A recent epidemiologic survey in the United States reported an overall CMV seroprevalence of 50.4% (8). Other studies have described seroprevalence rates as high as 100% in some populations (9). Seroprevalence rates vary depending on age (higher rates are observed among older persons), geography (higher rates in developing countries), and socioeconomic status (higher rates in economically depressed regions) (8, 9).
CMV is acquired from exposure to saliva, tears, urine, stool, breast milk, semen, and other bodily secretions of infected individuals (10). Contact with contaminated environmental surfaces containing viable virus may also transmit the virus (10). CMV can also be acquired through blood transfusion and organ transplantation from CMV-infected donors (1, 11, 12).
Primary CMV infection occurs most commonly during the first 2 decades of life (13). In immunocompetent individuals, primary CMV infection is generally asymptomatic. In some patients, a benign self-limited febrile illness may ensue and last for several days. This febrile illness may be accompanied by generalized lymphadenopathy and may mimic the clinical illness of infectious mononucleosis (14).
Primary CMV infection induces a robust cellular and humoral immune response (15). CMV immunoglobulin M (CMV-IgM) is initially secreted during early CMV infection, and the detection of CMV-IgM by serologic assays is indicative of active, acute, or recent infection (16). Weeks into the course of primary infection, CMV-IgG antibody is secreted, and this antibody persists for life (16). The detection of CMV-IgG is indicative of previous or past infection (16). Durable control of CMV infection is the function of a robust cell-mediated immunity, with generation of CMV-specific CD4+ and CD8+ T cells (15, 17–19). Suppression of the number and function of CMV-specific CD4+ and CD8+ T cells allows for reactivation of the virus from latency, leading to uncontrolled viral replication and clinical disease in immunocompromised patients, including SOT recipients (15, 17, 18).
Several studies have demonstrated complex immune evasion mechanisms that circumvent the ability of humoral and cell-mediated immunity to eliminate CMV (20). Accordingly, primary CMV infection results in the virus entering a state of latency. The latent CMV genome has been demonstrated to be present in numerous cells of the body, including macrophages, mononuclear cells, neutrophils, polymorphonuclear cells, epithelial and endothelial cells, fibroblasts, neuronal cells, and parenchymal cells, among others (21–23). The tissue distributions of cells harboring latent CMV are widespread and include the bone marrow, liver, kidney, gastrointestinal tract, lungs, and brain. The widespread cellular distribution of viral latency may account for the potentially multisystemic involvement of disease caused by this virus.
Cells harboring latent CMV serve as niduses of viral reactivation (3). Viral reactivation occurs intermittently throughout life, even in immunologically competent hosts (24). These intermittent viral reactivation events can pose considerable and recurrent burdens to the immune system (25). In healthy individuals, these events trigger immunologic memory, which effectively controls viral replication at a low subclinical level, without apparent clinical effects (20, 25). In individuals with impaired immune function, such as SOT recipients, viral reactivation is the initial step in the pathogenesis of a potentially severe CMV disease (3, 26). What controls the fate of CMV reactivation in these patients is the state of pathogen-specific immunity. Severely impaired CMV-specific immunity in a SOT recipient may permit uncontrolled viral replication, leading to high levels of viremia and clinically manifesting with systemic and, often, tissue-invasive illness (3, 27, 28).
Cells that harbor latent virus serve as primary vectors for transmission of CMV to susceptible individuals. Since latent CMV is widely distributed in almost every organ, its transmission through organ transplantation is very likely (3, 11, 12). In this context, primary infection occurs when a CMV-seropositive donor transmits the latent virus through organ donation to a susceptible CMV-seronegative transplant recipient (herein referred to as CMV D+/R−). The CMV D+/R− mismatch category occurs in an estimated 20 to 25% of all SOT recipients (3) and is the single most important risk factor for the development of CMV disease in SOT recipients (3, 29–35). Much less commonly, primary CMV infection may be acquired from exposure to infected individuals in the community (i.e., saliva and other body fluids) or through transfusion of blood from CMV-infected donors (3, 29). Because CMV-seronegative SOT recipients lack preexisting CMV-specific humoral and cell-mediated immunity, their ability to suppress viral reactivation (in the infected allograft) is nonexistent, thereby allowing for very rapid CMV replication dynamics. The growth rate has been calculated at 1.82 units/day (95% confidence interval [CI], 1.44 to 2.56 units/day) (36), which is commensurate to a doubling time of <2 days (Table 1) (3, 29–43). Clinically, this translates to a higher incidence and greater severity of CMV disease, characterized by high viral loads, among CMV D+/R− SOT recipients (3, 5, 36, 41–45).
Table 1.
Viral replication kinetics, incidence rates, and severity of CMV disease in solid organ transplant recipients
| Parameter | Value or descriptiona |
|||
|---|---|---|---|---|
| CMV D+/R− SOT recipients |
CMV R+ SOT recipients |
|||
| Viral replication kinetics (units/day) (mean [95% CI]) | 1.82 (1.44–2.56) | 0.61 (0.55–0.7) | ||
| Viral doubling time (days) (mean [range]) | 1.54 (0.5–5.5) | 2.67 (0.27–26.7) | ||
| Viral load at time of CMV disease diagnosis (absolute values dependent on specific viral load assay) | High to very high | Low to high | ||
| Severity of CMV disease | Often moderate to severe | Often mild to moderate | ||
| Incidence (%) of CMV disease in SOT organ | No prophylaxis | With prophylaxisb | No prophylaxis | With prophylaxisb |
| Kidney and/or pancreas | 45–65 | 6–38 | 8–20 | 1–2 |
| 17* | ||||
| Liver | 45–65 | 6–29 | 8–19 | 4–6 |
| Heart | 29–74 | 19–30 | 20–40 | 2 |
| Lung and lung-heart | 50–91 | 32 | 35–59 | 32 |
| 10* | <5–10* | |||
| 4** | 4** | |||
| Small bowel (intestinal) | LD | 7–37 | LD | 7–44 |
| Composite tissue (hand/face) | LD | 66–100 (LD) | LD | 45 (LD) |
The risk of CMV disease is higher (i.e., rates at the higher end of the reported range) for CMV D+/R+ patients than for CMV D−/R+ solid organ transplant recipients. Rates are estimates based on a review of clinical trials and retrospective and prospective clinical studies. LD, limited data are available for intestinal and composite tissue transplant recipients. Data were gathered from references 3 and 29 to 43.
Prophylaxis is given for a duration of 3 months unless otherwise indicated. *, 6 months of prophylaxis; **, 12 months of prophylaxis after lung transplantation. CMV disease in patients who receive prophylaxis generally occurs after the completion of antiviral prophylaxis (delayed-onset CMV disease).
Secondary CMV infection occurs in CMV-seropositive SOT recipients, as either reactivation or superinfection (reinfection) (3). Reactivation of endogenous latent CMV occurs after SOT in a CMV-seropositive patient, when CMV-specific immunity can be impaired by immunosuppressive drugs, especially T-lymphocyte-depleting compounds (17). In addition, superinfection (or reinfection) may occur when a CMV-seropositive recipient receives exogenous CMV from a CMV-seropositive donor, and subsequently, the circulating CMV may consist of both donor allograft-transmitted exogenous CMV and recipient-derived endogenous CMV (3). Differentiating reactivation from superinfection is not currently possible in routine clinical testing unless sophisticated genetic analyses are performed (46–48). Available studies indicate that superinfection occurs more commonly than reactivation of endogenous virus (46–49). CMV-seropositive recipients have preexisting CMV-specific humoral and cell-mediated immunity, and this immunologic memory can be mobilized during viral reactivation or reinfection. Such immunologic memory in CMV-seropositive transplant recipients dampens CMV replication dynamics to a much lower rate (with a calculated growth rate of 0.61 unit/day [95% CI, 0.55 to 0.7 unit/day]) (36, 43). This is indicated by a relatively lower viral load (and lower rate of rise), which clinically translates into a typically lower incidence and reduced severity of CMV disease (3, 43) (Table 1). However, CMV-seropositive SOT recipients who receive a high degree of immunosuppression, especially with agents that deplete T lymphocytes, may have much more rapid CMV replication dynamics, leading to a higher risk of disease (28, 50, 51).
CLINICAL DISEASE
CMV infection in SOT recipients exhibits a wide spectrum of clinical manifestations, from asymptomatic low-grade infection (typically associated with a low viral load) to severe, widely disseminated and potentially fatal CMV disease (characterized by a high viral load) (1, 3, 29). The clinical course of CMV infection and its presentation are influenced by several interrelated factors, including the degree of immunosuppression (1, 3, 29). In general, CMV disease manifests with greater severity in patients who lack or are deficient in CMV-specific immunity (17, 18, 27, 28, 52). Hence, CMV D+/R− SOT recipients have a higher incidence of infection and are predisposed to develop more severe forms of CMV disease (Table 1) (17, 18, 27, 28, 52). The predisposition to develop CMV disease is augmented by the use of intense pharmacologic immunosuppression, especially those agents that deplete T lymphocytes (27, 28, 53). CMV-seropositive SOT recipients who possess preexisting CMV-specific immunity have a relatively lower risk of CMV disease, and the occurrence of CMV disease in these patients is likely the result of the use of intense immunosuppression that severely impairs T cell function (17, 52). In all these cases, the risk of CMV disease and its severity can be correlated directly to viral load—in general, a higher viral load corresponds to a greater risk and severity of CMV disease (3, 5, 36, 41–45).
CMV infection in SOT recipients starts as local replication at sites that harbor latent virus. In CMV D+/R− SOT recipients, this typically means the transplanted allograft. Hence, allograft CMV infection in CMV D+/R− patients is not uncommon, and this may cause allograft dysfunction that can be mistaken as rejection (3, 28). In CMV-seropositive SOT recipients, local reactivation may occur anywhere, since the virus has widespread tissue distribution (54). If the virus is uncontrolled, local reactivation is followed by hematogenous dissemination (viremic phase). The vast majority of cases of CMV infection are diagnosed during this viremic phase, through viral load or antigen testing (3, 5, 36, 41–45).
SOT recipients with CMV disease may manifest with a syndrome characterized by fever, anorexia, myalgias, and arthralgias (3). This is often accompanied by leukopenia and thrombocytopenia (3). This clinical illness, termed CMV syndrome, is the most common clinical presentation of CMV disease in SOT recipients (Table 2) (3). The clinical diagnosis of CMV syndrome is confirmed by the demonstration of CMV in the blood (by either viral culture, antigen testing, or NAT) (3). In a smaller number of cases, CMV disease involves the invasion of end organs, and patients present with gastritis, enteritis, colitis, pneumonitis, encephalitis, hepatitis, nephritis, and carditis, among others. Virtually any organ system can be affected by CMV (1, 29, 55), but gastrointestinal involvement is the most common presentation of tissue-invasive CMV disease in SOT recipients (54). It can affect any segment of the gastrointestinal tract and manifests clinically as dysphagia, nausea, vomiting, abdominal pain, diarrhea, and gastrointestinal hemorrhage, depending on the site of involvement (54). Findings on endoscopy or colonoscopy include mucosal hyperemia, erosions, and ulcerations (54). Not uncommonly, CMV invasion may be localized to the transplanted allograft, such that pneumonitis, hepatitis, carditis, nephritis, and pancreatitis may be observed among lung, liver, heart, kidney, and pancreas transplant recipients, respectively (1, 29). Ideally, the diagnosis of tissue-invasive CMV disease should be supported by biopsy and histopathology. However, clinicians are sometimes hesitant to perform invasive procedures to obtain tissue for diagnosis. Hence, in the presence of appropriate signs and symptoms, the clinical diagnosis of tissue-invasive CMV disease may be suggested by the detection of CMV in the blood by culture, antigen testing, or NAT (3). The correlation between these blood tests and the diagnosis of CMV disease has led to a decline in obtaining tissue to confirm tissue-invasive CMV diseases (1, 29). However, the detection of CMV in the blood does not necessarily exclude the presence of copathogens or concomitant conditions, such as allograft rejection. Indeed, the clinical manifestations of CMV disease involving the transplanted allograft may be nonspecific and difficult to differentiate from allograft rejection (28). It therefore remains necessary for tissue biopsy to be performed for definitive diagnosis given clinical suspicion and unresponsiveness to antiviral treatment. Conversely, the absence of CMV in the blood of these patients does not totally exclude CMV disease as a diagnosis, as some cases of compartmentalized or localized CMV diseases have very low or transient periods of viremia (1, 3, 29).
Table 2.
Clinical manifestations and impact of CMV infection after solid organ transplantation
| Effecta |
|---|
| Direct effects |
| CMV syndrome—most common clinical manifestation |
| Tissue-invasive CMV disease |
| Gastrointestinal disease—most common organ involvement |
| Allograft infection |
| Hepatitis |
| Pneumonitis |
| Nephritis |
| Pancreatitis |
| Carditis |
| CNS disease |
| Retinitis (rare) |
| Others (any organ can be infected by CMV) |
| Multiorgan disease |
| Mortality |
| Indirect effects |
| Acute allograft rejection |
| Chronic allograft rejection |
| Bronchiolitis obliterans |
| Coronary vasculopathy |
| Tubulointerstitial fibrosis |
| Vanishing bile duct syndrome |
| Opportunistic and other infections |
| Fungal superinfection |
| Bacterial superinfection |
| Epstein-Barr virus-associated PTLD |
| Hepatitis C recurrence |
| Infections with other viruses (e.g., HHV-6 and HHV-7) |
| Increased risk of death |
CMV, cytomegalovirus; PTLD, posttransplant lymphoproliferative disease; HHV, human herpesvirus.
The impact of CMV on SOT also encompasses numerous indirect effects that are presumed to be the result of viral immunomodulation (Table 2) (1, 3, 29). In this regard, CMV is believed to increase the risk of acute and chronic allograft rejection (1, 3, 29, 56). CMV has also been associated with bronchiolitis obliterans syndrome after lung transplantation (57), accelerated coronary vasculopathy after heart transplantation (58–60), and interstitial fibrosis and chronic allograft nephropathy after kidney transplantation (61). In liver transplant recipients with chronic hepatitis C, the risk and severity of hepatitis C recurrence are greater in the presence of CMV infection and disease (62–64). CMV has been associated with an increased predisposition of SOT recipients to other opportunistic infections (3, 63, 65), including bacterial, fungal, and other viral infections. Functional exhaustion of CD4+ T lymphocytes occurs during CMV infection, and this immunologic dysfunction could account for the heightened risk of other opportunistic infections (63, 66). CMV-infected patients are also more likely to develop Epstein-Barr virus infection with subsequent posttransplant lymphoproliferative disease (67). Finally, CMV has been associated with poorer long-term allograft and patient survival outcomes (57, 68, 69).
LABORATORY METHODS OF CMV DIAGNOSIS
The clinical manifestations of CMV disease are nonspecific and can be mimicked by diseases caused by other infectious and noninfectious etiologies. Diagnosis of CMV infection and disease based on clinical grounds alone is often unreliable. Laboratory confirmation is essential in establishing the diagnosis of CMV infection.
The diagnosis of CMV infection is established by the demonstration (or isolation) of the virus in culture or the demonstration of viral antigen or nucleic acid in clinical samples (2). Table 3 lists the various methods for the diagnosis of CMV.
Table 3.
Laboratory tests for cytomegalovirus detection in solid organ transplant recipientsa
| Method | Principle | Sample type or processing details | Turnaround time | Results and clinical utility | Advantages | Disadvantages |
|---|---|---|---|---|---|---|
| Non-nucleic acid-based methods | ||||||
| Serology | Detection of antibody against CMV (IgG, IgM) | Serum | 6 h | CMV-IgG indicates past CMV infection (latent infection); CMV-IgM implies acute or recent infection | Prognostication and risk assignment of patients and their donors prior to transplantation (CMV D+/−, D+/R+, D−/R+, or D−/R−) | Not useful for CMV disease diagnosis in transplant recipients due to attenuated and delayed antibody production; not useful for guiding duration of treatment |
| Histopathology | Demonstration of CMV-infected cells (enlarged cells with nuclear inclusions) | Tissue microscopy with H&E stain; may need in situ hybridization and immunohistochemical staining to increase sensitivity and specificity | 24–48 h | Detection of CMV-infected cells indicates active tissue-invasive disease | Confirmatory test for tissue-invasive CMV disease; highly specific | Need for invasive method to obtain tissue specimen; not generally used to monitor treatment response or risk of relapse |
| Virus culture | ||||||
| Tube culture | Viral growth in human fibroblast cells is indicated by CPE | Cell culture facility; light microscopy | 2–4 weeks | Detection of characteristic CPE indicates presence of virus | Highly specific for CMV infection; the viral isolate can be tested for phenotypic susceptibility | Prolonged processing time is not clinically useful in real-time clinical management; poor sensitivity; requires viable CMV; not generally used to monitor treatment response or risk of relapse due to poor sensitivity and long processing time |
| Shell vial assay | Viral growth with detection using monoclonal antibodies against viral antigens | Cell culture facility; immunofluorescence detection | 16–48 h | Infectious foci detected by monoclonal antibody directed to immediate-early antigen of CMV (prior to the onset of CPE) | Highly specific for CMV infection; more sensitive and rapid than conventional tube cultures | Relatively low sensitivity compared to molecular methods; rapid decrease of CMV activity in clinical specimens; not generally used to monitor treatment response or risk of relapse |
| Antigenemia assay | Detection of pp65 antigen | Polymorphonuclear cells; processing within 4–6 h; light microscopy or immunofluorescence | 6 h | Number of CMV-infected cells per total number of cells (e.g., 5 × 104) | Rapid diagnosis of CMV infection; quantification (no. of positive cells) may indicate disease and infection severity, may be used as a guide for preemptive therapy, and may be used as a guide for treatment response and duration of treatment | Subjective interpretation of results; requires rapid processing; not useful in leukopenic patients; lack of standardization in no. of positive cells for various clinical indications |
| Nucleic acid-based molecular methods | ||||||
| Nucleic acid amplification tests | PCR amplification and detection of CMV DNA or RNA | Various clinical samples (blood, other body fluids); various assays (commercial and laboratory-developed tests) | Few hours | Standardized assay results reported as IU/ml; nonstandardized assay results reported as no. of CMV copies per volume of specimen or PCR | Highly sensitive and specific for rapid diagnosis of CMV; quantification (viral load) allows for individualized management of patients; used to indicate disease severity; used to guide preemptive therapy; used to assess the risk of CMV disease; used to guide duration of antiviral treatment; used as surrogate of disease relapse or infection with resistant virus | Currently without a widely accepted viral threshold for predicting CMV disease; lack of assay standardization limits portability of results—ongoing efforts at assay standardization may reduce this limitation; highly sensitive assay may detect latent CMV |
| NucliSens pp67 test | mRNA detection | Whole blood; samples must be processed within 24 h (or kept in lysis buffer at −80°C indefinitely) | 6 h | Qualitative assay for detection of active CMV infection | Highly specific for viral replication, since it measures the replicative intermediate; clinical utility for preemptive therapy; clinical utility for monitoring response to treatment | Qualitative assay; less sensitive than nucleic acid amplification tests |
| Hybrid capture assay | DNA-RNA hybrid | Whole blood; delayed processing is possible | 6 h | No. of CMV copies per ml (lowest limit of detection, 7 × 102 copies per ml of whole blood) | Highly specific for CMV infection; rapid diagnosis of CMV infection | Less sensitive than nucleic acid amplification tests |
CMV, cytomegalovirus; CPE, cytopathic effects; H&E, hematoxylin and eosin.
Histopathology
Principle, methods, and clinical applications.
Histopathology remains the reference standard for diagnosis of tissue-invasive CMV disease (2, 3, 54, 70). CMV infection is indicated in a tissue biopsy specimen by cellular and nuclear enlargement (cytomegalic cells) and the presence of amphophilic to basophilic cytoplasmic inclusions (aggregates of CMV nucleoproteins that are produced during viral replication) (2, 3, 54, 71). The severity of CMV infection can be assessed based on the degree of histological involvement. While these histopathologic findings are highly characteristic of CMV infection (72), atypical features may be present and may overlap in appearance both with reactive changes and with inclusions of other intracellular viruses. Hence, the diagnosis can be confirmed further by in situ hybridization (ISH) or immunohistochemical (IHC) testing (71). To facilitate histopathological identification of CMV-infected cells in tissue specimens, ISH uses CMV-specific cDNA probes that bind to viral DNA in the cellular material (71). Likewise, IHC uses monoclonal or polyclonal antibody against early CMV antigen; this process increases the sensitivity and specificity of histopathology in the diagnosis of CMV disease compared to standard hematoxylin and eosin staining (71, 73). The greatest value of ISH and IHC tests is in cases where results of routine histopathology are equivocal or nondiagnostic.
