Abstract
Pacidamycins (or uridyl peptide antibiotics) possess selective in vivo activity against Pseudomonas aeruginosa. An important limitation for the therapeutic use of pacidamycins with P. aeruginosa is the high frequency (10−6 to 10−7) at which resistant mutants emerge. To elucidate the mechanism(s) of this resistance, pacidamycin-resistant P. aeruginosa mutants were isolated. Two types of mutants were obtained. Type 1, or high-level resistance mutants with a pacidamycin MIC of 512 μg/ml, were more abundant, with a frequency of ∼2 × 10−6, and did not show cross-resistance with other antibiotics. Type 2, low-level resistance mutants, were isolated with a frequency of ∼10−8 and had a pacidamycin MIC of 64 μg/ml (the MIC for the wild-type strain was 4 to 16 μg/ml). These mutants were cross-resistant to levofloxacin, tetracycline, and erythromycin and were shown to overexpress either the MexAB-OprM or MexCD-OprJ multidrug resistance efflux pumps. High-level resistant mutants were isolated by transposon mutagenesis and one insertion was localized to oppB, one of two periplasmic binding protein components of an oligopeptide transport system which is encoded by the opp-fabI operon. The Opp system is required for uptake of pacidamycin across the inner membrane, since various opp, but not fabI, mutants were resistant to high levels of pacidamycin. Both of the two putative Opp periplasmic binding proteins, OppA and OppB, were required for pacidamycin uptake. Although both impaired uptake into and efflux from the cell can cause pacidamycin resistance in P. aeruginosa, our data suggest that impaired uptake is the primary reason for the high-frequency and high-level pacidamycin resistance.
INTRODUCTION
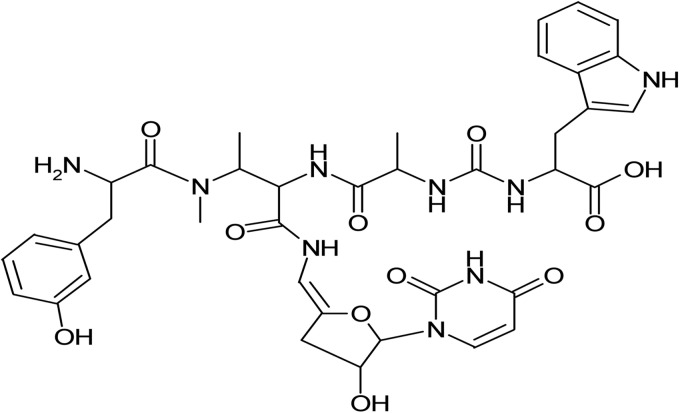
Pacidamycins are uridyl peptide antibiotics that were first discovered in 1989 (1–3) (pacidamycin 4, used in this study, is shown in Fig. 1). Since that time, at least 10 related compounds have been reported. Despite their discovery over 2 decades ago, the biosynthetic gene clusters for pacidamycin biosynthesis were only recently identified and characterized in Streptomyces coeruleorubidus (4, 5). The pacidamycins have an unusual spectrum of antibacterial activity. While they possess in vivo activity against Pseudomonas aeruginosa, one of the organisms most refractory to antibacterial therapy, they are not active against many other bacteria, for example, Escherichia coli and Staphylococcus aureus (2). Pacidamycins target MraY, or translocase I, an essential enzyme in peptidoglycan biosynthesis in most Gram-positive and Gram-negative bacteria (6). Located on the cytoplasmic face of the inner membrane, MraY catalyzes the formation of the first lipid intermediate, undecaprenylpyrophosphate-N-acetylmuramyl-pentapeptide (or lipid I), in cell wall synthesis. Although E. coli and S. aureus are intrinsically resistant to pacidamycin, MraY purified from either bacterium can be efficiently inhibited in vitro by pacidamycin (6). High-level intrinsic pacidamycin resistance in these bacteria could be explained by lack of uptake of the peptide antibiotic, efficient extrusion via efflux pumps, or a combination of these mechanisms. The intrinsic resistance of E. coli to mureidomycins was previously attributed to efflux by the AcrAB-TolC pump (7, 8). A significant limitation for the therapeutic use of pacidamycins with P. aeruginosa is the high frequency (10−6 to 10−7) at which resistant mutants emerge. In this paper, we report that both impaired uptake into and efflux from the cell are pacidamycin resistance determinants in this bacterium. However, the primary reason for high-frequency and high-level resistance is loss of uptake into the cell due to mutations in the opp operon encoding an oligopeptide transport system.