Histopathology requires an invasive procedure to obtain tissue samples for testing. Esophagogastroduodenoscopy and colonoscopy may be needed to demonstrate mucosal ulcerations and to obtain tissue samples from patients clinically suspected to have gastrointestinal tissue-invasive CMV disease (54). Bronchoscopy may be needed to obtain respiratory fluid and tissue for the diagnosis of CMV pneumonia (73). Allograft biopsy may be needed to document allograft tissue invasion by CMV and to rule out other causes of allograft dysfunction (3). Because these procedures are invasive, clinicians are often hesitant to perform them. Moreover, repeated biopsies are generally not performed serially to assess the response to treatment (54). Accordingly, many clinicians rely on the demonstration of CMV in the peripheral blood by NAT or antigen testing to support the clinical diagnosis of tissue-invasive CMV disease in patients with compatible clinical signs and symptoms. For example, SOT patients presenting with diarrhea are presumed to have tissue-invasive gastrointestinal CMV disease if a high CMV load is concomitantly demonstrated (3). Likewise, CMV pneumonia is probable in a patient with respiratory symptoms, radiographic findings, and presence of CMV in the blood, especially if a high viral load is observed (3).
Histopathology of an allograft biopsy specimen is highly recommended if allograft rejection is a diagnostic consideration in a patient with CMV viremia (3). It is also recommended when it is critical to distinguish CMV disease from other conditions or copathogens, especially when anti-CMV treatment does not lead to complete resolution of clinical symptoms (3, 74). Histopathology is also required in cases of compartmentalized or localized CMV disease when CMV testing of the blood is negative (3).
Serology
Principle and methods.
Serology relies on the sensitive detection of antibodies against CMV in the blood (3, 16). Several methodologies are available for antibody detection, but the most commonly used is the enzyme-linked immunosorbent assay (ELISA), for which there are various commercial products available (16).
Clinical applications.
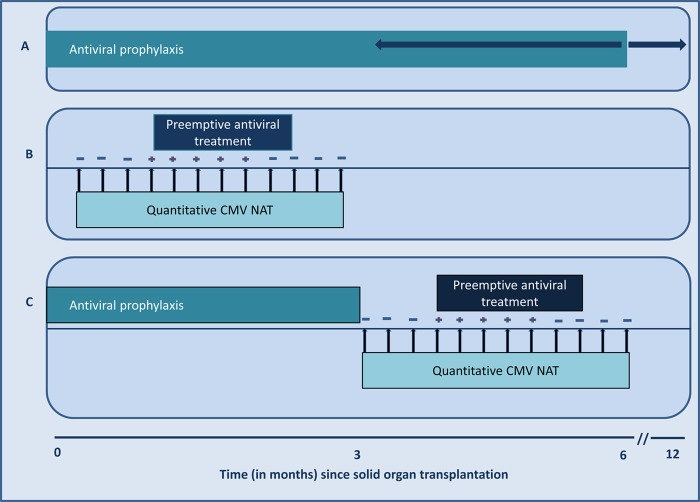
The main clinical utility of CMV serology in transplantation is in the pretransplant screening of organ (and blood) donors and transplant candidates (3). The detection of CMV-IgG is recommended, but IgM testing is discouraged due to frequent false-positive results. Knowledge of the CMV-IgG serostatus of the donor and recipient guides the stratification of SOT patients into different categories of CMV disease risk after transplantation (3). The high-risk category includes CMV D+/R− patients, the moderate-risk category includes CMV D+/R+ and CMV D−/R+ patients, and the low-risk category includes CMV D−/R− patients (3). Depending on the risk category, the recommended prevention measures vary, with either antiviral prophylaxis or preemptive therapy guided by antigen testing or CMV NAT.
CMV-IgM and -IgG antibody testing is generally not recommended for the diagnosis of active CMV infection or disease in SOT recipients (3). It is also not used to monitor the clinical course of infection or response to treatment. Because of drug-induced immune suppression, SOT recipients have a delayed or impaired ability to develop antibody responses, thereby limiting the clinical utility of serology for real-time diagnosis of acute CMV infection (3). Previous studies have shown the lack of timely serologic conversion in transplant recipients with CMV infection (70).
The clinical utility of CMV serologic testing to assess seroconversion (or lack thereof) in CMV D+/R− SOT recipients has been assessed as a potential predictor of late-onset CMV disease. Serocoversion at the completion of a 3-month antiviral prophylaxis in CMV D+/R− SOT recipients was not significantly associated with protection from late-onset CMV disease (75). In contrast, the detection of CMV-IgG at 6 months (in patients who received 3 months of antiviral prophylaxis) was associated with a lower risk of late-onset CMV disease, although the clinical benefit for this is tempered by the fact that most CMV disease cases occurred prior to the 6th month after transplantation (75). Accordingly, current guidelines do not strongly recommend the use of CMV serology during the posttransplant period to guide treatment duration or discontinuation of antiviral prophylaxis, as there are currently no solid data to support this clinical application (3).
Culture
Principle, assay characteristics, clinical applications, and limitations.
Viral culture is highly specific for the diagnosis of CMV infection in SOT recipients (3, 70). Culture can be performed using the conventional plaque assay or the more rapid shell vial centrifugation culture system (2). Viral culture can be performed on blood, respiratory secretions (including bronchoalveolar lavage [BAL] fluid), saliva, urine, stool, cerebrospinal fluid (CSF), and tissue biopsy specimens. Isolation of CMV from most clinical samples (other than urine, saliva, and stool) is highly predictive of the diagnosis of CMV disease or the risk of progression from CMV infection into clinical illness (2, 76, 77). In contrast, the use of urine, saliva, and stool samples for CMV culture is of limited clinical utility because viral shedding may be detected in these specimens in CMV-seropositive SOT recipients even in the absence of clinical illness (3). Urine, stool, and saliva are therefore not recommended as clinical samples for diagnostic purposes for most patients (3). For CMV-seronegative patients (seen most commonly in pediatric age groups), however, the isolation of CMV in urine (and other samples) may be clinically relevant, since it is suggestive of active primary infection (instead of shedding) (3). Indeed, the detection of CMV in the urine has been associated with a 2-fold higher risk of developing clinical CMV disease (76, 77).
The major drawbacks to viral culture are its low to modest sensitivity and long turnaround time (2, 3). Traditional tissue tube culture (plaque assay) requires the growth of CMV in human fibroblast cultures (MRC-5 cells). The presence of CMV is indicated by its typical cytopathic effect (CPE), characterized by foci of flat, swollen cells (2, 3). Notably, the time to the development of CPE has been correlated directly with the titer of CMV present in the sample. CPE has been detected as early as 2 days (in patients with high viral titers) or as late as 21 days (in patients with low-titer infections) after the start of culture. Because tube culture is very laborious and may take weeks for viral isolation and detection, its utility is very limited in contemporary clinical practice (2, 3). Its subsequent modification using the shell vial centrifugation technique has resulted in a shorter turnaround time (i.e., 48 h). Using the shell vial technique, cultured cells are stained by monoclonal antibodies to detect the presence of immediate-early antigens produced during viral replication (2, 3). This modification has allowed for the detection of CMV in cell culture prior to the development of CPE. Despite this modification, the test remains significantly much less sensitive than antigen detection and molecular assays (78–81). Accordingly, the clinical use of viral culture is minimal in the contemporary era, when molecular assays are most commonly used in the clinical setting (2, 3). The remaining major clinical use of viral culture is in the diagnosis of CMV infection by use of samples that have not been validated or optimized for molecular testing (2, 3). Viral culture may also be required when phenotypic antiviral drug resistance testing is needed, although advances in molecular genotypic assays have now emerged as methods of choice for detecting antiviral drug resistance (82).
Antigen Testing
Principle and assay characteristics.
CMV antigen detection in the blood is the most commonly used phenotypic (nonmolecular) method for the rapid and sensitive diagnosis of CMV infection in SOT recipients (2, 3). CMV antigenemia assay uses monoclonal antibodies to detect the CMV lower matrix phosphoprotein pp65 antigen (encoded by UL83), a structural late protein expressed in CMV-infected leukocytes during the early phase of the CMV replication process (2, 3). The result of the test is reported as the number of positive cells per total number of cells counted (2, 3). Because pp65 is secreted during viral replication, its detection in peripheral blood leukocytes (PBL) generally signifies active CMV infection (83, 84).
The CMV antigen assay is a rapid and easy test to perform and has a higher sensitivity than that of virus culture (80, 85, 86). It is able to detect CMV infection earlier than virus culture, with some studies reporting the detection of antigenemia an average of 5 to 14 days before the onset of CMV disease (87, 88). In some studies, the sensitivity of pp65 antigenemia testing for the diagnosis of CMV infection was comparable to that of CMV NAT by PCR (78, 89–93). One of these studies reported a strong correlation between pp65 antigenemia and CMV PCR performed on whole-blood specimens by use of an in-house laboratory-developed test (LDT) (91). Other studies, however, have reported a significantly lower sensitivity of antigenemia testing than those of molecular tests (80, 88, 94–99). In one study, the sensitivity of pp65 antigenemia was lower (39%) than that of an ultrasensitive PCR-based assay performed on plasma (67%) (96). Moreover, the LDT plasma PCR assay detected CMV infection 12 days earlier than the antigenemia test (96). Another study reported a significantly lower sensitivity (26%) of pp65 antigenemia testing than those of two PCR-based assays (COBAS Amplicor CMV Monitor test [48.6%] and an in-house LDT [performed on a LightCycler] [54%]) (98). Similar observations were reported in a study that compared pp65 antigenemia testing with three different CMV PCR assays (each using different primer sets), using cell-free plasma samples (99).
Clinical applications.
Detection of pp65-positive cells in the blood of a patient with compatible symptoms confirms the diagnosis of CMV disease (100, 101). The quantitative ability of pp65 antigenemia testing (i.e., the ability to quantify the viral burden based on the number of positive cells) may indicate the severity of CMV infection (84): the higher the number of pp65 antigen-positive cells, the greater is the disease severity or risk of progression to CMV disease.
CMV antigenemia can be used to detect early CMV replication and to guide the initiation of preemptive therapy. On average, antigenemia can be detected 5 to 14 days before the onset of CMV disease (102–105). Using this preemptive therapy approach for CMV disease prevention, SOT recipients are monitored regularly (usually once weekly) for the presence of pp65 antigen-positive cells in the blood. Once pp65 antigen-positive cells are detected at a predefined threshold (which varies depending on the institution and patient population), and prior to the onset of clinical symptoms, SOT patients are treated with antiviral drugs (most commonly with valganciclovir) in order to prevent the progression of asymptomatic CMV infection into clinical disease. In general, the degree of pp65 antigenemia correlates with the risk of subsequent CMV disease. However, there is a lack of consensus as to the threshold of pp65-positive cells that should trigger the initiation of antiviral therapy. One study recommended initiation of preemptive therapy once the number of pp65 antigen-positive cells exceeded 10 per 2 × 105 cells counted. However, some studies have demonstrated that a patient with a small number of pp65-positive cells may still develop CMV disease, while some patients with larger numbers of positive cells resolve their infection spontaneously, without antiviral therapy (106). These divergent observations and the lack of a defined standardized threshold are likely due to the various risks among SOT recipients (highest among lung recipients and lower among kidney recipients), their preexisting CMV-specific immunity (highest among CMV D+/R− patients compared to others), and the severity of pharmacologic immunosuppression (highest with lymphocyte-depleting drugs) (83, 84, 88, 106–109).
CMV antigenemia has been used to guide antiviral treatment of CMV disease and to determine treatment endpoints (3, 95, 105). The number of pp65-positive leukocytes declines during the course of effective antiviral treatment (3). A few studies, however, have demonstrated that there may be intermittent rises in the level of antigenemia during the first 2 weeks of antiviral treatment (95, 105). The mechanism for this intermittent rise in antigenemia is not completely understood, but it does not necessarily suggest treatment failure as long the patient is clinically improving. Antiviral treatment is continued until pp65 antigenemia is no longer detected in the blood or has declined below a predefined threshold (3). A persistence or rise in the number of pp65-positive cells may indicate drug-resistant virus or the need to reduce immunosuppression as a component of antiviral therapy (3).
Limitations.
The disadvantages of CMV antigenemia testing are its labor-intensive and manual nature (2). The interpretation of the test is subjective, and there is limited interlaboratory standardization of thresholds of positive cell counts to guide various clinical actions (2). Blood samples being subjected to pp65 antigenemia testing should be processed rapidly (ideally within 6 h) to optimize sensitivity, since test results depend on the life span of leukocytes ex vivo. Delays in the processing of a sample for longer than 24 h may lead to a significant decrease in the number of detectable pp65-positive cells in the blood (110, 111). Since the test relies on a sufficient number of polymorphonuclear leukocytes, the pp65 antigenemia assay has limited utility and may be falsely negative for patients with severe leukopenia, and it is not useful in relatively acellular samples, although it has been used to demonstrate the presence of CMV in BAL fluid (100, 101). Moreover, the use of NAT has continued to increase, displacing the use of antigenemia testing. Improvements in NAT, including efforts at standardization, may further increase its appeal, potentially leading to a continued decline in the clinical use of pp65 antigenemia testing.
Nucleic Acid-Based Methods
Nucleic acid amplification tests have emerged as the preferred methods for the rapid diagnosis of CMV after SOT (3). NAT assays are considered the most sensitive methods for CMV diagnosis, typically relying on PCR technology to detect minute amounts of viral nucleic acid in clinical samples. However, CMV persists in latent form in many nucleated cells; therefore, NAT has the risk of detecting and quantifying inactive, nonreplicating CMV. Laboratories should develop strategies aimed at distinguishing active viral infection from latent viral DNA detection. In the absence of such a discriminating test, the clinician is left to rely on clinical judgement in interpreting assay results and differentiating true infection from viral latency.
Several platforms are available for CMV NAT, and one has been approved by the U.S. Food and Drug Administration (CAP/CTM CMV test [Roche]) for viral load monitoring in patients receiving antiviral treatment for CMV disease (4, 5). The vast majority of CMV NAT assays are developed in-house (LDTs). LDTs are developed, optimized, and validated by each performing laboratory, and each has unique assay characteristics, such as the upper and lower limits of detection, linear range of detection, precision, and accuracy. The protocols for CMV NAT assays differ in many other aspects, including specimen types (blood, urine, CSF, BAL fluid, and others), blood sample preparations (whole blood, plasma, serum, and leukocytes), nucleic acid extraction methods, primers and targets (various CMV genes [DNA polymerase gene, glycoprotein B gene, immediate-early gene, major immediate-early gene, UL83, and others], DNA versus RNA), quantitation standards and controls (versus qualitative assays), reaction and amplification protocols (e.g., number of cycles), signal generation systems, and methods for calculating copy numbers and reporting of results (2, 6, 97). In other words, all available CMV NAT assays were not created similarly, and their results are not interchangeable in the absence of standardization.
CMV DNA versus RNA as a target.
As an enveloped double-stranded DNA virus, CMV produces viral mRNA transcripts during its replication cycle. In most CMV NAT assays, the target nucleic acid is DNA (2), although several assay have been developed to detect RNA through reverse transcriptase PCR (112).
Studies have consistently demonstrated that NAT to detect CMV DNA is a highly sensitive method of detecting CMV infection (2, 113). Because of its property of target amplification by the polymerase enzyme, CMV PCR has the ability to rapidly detect and quantify even small amounts of viral nucleic acid in clinical samples (113). CMV DNA is stable in clinical specimens over time, and delayed sample processing has not been associated with any major impact on CMV DNA quantification (111). A recent study demonstrated the stability of CMV DNA in EDTA-blood samples that had been stored at 4°C for 14 days (114). While it is a highly sensitive indicator of the presence of CMV in clinical samples, the detection of CMV DNA is a relatively less specific indicator (compared to RNA testing) of active CMV replication, as a highly sensitive CMV DNA test may detect inactive latent viral DNA (113). Indeed, CMV DNA has been detected by sensitive PCR in blood from otherwise healthy seropositive individuals (113). Detection of latent DNA may therefore lead to unnecessary antiviral treatment of patients without active CMV infection. Several CMV DNA targets have been used in various NAT assays, including the DNA polymerase gene and the glycoprotein B gene, among others. The amplification efficiencies of these DNA targets vary, resulting in noncomparable viral load results (6).
The concern of detecting latent CMV DNA has led to the development of assays that detect viral RNA targets. Because RNA intermediates are generally produced mainly during CMV replication and serve as the biologic link between the CMV genome and gene expression, their detection should indicate active viral infection (115–119). Reverse transcriptase PCR is the method used to selectively detect viral mRNA transcripts in blood and other clinical specimens. However, RNA molecules are readily degraded, and their degradation in vitro can lead to false-negative results (115, 116). It is therefore important to safely transport and process clinical samples promptly. Compared to that of NAT detection of CMV DNA, the sensitivity of CMV RNA testing is lower (112). There have been several studies evaluating CMV mRNA in clinical samples as an indicator of CMV disease or risk of disease (94, 115, 116, 120). One study reported the presence of CMV immediate-early mRNA in leukocytes of transplant patients with active CMV infection (121). In another study, the presence of pp67 mRNA had 100% specificity for CMV disease, but it was detected only in patients with very high viral loads (94). Another study demonstrated that the presence of mRNA was more specific for CMV disease than CMV DNA detection (116). However, CMV RNA detection had a lower sensitivity than that of CMV DNA testing (122), and in some instances, it even showed a lower sensitivity than that of antigenemia testing (123).
Qualitative and quantitative assays.
NAT assays can be developed as qualitative (reported as positive or negative) or quantitative (reported as the amount of virus, typically normalized to the volume of the input specimen) assays. Qualitative CMV DNA tests are highly sensitive for the diagnosis of CMV infection in SOT recipients (2, 124). In one study, the sensitivity of qualitative nested PCR in detecting CMV infection was reported to be as high as 95%, compared to only 83% for the TaqMan-based quantitative assay, with a threshold of >125,000 copies/2 × 106 peripheral blood leukocytes (93). However, the specificity of qualitative NAT was dismal, and its positive predictive value was low compared to that of the quantitative assay (47% versus 68%) (93).
Qualitative CMV DNA tests do not reliably distinguish latent DNA from active viral replication. A qualitative CMV RNA test can offset this limitation, since detection of RNA intermediates is generally indicative of a replicating virus (112, 117, 118). Qualitative CMV DNA or RNA tests do not quantify viral loads, and hence they are not able to assess the severity of infection (2, 124). These tests are not able to differentiate low-level infection (associated with asymptomatic infection) from high-level viral replication (associated with CMV disease) (2, 124). Qualitative tests therefore have very limited clinical utility in disease prognostication and in monitoring antiviral treatment responses (2, 3). Qualitative tests cannot reliably be used to assess trends in the rise or decline of infection (2, 3); in one study, a qualitative CMV DNA test remained positive long after antiviral treatment was no longer required (93).
To increase the specificity of CMV DNA tests, laboratories have developed quantitative NAT (QNAT) and commonly report results in absolute values per volume of specimen or per PCR (2). Quantification of CMV DNA has correlated disease and infection severity with the degree of viral replication (i.e., viral load) (2, 3, 5, 36, 41–45). Active CMV replication is indicated by high absolute viral load values or a rising trend in viral loads, while low-level viral loads may indicate detection of latent viral DNA (2, 3, 5, 36, 41–45).
Clinical samples.
(i) Blood compartments.
NAT can detect CMV nucleic acid in various clinical specimens, although this is most commonly performed on peripheral blood samples (2, 114, 125–127). As discussed above, the pathogenesis of CMV infection results in its systemic spread through the blood (viremic phase). Hence, the majority of CMV disease can be diagnosed by demonstrating CMV nucleic acid in the blood (3).
CMV NAT using blood samples is highly sensitive for diagnosis of CMV infection (2, 114, 125–127). Different compartments of the blood have been used in the diagnosis of CMV infection, including leukocyte preparations, whole blood, plasma, and serum (2, 114, 125–127). Which of the various blood compartments is optimal for CMV DNA detection has been the subject of several studies (91, 126, 128). Overall, these studies have demonstrated that whole blood and leukocyte samples have the highest sensitivity for CMV DNA detection compared to plasma and serum (80, 91, 126, 128, 129). Whole blood is easy to process because it does not require complex sample preparation compared to preparation of leukocyte subpopulations (126). While studies have shown good correlations in viral load values among the various compartments, significantly higher viral load levels have been detected in whole blood (91, 126, 128, 130, 131). Accordingly, many authorities have advocated the use of whole blood for CMV DNA detection due to its higher sensitivity and ability to detect low-copy-number viral reactivation. In a study that compared 170 plasma and whole-blood samples obtained from 61 transplant recipients, 14% of the samples had discordant results (positive viral load in whole blood but negative result for plasma) (128). The majority of the discordant samples were observed at low viral load copy levels, implying the higher sensitivity of whole blood in detecting low-level viral loads (128). Some have suggested that using a highly sensitive sample will identify CMV disease in patients with low viral loads, but the specificity of detecting low-level CMV DNA in whole blood for predicting CMV disease is only modest (as some tests may detect latent virus). Moreover, many patients with low viral load values have a transient viremia that resolves spontaneously, and their detection may lead to unnecessary treatment. The use of a highly sensitive whole-blood PCR test may also lead to a longer course of antiviral therapy, since treatment is usually continued until CMV DNA is undetectable (128).