Fig 1.
Structure of the uridylpeptide antibiotic pacidamycin 4 employed in this study.
(Portions of this work were previously presented in part as a poster at the 101st General Meeting of the American Society for Microbiology, Orlando, FL, 20 to 24 May 2001 [9].)
MATERIALS AND METHODS
Bacterial strains and media.
Pseudomonas aeruginosa strains used in this study are listed in Table 1. E. coli strains used for cloning were DH5α (15) and HPS1 (16). Bacteria were generally cultivated in Lennox LB broth base or agar (Life Technologies, Grand Island, NY). Other growth media used in this study were super optimal broth (SOB) (17), peptone tryptic soy broth (PTSB) (18), and M9 minimal medium (28) supplemented with 1% Difco Casitone (Becton, Dickinson and Company, Franklin Lakes, NJ). For plasmid maintenance, media were supplemented with 100 μg/ml of ampicillin or 15 μg/ml gentamicin for E. coli and 200 μg/ml carbenicillin or 30 μg/ml gentamicin for P. aeruginosa, unless noted otherwise.
Table 1.
Pseudomonas aeruginosa strains and plasmids used in this study
| Strain/plasmid | Characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| PAO1 | Prototype strain (PAM1020); obtained from N. Gotoh | 10 |
| PAO1-UW | Sequenced prototype PAO1 strain | 11 |
| PAM1032 | nalB (MexAB-OprM up) | 12 |
| PAM1033 | nfxB (MexCD-OprJ up) | 12 |
| PAM1034 | nfxC (MexEF-OprN up) | 12 |
| PAM1043 | nalB-type (MexAB-OprM up) | This study |
| PAM1044 | nfxB-type (MexCD-OprJ up) | This study |
| PAM1135 | PAO1 pac-1 | This study |
| PAM1154 | PAO1 oprM::Hgra | This study |
| PAM1194 | PAO1 pac::mini-D171b | This study |
| PAM1316 | PAM1194 oprM::Hgr | This study |
| PAM1348 | PAM1135 oprM::Hgr | This study |
| PAM2386 | PAO1 pac::mini-D171b,c | This study |
| PAO235 | PAO1 fabI::FRT | This study |
| PAO315 | PAO1 oppE::FRT | This study |
| PAO449 | PAO1 oppA::FRT | This study |
| PAO306 | PAO1 with chromosomally integrated fabI′-lacZ | This study |
| PW4183 | PAO1-UW oppA::ISlacZ/hah | 13 |
| PW4184 | PAO1-UW oppA::ISlacZ/hah | 13 |
| PAO1284 | PAO1 ΔoppB::FRT | This study |
| Plasmids | ||
| pCR2.1 | TA cloning vector for PCR fragments; Ampr Kanr | Life Technologies |
| pEX18Ap | Gene replacement vector; Ampr | 14 |
| pPS856 | Source for GENr cassette | 14 |
| pOPPA1 | pCR2.1 with 1,852-bp oppA fragment | This study |
| pOPPA2 | pOPPA1 with GENr cassette from pPS856 | This study |
| pOPPA3 | pEX18Ap with oppB::GENr fragment from pOPPA2 | This study |
| pOPPB1 | pCR2.1 with 834-bp oppB fragment | This study |
| pOPPB2 | pOPPB1 with GENr cassette from pPS856 | This study |
| pOPPB3 | pEX18Ap with oppB::GENr fragment from pOPPB2 | This study |
| pOPPE1 | pCR2.1 with 1,745-bp oppE fragment | This study |
| pOPPE2 | pOPPE1 with GENr cassette from pPS856 | This study |
| pOPPE3 | pEX18Ap with oppE::GENr fragment from pOPPE2 | This study |
Hgr, mercury resistance determinant from Tn501.