Because latent CMV may be detected and amplified in leukocyte-containing blood samples, the use of cell-free plasma or serum has been advocated by some as more indicative of active CMV infection in SOT recipients. There have been several studies showing a correlation between CMV infection and the viral load present in cell-free serum (132, 133) or plasma (80, 99, 126, 134). The source of CMV in plasma samples may be lysis of infected leukocytes or release from other infected sites, such as parenchymal and endothelial cells. Since CMV in plasma or serum may be due primarily to release from actively infected cells, it has been suggested that the detection of CMV in these samples is more specific for CMV infection than its detection in whole blood or peripheral blood leukocytes (whose CMV levels may be due to cell-associated latent CMV). Indeed, studies have demonstrated that detection of CMV DNA in plasma is highly associated with CMV disease (132, 133, 135).
The detection of CMV in blood specimens may be affected by the volume of whole blood, cells, or plasma samples used for processing and nucleic acid extraction. Theoretically, the sensitivity of NAT may vary among assays that utilize nucleic acid extracted from 0.2 ml of plasma or whole blood compared to larger sample volumes, especially when the viral load level is low. Such variability in viral load values based on sample volume may also be observed for other body fluids and tissues.
(ii) Cerebrospinal fluid.
CSF analysis for CMV is indicated for patients presenting with compatible clinical symptoms of encephalitis, meningitis, polyradiculopathy, and others. CSF is a relatively acellular specimen, and the detection of CMV DNA by either qualitative or quantitative assay is highly suggestive of CMV central nervous system (CNS) disease (136–138). However, there should be cautious interpretation of CMV DNA results of qualitative tests, since significant pleocytosis (from inflammatory causes other than CMV disease) may result in falsely positive results due to detection of latent CMV in CSF leukocytes. Often, however, there are other clinical clues to the diagnosis of CMV CNS disease, such as magnetic resonance imaging (MRI) findings of periventricular enhancement. As in other cases, quantification has been advocated to assess the severity of clinical disease (in a study conducted in patients with AIDS) (136–138).
(iii) Aqueous and vitreous humor fluid.
A detailed funduscopic examination by an experienced ophthalmologist can reliably diagnose CMV retinitis, which is characterized by retinal hemorrhages and a whitish granular appearance to the retina. The detection of CMV DNA in aqueous and vitreous fluid in these patients confirms the clinical diagnosis (139, 140). Obtaining vitreous fluid is also needed to exclude other potential etiologies, while others have used CMV NAT on vitreous fluid to monitor the efficacy of antiviral treatment responses (140).
(iv) Respiratory samples.
The detection of CMV in BAL fluid may or may not be indicative of CMV pneumonia (68, 73, 141–145). Shedding of CMV in saliva and respiratory secretions is not uncommon, and the demonstration of CMV DNA in these respiratory samples in the absence of compatible clinical signs and symptoms (or in the absence of biopsy confirmation) is of unclear significance and does not necessarily indicate CMV pneumonia (141). In the presence of compatible clinical symptoms, however, the demonstration of CMV DNA may be helpful and may obviate risky lung biopsy in certain situations. Contamination by CMV that is shed in the saliva is a theoretical concern in the interpretation of BAL fluid NAT results, but a study of 76 simultaneously collected BAL fluid and throat wash samples from lung transplant recipients indicated that such contamination is unlikely and that demonstration of CMV DNA in the BAL fluid is highly representative of virus replication in the lung (144).
There have also been investigations to determine whether the degree of viral load is correlated with respiratory disease (141, 146). In a study of 27 lung transplant recipients, CMV loads of >500,000 copies/ml of BAL fluid were highly correlated with biopsy-proven CMV pneumonitis (141). The viral load in the BAL fluid was also correlated with the severity of CMV lung tissue involvement, as measured by histopathologic findings and IHC staining (73). CMV NAT of BAL fluid has been suggested as a better marker of CMV pneumonia in lung recipients than CMV NAT of the blood, since sporadic cases of “compartmentalized” pneumonia without concomitant viremia have been reported (147). While CMV NAT of BAL fluid has been correlated highly with CMV disease in the lungs by some (148), others have found no correlation (149). Such discrepant results may be due to a lack of standardization in various aspects of sample collection and testing, including the amount of fluid effluent, sampling error, and assay performance characteristics. In addition, differences in patient risk profiles may account for the discrepant findings.
(v) Urine and other specimens.
CMV can also be detected in urine and stool, although these are generally not recommended as samples for CMV disease diagnosis (3, 150, 151). Approximately 50% of transplant recipients excrete CMV in body secretions such as urine and stool at some stage after transplantation (152). Detection of CMV in these samples is therefore of minimal to modest clinical significance, as it does not necessarily indicate a high risk of clinical disease, particularly in CMV-seropositive transplant recipients. Detection of CMV DNA in urine is not specific for CMV disease of the genitourinary tract in adults, since CMV DNA is shed in the genitourinary system even in healthy individuals (3, 152).
The clinical utility of CMV DNA testing in the urine is in young infants, for whom serology may be difficult to interpret (due to the presence of maternal antibodies). The demonstration of CMV DNA in the urine of infants and children should indicate prior infection (and latency) or active infection (3). Demonstration of CMV DNA in the urine of any CMV D+/R− SOT recipient is also clinically significant, as it indicates true infection (3).
Assay variability, calibration, and standardization.
One of the central issues that have emerged during the widespread application of CMV NAT in clinical practice is the significant interassay variability in viral load detection and quantification (4, 6, 97). Assays have differed in every aspect of design, including instrumentation, genetic detection targets (e.g., polymerase gene and glycoprotein B gene, among others), sample types (whole blood, plasma, and leukocytes), nucleic acid extraction methods, cycling parameters, detection chemistries and reagents, and reporting parameters (4, 6, 97).
Numerous investigators have highlighted the disparity of various CMV DNA tests, as exemplified by a few studies discussed here (2, 125, 153, 154). A comparative study of a TaqMan-based assay and another commercial real-time PCR assay (COBAS Amplicor CMV Monitor test [Roche]) among 27 kidney and liver transplant patients demonstrated that while the results of the two assays were highly correlated, the TaqMan assay was more sensitive (92% versus 80% detection of all positive samples) and yielded higher viral load results (155). Another study compared the commercial COBAS Amplicor CMV Monitor test (targeting the CMV DNA polymerase gene) and an LDT using a LightCycler intrument (targeting the glycoprotein B gene) and observed that viral load values from the LightCycler assay were significantly higher (98). In contrast, another group of investigators compared the same PCR systems but used a different target for the LDT LightCycler system, and they observed higher viral load values with the COBAS Amplicor CMV Monitor test (125). The findings were consistent across blood compartments, with the COBAS Amplicor CMV Monitor test reporting higher viral load results than the LDT for whole blood, plasma, peripheral blood leukocytes, and mononuclear cells (125). Another study comparing two generations of the LightCycler platform (version 2.0 and model 480 real-time PCR) further demonstrated that while the viral load results were highly correlated, there were statistically significant differences in the absolute viral load values reported by the instruments (156). The assays differed in both detection platform and probe chemistry (156), again suggesting factors that may play a role in interassay result variability.
The significant interassay and interlaboratory variability in viral load detection and reporting was highlighted by three recent multicenter trials that compared various CMV NAT assays. Potentially due to differences in assay platforms, clinical samples, calibrator standards, gene targets, extraction techniques, and other factors (Table 4) (6), there was up to a 3-log10 variation in viral loads reported among different CMV QNAT assays using common samples (97). In a multicenter study conducted across 33 laboratories in Europe and North America, variability in viral load results for individual samples ranged from 2.0 log10 copies/ml to 4.3 log10 copies/ml (97). For example, a clinical sample with 100 copies/ml of CMV reported by one assay may have shown 100,000 copies/ml when tested by another method (97). There was also significant intralaboratory variability, although to a lesser degree than that between laboratories (97). Likewise, another multisite assessment of CMV NATs in 23 laboratories (including 22 which used LDTs on a wide variety of platforms) showed significant interassay quantitative variability in viral load reporting (157). Ten of the laboratories reported viral load values that were significantly different from the expected values (with bias ranging from −0.82 to 1.4 log) (157). This study further determined that changes in reported viral loads of <3- to 5-fold for similar assays may not be significantly different (157). These studies indicated that standardization of NAT methodologies and the presence of a common CMV DNA reference standard are needed to allow laboratories to achieve comparable numeric results (97, 157).
Table 4.
Selected factors contributing to interassay variability in viral load reporting among quantitative nucleic acid amplification testsa
| Major factor | Specific variable(s) | Comments |
|---|---|---|
| Sample selection and specimen volume | Blood compartments | Higher viral load obtained with whole blood and cell-containing compartments than with plasma or serum |
| Pipetting technique | Automated vs manual pipetting devices | |
| Nucleic acid extraction | Liquid-phase magnetic beads, silica membrane/column | Viral load reporting may depend on the efficiency of viral nucleic acid extraction; in one study, the lowest level of variability in viral load reporting was observed for those methods using liquid-phase extraction compared to magnetic bead and silica membrane extraction (see the text); the volume of sample for nucleic acid extraction may also affect viral load reporting |
| PCR instrument | TaqMan, LightCycler or iCycler, others | Multiple studies have demonstrated differences in viral load reporting depending on the assay platform (see the text) |
| Molecular amplification targets | CMV polymerase gene, glycoprotein B gene, immediate-early gene, others | Differences in amplification efficiency have been demonstrated among various gene targets; the presence of gene polymorphisms in the gene target can reduce sensitivity and quantitative results (see the text) |
| Probe chemistry | Hydrolysis (TaqMan), fluorescence resonance energy transfer (FRET) | |
| Detection reagents | Real-time EIA Southern blot | One study identified detection reagents as the biggest contributor to overall assay variability |
| Calibrators | Abbott, Acrometrix, Advanced Biotechnologies, Qiagen, Roche | One of the most significant variables associated with interassay variability; different quantitative standards can have dramatic effects on viral load results; the introduction of the first WHO international CMV reference standard and the U.S. National Institute of Standards and Technology standard reference material can be expected to address this issue |
| Interaction among variables | The interaction among any of the above variables may compound and increase the variability of viral load reporting |
Data were gathered mostly from a large study of laboratory proficiency testing (85). See the text for further discussion of these variables.
Based on such findings, in November 2010, the WHO released the first international reference standard (NIBSC 09/162) for the quantification of CMV nucleic acid by NAT (7). This reference standard comprises a whole-virus preparation of the human CMV Merlin strain, formulated in a universal buffer comprising Tris-HCl and human serum albumin. This standard material was evaluated in a worldwide collaborative study of 32 laboratories performing NAT-based assay of CMV (7). When reconstituted in 1 ml of nuclease-free water, the material has been assigned a concentration of 5 × 106 international units (IU). The U.S. National Institute of Standards and Technology also produced a standard reference material (SRM2366) for CMV that is appropriate for establishing metrological traceability of assay calibrants (158). Availability of these standards should allow common calibration of both commercially developed CMV NAT and LDTs. To date, the only commercial test that has been approved by the U.S. FDA for CMV disease monitoring is the COBAS AmpliPrep/COBAS TaqMan CMV test (CAP/CTM CMV Test; Roche Molecular Diagnostics), which has been calibrated based on the WHO standard and produces results in IU/ml (5). In a study that compared the performances of the CAP/CTM CMV test across five centers in the United States and Europe, there was a high level of quantitative agreement in the reported viral loads across different test centers (4). However, there was high quantitative variability observed for the CAP/CTM CMV test at lower viral load values, at or near the lower limit of detection (i.e., 2.8 log10 copies/ml) (4). With such low viral loads, the CAP/CTM CMV test detected all positive samples but was able to provide quantitative results for only 83% of samples (4). The most stable viral load results, with the lowest interlaboratory variability, were those within the middle range of the assay (4). Interestingly, the samples were also tested by the existing noncalibrated CMV NAT assays in the five laboratories, which showed high interassay quantitative variability (4). For three of the five assays (real-time PCR based on Artus reagents [Qiagen] and LDTs targeting UL111a and pp65 gene targets), reported values were higher than those from the CAP/CTM CMV test, while differences were variable across the quantitative testing range for the other two tests (Affigene real-time PCR test [Cepheid] and COBAS Amplicor Monitor CMV test [Roche]). This study again emphasized the high variability among assays that have not been calibrated to the WHO standard (4). Another study recently compared the performance of an LDT and the commercially available RealTime CMV assay (Abbott) in 513 samples obtained from 37 transplant patients (159). There was significant correlation between the two assays, but despite standardized reporting in international units, using the WHO reference standard, there were discordant results for 23% of samples (positive by the Abbott assay and negative by the LDT). These studies emphasize that even in the presence of an international reference standard, there are still potential differences in viral load test results, based on other variables, such as the assay's performance characteristics and limits of detection (159).
Indeed, while the availability of the international reference materials may significantly harmonize viral load reporting (i.e., in IU/ml) (4), there remains assay-specific variability due to other differences in test characteristics. Differences in nucleic acid extraction methods, type and volume of clinical samples, selection of primers and probes, target-specific amplification efficiencies, detection chemistries and reagents, instrumentation, and operator-dependent variability may independently or collectively account for assay-specific variability (Table 4) (6). One study reported that differences in nucleic acid extraction efficiency over a wide range of plasma CMV DNA loads could account for differences in viral load reporting (160). Another study showed that among three methods of nucleic acid extraction used for CMV NAT, the lowest variability was observed for liquid-phase compared to silica membrane/column and magnetic bead methods (6).
Differences in viral load reporting may be observed if the assay is run on different platforms (such as Abbott, Roche LightCycler, and Nanogen PCR platforms) (160). There are also various amplification efficiencies based on the primers used (161). For example, in a study that compared three different primer sets (UL125 alone, UL126 alone, and UL55/UL123-exon 4), the double-primer assay demonstrated the highest sensitivity, specificity, and predictive values (99). In a large study of laboratory proficiency testing, amplification of the CMV DNA polymerase gene yielded lower mean viral loads than those obtained using glycoprotein B, immediate-early, major immediate-early, and other genes (6). One factor that could partly account for the various efficiencies observed is the presence of genetic mutations or polymorphisms in the chosen gene target. The presence of genetic polymorphisms or mutations in target genes can impede primer binding, reduce PCR efficiency, and reduce sensitivity. Interactions among several apparently independent assay designs and characteristics have also been observed to increase the variability in viral load reporting (6). Thus, standardization of quantitative calibrators, while a great step forward, will not completely eliminate variability of results. Other issues will need to be addressed over time to ensure uniformity in CMV load reporting. The availability of automated commercial PCR tests that encompass all aspects of specimen preparation and testing may be another significant step in this direction (6).
In addition, preanalytical considerations may affect the performance of these tests (Table 4). These include specimen selection and volume, collection, and transport. Storage conditions of clinical samples for PCR testing may affect viral load results, and while one study suggested that storage should not last longer than 72 h (162), another showed stability for up to 14 days (114). As noted earlier in this review, the type of clinical sample may play a role, with whole-blood samples yielding higher values (about a 1-log10 difference) than those obtained with plasma samples (126, 128).
Several other test characteristics are important to consider, including the upper and lower limits of detection and quantification (i.e., the highest and lowest concentrations of DNA that can be detected and quantified in 95% of replicates, respectively), linear range, precision, and accuracy. Finally, variations in patient populations being studied and their level of immunosuppression may account for viral load variability, and thus, viral load threshold recommendations may need to be specific for every type of organ transplant, risk stratum, and level of immunosuppression (3).
Non-PCR amplification methods.
There are several non-PCR methodologies for CMV diagnosis based on signal amplification methods rather than on direct detection of CMV DNA or RNA by target amplification methods such as PCR (Table 3). Examples of signal amplification methods include the branched DNA (bDNA) assay (Chiron Corporation, Emeryville, CA) and the hybrid capture assay (HCA; Qiagen, Hilden, Germany [previously Digene Diagnostics]). The bDNA signal amplification assay measures viral nucleic acid from clinical specimens by using bDNA amplifiers to boost the reporter signal rather than amplifying the target sequences. The target nucleic acid binds to the bDNA molecule, which contains multiple binding sites for an enzyme labeled probe, and the target-bDNA complex is detected with a chemoluminescent substrate. The intensity of the light output is directly proportional to the amount of DNA in the clinical sample. Because it does not amplify the signal, it has been considered to be less sensitive than PCR-based assays (163).
The HCA is a solution hybridization antibody capture assay that uses a cRNA probe to hybridize with the CMV DNA target (reported to be 17% of the CMV genome) (141, 164–166). The target-probe complex (termed a DNA-RNA hybrid) is captured by antibodies specific to the hybrid, and the resulting signal (chemiluminescence) is then measured by a luminometer. The amount of light emitted is quantified and is proportional to the amount of DNA in the clinical sample. The clinical application of HCA has been demonstrated in several studies conducted on SOT recipients (141, 167–169), and these studies suggested the correlation between high viral load and the risk of CMV disease. Few studies have reported lower sensitivities than those of antigenemia and PCR-based assays (165, 170).
Another methodology that has been used for CMV diagnosis in SOT recipients is nucleic acid sequence-based amplification (NASBA), which is a specific isothermal technique of amplification (163). One example of a specific test that has been developed based on NASBA technology is the Nuclisens pp67 test (Organon Teknika), which monitors CMV late pp67 mRNA expression. The Nuclisens pp67 assay has been tested in a few studies of SOT recipients (171), although its clinical use is not as widespread as that of PCR-based assays. In a study that assessed its performance in comparison with antigenemia and PCR-based testing, the Nuclisens pp67 test had the lowest sensitivity (20% versus 65% and 95%, respectively) (94). However, it has the highest specificity for CMV disease (93%). The pp67 mRNA assay was highly effective in identifying those with very high CMV loads (94). However, in another study, the pp67 mRNA test was not able to detect the virus in 4 of 11 patients who developed CMV disease (163). Its low to modest sensitivity has limited its use and application in clinical practice. Development of a more sensitive assay, and preferably one with quantitative capability, is needed (172).
CLINICAL CORRELATION
CMV NAT has revolutionized the contemporary management of CMV disease in SOT recipients. Not only does CMV NAT allow for the rapid diagnosis of CMV infection, but its quantitative capabilities have introduced several principles that are relevant to CMV prevention and treatment, including risk stratification, early detection for preemptive therapy, assessment of antiviral treatment responses, and assessment of disease relapse (Table 5). Multiple studies conducted over the past 2 decades have highlighted these clinical indications for CMV load testing. However, due to the lack of standardization until recently, there have been no widely accepted viral load thresholds that can be applied universally for these various clinical indications across centers. With the attempt to standardize reporting, there are now emerging data to suggest such viral thresholds, as discussed below.
Table 5.
Clinical utility of viral load in solid organ transplant recipients
| General indication of use | Specific clinical utility | Commentsa |
|---|---|---|
| Prognostication | Assessment of CMV disease risk | Higher initial and peak viral loads are associated with a higher risk of CMV disease development (see the text); a rapid rise or upward trend in viral load is associated with a higher risk of CMV disease development (see the text) |
| Assessment of CMV disease severity | Higher viral loads are associated with more severe CMV disease, tissue-invasive CMV disease, and potential multiorgan involvement (see the text) | |
| Prevention | Guidance in the initiation and duration of preemptive therapy | Detection of a viral load above a predefined assay-specific threshold is an indication for the initiation of preemptive antiviral therapy; weekly NAT will guide in determining duration of preemptive therapy (see treatment below) |
| Diagnosis | Rapid and sensitive diagnosis of CMV infection | Rapid and sensitive detection of CMV in the blood or tissue of patients with compatible clinical symptoms confirms the clinical suspicion of CMV disease; higher viral loads are associated with more severe CMV disease, tissue-invasive CMV disease, and potential multiorgan involvement; a negative CMV NAT in blood does not completely rule out CMV disease if the clinical suspicion is high—in this case, pursue other diagnostic tests |
| Treatment | Assessment of efficacy of antiviral therapy | Weekly NAT will indicate the decline in viral load as a measure of the efficacy of antiviral therapy |
| Assessment of treatment duration | Higher pretreatment viral loads (e.g., >18,200 [4.3 log10] IU/ml) have lower rates of CMV disease resolution; viral load suppression (e.g., <137 IU/ml or <2.1 log10 IU/ml) is associated with a shorter time to clinical CMV disease resolution | |
| Assessment of risk of relapse | Detection of a viral load above a threshold at the end of antiviral treatment is significantly associated with clinical and virological relapse; viral decay (t1/2) in patients with successful antiviral treatment was 3.17 days, compared to 8.8 days for patients with subsequent CMV relapse | |
| Assessment of risk of antiviral resistance | Persistent detection (or rise) of viral load in a patient receiving a prolonged course of antiviral therapy is a surrogate for the presence of a drug-resistant virus |
The viral load thresholds for most clinical indications have not been defined due to the lack of a standardized assay in most studies that have reported these clinical uses. See the text for examples of viral load thresholds specific to assays used and populations studied.
Risk Stratification
From the sites of initial reactivation, CMV disseminates in the blood and may spread to other organs. There is strong evidence that the degree of CMV dissemination in the blood, as measured directly by absolute CMV load values, is a significant risk factor for progression from CMV infection to symptomatic disease (173). The initial viral load has been correlated significantly with peak CMV load and with CMV disease in liver (odds ratio [OR], 1.82 [95% CI, 1.11 to 2.98]; P = 0.02) and kidney (OR, 1.34 [1.07 to 1.68]; P = 0.01) transplant recipients (43). In a study of liver transplant recipients, the risk of CMV disease was 8.8-fold and 51.5-fold higher among those with a detectable CMV load but <2,860 copies/106 PBL and those with >2,860 copies/106 PBL, respectively, than among those with an undetectable viral load (127). Several other studies have confirmed these observations by demonstrating that transplant recipients predisposed to develop CMV disease have significantly higher viral loads than those in patients without symptoms (151, 167, 173–177). These observations collectively serve as the basis for the use of CMV NAT in the blood as a prognostic marker of risk for CMV disease.
However, the numerical viral load values that reliably predict CMV disease risk compared to asymptomatic infection have not been defined, and they vary among various tests (due to a lack of assay standardization). Moreover, the viral load values are likely influenced by a patient's underlying level of immunity. Higher viral loads are expected in CMV D+/R− SOT patients with CMV disease than in CMV-seropositive patients, even using the same assay (Table 1) (178). Lung transplant recipients, who are likely more immunosuppressed, are likely to have higher viral loads than those of liver or kidney transplant recipients. Although there is considerable overlap, the viral load is generally higher in patients with tissue-invasive and disseminated disease than in those with CMV syndrome (155, 178). In a recent study using a CMV test that has been calibrated to the WHO international standard, the mean baseline viral load of patients with CMV syndrome was 9,120 (3.96 log10) IU/ml, compared to 20,893 (4.32 log10) IU/ml in patients with tissue-invasive CMV disease (5). A contribution of viral genotype to the level of viral load has also been suggested, although recent studies indicate that this may not be the case (179, 180).
In addition to absolute viral load values (i.e., initial and peak viral loads), trends (or changes over time) in viral load have also been used for CMV disease risk stratification (36, 41–43, 178). The risk of CMV disease is significantly higher in SOT recipients, with a faster and steeper rise in viral load over time (36, 41–43, 178). In one study, the rate of increase in CMV load between the last PCR-negative and first PCR-positive sample was significantly higher in patients who developed CMV disease than in those who did not (0.33 log10 versus 0.19 log10 genomes/ml daily; P < 0.001). In multivariate regression analyses, the rate of viral load increase was an independent risk factor for CMV disease (OR, 1.52 [1.06 to 2.17] [P = 0.02], per 0.1-log10 increase in CMV load/ml daily) (43). However, the exact log change in viral load that is clinically relevant based on CMV biology in SOT recipients has yet to be defined. While a 1-log change is considered significant for most viral infections, the significant change in value may be higher for CMV, especially at lower viral load levels (4, 159). One study recently suggested that a change in viral load of <3- to 5-fold might not be significantly different (157).
Preemptive Therapy
Preemptive therapy is one of the two major strategies for preventing CMV disease in SOT recipients (Table 6) (3). In contrast to the strategy of antiviral prophylaxis (34, 38), wherein all at-risk SOT patients receive antiviral drugs, most commonly valganciclovir, for a defined period after transplantation, preemptive therapy provides antiviral drugs only to those with evidence of asymptomatic CMV infection, as detected by CMV NAT (Fig. 1) (3).
Table 6.
Prevention of CMV disease in solid organ transplant recipients
| Parameter | Commentsa |
|
|---|---|---|
| Preemptive therapy | Antiviral prophylaxis | |
| Strategy | Monitor patients for CMV reactivation by using highly sensitive and predictive laboratory assays (such as nucleic acid amplification tests) | Administer antiviral drug (valganciclovir) for a defined duration to all patients at risk for CMV disease (duration varies depending on the organ transplant and the risk profile of patients) |
| Laboratory monitoring is performed once weekly. More frequent monitoring may be considered for patients at very high risk of CMV disease | Laboratory monitoring (NAT) for CMV reactivation is not routinely recommended for patients receiving antiviral prophylaxis | |
| Administer antiviral therapy (valganciclovir) only to patients who develop CMV reactivation above a predefined viral load threshold | ||
| Monitor patients by use of CMV NAT once weekly to guide treatment duration; treatment is continued until the viral load falls to an undetectable level or below a predefined threshold | ||
| Recommended populations | CMV-seropositive (CMV D+/R+ and CMV D−/R+) heart, liver, kidney, and pancreas transplant recipients | All CMV D+/R− solid organ transplant recipients |
| Considered but less preferred for CMV D+/R− heart, liver, kidney, and pancreas transplant recipients due to potential risk of breakthrough disease as a result of rapid replication dynamics | All CMV D+/R− and CMV R+ lung, intestinal, and composite tissue allograft transplant recipients | |
| Not recommended for lung, intestinal, and composite tissue allograft transplant recipients | All CMV-seropositive (CMV D+/R+ and CMV D−/R+) heart, liver, kidney, and pancreas transplant recipients | |
| All transplant patients in centers that do not have an available assay for sensitive detection of CMV reactivation | ||
| Advantages | Reduced no. of patients exposed to antiviral drugs | Prevents reactivation of other herpesviruses (i.e., herpes simplex virus, HHV-6) |
| Reduced direct antiviral drug costs | Does not rely on a highly sensitive and predictive assay for CMV detection | |
| Reduced duration of antiviral drug use | Reduces incidence of indirect CMV effects | |
| Reduced toxicity related to antiviral drugs | ||
| Lower risk of antiviral drug resistance | ||
| Disadvantages | Requires a predictive test for early identification of patients at risk of CMV disease | Prolonged antiviral drug use may lead to emergence of antiviral drug resistance |
| Requires patients to comply with stringent laboratory surveillance schedule | Prolonged antiviral drug use may lead to higher incidence of adverse drug effects | |
| No widely accepted viral load threshold for initiation of antiviral therapy | Use of expensive drugs by all patients, including those who may not have viral reactivation | |
| Increased cost of diagnostic surveillance testing | ||
| May not identify all patients at risk of CMV disease (breakthrough infection may occur in high-risk CMV D+/R− patients between scheduled weekly testing) | ||
| CMV-selective nature does not prevent reactivation of other herpesviruses | ||
| Prolonged duration of preemptive antiviral therapy may be associated with drug resistance | ||
CMV, cytomegalovirus; HHV, human herpesvirus.
Fig 1.
Strategies for prevention of cytomegalovirus disease in transplant recipients. (A) Antiviral prophylaxis. An antiviral drug, most commonly valganciclovir, is given to all at-risk patients for a defined period after transplantation. In general, the duration is 3 to 6 months, although it can be shortened (backward arrow) or prolonged (forward arrow) depending on the risk profile. (B) Preemptive therapy. This strategy entails routine cytomegalovirus surveillance by nucleic acid testing (often on a weekly basis, as indicated by arrows). Upon detection of a positive viral load threshold, antiviral treatment is initiated and continued until the viral level falls below the clinically relevant threshold. Viral load monitoring for patients is usually conducted during the first 3 months after transplantation. (C) Hybrid approach, wherein antiviral prophylaxis is followed by a preemptive strategy. This is an approach to reduce the incidence of late-onset cytomegalovirus disease in high-risk transplant patients who start off with antiviral prophylaxis as the primary method of cytomegalovirus prevention.
This strategy of preemptive therapy takes advantage of one of the hallmarks of CMV disease pathogenesis, i.e., viral dissemination in the blood. Detection of CMV reactivation early in its course, as it starts to disseminate in the blood, is therefore a prerequisite for successful preemptive therapy (3, 181). Use of a rapid and sensitive diagnostic test such as NAT has enabled clinicians to detect CMV replication early in its course and prior to the onset of clinical disease (3, 124, 178).
With this preemptive therapy approach, SOT recipients undergo serial CMV surveillance during the period at highest risk of CMV reactivation (Fig. 1) (3, 166). The traditional at-risk period is the first 3 months after SOT; hence, guidelines recommend performance of weekly CMV surveillance for 12 weeks after transplantation. In the current era, preemptive therapy has also been applied as part of a hybrid approach to prevent late-onset CMV disease (182, 183). In this regard, high-risk CMV D+/R− SOT recipients who receive antiviral prophylaxis as a primary strategy for CMV prevention undergo CMV surveillance by NAT after completion of CMV prophylaxis (182, 183). At least two studies have assessed the efficacy of this approach (182, 183). One study evaluated the approach of performing CMV NAT every 2 weeks for 3 months after completion of prophylaxis, while another performed CMV NAT once weekly for 8 weeks after completion of antiviral prophylaxis. In both studies, the clinical utility of this hybrid approach was considered modest, since breakthrough CMV disease may still occur (if the patient is tested less frequently) (182), and CMV disease may still occur beyond the period of active CMV surveillance (if the period of surveillance is not long enough) (183).
Once-weekly CMV surveillance is the recommended frequency of laboratory testing for preemptive therapy. The majority of published studies on preemptive therapy have performed once-weekly CMV surveillance during the first 3 months after SOT (3, 124, 127). Some centers have performed CMV surveillance every 2 weeks (102, 103, 184), while others have observed that even once-weekly testing may not be frequent enough for high-risk groups (and hence they recommend twice-weekly monitoring) (124, 127, 178). In a cohort of liver transplant recipients undergoing once-weekly NAT surveillance for preemptive therapy, a significant number of CMV D+/R− patients developed clinical symptoms during the period between once-weekly testing, or at the time of first PCR positivity (124, 127). The failure to detect CMV early in its course was likely due to the rapid CMV replication dynamics in CMV D+/R− patients (36). Hence, the frequency of CMV testing may be tailored according to the underlying risk profile, with more frequent testing recommended for the high-risk CMV D+/R− group. In one study, the rate of increase in viral load in whole blood was significantly higher for CMV D+/R− patients, with a median doubling time of 1.54 days (range, 0.5 to 5.5 days), compared to 2.67 days (range, 0.27 to 26.7 days) for the CMV D+/R+ group (178). Because of the rapid CMV replication dynamics in CMV D+/R− SOT recipients, antiviral prophylaxis has been recommended for these patients, especially when frequent CMV surveillance is not logistically possible (Table 6) (3). Alternatively, others have recommended and performed more frequent CMV monitoring (i.e., twice weekly) for this high-risk group (178).
Upon CMV detection above a predefined center-specific viral load threshold, antiviral therapy (usually oral valganciclovir and, less commonly, intravenous ganciclovir) is initiated preemptively to prevent the progression of asymptomatic CMV infection into clinical disease (3, 185, 186). In one study, this approach reduced the incidence of CMV disease from 12% to 0% in a cohort of liver transplant recipients (124). What is still undefined is a widely accepted viral load threshold that can be applied universally (across different institutions) for preemptive therapy. It is therefore emphasized that previously reported thresholds should be interpreted with caution, depending on the assay used, specimen tested, and population studied (3). One center suggested a viral load of 1,000 to 5,000 copies/ml of plasma as the optimal threshold for guiding preemptive therapy in SOT recipients. Another center validated a viral load threshold of 3,000 genomes/ml of whole blood as the level that should trigger preemptive therapy, with the aim of preventing viral replication above 37,000 genomes/ml—a level associated with tissue-invasive CMV disease (178). Using CMV NAT to monitor 689 liver and kidney transplant recipients, CMV infection was demonstrated in 43% of recipients during the first 90 days after transplantation, but only 21% of patients reached viral loads above 3,000 genomes/ml. The majority of the patients who developed viremia that required antiviral therapy belonged to the high-risk CMV D+/R− group (viremia in this group developed in 78% of cases, and antiviral therapy was required in 69% of cases) (178). This was followed by the D+/R+ group (54% developed viremia, and 23% required antiviral therapy), while the D−/R+ group had the lowest rates (40% and 13%, respectively) (178). Based on these observations, the clinical utility of preemptive therapy may relate to the specific population being treated. Another center developed risk-specific viral load thresholds for preemptive therapy. In its report, viral load thresholds (as measured using a TaqMan assay [ABI Prism 7700 sequence detection system]) of 1,000 copies, 4,000 copies, and 10,000 copies/ml of whole blood were suggested as indicators for initiating preemptive antiviral therapy in high-risk, moderate-risk, and low-risk transplant groups, respectively (89). A more recent prospective study that quantified viral load by using standardized CMV NAT suggested 3,983 IU/ml of plasma as the threshold for initiation of preemptive therapy in moderate-risk, CMV-seropositive liver, kidney, and heart transplant recipients (187). That cutoff value, which was first defined in a study of 141 CMV-seropositive SOT recipients (derivation cohort) and validated in a prospective study of 252 transplant recipients (validation cohort), had an 89.9% sensitivity and 88.9% specificity in correctly identifying the need for preemptive therapy, with a negative predictive value of 99.6% (187). Similar studies must be performed to confirm this finding and to derive and validate viral thresholds for preemptive therapy in high-risk CMV D+/R− SOT recipients and other transplant populations.
Overall, the use of preemptive antiviral therapy, as guided by CMV NAT, in patients with asymptomatic CMV infection has resulted in a significant reduction in the incidence of CMV disease in SOT recipients (181). Individual clinical trials and collective data from meta-analyses have clearly shown the benefits of NAT-guided preemptive therapy for CMV disease prevention (124, 188). Based on these studies, NAT-directed preemptive therapy is recommended as a strategy for CMV disease prevention in CMV-seropositive heart, liver, kidney, and pancreas transplant recipients (3). While this approach has also been demonstrated to be effective with CMV D+/R− SOT recipients, the current guidelines recommend antiviral prophylaxis for this high-risk patient group, if frequent CMV surveillance is not logistically feasible, due to the high rate of viral replication in these patients (3). Likewise, lung, intestinal, and composite tissue allograft transplant recipients should receive antiviral prophylaxis rather than preemptive therapy, because of the high risk of CMV disease in these patient groups (3) (Table 6).
Rapid Diagnosis
The earliest clinical use of CMV NAT was for the rapid and sensitive diagnosis of CMV disease in SOT recipients (2). Clinicians use NAT to assess the likelihood that a patient with compatible clinical symptoms has active CMV disease. During most CMV infections, there is hematogenous viral dissemination which can be captured by peripheral blood NAT. Quantitative results from such testing (viral loads) have been used as an objective measure of CMV disease activity and severity.
The clinical diagnosis of CMV syndrome in SOT patients with fever, arthralgias, myalgias, and bone marrow suppression is confirmed if CMV DNA is demonstrated in the blood. The diagnosis is generally confirmed if the CMV load is high and no other pathogens can be demonstrated. In a study of kidney transplant recipients who developed late-onset CMV disease, the median peak viral load in symptomatic patients was 13,500 copies/ml and ranged from as low as 400 copies/ml (lowest limit of detection for the assay) to as high as 2,831,000 copies/ml (189). The wide variability in the viral loads reported in this study illustrates the difficulty in defining the clinically relevant value. The viral load value is likely influenced by the patient's immune status and the clinical stage of the infection or disease. For example, viral load values are higher for patients with CMV disease than for those with asymptomatic infection (190) and for those with severe disease than for those with milder infections (138). On the other hand, a negative CMV NAT result for a febrile patient is a strong argument against CMV syndrome, and clinicians should search for alternative diagnoses (with few exceptions, as noted below) (191, 192).
The clinical diagnosis of tissue-invasive CMV disease is also supported by the demonstration of a high peripheral blood CMV load (3, 189, 190). Ideally, CMV should be demonstrated at the site of tissue invasion, such as BAL fluid for CMV pneumonia, CSF for CMV encephalitis and meningitis, and vitreous humor for CMV retinitis, among others (3, 138). The degree of viral load in these clinical specimens, including BAL fluid and CSF, has been associated directly with the severity of infection (73, 138, 141). However, clinicians are hesitant to perform such invasive biopsy procedures on immunocompromised and often critically ill patients. Hence, in patients with compatible signs and symptoms of tissue dysfunction, the clinical suspicion of tissue-invasive CMV disease can be supported by peripheral blood CMV NAT (3). Such testing sometimes obviates the need for histopathologic diagnosis of tissue-invasive CMV disease (54). In clinical practice, the diagnosis of gastrointestinal tissue-invasive CMV disease may be suggested by a high-copy-number blood viral load in a patient with diarrhea and other symptoms (54). Likewise, the diagnosis of CMV pneumonitis may be suggested for a patient with pulmonary symptoms, radiographic findings, and a high viral load in the peripheral blood (141). High viral loads are often seen in patients with multiorgan tissue-invasive disease, and lower values may be seen in those with localized or compartmentalized CMV disease. A response to antiviral therapy, as indicated by a decline in viral load and resolution of clinical symptoms, further supports the diagnosis of tissue-invasive disease. Unresponsiveness to antiviral therapy, as evidenced by a nondeclining or rising viral load, may warrant a further investigation for alternative etiologic agents or raise the possibility of antiviral resistance (3, 74).
Peripheral blood CMV NAT may be negative for a proportion of patients with CMV disease (3, 5). In a cohort of 267 patients with CMV disease, 11.7% had no quantifiable viral load (i.e., loads were lower than the assay reporting value of 137 [2.14 log10] IU/ml) (5). There are several possible reasons for a negative CMV NAT result in the presence of CMV disease. The infection may be a form of compartmentalized tissue-invasive disease, wherein the infection is localized in an organ with no systemic dissemination. A number of CMV-seropositive SOT patients with gastrointestinal CMV disease have no detectable CMV viremia (193). These patients often present late, generally many years after transplantation (193). Diagnosis in these cases is generally confirmed by tissue biopsy and histology with ISH (193). Rare cases of CMV retinitis in SOT recipients, especially those that occur late, also generally present without concomitant viremia (139). Diagnosis of CMV retinitis is generally confirmed by detailed funduscopic examination and demonstration of CMV in vitreous fluid by NAT (139). CMV NAT may also give a falsely negative result if the viral load is below the limit of assay detection for the assay being used. Peripheral blood CMV NAT may be negative if sampling occurs prior to the onset of viremia or after viremia has subsided, as in cases of delayed diagnosis, although this is rare. Finally, CMV NAT may be negative (or yield a falsely low viral load) if the virus has genetic polymorphisms in the PCR target gene, reducing the efficiency of viral amplification (6). To confirm the clinical suspicion of tissue-invasive CMV disease in these nonviremic situations, one may repeat the NAT at a later time point, use another NAT assay with higher sensitivity or a different genetic target, or obtain tissue for histopathologic diagnosis (54). In such cases, one should strongly consider sampling body fluid or tissue from the suspected site of infection, such as the aqueous and vitreous humor fluid for patients suspected to have CMV retinitis (139, 194), BAL fluid for patients suspected to have CMV pneumonia (142, 143), and CSF for patients suspected to have CMV encephalitis, meningitis, or polyradiculopathy (147, 195, 196).
Treatment Response
CMV load may be used to assess the response to antiviral treatment and to determine antiviral treatment endpoints. In this regard, three different measures have been proposed: (i) initial viral load at diagnosis, (ii) viral load decline, and (iii) viral load suppression. These three principles apply to antiviral treatment of established CMV disease and to preemptive treatment of asymptomatic CMV infection.
Viral load at time of diagnosis.