Insertion mapping revealed that in this strain, pac is identical to oppB.
The mini-D171 element was transferred from PAM1194 to PAO1 by F116L-mediated transduction selecting tetracycline resistance encoded by mini-D171.
MIC determinations.
Antimicrobial susceptibilities were assessed by determining MICs using the 2-fold broth microdilution method by following Clinical and Laboratory Standards Institute guidelines (20).
Selection of pacidamycin-resistant mutants.
Spontaneous pacidamycin-resistant mutants of wild-type strain PAO1 (PAM1020) were selected on LB agar plates containing pacidamycin at either 50 μg/ml (4× MIC) or 200 μg/ml (16× MIC) using overnight cultures grown in LB. The potential efflux-mediated multidrug resistance phenotype was assessed by replica plating on LB plates containing levofloxacin at 0.5 μg/ml (4× MIC). Levofloxacin was used since it is a substrate of major Mex pumps from P. aeruginosa (12). Pacidamycin-resistant mutants that were also resistant to levofloxacin were selected on plates contacting pacidamycin at 50 μg/ml. Such mutants were much less (50- to 100-fold) frequent than mutants resistant to pacidamycin alone. In order to carefully estimate the frequency of emergence of pacidamycin-resistant mutants that exhibited a multidrug resistance phenotype, such mutants were also selected directly on plates containing both pacidamycin (at 50 μg/ml) and levofloxacin (at 0.5 μg/ml). Frequency of resistance emergence was calculated as a ratio of viable colonies from antibiotic-containing and antibiotic-free plates.
Transposon mutagenesis and transduction.
Transposon mutagenesis of PAM1020 to identify pacidamycin resistance mutants was performed with the mini-D3112 bacteriophage transposable element mini-D171 (encoding tetracycline resistance) (21). Mutants were selected on LB agar plates containing 100 μg/ml tetracycline and 200 μg/ml pacidamycin. Transductions into P. aeruginosa recipient strains were performed with phage F116L (22).
Inverse PCR.
Inverse PCR was used to localize mini-D171 insertions resulting in pacidamycin resistance. Chromosomal DNA was isolated from the mutant PAM1194 (PAM1020 pac::TET-16), digested with HindIII, ligated, and used as a template for PCR with primer 1 (primer sequences are shown in Table 2), which anneals close to the left end of mini-D171), and primer 2, annealing upstream of the HindIII site in mini-D171. The resulting ∼600-bp PCR fragment was cloned into pCR2.1 (Life Technologies) and sequenced using M13 reverse and M13 forward (−40) primers.
Table 2.
Oligonucleotides used in this study
| Name | Sequencea |
|---|---|
| Primer 1 | 5′-GCAGCTCTTGCTCAATCC |
| Primer 2 | 5′-CAAGCGACTTATGGGAGG |
| oppE#1 | 5′-CTGCTGGAGCTCATCGGCGAAGC |
| oppE#2 | 5′-ATGGAGAGCTCGCTGGCGACTCC |
| oppB-up | 5′-GGAGCTCGACAAGGTGGTCGC |
| oppB-down | 5′-GGAGCTCAGTGGCTGGCCGTC |
| oppA-up | 5′-CAAGCTTGGCCAGAACCCCGG |
| oppA-down | 5′-AGGTACCAGTTGGGAATGCTGTAG |
| FabI-2 | 5′-CGCGGGTGGTCACCGCGG |
| OppE-1 | 5′-ATGGCCCCGGCAAGCGCG |
| P244 | 5′-CTGCGGCAACTGGAGGGCAAG |
| P245 | 5′-CCGGGCCCTGAGCTTGTCGTT |
| lacZ-148 | 5′-GGGTAACGCCAGGGTTTTCC |
| Pser-down | 5′-AGTTCGGCCTGGTGGAACAACTCG |
Newly introduced SacI (oppE#1 and oppE#2), HindIII (oppA-up), KpnI (oppA-down), and ApaI (P245) restriction sites are underlined.
PAβN uptake assay.