Viral load at the time of diagnosis has been correlated with antiviral treatment efficacy (197). In a large clinical trial that compared oral valganciclovir and intravenous ganciclovir for the treatment of CMV disease in 321 SOT recipients, baseline (pretreatment) viral load was the only predictor that was significantly associated with viral load eradication at the end of treatment (197). SOT recipients with viral loads of <10,000 copies/ml had a 6.4-fold higher chance of eradicating the virus after 21 days of induction treatment and a 2.5-fold higher chance of eradicating the virus at day 49 of treatment than those with viral loads of >10,000 copies/ml of plasma (197). In general, a higher viral load at diagnosis has been associated with a longer course of treatment and a higher risk of treatment relapse. In another study of kidney and liver transplant recipients, the total duration of antiviral treatment was associated with peak viral load at the start of treatment (178). Each 1-log-higher peak viral load was associated with a 31-day longer (95% CI, 19 to 43 days) total treatment time (178). In a more recent study of 267 SOT recipients who received intravenous ganciclovir and valganciclovir for treatment of CMV disease, CMV disease resolution was again correlated with pretreatment viral load. Using an assay that has been calibrated to the WHO international standard, SOT recipients with a pretreatment CMV load of <18,200 (4.3 log10) IU/ml had a shorter time to clinical CMV disease resolution than those with a higher CMV load (adjusted hazard ratio [AHR], 1.56; P = 0.001) (5). The median time to clinical resolution was 5 days longer for those with a higher viral load (12 days versus 7 days) (5).
Viral load decline.
Viral load decline over time has been suggested as a measure of therapeutic efficacy. Thus, SOT patients should undergo CMV surveillance during antiviral treatment (197). In a study of patients with asymptomatic CMV infection, the viral decay (also known as half-life) for 22 patients receiving preemptive valganciclovir treatment was 2.16 days, which was comparable to the viral decay of 1.73 days for 23 patients who received preemptive intravenous ganciclovir (198). In another study of patients with CMV disease and subclinical CMV viremia, the median half-life was 4.78 days in plasma, 5.78 days in whole blood, 4.42 days in peripheral blood mononuclear cells, and 5.10 days in peripheral blood leukocytes during treatment with intravenous ganciclovir (126). During treatment of established CMV disease, one study demonstrated a viral half-life of 5 to 8 days (44). In a large cohort of SOT patients with established CMV disease, the viral load half-life was 11.5 days (range, 8.3 to 16.5 days) for patients receiving oral valganciclovir treatment and 10.4 days (range, 7.9 to 14.5 days) for patients receiving intravenous ganciclovir (197). Based on these studies, once-weekly CMV monitoring is recommended to assess therapeutic responses (3). Assessing the viral load response sooner (before 7 days) or more frequently is generally not recommended, since there is often a lag time before the reduction in viral load is observed (197). The mean time to clinically relevant viral load decline was 6.1 (±4.5) days for patients treated with oral valganciclovir and 6.6 (±4.7) days for those treated with intravenous ganciclovir (197).
A decline in the peripheral blood viral load generally indicates that a patient is responding appropriately to antiviral treatment (80, 197, 199). In a study of 19 episodes of CMV disease, the viral load declined from the pretreatment mean value of 45,412 to 8,721 copies/ml of whole blood after 1 week of intravenous ganciclovir treatment (126). A similar pattern of viral decline was observed in plasma, peripheral blood mononuclear cells, and peripheral blood leukocytes (126). However, a recent study that utilized the WHO international standard for viral load reporting did not find a significant association between relative CMV load reductions from baseline of predefined magnitudes (1.0-, 1.5-, 2.0-, and 2.5-log declines) and faster disease resolution (5). It is possible that the association between clinical response and the degree of viral load decline is complex, and it may vary depending on the severity of clinical disease, degree of initial viremia, degree of peak viremia, and underlying level of immunosuppression.
The majority of SOT recipients demonstrate a decline in viral load during the first week of antiviral treatment. However, in some cases, the viral load may rise during the first few days to up to 2 weeks of antiviral treatment (200). This intermittent rise or fluctuations in some patients are often benign and will be followed eventually by a satisfactory decline in viral load over ensuing weeks (this pattern has also been observed with antigenemia testing). While this may be a natural course in these patients, the rise in viral load should raise the potential for treatment failure, recurrent disease, or drug-resistant CMV disease.
Viral load suppression.
Antiviral treatment is generally continued until clinical resolution is achieved and viral eradication is demonstrated. Clinical response appears earlier than viral clearance. In a clinical trial that assessed antiviral treatment of CMV disease, the mean time to clinical resolution of symptoms was 15 days of antiviral treatment (range, 13 to 17 days) (197). However, viral eradication (<600 copies/ml) at treatment day 21 was achieved in only 45% of patients receiving valganciclovir and 48% of those treated with intravenous ganciclovir (197). In a recent study of 267 SOT recipients who received intravenous ganciclovir and valganciclovir for treatment of CMV disease, those with CMV load suppression (<137 IU/ml or <2.1 log10 IU/ml), as measured by a CMV NAT calibrated to the WHO international reference standard, at days 7, 14, and 21 had shorter times to clinical disease resolution than those without viral suppression, even after adjusting for potential confounders (AHR = 1.61, 1.73, and 1.64, respectively [P values of 0.005, <0.001, and <0.001, respectively]) (5). This finding translates to 5 days, 5 days, and 7 days longer for clinical disease resolution among patients with detectable virus at 7, 14, and 21 days of antiviral treatment (5). This recent report supports the current treatment guidelines recommending viral eradication from blood prior to discontinuing antiviral therapy. The duration of antiviral therapy may vary depending on the sensitivity of the NAT assay used for monitoring. Use of a highly sensitive PCR test for monitoring response may result in a longer course of treatment, while those monitored by less sensitive assays may be treated for shorter periods. Standardization of CMV NAT assays for monitoring is therefore needed, and viral values considered safe for cessation of antiviral therapy must still be defined. In one study, the risk of CMV disease relapse was not significantly different between patients with discordant positive whole-blood and negative plasma PCRs and those with negative whole-blood and negative plasma PCRs. This observation suggests that the higher sensitivity of whole-blood PCR assays may not be highly clinically relevant for assessing treatment responses (128). In these situations, antiviral therapy should be continued until the viral load is suppressed to levels below what is yet to be defined as a “safe threshold” for treatment cessation.
The total duration of antiviral treatment may also be influenced by the underlying immunologic risk profile of patients. The duration of viremia (and consequently the number of days needed for treatment) is generally longer for CMV D+/R− liver transplant patients (32 days; range, 7 to 87 days) than for D+/R+ (14 days; range, 1 to 111 days) and D−/R+ (5.5 days; range, 1 to 53 days) patients (178). A similar pattern was observed in kidney transplant recipients (178). Consequently, antiviral treatment is continued longer for D+/R− patients (43 days; range, 18 to 102 days) than for D+/R+ (25 days; range, 6 to 141 days) and D−/R+ (23 days; range not provided) liver transplant patients (178).
Relapse and Resistance
Viral load is used clinically as an objective measure of CMV disease activity (37). Hence, it is generally recommended that antiviral treatment be continued until the viral load is no longer detectable in the blood or has declined to a predefined threshold where it is safe to discontinue antiviral treatment (37). These recommendations are based on studies that have significantly associated relapse with the presence of detectable virus in the blood (37). These are further supported by a recent study, using an assay that has been calibrated to the WHO international reference standard, showing that viral load suppression to levels below 137 (2.14 log10) IU/ml at the end of treatment is significantly associated with CMV disease resolution (5).
In a study of 24 SOT recipients with CMV disease, eight patients developed relapsing CMV infection following a 14-day course of intravenous ganciclovir treatment. Relapsing CMV infection was characterized by a significantly higher pretreatment (or baseline) viral load and the persistence of viral load in the blood at the end of treatment (45). In another study, recurrent CMV disease was observed in 12 (23%) of 50 SOT patients with CMV disease. The two factors that were associated with recurrent disease were (i) a longer time to viral clearance and (ii) a lack of viral eradication at the end of treatment (44). The viral load half-life was 8.8 days among patients with CMV recurrence, compared to only 3.17 days among patients who resolved their CMV disease without recurrence (44). In a recent study of 321 SOT recipients with CMV disease who received 21 days of intravenous ganciclovir or oral valganciclovir followed by valganciclovir maintenance for 4 weeks, recurrence of clinical disease was observed in 15.1% of patients, and virological recurrence was seen in 30.0% of patients. The only independent predictor of clinical and virological recurrence was a failure to eradicate viremia at day 21 (for clinical recurrence, the OR was 3.9 [1.3 to 11.3] [P = 0.012]; and for virological recurrence, the OR was 5.6 [2.5 to 12.6] [P < 0.0001]) (37). The vast majority of recurrent CMV infection and disease are due to ganciclovir-susceptible viruses, and these viruses should remain susceptible and respond well to retreatment with oral valganciclovir or intravenous ganciclovir (3, 201, 202). In a few cases, recurrence of CMV infection heralds the onset of disease due to drug-resistant virus (3, 201, 202).
To reduce the risk of CMV disease recurrence, some experts have recommended the use of secondary antiviral prophylaxis, wherein valganciclovir is given at prophylaxis doses following completion of induction antiviral therapy (3). However, the clinical utility of this approach is much debated. Several studies have demonstrated that this does not affect the rates of clinical and virological recurrence. In a study of gastrointestinal CMV disease treatment, the rates of recurrence did not differ significantly between those who received and those who did not receive secondary prophylaxis (54). Nonetheless, current guidelines do recommend such an approach for transplant patients who are considered at high risk of recurrence. For example, those patients with severe gastrointestinal involvement at the time of CMV disease diagnosis are at higher risk of relapse (54). Higher viral loads at diagnosis and peak viral load measures have also been associated with the risk of recurrence, albeit inconsistently (45, 54). However, there are currently no solid data to support the use of viral load measures to guide the need for secondary antiviral prophylaxis.
Viral load can be used to predict the potential risk for ganciclovir-resistant CMV disease. Using simple mathematical modeling, the efficacy of intravenous ganciclovir against susceptible wild-type CMV was found to be significantly higher than that against ganciclovir-resistant virus (91.5% [95% CI, 89 to 94%] versus 62% [95% CI, 57 to 66%]) (40). The persistence of CMV load despite sufficient treatment is the most common indicator of antiviral drug resistance (3, 201, 203, 204). Often, the virus population during CMV disease is a mixture of wild-type and mutant viruses (40). Hence, one typically observes a decline in viral load during the initial phase of antiviral treatment, corresponding to the decline in the susceptible virus population. Thereafter, the drug-resistant virus predominates and becomes detected persistently in the blood. The incidence of ganciclovir-resistant CMV infection in SOT recipients remains low, at less than 1% (205, 206). However, ganciclovir resistance should be suspected when (i) the viral load persists, plateaus, or rises during treatment; (ii) the patient is highly immunocompromised from use of lymphocyte-depleting agents; (iii) the patient is a high-risk CMV D+/R− SOT recipient; and (iv) there is a history of prolonged antiviral exposure, through either antiviral prophylaxis, preemptive therapy, or antiviral treatment (3). Lung transplant recipients are generally at higher risk of drug-resistant CMV than the other SOT populations (202, 207, 208).
Patients suspected to have infection due to drug-resistant virus should have viral genotype analysis to detect mutations in UL97 and UL54 to define the phenotype of ganciclovir resistance or possible cross-resistance to other antiviral drugs (foscarnet and cidofovir) (209–211). Laboratory testing for drug-resistant CMV was recently reviewed in this journal (209). The clinical impact of drug-resistant CMV is enormous, as it has led to high rates of tissue-invasive disease, poor allograft survival, and heightened patient mortality (201, 212). Treatment is very limited, and depending on the genetic profile, one may use highly nephrotoxic drugs, such as foscarnet and cidofovir (for UL97-exclusive mutants), or off-label and investigational drugs, such as leflunomide, maribavir, letermovir, and CMX-001 (201, 213–215).
CONCLUSIONS
CMV load testing has contributed to contemporary principles for the diagnosis, prevention, and treatment of CMV disease in the SOT population. CMV NAT assays are helpful clinically in assessing the risk of disease, ensuring sensitive and rapid diagnosis, and monitoring therapeutic responses. Despite the large amount of published research data, the interpretation of viral load results remains highly complex. This is due to the heterogeneity of the SOT population, variability of the risks within each SOT group, differing patient immune profiles and immunosuppressive regimens, and nonuniformity and lack of standardization among the many molecular assays used for CMV DNA detection and quantification. Without standardized tests available across different laboratories, clinicians must establish assay- and institution-specific viral load thresholds, correlating to the test used, population tested, and disease characteristics present in any given medical center. The recently released WHO standard is a significant advance that will help to harmonize viral load reporting and assist the medical community in attaining the goal of standardized practice. However, there remain other variables that influence the viral load threshold. Reducing the variability of NAT assays will require a multifaceted approach to improving the performance and comparability of these tests. A better understanding of the variables involved should help us to better interpret the test results and, more importantly, better appreciate the complexity of our patients and the individuality that is needed for the management of every single transplant recipient.
Biographies

Raymund R. Razonable is Professor of Medicine at the College of Medicine at Mayo Clinic. He also serves as Chair of Transplant Infectious Diseases, Associate Chair for Faculty Development, Program Director of the Transplant ID Fellowship Program, and Associate Program Director of the Infectious Disease Fellowship Program in the Division of Infectious Diseases at the Mayo Clinic. Dr. Razonable's clinical and research interests are infections in immunocompromised hosts. He has authored more than 150 peer-reviewed original and review articles and book chapters on these topics, and he has given numerous presentations at regional, national, and international meetings. He is the author of the American Society of Transplantation guideline for CMV in solid organ transplantation. Dr. Razonable is an editor of the journal Transplant Infectious Disease, and he serves on the editorial boards of several journals. He is a fellow of the Infectious Diseases Society of America and a member of numerous professional societies.

Randall T. Hayden is Director of Clinical and Molecular Microbiology and Member in the Department of Pathology at St. Jude Children's Research Hospital, Memphis, TN. He joined the faculty there in 2000, following postdoctoral training in microbiology and molecular microbiology at the Mayo Clinic and in surgical pathology at the M. D. Anderson Cancer Center. He is board certified in anatomic and clinical pathology, with a subspecialty certification in medical microbiology. His research interests focus on the application of molecular methods to diagnostic challenges in clinical microbiology, with particular emphasis on the diagnosis of infections in the immunocompromised host. Work in his laboratory has included development of several assays for the quantitative detection of systemic viral disease in hematopoietic stem cell transplant recipients. He has also worked on several new methods for enhancing detection of fungal infections, including the use of both antigen detection and molecular amplification assays for this purpose. He is editor-in-chief of Diagnostic Microbiology of the Immunocompromised Host and coeditor of the second edition of the widely read Molecular Microbiology, Diagnostic Principles and Practice, both from ASM Press.
REFERENCES
- 1.Beam E, Razonable RR. 2012. Cytomegalovirus in solid organ transplantation: epidemiology, prevention, and treatment. Curr. Infect. Dis. Rep. 14:633–641 [DOI] [PubMed] [Google Scholar]
- 2.Razonable RR, Paya CV, Smith TF. 2002. Role of the laboratory in diagnosis and management of cytomegalovirus infection in hematopoietic stem cell and solid-organ transplant recipients. J. Clin. Microbiol. 40:746–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Razonable R, Humar A. 2013. Cytomegalovirus in solid organ transplant recipients. Am. J. Transplant. 13:93–106 [DOI] [PubMed] [Google Scholar]
- 4.Hirsch HH, Lautenschlager I, Pinsky BA, Cardenoso L, Aslam S, Cobb B, Vilchez RA, Valsamakis A. 2013. An international multicenter performance analysis of cytomegalovirus load tests. Clin. Infect. Dis. 56:367–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Razonable R, Asberg A, Rollag H, Duncan J, Boisvert D, Yao JD, Caliendo AM, Humar A, Do T. 2013. Viral load suppression as measured by a CMV test calibrated to the WHO international standard is predictive of CMV disease resolution in transplant recipients. Clin. Infect. Dis. 56:1546–1553 [DOI] [PubMed] [Google Scholar]
- 6.Hayden RT, Yan X, Wick MT, Rodriguez AB, Xiong X, Ginocchio CC, Mitchell MJ, Caliendo AM. 2012. Factors contributing to variability of quantitative viral PCR results in proficiency testing samples: a multivariate analysis. J. Clin. Microbiol. 50:337–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freyer JF, Heath AB, Anderson R, Minor PD. 2010. Collaborative study to evaluate the proposed 1st WHO international standard for human cytomegalovirus for nucleic acid amplification-based assays. WHO/BS/10.2138. WHO, Geneva, Switzerland [Google Scholar]
- 8.Bate SL, Dollard SC, Cannon MJ. 2010. Cytomegalovirus seroprevalence in the United States: the national health and nutrition examination surveys, 1988–2004. Clin. Infect. Dis. 50:1439–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cannon MJ, Schmid DS, Hyde TB. 2010. Review of cytomegalovirus seroprevalence and demographic characteristics associated with infection. Rev. Med. Virol. 20:202–213 [DOI] [PubMed] [Google Scholar]
- 10.Stowell JD, Forlin-Passoni D, Din E, Radford K, Brown D, White A, Bate SL, Dollard SC, Bialek SR, Cannon MJ, Schmid DS. 2012. Cytomegalovirus survival on common environmental surfaces: opportunities for viral transmission. J. Infect. Dis. 205:211–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho M, Suwansirikul S, Dowling JN, Youngblood LA, Armstrong JA. 1975. The transplanted kidney as a source of cytomegalovirus infection. N. Engl. J. Med. 293:1109–1112 [DOI] [PubMed] [Google Scholar]
- 12.Suwansirikul S, Rao N, Dowling JN, Ho M. 1977. Primary and secondary cytomegalovirus infection. Arch. Intern. Med. 137:1026–1029 [PubMed] [Google Scholar]
- 13.Joseph SA, Beliveau C, Muecke CJ, Rahme E, Soto JC, Flowerdew G, Johnston L, Langille D, Gyorkos TW. 2006. Cytomegalovirus as an occupational risk in daycare educators. Paediatr. Child Health 11:401–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Escobar MR, Allison MJ, Dalton HP. 1970. Etiology and laboratory diagnosis of infectious mononucleosis. II. Cytomegalovirus (CMV) mononucleosis. Va. Med. Mon. (1918) 97:191–192 [PubMed] [Google Scholar]
- 15.Watkins RR, Lemonovich TL, Razonable RR. 2012. Immune response to CMV in solid organ transplant recipients: current concepts and future directions. Expert Rev. Clin. Immunol. 8:383–393 [DOI] [PubMed] [Google Scholar]
- 16.Vauloup-Fellous C, Berth M, Heskia F, Dugua JM, Grangeot-Keros L. 2013. Re-evaluation of the VIDAS((R)) cytomegalovirus (CMV) IgG avidity assay: determination of new cut-off values based on the study of kinetics of CMV-IgG maturation. J. Clin. Virol. 56:118–123 [DOI] [PubMed] [Google Scholar]
- 17.Eid AJ, Brown RA, Arthurs SK, Lahr BD, Eckel-Passow JE, Larson TS, Razonable RR. 2010. A prospective longitudinal analysis of cytomegalovirus (CMV)-specific CD4+ and CD8+ T cells in kidney allograft recipients at risk of CMV infection. Transpl. Int. 23:506–513 [DOI] [PubMed] [Google Scholar]
- 18.Manuel O, Husain S, Kumar D, Zayas C, Mawhorter S, Levi ME, Kalpoe J, Lisboa L, Ely L, Kaul DR, Schwartz BS, Morris MI, Ison MG, Yen-Lieberman B, Sebastian A, Assi M, Humar A. 2013. Assessment of cytomegalovirus-specific cell-mediated immunity for the prediction of cytomegalovirus disease in high-risk solid-organ transplant recipients: a multicenter cohort study. Clin. Infect. Dis. 56:817–824 [DOI] [PubMed] [Google Scholar]
- 19.Lisboa LF, Kumar D, Wilson LE, Humar A. 2012. Clinical utility of cytomegalovirus cell-mediated immunity in transplant recipients with cytomegalovirus viremia. Transplantation 93:195–200 [DOI] [PubMed] [Google Scholar]
- 20.Engel P, Angulo A. 2012. Viral immunomodulatory proteins: usurping host genes as a survival strategy. Adv. Exp. Med. Biol. 738:256–276 [DOI] [PubMed] [Google Scholar]
- 21.Schafer P, Tenschert W, Cremaschi L, Gutensohn K, Laufs R. 1998. Utility of major leukocyte subpopulations for monitoring secondary cytomegalovirus infections in renal-allograft recipients by PCR. J. Clin. Microbiol. 36:1008–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerna G, Zipeto D, Percivalle E, Parea M, Revello MG, Maccario R, Peri G, Milanesi G. 1992. Human cytomegalovirus infection of the major leukocyte subpopulations and evidence for initial viral replication in polymorphonuclear leukocytes from viremic patients. J. Infect. Dis. 166:1236–1244 [DOI] [PubMed] [Google Scholar]
- 23.Grefte A, van der Giessen M, van Son W, The TH. 1993. Circulating cytomegalovirus (CMV)-infected endothelial cells in patients with an active CMV infection. J. Infect. Dis. 167:270–277 [DOI] [PubMed] [Google Scholar]
- 24.Landini MP, Michelson S. 1988. Human cytomegalovirus proteins. Prog. Med. Virol. 35:152–185 [PubMed] [Google Scholar]
- 25.Dunn HS, Haney DJ, Ghanekar SA, Stepick-Biek P, Lewis DB, Maecker HT. 2002. Dynamics of CD4 and CD8 T cell responses to cytomegalovirus in healthy human donors. J. Infect. Dis. 186:15–22 [DOI] [PubMed] [Google Scholar]
- 26.Asberg A, Jardine AG, Bignamini AA, Rollag H, Pescovitz MD, Gahlemann CC, Humar A, Hartmann A. 2010. Effects of the intensity of immunosuppressive therapy on outcome of treatment for CMV disease in organ transplant recipients. Am. J. Transplant. 10:1881–1888 [DOI] [PubMed] [Google Scholar]
- 27.Razonable RR, Rivero A, Rodriguez A, Wilson J, Daniels J, Jenkins G, Larson T, Hellinger WC, Spivey JR, Paya CV. 2001. Allograft rejection predicts the occurrence of late-onset cytomegalovirus (CMV) disease among CMV-mismatched solid organ transplant patients receiving prophylaxis with oral ganciclovir. J. Infect. Dis. 184:1461–1464 [DOI] [PubMed] [Google Scholar]
- 28.Razonable RR, Paya CV. 2005. Infections and allograft rejection-intertwined complications of organ transplantation. Swiss Med. Wkly. 135:571–573 [DOI] [PubMed] [Google Scholar]
- 29.Eid AJ, Razonable RR. 2010. New developments in the management of cytomegalovirus infection after solid organ transplantation. Drugs 70:965–981 [DOI] [PubMed] [Google Scholar]
- 30.Bueno J, Green M, Kocoshis S, Furukawa H, Abu-Elmagd K, Yunis E, Irish W, Todo S, Reyes J, Starzl TE. 1997. Cytomegalovirus infection after intestinal transplantation in children. Clin. Infect. Dis. 25:1078–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Finlen Copeland CA, Davis WA, Snyder LD, Banks M, Avery R, Davis RD, Palmer SM. 2011. Long-term efficacy and safety of 12 months of valganciclovir prophylaxis compared with 3 months after lung transplantation: a single-center, long-term follow-up analysis from a randomized, controlled cytomegalovirus prevention trial. J. Heart Lung Transplant. 30:990–996 [DOI] [PubMed] [Google Scholar]
- 32.Florescu DF, Langnas AN, Grant W, Mercer DF, Botha J, Qiu F, Shafer L, Kalil AC. 2012. Incidence, risk factors, and outcomes associated with cytomegalovirus disease in small bowel transplant recipients. Pediatr. Transplant. 16:294–301 [DOI] [PubMed] [Google Scholar]
- 33.Knoll BM, Hammond SP, Koo S, Issa NC, Tullius SG, Baden LR, Pomahac B, Marty FM. 2013. Infections following facial composite tissue allotransplantation—single center experience and review of the literature. Am. J. Transplant. 13:770–779 [DOI] [PubMed] [Google Scholar]
- 34.Palmer SM, Limaye AP, Banks M, Gallup D, Chapman J, Lawrence EC, Dunitz J, Milstone A, Reynolds J, Yung GL, Chan KM, Aris R, Garrity E, Valentine V, McCall J, Chow SC, Davis RD, Avery R. 2010. Extended valganciclovir prophylaxis to prevent cytomegalovirus after lung transplantation: a randomized, controlled trial. Ann. Intern. Med. 152:761–769 [DOI] [PubMed] [Google Scholar]
- 35.Schneeberger S, Lucchina S, Lanzetta M, Brandacher G, Bosmuller C, Steurer W, Baldanti F, Dezza C, Margreiter R, Bonatti H. 2005. Cytomegalovirus-related complications in human hand transplantation. Transplantation 80:441–447 [DOI] [PubMed] [Google Scholar]
- 36.Emery VC, Cope AV, Bowen EF, Gor D, Griffiths PD. 1999. The dynamics of human cytomegalovirus replication in vivo. J. Exp. Med. 190:177–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Asberg A, Humar A, Jardine AG, Rollag H, Pescovitz MD, Mouas H, Bignamini A, Toz H, Dittmer I, Montejo M, Hartmann A. 2009. Long-term outcomes of CMV disease treatment with valganciclovir versus IV ganciclovir in solid organ transplant recipients. Am. J. Transplant. 9:1205–1213 [DOI] [PubMed] [Google Scholar]
- 38.Humar A, Lebranchu Y, Vincenti F, Blumberg EA, Punch JD, Limaye AP, Abramowicz D, Jardine AG, Voulgari AT, Ives J, Hauser IA, Peeters P. 2010. The efficacy and safety of 200 days valganciclovir cytomegalovirus prophylaxis in high-risk kidney transplant recipients. Am. J. Transplant. 10:1228–1237 [DOI] [PubMed] [Google Scholar]
- 39.Humar A, Limaye AP, Blumberg EA, Hauser IA, Vincenti F, Jardine AG, Abramowicz D, Ives JA, Farhan M, Peeters P. 2010. Extended valganciclovir prophylaxis in D+/R− kidney transplant recipients is associated with long-term reduction in cytomegalovirus disease: two-year results of the IMPACT study. Transplantation 90:1427–1431 [DOI] [PubMed] [Google Scholar]
- 40.Emery VC, Griffiths PD. 2000. Prediction of cytomegalovirus load and resistance patterns after antiviral chemotherapy. Proc. Natl. Acad. Sci. U. S. A. 97:8039–8044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emery VC, Hassan-Walker AF, Burroughs AK, Griffiths PD. 2002. Human cytomegalovirus (HCMV) replication dynamics in HCMV-naive and -experienced immunocompromised hosts. J. Infect. Dis. 185:1723–1728 [DOI] [PubMed] [Google Scholar]
- 42.Emery VC, Manuel O, Asberg A, Pang X, Kumar D, Hartmann A, Preiksaitis JK, Pescovitz MD, Rollag H, Jardine AG, Gahlemann CG, Humar A. 2012. Differential decay kinetics of human cytomegalovirus glycoprotein B genotypes following antiviral chemotherapy. J. Clin. Virol. 54:56–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Emery VC, Sabin CA, Cope AV, Gor D, Hassan-Walker AF, Griffiths PD. 2000. Application of viral-load kinetics to identify patients who develop cytomegalovirus disease after transplantation. Lancet 355:2032–2036 [DOI] [PubMed] [Google Scholar]
- 44.Humar A, Kumar D, Boivin G, Caliendo AM. 2002. Cytomegalovirus (CMV) virus load kinetics to predict recurrent disease in solid-organ transplant patients with CMV disease. J. Infect. Dis. 186:829–833 [DOI] [PubMed] [Google Scholar]
- 45.Sia IG, Wilson JA, Groettum CM, Espy MJ, Smith TF, Paya CV. 2000. Cytomegalovirus (CMV) DNA load predicts relapsing CMV infection after solid organ transplantation. J. Infect. Dis. 181:717–720 [DOI] [PubMed] [Google Scholar]
- 46.Chou SW. 1986. Acquisition of donor strains of cytomegalovirus by renal-transplant recipients. N. Engl. J. Med. 314:1418–1423 [DOI] [PubMed] [Google Scholar]
- 47.Chou SW. 1987. Cytomegalovirus infection and reinfection transmitted by heart transplantation. J. Infect. Dis. 155:1054–1056 [DOI] [PubMed] [Google Scholar]
- 48.Chou SW. 1989. Reactivation and recombination of multiple cytomegalovirus strains from individual organ donors. J. Infect. Dis. 160:11–15 [DOI] [PubMed] [Google Scholar]
- 49.Manuel O, Pang XL, Humar A, Kumar D, Doucette K, Preiksaitis JK. 2009. An assessment of donor-to-recipient transmission patterns of human cytomegalovirus by analysis of viral genomic variants. J. Infect. Dis. 199:1621–1628 [DOI] [PubMed] [Google Scholar]
- 50.Boland GJ, Hene RJ, Ververs C, de Haan MA, de Gast GC. 1993. Factors influencing the occurrence of active cytomegalovirus (CMV) infections after organ transplantation. Clin. Exp. Immunol. 94:306–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krogsgaard K, Boesgaard S, Aldershvile J, Arendrup H, Mortensen SA, Petterson G. 1994. Cytomegalovirus infection rate among heart transplant patients in relation to anti-thymocyte immunoglobulin induction therapy. Copenhagen Heart Transplant Group. Scand. J. Infect. Dis. 26:239–247 [DOI] [PubMed] [Google Scholar]
- 52.Kumar D, Chernenko S, Moussa G, Cobos I, Manuel O, Preiksaitis J, Venkataraman S, Humar A. 2009. Cell-mediated immunity to predict cytomegalovirus disease in high-risk solid organ transplant recipients. Am. J. Transplant. 9:1214–1222 [DOI] [PubMed] [Google Scholar]
- 53.Portela D, Patel R, Larson-Keller JJ, Ilstrup DM, Wiesner RH, Steers JL, Krom RA, Paya CV. 1995. OKT3 treatment for allograft rejection is a risk factor for cytomegalovirus disease in liver transplantation. J. Infect. Dis. 171:1014–1018 [DOI] [PubMed] [Google Scholar]
- 54.Eid AJ, Arthurs SK, Deziel PJ, Wilhelm MP, Razonable RR. 2010. Clinical predictors of relapse after treatment of primary gastrointestinal cytomegalovirus disease in solid organ transplant recipients. Am. J. Transplant. 10:157–161 [DOI] [PubMed] [Google Scholar]
- 55.Drage M, Reid A, Callaghan CJ, Baber Y, Freeman S, Huguet E, Watson CJ. 2009. Acute cytomegalovirus cholecystitis following renal transplantation. Am. J. Transplant. 9:1249–1252 [DOI] [PubMed] [Google Scholar]
- 56.Smith JM, Corey L, Bittner R, Finn LS, Healey PJ, Davis CL, McDonald RA. 2010. Subclinical viremia increases risk for chronic allograft injury in pediatric renal transplantation. J. Am. Soc. Nephrol. 21:1579–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paraskeva M, Bailey M, Levvey BJ, Griffiths AP, Kotsimbos TC, Williams TP, Snell G, Westall G. 2011. Cytomegalovirus replication within the lung allograft is associated with bronchiolitis obliterans syndrome. Am. J. Transplant. 11:2190–2196 [DOI] [PubMed] [Google Scholar]
- 58.Valantine H. 2004. Cardiac allograft vasculopathy after heart transplantation: risk factors and management. J. Heart Lung Transplant. 23:S187–S193 [DOI] [PubMed] [Google Scholar]
- 59.Valantine H. 2005. Prevention of cardiac allograft vasculopathy with Certican (everolimus): the Stanford University experience within the Certican phase III clinical trial. J. Heart Lung Transplant. 24:S191–S195 [DOI] [PubMed] [Google Scholar]
- 60.Valantine HA. 2004. The role of viruses in cardiac allograft vasculopathy. Am. J. Transplant. 4:169–177 [DOI] [PubMed] [Google Scholar]
- 61.Arthurs SK, Eid AJ, Pedersen RA, Kremers WK, Cosio FG, Patel R, Razonable RR. 2008. Delayed-onset primary cytomegalovirus disease and the risk of allograft failure and mortality after kidney transplantation. Clin. Infect. Dis. 46:840–846 [DOI] [PubMed] [Google Scholar]
- 62.Burak KW, Kremers WK, Batts KP, Wiesner RH, Rosen CB, Razonable RR, Paya CV, Charlton MR. 2002. Impact of cytomegalovirus infection, year of transplantation, and donor age on outcomes after liver transplantation for hepatitis C. Liver Transpl. 8:362–369 [DOI] [PubMed] [Google Scholar]
- 63.Bosch W, Heckman MG, Pungpapong S, Diehl NN, Shalev JA, Hellinger WC. 2012. Association of cytomegalovirus infection and disease with recurrent hepatitis C after liver transplantation. Transplantation 93:723–728 [DOI] [PubMed] [Google Scholar]
- 64.Razonable RR, Burak KW, van Cruijsen H, Brown RA, Charlton MR, Smith TF, Espy MJ, Kremers W, Wilson JA, Groettum C, Wiesner R, Paya CV. 2002. The pathogenesis of hepatitis C virus is influenced by cytomegalovirus. Clin. Infect. Dis. 35:974–981 [DOI] [PubMed] [Google Scholar]
- 65.Munoz-Price LS, Slifkin M, Ruthazer R, Poutsiaka DD, Hadley S, Freeman R, Rohrer R, Angelis M, Cooper J, Fairchild R, Barefoot L, Bloom J, Fitzmaurice S, Snydman DR. 2004. The clinical impact of ganciclovir prophylaxis on the occurrence of bacteremia in orthotopic liver transplant recipients. Clin. Infect. Dis. 39:1293–1299 [DOI] [PubMed] [Google Scholar]
- 66.Antoine P, Olislagers V, Huygens A, Lecomte S, Liesnard C, Donner C, Marchant A. 2012. Functional exhaustion of CD4+ T lymphocytes during primary cytomegalovirus infection. J. Immunol. 189:2665–2672 [DOI] [PubMed] [Google Scholar]
- 67.Walker RC, Marshall WF, Strickler JG, Wiesner RH, Velosa JA, Habermann TM, McGregor CG, Paya CV. 1995. Pretransplantation assessment of the risk of lymphoproliferative disorder. Clin. Infect. Dis. 20:1346–1353 [DOI] [PubMed] [Google Scholar]
- 68.Kerschner H, Jaksch P, Karigl G, Popow-Kraupp T, Klepetko W, Puchhammer-Stockl E. 2009. Cytomegalovirus DNA load patterns developing after lung transplantation are significantly correlated with long-term patient survival. Transplantation 87:1720–1726 [DOI] [PubMed] [Google Scholar]
- 69.Bosch W, Heckman MG, Diehl NN, Shalev JA, Pungpapong S, Hellinger WC. 2011. Association of cytomegalovirus infection and disease with death and graft loss after liver transplant in high-risk recipients. Am. J. Transplant. 11:2181–2189 [DOI] [PubMed] [Google Scholar]
- 70.Paya CV, Smith TF, Ludwig J, Hermans PE. 1989. Rapid shell vial culture and tissue histology compared with serology for the rapid diagnosis of cytomegalovirus infection in liver transplantation. Mayo Clin. Proc. 64:670–675 [DOI] [PubMed] [Google Scholar]
- 71.Paya CV, Holley KE, Wiesner RH, Balasubramaniam K, Smith TF, Espy MJ, Ludwig J, Batts KP, Hermans PE, Krom RA. 1990. Early diagnosis of cytomegalovirus hepatitis in liver transplant recipients: role of immunostaining, DNA hybridization and culture of hepatic tissue. Hepatology 12:119–126 [DOI] [PubMed] [Google Scholar]
- 72.Mattes FM, McLaughlin JE, Emery VC, Clark DA, Griffiths PD. 2000. Histopathological detection of owl's eye inclusions is still specific for cytomegalovirus in the era of human herpesviruses 6 and 7. J. Clin. Pathol. 53:612–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chemaly RF, Yen-Lieberman B, Castilla EA, Reilly A, Arrigain S, Farver C, Avery RK, Gordon SM, Procop GW. 2004. Correlation between viral loads of cytomegalovirus in blood and bronchoalveolar lavage specimens from lung transplant recipients determined by histology and immunohistochemistry. J. Clin. Microbiol. 42:2168–2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Arslan H, Inci EK, Azap OK, Karakayali H, Torgay A, Haberal M. 2007. Etiologic agents of diarrhea in solid organ recipients. Transpl. Infect. Dis. 9:270–275 [DOI] [PubMed] [Google Scholar]
- 75.Humar A, Mazzulli T, Moussa G, Razonable RR, Paya CV, Pescovitz MD, Covington E, Alecock E. 2005. Clinical utility of cytomegalovirus (CMV) serology testing in high-risk CMV D+/R− transplant recipients. Am. J. Transplant. 5:1065–1070 [DOI] [PubMed] [Google Scholar]
- 76.Pillay D, Ali AA, Liu SF, Kops E, Sweny P, Griffiths PD. 1993. The prognostic significance of positive CMV cultures during surveillance of renal transplant recipients. Transplantation 56:103–108 [DOI] [PubMed] [Google Scholar]
- 77.Pillay D, Charman H, Burroughs AK, Smith M, Rolles K, Griffiths PD. 1992. Surveillance for CMV infection in orthotopic liver transplant recipients. Transplantation 53:1261–1265 [DOI] [PubMed] [Google Scholar]
- 78.Piiparinen H, Helantera I, Lappalainen M, Suni J, Koskinen P, Gronhagen-Riska C, Lautenschlager I. 2005. Quantitative PCR in the diagnosis of CMV infection and in the monitoring of viral load during the antiviral treatment in renal transplant patients. J. Med. Virol. 76:367–372 [DOI] [PubMed] [Google Scholar]
- 79.Gerna G, Furione M, Baldanti F, Percivalle E, Comoli P, Locatelli F. 1995. Quantitation of human cytomegalovirus DNA in bone marrow transplant recipients. Br. J. Haematol. 91:674–683 [DOI] [PubMed] [Google Scholar]
- 80.Gerna G, Furione M, Baldanti F, Sarasini A. 1994. Comparative quantitation of human cytomegalovirus DNA in blood leukocytes and plasma of transplant and AIDS patients. J. Clin. Microbiol. 32:2709–2717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gerna G, Revello MG, Percivalle E, Zavattoni M, Parea M, Battaglia M. 1990. Quantification of human cytomegalovirus viremia by using monoclonal antibodies to different viral proteins. J. Clin. Microbiol. 28:2681–2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hakki M, Chou S. 2011. The biology of cytomegalovirus drug resistance. Curr. Opin. Infect. Dis. 24:605–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lo CY, Ho KN, Yuen KY, Lui SL, Li FK, Chan TM, Lo WK, Cheng IK. 1997. Diagnosing cytomegalovirus disease in CMV seropositive renal allograft recipients: a comparison between the detection of CMV DNAemia by polymerase chain reaction and antigenemia by CMV pp65 assay. Clin. Transplant. 11:286–293 [PubMed] [Google Scholar]
- 84.Niubo J, Perez JL, Martinez-Lacasa JT, Garcia A, Roca J, Fabregat J, Gil-Vernet S, Martin R. 1996. Association of quantitative cytomegalovirus antigenemia with symptomatic infection in solid organ transplant patients. Diagn. Microbiol. Infect. Dis. 24:19–24 [DOI] [PubMed] [Google Scholar]
- 85.Landry ML, Ferguson D. 1993. Comparison of quantitative cytomegalovirus antigenemia assay with culture methods and correlation with clinical disease. J. Clin. Microbiol. 31:2851–2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.van der Bij W, Schirm J, Torensma R, van Son WJ, Tegzess AM, The TH. 1988. Comparison between viremia and antigenemia for detection of cytomegalovirus in blood. J. Clin. Microbiol. 26:2531–2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Boeckh M, Bowden RA, Goodrich JM, Pettinger M, Meyers JD. 1992. Cytomegalovirus antigen detection in peripheral blood leukocytes after allogeneic marrow transplantation. Blood 80:1358–1364 [PubMed] [Google Scholar]
- 88.The TH, van der Ploeg M, van den Berg AP, Vlieger AM, van der Giessen M, van Son WJ. 1992. Direct detection of cytomegalovirus in peripheral blood leukocytes—a review of the antigenemia assay and polymerase chain reaction. Transplantation 54:193–198 [DOI] [PubMed] [Google Scholar]
- 89.Li H, Dummer JS, Estes WR, Meng S, Wright PF, Tang YW. 2003. Measurement of human cytomegalovirus loads by quantitative real-time PCR for monitoring clinical intervention in transplant recipients. J. Clin. Microbiol. 41:187–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hebart H, Muller C, Loffler J, Jahn G, Einsele H. 1996. Monitoring of CMV infection: a comparison of PCR from whole blood, plasma-PCR, pp65-antigenemia and virus culture in patients after bone marrow transplantation. Bone Marrow Transplant. 17:861–868 [PubMed] [Google Scholar]
- 91.Garrigue I, Boucher S, Couzi L, Caumont A, Dromer C, Neau-Cransac M, Tabrizi R, Schrive MH, Fleury H, Lafon ME. 2006. Whole blood real-time quantitative PCR for cytomegalovirus infection follow-up in transplant recipients. J. Clin. Virol. 36:72–75 [DOI] [PubMed] [Google Scholar]
- 92.Martin-Davila P, Fortun J, Gutierrez C, Marti-Belda P, Candelas A, Honrubia A, Barcena R, Martinez A, Puente A, de Vicente E, Moreno S. 2005. Analysis of a quantitative PCR assay for CMV infection in liver transplant recipients: an intent to find the optimal cut-off value. J. Clin. Virol. 33:138–144 [DOI] [PubMed] [Google Scholar]
- 93.Meyer-Koenig U, Weidmann M, Kirste G, Hufert FT. 2004. Cytomegalovirus infection in organ-transplant recipients: diagnostic value of pp65 antigen test, qualitative polymerase chain reaction (PCR) and quantitative Taqman PCR. Transplantation 77:1692–1698 [DOI] [PubMed] [Google Scholar]
- 94.Mengoli C, Cusinato R, Biasolo MA, Cesaro S, Parolin C, Palu G. 2004. Assessment of CMV load in solid organ transplant recipients by pp65 antigenemia and real-time quantitative DNA PCR assay: correlation with pp67 RNA detection. J. Med. Virol. 74:78–84 [DOI] [PubMed] [Google Scholar]
- 95.Boeckh M, Gooley TA, Myerson D, Cunningham T, Schoch G, Bowden RA. 1996. Cytomegalovirus pp65 antigenemia-guided early treatment with ganciclovir versus ganciclovir at engraftment after allogeneic marrow transplantation: a randomized double-blind study. Blood 88:4063–4071 [PubMed] [Google Scholar]
- 96.Hadaya K, Wunderli W, Deffernez C, Martin PY, Mentha G, Binet I, Perrin L, Kaiser L. 2003. Monitoring of cytomegalovirus infection in solid-organ transplant recipients by an ultrasensitive plasma PCR assay. J. Clin. Microbiol. 41:3757–3764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pang XL, Fox JD, Fenton JM, Miller GG, Caliendo AM, Preiksaitis JK. 2009. Interlaboratory comparison of cytomegalovirus viral load assays. Am. J. Transplant. 9:258–268 [DOI] [PubMed] [Google Scholar]
- 98.Pang XL, Chui L, Fenton J, LeBlanc B, Preiksaitis JK. 2003. Comparison of LightCycler-based PCR, COBAS Amplicor CMV Monitor, and pp65 antigenemia assays for quantitative measurement of cytomegalovirus viral load in peripheral blood specimens from patients after solid organ transplantation. J. Clin. Microbiol. 41:3167–3174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Boeckh M, Huang M, Ferrenberg J, Stevens-Ayers T, Stensland L, Nichols WG, Corey L. 2004. Optimization of quantitative detection of cytomegalovirus DNA in plasma by real-time PCR. J. Clin. Microbiol. 42:1142–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Solans EP, Yong S, Husain AN, Eichorst M, Gattuso P. 1997. Bronchioloalveolar lavage in the diagnosis of CMV pneumonitis in lung transplant recipients: an immunocytochemical study. Diagn. Cytopathol. 16:350–352 [DOI] [PubMed] [Google Scholar]
- 101.Solans EP, Garrity ER, Jr, McCabe M, Martinez R, Husain AN. 1995. Early diagnosis of cytomegalovirus pneumonitis in lung transplant patients. Arch. Pathol. Lab. Med. 119:33–35 [PubMed] [Google Scholar]
- 102.Singh N, Paterson DL, Gayowski T, Wagener MM, Marino IR. 2000. Cytomegalovirus antigenemia directed pre-emptive prophylaxis with oral versus I.V. ganciclovir for the prevention of cytomegalovirus disease in liver transplant recipients: a randomized, controlled trial. Transplantation 70:717–722 [DOI] [PubMed] [Google Scholar]
- 103.Singh N. 2001. Preemptive therapy versus universal prophylaxis with ganciclovir for cytomegalovirus in solid organ transplant recipients. Clin. Infect. Dis. 32:742–751 [DOI] [PubMed] [Google Scholar]
- 104.Kusne S, Grossi P, Irish W, St George K, Rinaldo C, Rakela J, Fung J. 1999. Cytomegalovirus PP65 antigenemia monitoring as a guide for preemptive therapy: a cost effective strategy for prevention of cytomegalovirus disease in adult liver transplant recipients. Transplantation 68:1125–1131 [DOI] [PubMed] [Google Scholar]
- 105.Grossi P, Kusne S, Rinaldo C, St George K, Magnone M, Rakela J, Fung J, Starzl TE. 1996. Guidance of ganciclovir therapy with pp65 antigenemia in cytomegalovirus-free recipients of livers from seropositive donors. Transplantation 61:1659–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yang CW, Kim YO, Kim YS, Kim SY, Moon IS, Ahn HJ, Koh YB, Bang BK. 1998. Clinical course of cytomegalovirus (CMV) viremia with and without ganciclovir treatment in CMV-seropositive kidney transplant recipients. Longitudinal follow-up of CMV pp65 antigenemia assay. Am. J. Nephrol. 18:373–378 [DOI] [PubMed] [Google Scholar]
- 107.Iberer F, Tscheliessnigg K, Halwachs G, Rehak P, Wasler A, Petutschnigg B, Schreier G, Muller H, Allmayer T, Freigassner M, Prenner G, Hipmair G, Grasser B. 1996. Definitions of cytomegalovirus disease after heart transplantation: antigenemia as a marker for antiviral therapy. Transpl. Int. 9:236–242 [DOI] [PubMed] [Google Scholar]
- 108.Gaeta A, Nazzari C, Angeletti S, Lazzarini M, Mazzei E, Mancini C. 1997. Monitoring for cytomegalovirus infection in organ transplant recipients: analysis of pp65 antigen, DNA and late mRNA in peripheral blood leukocytes. J. Med. Virol. 53:189–195 [DOI] [PubMed] [Google Scholar]
- 109.Murray BM, Amsterdam D, Gray V, Myers J, Gerbasi J, Venuto R. 1997. Monitoring and diagnosis of cytomegalovirus infection in renal transplantation. J. Am. Soc. Nephrol. 8:1448–1457 [DOI] [PubMed] [Google Scholar]
- 110.Boeckh M, Woogerd PM, Stevens-Ayers T, Ray CG, Bowden RA. 1994. Factors influencing detection of quantitative cytomegalovirus antigenemia. J. Clin. Microbiol. 32:832–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Schafer P, Tenschert W, Gutensohn K, Laufs R. 1997. Minimal effect of delayed sample processing on results of quantitative PCR for cytomegalovirus DNA in leukocytes compared to results of an antigenemia assay. J. Clin. Microbiol. 35:741–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hebart H, Lengerke C, Ljungman P, Paya CV, Klingebiel T, Loeffler J, Pfaffenrath S, Lewensohn-Fuchs I, Barkholt L, Tomiuk J, Meisner C, Lunenberg J, Top B, Razonable RR, Patel R, Litzow MR, Jahn G, Einsele H. 2011. Prospective comparison of PCR-based vs late mRNA-based preemptive antiviral therapy for HCMV infection in patients after allo-SCT. Bone Marrow Transplant. 46:408–415 [DOI] [PubMed] [Google Scholar]
- 113.Larsson S, Soderberg-Naucler C, Wang FZ, Moller E. 1998. Cytomegalovirus DNA can be detected in peripheral blood mononuclear cells from all seropositive and most seronegative healthy blood donors over time. Transfusion 38:271–278 [DOI] [PubMed] [Google Scholar]
- 114.Abdul-Ali D, Kraft CS, Ingersoll J, Frempong M, Caliendo AM. 2011. Cytomegalovirus DNA stability in EDTA anti-coagulated whole blood and plasma samples. J. Clin. Virol. 52:222–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gozlan J, Salord JM, Chouaid C, Duvivier C, Picard O, Meyohas MC, Petit JC. 1993. Human cytomegalovirus (HCMV) late-mRNA detection in peripheral blood of AIDS patients: diagnostic value for HCMV disease compared with those of viral culture and HCMV DNA detection. J. Clin. Microbiol. 31:1943–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Randhawa PS, Manez R, Frye B, Ehrlich GD. 1994. Circulating immediate-early mRNA in patients with cytomegalovirus infections after solid organ transplantation. J. Infect. Dis. 170:1264–1267 [DOI] [PubMed] [Google Scholar]
- 117.Gerna G, Baldanti F, Lilleri D, Parea M, Alessandrino E, Pagani A, Locatelli F, Middeldorp J, Revello MG. 2000. Human cytomegalovirus immediate-early mRNA detection by nucleic acid sequence-based amplification as a new parameter for preemptive therapy in bone marrow transplant recipients. J. Clin. Microbiol. 38:1845–1853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gerna G, Lilleri D, Baldanti F, Torsellini M, Giorgiani G, Zecca M, De Stefano P, Middeldorp J, Locatelli F, Revello MG. 2003. Human cytomegalovirus immediate-early mRNAemia versus pp65 antigenemia for guiding pre-emptive therapy in children and young adults undergoing hematopoietic stem cell transplantation: a prospective, randomized, open-label trial. Blood 101:5053–5060 [DOI] [PubMed] [Google Scholar]
- 119.Revello MG, Lilleri D, Zavattoni M, Stronati M, Bollani L, Middeldorp JM, Gerna G. 2001. Human cytomegalovirus immediate-early messenger RNA in blood of pregnant women with primary infection and of congenitally infected newborns. J. Infect. Dis. 184:1078–1081 [DOI] [PubMed] [Google Scholar]
- 120.Gozlan J, Salord JM, Roullet E, Baudrimont M, Caburet F, Picard O, Meyohas MC, Duvivier C, Jacomet C, Petit JC. 1992. Rapid detection of cytomegalovirus DNA in cerebrospinal fluid of AIDS patients with neurologic disorders. J. Infect. Dis. 166:1416–1421 [DOI] [PubMed] [Google Scholar]
- 121.Lam KM, Oldenburg N, Khan MA, Gaylore V, Mikhail GW, Strouhal PD, Middeldorp JM, Banner N, Yacoub M. 1998. Significance of reverse transcription polymerase chain reaction in the detection of human cytomegalovirus gene transcripts in thoracic organ transplant recipients. J. Heart Lung Transplant. 17:555–565 [PubMed] [Google Scholar]
- 122.Patel R, Smith TF, Espy M, Portela D, Wiesner RH, Krom RA, Paya CV. 1995. A prospective comparison of molecular diagnostic techniques for the early detection of cytomegalovirus in liver transplant recipients. J. Infect. Dis. 171:1010–1014 [DOI] [PubMed] [Google Scholar]
- 123.Meyer-Konig U, Serr A, von Laer D, Kirste G, Wolff C, Haller O, Neumann-Haefelin D, Hufert FT. 1995. Human cytomegalovirus immediate early and late transcripts in peripheral blood leukocytes: diagnostic value in renal transplant recipients. J. Infect. Dis. 171:705–709 [DOI] [PubMed] [Google Scholar]
- 124.Paya CV, Wilson JA, Espy MJ, Sia IG, DeBernardi MJ, Smith TF, Patel R, Jenkins G, Harmsen WS, Vanness DJ, Wiesner RH. 2002. Preemptive use of oral ganciclovir to prevent cytomegalovirus infection in liver transplant patients: a randomized, placebo-controlled trial. J. Infect. Dis. 185:854–860 [DOI] [PubMed] [Google Scholar]
- 125.Razonable RR, Brown RA, Espy MJ, Rivero A, Kremers W, Wilson J, Groettum C, Smith TF, Paya CV. 2001. Comparative quantitation of cytomegalovirus (CMV) DNA in solid organ transplant recipients with CMV infection by using two high-throughput automated systems. J. Clin. Microbiol. 39:4472–4476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Razonable RR, Brown RA, Wilson J, Groettum C, Kremers W, Espy M, Smith TF, Paya CV. 2002. The clinical use of various blood compartments for cytomegalovirus (CMV) DNA quantitation in transplant recipients with CMV disease. Transplantation 73:968–973 [DOI] [PubMed] [Google Scholar]
- 127.Razonable RR, van Cruijsen H, Brown RA, Wilson JA, Harmsen WS, Wiesner RH, Smith TF, Paya CV. 2003. Dynamics of cytomegalovirus replication during preemptive therapy with oral ganciclovir. J. Infect. Dis. 187:1801–1808 [DOI] [PubMed] [Google Scholar]
- 128.Lisboa LF, Asberg A, Kumar D, Pang X, Hartmann A, Preiksaitis JK, Pescovitz MD, Rollag H, Jardine AG, Humar A. 2011. The clinical utility of whole blood versus plasma cytomegalovirus viral load assays for monitoring therapeutic response. Transplantation 91:231–236 [DOI] [PubMed] [Google Scholar]
- 129.Saltzman RL, Quirk MR, Jordan MC. 1992. High levels of circulating cytomegalovirus DNA reflect visceral organ disease in viremic immunosuppressed patients other than marrow recipients. J. Clin. Invest. 90:1832–1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Garrigue I, Doussau A, Asselineau J, Bricout H, Couzi L, Rio C, Merville P, Fleury H, Lafon ME, Thiebaut R. 2008. Prediction of cytomegalovirus (CMV) plasma load from evaluation of CMV whole-blood load in samples from renal transplant recipients. J. Clin. Microbiol. 46:493–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Koidl C, Bozic M, Marth E, Kessler HH. 2008. Detection of CMV DNA: is EDTA whole blood superior to EDTA plasma? J. Virol. Methods 154:210–212 [DOI] [PubMed] [Google Scholar]
- 132.Cunningham R, Harris A, Frankton A, Irving W. 1995. Detection of cytomegalovirus using PCR in serum from renal transplant recipients. J. Clin. Pathol. 48:575–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Eckart P, Brouard J, Vabret A, Freymuth F, Guillot M, Ryckelynck JP, Hurault de Ligny B. 1997. Detection of human cytomegalovirus in renal transplantation: comparison of four diagnostic methods: DNA in sera by polymerase chain reaction (PCR), DNA in leukocyte by PCR, pp65 leukocytic antigenemia, and viremia. Transplant Proc. 29:2387–2389 [DOI] [PubMed] [Google Scholar]
- 134.Preiser W, Rabenau HF, Vogel JU, Brixner V, Doerr HW. 2002. Performance characteristics of an automated PCR assay for the quantification of cytomegalovirus DNA in plasma. J. Virol. Methods 101:149–157 [DOI] [PubMed] [Google Scholar]
- 135.Evans PC, Soin A, Wreghitt TG, Alexander GJ. 1998. Qualitative and semiquantitative polymerase chain reaction testing for cytomegalovirus DNA in serum allows prediction of CMV related disease in liver transplant recipients. J. Clin. Pathol. 51:914–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Revello MG, Percivalle E, Sarasini A, Baldanti F, Furione M, Gerna G. 1994. Diagnosis of human cytomegalovirus infection of the nervous system by pp65 detection in polymorphonuclear leukocytes of cerebrospinal fluid from AIDS patients. J. Infect. Dis. 170:1275–1279 [DOI] [PubMed] [Google Scholar]
- 137.Shinkai M, Spector SA. 1995. Quantitation of human cytomegalovirus (HCMV) DNA in cerebrospinal fluid by competitive PCR in AIDS patients with different HCMV central nervous system diseases. Scand. J. Infect. Dis. 27:559–561 [DOI] [PubMed] [Google Scholar]
- 138.Arribas JR, Clifford DB, Fichtenbaum CJ, Commins DL, Powderly WG, Storch GA. 1995. Level of cytomegalovirus (CMV) DNA in cerebrospinal fluid of subjects with AIDS and CMV infection of the central nervous system. J. Infect. Dis. 172:527–531 [DOI] [PubMed] [Google Scholar]
- 139.Eid AJ, Bakri SJ, Kijpittayarit S, Razonable RR. 2008. Clinical features and outcomes of cytomegalovirus retinitis after transplantation. Transpl. Infect. Dis. 10:13–18 [DOI] [PubMed] [Google Scholar]
- 140.Harper TW, Miller D, Schiffman JC, Davis JL. 2009. Polymerase chain reaction analysis of aqueous and vitreous specimens in the diagnosis of posterior segment infectious uveitis. Am. J. Ophthalmol. 147:140–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Chemaly RF, Yen-Lieberman B, Chapman J, Reilly A, Bekele BN, Gordon SM, Procop GW, Shrestha N, Isada CM, Decamp M, Avery RK. 2005. Clinical utility of cytomegalovirus viral load in bronchoalveolar lavage in lung transplant recipients. Am. J. Transplant. 5:544–548 [DOI] [PubMed] [Google Scholar]
- 142.Kotsimbos AT, Sinickas V, Glare EM, Esmore DS, Snell GI, Walters EH, Williams TJ. 1997. Quantitative detection of human cytomegalovirus DNA in lung transplant recipients. Am. J. Respir. Crit. Care Med. 156:1241–1246 [DOI] [PubMed] [Google Scholar]
- 143.Stephan F, Fajac A, Grenet D, Honderlick P, Ricci S, Frachon I, Friard S, Caubarrere I, Bernaudin JF, Stern M. 1997. Predictive value of cytomegalovirus DNA detection by polymerase chain reaction in blood and bronchoalveolar lavage in lung transplant patients. Transplantation 63:1430–1435 [DOI] [PubMed] [Google Scholar]
- 144.Kerschner H, Jaksch P, Zweytick B, Puchhammer-Stockl E. 2010. Detection of human cytomegalovirus in bronchoalveolar lavage fluid of lung transplant recipients reflects local virus replication and not contamination from the throat. J. Clin. Microbiol. 48:4273–4274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Costa C, Delsedime L, Solidoro P, Curtoni A, Bergallo M, Libertucci D, Baldi S, Rinaldi M, Cavallo R. 2010. Herpesviruses detection by quantitative real-time polymerase chain reaction in bronchoalveolar lavage and transbronchial biopsy in lung transplant: viral infections and histopathological correlation. Transplant. Proc. 42:1270–1274 [DOI] [PubMed] [Google Scholar]
- 146.Boivin G, Olson CA, Quirk MR, Kringstad B, Hertz MI, Jordan MC. 1996. Quantitation of cytomegalovirus DNA and characterization of viral gene expression in bronchoalveolar cells of infected patients with and without pneumonitis. J. Infect. Dis. 173:1304–1312 [DOI] [PubMed] [Google Scholar]
- 147.Bauer CC, Jaksch P, Aberle SW, Haber H, Lang G, Klepetko W, Hofmann H, Puchhammer-Stockl E. 2007. Relationship between cytomegalovirus DNA load in epithelial lining fluid and plasma of lung transplant recipients and analysis of coinfection with Epstein-Barr virus and human herpesvirus 6 in the lung compartment. J. Clin. Microbiol. 45:324–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Storch GA, Ettinger NA, Ockner D, Wick MR, Gaudreault-Keener M, Rossiter J, Trulock EP, Cooper JD. 1993. Quantitative cultures of the cell fraction and supernatant of bronchoalveolar lavage fluid for the diagnosis of cytomegalovirus pneumonitis in lung transplant recipients. J. Infect. Dis. 168:1502–1506 [DOI] [PubMed] [Google Scholar]
- 149.Slavin MA, Gleaves CA, Schoch HG, Bowden RA. 1992. Quantification of cytomegalovirus in bronchoalveolar lavage fluid after allogeneic marrow transplantation by centrifugation culture. J. Clin. Microbiol. 30:2776–2779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Evans PC, Gray J, Wreghitt TG, Alexander GJ. 1998. Optimisation of the polymerase chain reaction and dot-blot hybridisation for detecting cytomegalovirus DNA in urine: comparison with detection of early antigen fluorescent foci and culture. J. Virol. Methods 73:41–52 [DOI] [PubMed] [Google Scholar]
- 151.Cope AV, Sweny P, Sabin C, Rees L, Griffiths PD, Emery VC. 1997. Quantity of cytomegalovirus viruria is a major risk factor for cytomegalovirus disease after renal transplantation. J. Med. Virol. 52:200–205 [PubMed] [Google Scholar]
- 152.Sia IG, Patel R. 2000. New strategies for prevention and therapy of cytomegalovirus infection and disease in solid-organ transplant recipients. Clin. Microbiol. Rev. 13:83–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Yerly S, Perrin L, Van Delden C, Schaffer S, Thamm S, Wunderli W, Kaiser L. 2007. Cytomegalovirus quantification in plasma by an automated real-time PCR assay. J. Clin. Virol. 38:298–303 [DOI] [PubMed] [Google Scholar]
- 154.Gouarin S, Vabret A, Scieux C, Agbalika F, Cherot J, Mengelle C, Deback C, Petitjean J, Dina J, Freymuth F. 2007. Multicentric evaluation of a new commercial cytomegalovirus real-time PCR quantitation assay. J. Virol. Methods 146:147–154 [DOI] [PubMed] [Google Scholar]
- 155.Piiparinen H, Hockerstedt K, Gronhagen-Riska C, Lautenschlager I. 2004. Comparison of two quantitative CMV PCR tests, Cobas Amplicor CMV Monitor and TaqMan assay, and pp65-antigenemia assay in the determination of viral loads from peripheral blood of organ transplant patients. J. Clin. Virol. 30:258–266 [DOI] [PubMed] [Google Scholar]
- 156.Przybylski M, Dzieciatkowski T, Les K, Mucha MA, Wroblewska M, Mlynarczyk G. 2012. Comparison of real-time PCR quantitative analysis of the cytomegalovirus DNA level using LightCycler 2.0 and LightCycler 480 instruments. J. Clin. Virol. 55:270–273 [DOI] [PubMed] [Google Scholar]
- 157.Wolff DJ, Heaney DL, Neuwald PD, Stellrecht KA, Press RD. 2009. Multi-site PCR-based CMV viral load assessment-assays demonstrate linearity and precision, but lack numeric standardization: a report of the association for molecular pathology. J. Mol. Diagn. 11:87–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Haynes RJ, Kline MC, Toman B, Scott C, Wallace P, Butler JM, Holden MJ. 2013. Standard reference material 2366 for measurement of human cytomegalovirus DNA. J. Mol. Diagn. 15:177–185 [DOI] [PubMed] [Google Scholar]
- 159.Furione M, Rognoni V, Cabano E, Baldanti F. 2012. Kinetics of human cytomegalovirus (HCMV) DNAemia in transplanted patients expressed in international units as determined with the Abbott RealTime CMV assay and an in-house assay. J. Clin. Virol. 55:317–322 [DOI] [PubMed] [Google Scholar]
- 160.Bravo D, Clari MA, Costa E, Munoz-Cobo B, Solano C, Jose Remigia M, Navarro D. 2011. Comparative evaluation of three automated systems for DNA extraction in conjunction with three commercially available real-time PCR assays for quantitation of plasma cytomegalovirus DNAemia in allogeneic stem cell transplant recipients. J. Clin. Microbiol. 49:2899–2904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Mendez JC, Espy MJ, Smith TF, Wilson JA, Paya CV. 1998. Evaluation of PCR primers for early diagnosis of cytomegalovirus infection following liver transplantation. J. Clin. Microbiol. 36:526–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Roberts TC, Buller RS, Gaudreault-Keener M, Sternhell KE, Garlock K, Singer GG, Brennan DC, Storch GA. 1997. Effects of storage temperature and time on qualitative and quantitative detection of cytomegalovirus in blood specimens by shell vial culture and PCR. J. Clin. Microbiol. 35:2224–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Pellegrin I, Garrigue I, Ekouevi D, Couzi L, Merville P, Merel P, Chene G, Schrive MH, Trimoulet P, Lafon ME, Fleury H. 2000. New molecular assays to predict occurrence of cytomegalovirus disease in renal transplant recipients. J. Infect. Dis. 182:36–42 [DOI] [PubMed] [Google Scholar]
- 164.Mazzulli T, Drew LW, Yen-Lieberman B, Jekic-McMullen D, Kohn DJ, Isada C, Moussa G, Chua R, Walmsley S. 1999. Multicenter comparison of the Digene hybrid capture CMV DNA assay (version 2.0), the pp65 antigenemia assay, and cell culture for detection of cytomegalovirus viremia. J. Clin. Microbiol. 37:958–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Mazzulli T, Wood S, Chua R, Walmsley S. 1996. Evaluation of the Digene hybrid capture system for detection and quantitation of human cytomegalovirus viremia in human immunodeficiency virus-infected patients. J. Clin. Microbiol. 34:2959–2962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Imbert-Marcille BM, Cantarovich D, Ferre-Aubineau V, Richet B, Soulillou JP, Billaudel S. 1997. Usefulness of DNA viral load quantification for cytomegalovirus disease monitoring in renal and pancreas/renal transplant recipients. Transplantation 63:1476–1481 [DOI] [PubMed] [Google Scholar]
- 167.Kuhn JE, Wendland T, Schafer P, Mohring K, Wieland U, Elgas M, Eggers HJ. 1994. Monitoring of renal allograft recipients by quantitation of human cytomegalovirus genomes in peripheral blood leukocytes. J. Med. Virol. 44:398–405 [DOI] [PubMed] [Google Scholar]
- 168.Macartney M, Gane EJ, Portmann B, Williams R. 1997. Comparison of a new quantitative cytomegalovirus DNA assay with other detection methods. Transplantation 63:1803–1807 [DOI] [PubMed] [Google Scholar]
- 169.Rollag H, Sagedal S, Holter E, Degre M, Ariansen S, Nordal KP. 1998. Diagnosis of cytomegalovirus infection in kidney transplant recipients by a quantitative RNA-DNA hybrid capture assay for cytomegalovirus DNA in leukocytes. Eur. J. Clin. Microbiol. Infect. Dis. 17:124–127 [DOI] [PubMed] [Google Scholar]
- 170.Lazzarotto T, Campisi T, Dal Monte P, Galli S, Spezzacatena P, Guglielmi P, Landini MP. 1996. A quantitative test (HCMV-hybrid-capture(TM)) to detect human cytomegalovirus DNA in the blood of immunocompromised patients compared with antigenemia and polymerase chain reaction. New Microbiol. 19:193–201 [PubMed] [Google Scholar]
- 171.Blok MJ, Goossens VJ, Vanherle SJ, Top Tacken BN, Middeldorp JM, Christiaans MH, van Hooff JP, Bruggeman CA. 1998. Diagnostic value of monitoring human cytomegalovirus late pp67 mRNA expression in renal-allograft recipients by nucleic acid sequence-based amplification. J. Clin. Microbiol. 36:1341–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Greijer AE, Verschuuren EA, Harmsen MC, Dekkers CA, Adriaanse HM, The TH, Middeldorp JM. 2001. Direct quantification of human cytomegalovirus immediate-early and late mRNA levels in blood of lung transplant recipients by competitive nucleic acid sequence-based amplification. J. Clin. Microbiol. 39:251–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Humar A, Gregson D, Caliendo AM, McGeer A, Malkan G, Krajden M, Corey P, Greig P, Walmsley S, Levy G, Mazzulli T. 1999. Clinical utility of quantitative cytomegalovirus viral load determination for predicting cytomegalovirus disease in liver transplant recipients. Transplantation 68:1305–1311 [DOI] [PubMed] [Google Scholar]
- 174.Cope AV, Sabin C, Burroughs A, Rolles K, Griffiths PD, Emery VC. 1997. Interrelationships among quantity of human cytomegalovirus (HCMV) DNA in blood, donor-recipient serostatus, and administration of methylprednisolone as risk factors for HCMV disease following liver transplantation. J. Infect. Dis. 176:1484–1490 [DOI] [PubMed] [Google Scholar]
- 175.Fox JC, Kidd IM, Griffiths PD, Sweny P, Emery VC. 1995. Longitudinal analysis of cytomegalovirus load in renal transplant recipients using a quantitative polymerase chain reaction: correlation with disease. J. Gen. Virol. 76:309–319 [DOI] [PubMed] [Google Scholar]
- 176.Mutimer D, Matyi-Toth A, Shaw J, Elias E, O'Donnell K, Stalhandske P. 1997. Patterns of viremia in liver transplant recipients with symptomatic cytomegalovirus infection. Transplantation 63:68–73 [DOI] [PubMed] [Google Scholar]
- 177.Toyoda M, Carlos JB, Galera OA, Galfayan K, Zhang X, Sun Z, Czer LS, Jordan SC. 1997. Correlation of cytomegalovirus DNA levels with response to antiviral therapy in cardiac and renal allograft recipients. Transplantation 63:957–963 [DOI] [PubMed] [Google Scholar]
- 178.Atabani SF, Smith C, Atkinson C, Aldridge RW, Rodriguez-Peralvarez M, Rolando N, Harber M, Jones G, O'Riordan A, Burroughs AK, Thorburn D, O'Beirne J, Milne RS, Emery VC, Griffiths PD. 2012. Cytomegalovirus replication kinetics in solid organ transplant recipients managed by preemptive therapy. Am. J. Transplant. 12:2457–2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Manuel O, Asberg A, Pang X, Rollag H, Emery VC, Preiksaitis JK, Kumar D, Pescovitz MD, Bignamini AA, Hartmann A, Jardine AG, Humar A. 2009. Impact of genetic polymorphisms in cytomegalovirus glycoprotein B on outcomes in solid-organ transplant recipients with cytomegalovirus disease. Clin. Infect. Dis. 49:1160–1166 [DOI] [PubMed] [Google Scholar]
- 180.Humar A, Kumar D, Gilbert C, Boivin G. 2003. Cytomegalovirus (CMV) glycoprotein B genotypes and response to antiviral therapy, in solid-organ-transplant recipients with CMV disease. J. Infect. Dis. 188:581–584 [DOI] [PubMed] [Google Scholar]
- 181.Sun HY, Cacciarelli TV, Wagener MM, Singh N. 2010. Preemptive therapy for cytomegalovirus based on real-time measurement of viral load in liver transplant recipients. Transpl. Immunol. 23:166–169 [DOI] [PubMed] [Google Scholar]
- 182.Humar A, Paya C, Pescovitz MD, Dominguez E, Washburn K, Blumberg E, Alexander B, Freeman R, Heaton N, Mueller B. 2004. Clinical utility of cytomegalovirus viral load testing for predicting CMV disease in D+/R− solid organ transplant recipients. Am. J. Transplant. 4:644–649 [DOI] [PubMed] [Google Scholar]
- 183.Lisboa LF, Preiksaitis JK, Humar A, Kumar D. 2011. Clinical utility of molecular surveillance for cytomegalovirus after antiviral prophylaxis in high-risk solid organ transplant recipients. Transplantation 92:1063–1068 [DOI] [PubMed] [Google Scholar]
- 184.Singh N, Yu VL, Mieles L, Wagener MM, Miner RC, Gayowski T. 1994. High-dose acyclovir compared with short-course preemptive ganciclovir therapy to prevent cytomegalovirus disease in liver transplant recipients. A randomized trial. Ann. Intern. Med. 120:375–381 [DOI] [PubMed] [Google Scholar]
- 185.Singh N, Wannstedt C, Keyes L, Mayher D, Tickerhoof L, Akoad M, Wagener MM, Cacciarelli TV. 2008. Valganciclovir as preemptive therapy for cytomegalovirus in cytomegalovirus-seronegative liver transplant recipients of cytomegalovirus-seropositive donor allografts. Liver Transpl. 14:240–244 [DOI] [PubMed] [Google Scholar]
- 186.Singh N, Wannstedt C, Keyes L, Gayowski T, Wagener MM, Cacciarelli TV. 2005. Efficacy of valganciclovir administered as preemptive therapy for cytomegalovirus disease in liver transplant recipients: impact on viral load and late-onset cytomegalovirus disease. Transplantation 79:85–90 [DOI] [PubMed] [Google Scholar]
- 187.Martin-Gandul C, Perez-Romero P, Sanchez M, Bernal G, Suarez G, Sobrino M, Merino L, Cisneros JM, Cordero E. 2013. Determination, validation and standardization of a CMV DNA cut-off value in plasma for preemptive treatment of CMV infection in solid organ transplant recipients at lower risk for CMV infection. J. Clin. Virol. 56:13–18 [DOI] [PubMed] [Google Scholar]
- 188.Strippoli GF, Hodson EM, Jones C, Craig JC. 2006. Preemptive treatment for cytomegalovirus viremia to prevent cytomegalovirus disease in solid organ transplant recipients. Transplantation 81:139–145 [DOI] [PubMed] [Google Scholar]
- 189.Helantera I, Kyllonen L, Lautenschlager I, Salmela K, Koskinen P. 2010. Primary CMV infections are common in kidney transplant recipients after 6 months valganciclovir prophylaxis. Am. J. Transplant. 10:2026–2032 [DOI] [PubMed] [Google Scholar]
- 190.Levitsky J, Freifeld AG, Puumala S, Bargenquast K, Hardiman P, Gebhart C, Wrenshall L, Langnas A, Kalil AC. 2008. Cytomegalovirus viremia in solid organ transplantation: does the initial viral load correlate with risk factors and outcomes? Clin. Transplant. 22:222–228 [DOI] [PubMed] [Google Scholar]
- 191.Manez R, Kusne S, Green M, Abu-Elmagd K, Irish W, Reyes J, Furukawa H, Tzakis A, Fung JJ, Todo S, Starzl TE. 1995. Incidence and risk factors associated with the development of cytomegalovirus disease after intestinal transplantation. Transplantation 59:1010–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Peiris JS, Taylor CE, Main J, Graham K, Madeley CR. 1995. Diagnosis of cytomegalovirus (CMV) disease in renal allograft recipients: the role of semiquantitative polymerase chain reaction (PCR). Nephrol. Dial. Transplant. 10:1198–1205 [PubMed] [Google Scholar]
- 193.Cummins NW, Deziel PJ, Abraham RS, Razonable RR. 2009. Deficiency of cytomegalovirus (CMV)-specific CD8+ T cells in patients presenting with late-onset CMV disease several years after transplantation. Transpl. Infect. Dis. 11:20–27 [DOI] [PubMed] [Google Scholar]
- 194.Gerna G, Baldanti F, Sarasini A, Furione M, Percivalle E, Revello MG, Zipeto D, Zella D. 1994. Effect of foscarnet induction treatment on quantitation of human cytomegalovirus (HCMV) DNA in peripheral blood polymorphonuclear leukocytes and aqueous humor of AIDS patients with HCMV retinitis. The Italian Foscarnet Study Group. Antimicrob. Agents Chemother. 38:38–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Cinque P, Vago L, Brytting M, Castagna A, Accordini A, Sundqvist VA, Zanchetta N, Monforte AD, Wahren B, Lazzarin A, Linde A. 1992. Cytomegalovirus infection of the central nervous system in patients with AIDS: diagnosis by DNA amplification from cerebrospinal fluid. J. Infect. Dis. 166:1408–1411. [DOI] [PubMed] [Google Scholar]
- 196.Weinberg A, Spiers D, Cai GY, Long CM, Sun R, Tevere V. 1998. Evaluation of a commercial PCR kit for diagnosis of cytomegalovirus infection of the central nervous system. J. Clin. Microbiol. 36:3382–3384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Asberg A, Humar A, Rollag H, Jardine AG, Mouas H, Pescovitz MD, Sgarabotto D, Tuncer M, Noronha IL, Hartmann A. 2007. Oral valganciclovir is noninferior to intravenous ganciclovir for the treatment of cytomegalovirus disease in solid organ transplant recipients. Am. J. Transplant. 7:2106–2113 [DOI] [PubMed] [Google Scholar]
- 198.Mattes FM, Hainsworth EG, Hassan-Walker AF, Burroughs AK, Sweny P, Griffiths PD, Emery VC. 2005. Kinetics of cytomegalovirus load decrease in solid-organ transplant recipients after preemptive therapy with valganciclovir. J. Infect. Dis. 191:89–92 [DOI] [PubMed] [Google Scholar]
- 199.Gerna G, Zipeto D, Parea M, Revello MG, Silini E, Percivalle E, Zavattoni M, Grossi P, Milanesi G. 1991. Monitoring of human cytomegalovirus infections and ganciclovir treatment in heart transplant recipients by determination of viremia, antigenemia, and DNAemia. J. Infect. Dis. 164:488–498 [DOI] [PubMed] [Google Scholar]
- 200.van den Berg AP, van Son WJ, Haagsma EB, Klompmaker IJ, Tegzess AM, Schirm J, Dijkstra G, van der Giessen M, Slooff MJ, The TH. 1993. Prediction of recurrent cytomegalovirus disease after treatment with ganciclovir in solid-organ transplant recipients. Transplantation 55:847–851 [DOI] [PubMed] [Google Scholar]
- 201.Eid AJ, Arthurs SK, Deziel PJ, Wilhelm MP, Razonable RR. 2008. Emergence of drug-resistant cytomegalovirus in the era of valganciclovir prophylaxis: therapeutic implications and outcomes. Clin. Transplant. 22:162–170 [DOI] [PubMed] [Google Scholar]
- 202.Limaye AP, Corey L, Koelle DM, Davis CL, Boeckh M. 2000. Emergence of ganciclovir-resistant cytomegalovirus disease among recipients of solid-organ transplants. Lancet 356:645–649 [DOI] [PubMed] [Google Scholar]
- 203.Razonable R. 2010. Direct and indirect effects of cytomegalovirus: can we prevent them? Enferm. Infecc. Microbiol. Clin. 28:1–5 [DOI] [PubMed] [Google Scholar]
- 204.Razonable RR, Emery VC. 2004. Management of CMV infection and disease in transplant patients. 27–29 February 2004 Herpes 11:77–86 [PubMed] [Google Scholar]
- 205.Boivin G, Goyette N, Farhan M, Ives J, Elston R. 2012. Incidence of cytomegalovirus UL97 and UL54 amino acid substitutions detected after 100 or 200 days of valganciclovir prophylaxis. J. Clin. Virol. 53:208–213 [DOI] [PubMed] [Google Scholar]
- 206.Boivin G, Goyette N, Gilbert C, Roberts N, Macey K, Paya C, Pescovitz MD, Humar A, Dominguez E, Washburn K, Blumberg E, Alexander B, Freeman R, Heaton N, Covington E. 2004. Absence of cytomegalovirus-resistance mutations after valganciclovir prophylaxis, in a prospective multicenter study of solid-organ transplant recipients. J. Infect. Dis. 189:1615–1618 [DOI] [PubMed] [Google Scholar]
- 207.Limaye AP, Raghu G, Koelle DM, Ferrenberg J, Huang ML, Boeckh M. 2002. High incidence of ganciclovir-resistant cytomegalovirus infection among lung transplant recipients receiving preemptive therapy. J. Infect. Dis. 185:20–27 [DOI] [PubMed] [Google Scholar]
- 208.Bhorade SM, Lurain NS, Jordan A, Leischner J, Villanueva J, Durazo R, Creech S, Vigneswaran WT, Garrity ER. 2002. Emergence of ganciclovir-resistant cytomegalovirus in lung transplant recipients. J. Heart Lung Transplant. 21:1274–1282 [DOI] [PubMed] [Google Scholar]
- 209.Lurain NS, Chou S. 2010. Antiviral drug resistance of human cytomegalovirus. Clin. Microbiol. Rev. 23:689–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Avery RK, Marty FM, Strasfeld L, Lee I, Arrieta A, Chou S, Tatarowicz W, Villano S. 2010. Oral maribavir for treatment of refractory or resistant cytomegalovirus infections in transplant recipients. Transpl. Infect. Dis. 12:489–496 [DOI] [PubMed] [Google Scholar]
- 211.Avery RK, Mossad SB, Poggio E, Lard M, Budev M, Bolwell B, Waldman WJ, Braun W, Mawhorter SD, Fatica R, Krishnamurthi V, Young JB, Shrestha R, Stephany B, Lurain N, Yen-Lieberman B. 2010. Utility of leflunomide in the treatment of complex cytomegalovirus syndromes. Transplantation 90:419–426 [DOI] [PubMed] [Google Scholar]
- 212.Boivin G, Goyette N, Gilbert C, Humar A, Covington E. 2005. Clinical impact of ganciclovir-resistant cytomegalovirus infections in solid organ transplant patients. Transpl. Infect. Dis. 7:166–170 [DOI] [PubMed] [Google Scholar]
- 213.Goldner T, Hewlett G, Ettischer N, Ruebsamen-Schaeff H, Zimmermann H, Lischka P. 2011. The novel anticytomegalovirus compound AIC246 (letermovir) inhibits human cytomegalovirus replication through a specific antiviral mechanism that involves the viral terminase. J. Virol. 85:10884–10893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Marschall M, Stamminger T, Urban A, Wildum S, Ruebsamen-Schaeff H, Zimmermann H, Lischka P. 2012. In vitro evaluation of the activities of the novel anticytomegalovirus compound AIC246 (letermovir) against herpesviruses and other human pathogenic viruses. Antimicrob. Agents Chemother. 56:1135–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Price NB, Prichard MN. 2011. Progress in the development of new therapies for herpesvirus infections. Curr. Opin. Virol. 1:548–554 [DOI] [PMC free article] [PubMed] [Google Scholar]