Opp transport activity was assessed utilizing Phe-Arg-β-naphthylamide (PAβN) as the substrate. PAβN is nonfluorescent in solution but is cleaved enzymatically inside cells to produce highly fluorescent β-naphthylamine (23). Since PAβN is effluxed via the MexAB-OprM system, mutants defective in this system were used for the uptake assays. Cultures of P. aeruginosa were grown in LB broth to an optical density (at 600 nm) of ∼1, washed, and resuspended in 50 mM K2HPO4 (pH 7.0), 1 mM MgSO4, 0.4% glucose. Assays were performed in 96-well plates in a final volume of 200 μl and were initiated by addition of 250 μg/ml PAβN (Sigma, St. Louis, MO) to suspensions of intact cells. Fluorescence was measured using excitation at 320 nm and emission at 460 nm.
Construction of chromosomal opp mutants by allelic exchange.
Using standard methods for PCR amplification of GC-rich templates (including 5% [vol/vol] dimethyl sulfoxide [DMSO]) and Taq DNA polymerase (Life Technologies), a 1,745-bp oppE gene fragment was PCR amplified from ∼100 ng of PAO1 genomic DNA using primers oppE#1 and oppE#2, which introduced SacI sites flanking the oppE gene fragment. Cycle conditions were an initial 5 min of denaturation at 96°C, followed by 35 cycles of 96°C for 45 s, 60°C for 45 s, 72°C for 1 min, and a final extension step for 10 min at 72°C. The resulting 1,745-bp fragment was cloned into the TA cloning vector pCR2.1 to form pOPPE1. A plasmid-borne oppE insertional mutant was generated by ligation of a 853-bp gentamicin resistance (GENr) cassette from pPS856 (14) into the single SmaI site located within the oppE gene fragment contained on pOPPE1. This step generated pOPPE2, whose oppE::GENr fragment was then cloned into the gene replacement vector pEX18Ap (14) to yield pOPPE3. For transfer to the P. aeruginosa chromosome, pOPPE3 was transformed into E. coli mobilizer strain SM10 (24) and conjugally transferred to PAO1, and gentamicin-resistant merodiploids were selected on VBMM plates containing 30 μg/ml gentamicin (14). Merodiploids were resolved by sucrose counterselection, and the gentamicin resistance marker was excised from the chromosome using Flp recombinase as previously described (14) to yield the unmarked oppE mutant strain PAO315. Insertional mutagenesis of oppE was verified by genomic Southern analysis using the biotin-labeled 1,745-bp oppE DNA fragment as the probe by following previously described procedures (14).
Using similar procedures and primers oppB-up and oppB-down, a 834-bp partial oppB fragment was amplified from PAO1 genomic DNA and cloned into pCR2.1 to form pOPPB1. pOPPB2 then was obtained by replacing a 204-bp NruI fragment from within the partial oppB coding sequence with a 1,077-bp gentamicin resistance marker-containing SmaI fragment from pPS856. Finally, a 1,811-bp HindIII-XhoI fragment of pOPPB2 was cloned between the HindIII-SalI sites of pEX18Ap to form pOPPB3. After transformation into SM10, pOPPB3 was conjugally transferred into PAO1 and the unmarked ΔoppB mutant PAO1284 was derived by utilizing the procedures described above for oppE. The presence of the correct oppB mutation was verified by PCR and DNA sequence analysis.
For isolation of an oppA mutant, this gene was amplified from PAO1 using primers oppA-down and oppA-up, and a 1,852-bp oppA fragment was cloned into pCR2.1 to form pOPPA1. pOPPA2 next was obtained by inserting a blunt-ended 1,054-bp GENr SacI fragment from pPS856 into an NruI site located within oppA (although we intended to delete three NruI fragments encompassing 1,044 bp of oppA, we inadvertently ended up isolating a GENr insertion in the third, most distal NruI site). Finally, a 2,902-bp HindIII-KpnI fragment of pOPPA2 was cloned between the same sites of pEX18Ap to form pOPPA3. The unmarked oppA::FRT mutant PAO449 was derived by utilizing the procedures described above for oppE, and PCR analysis and DNA sequencing were used to verify the presence of the oppA insertion.
P. aeruginosa oppA transposon mutants.
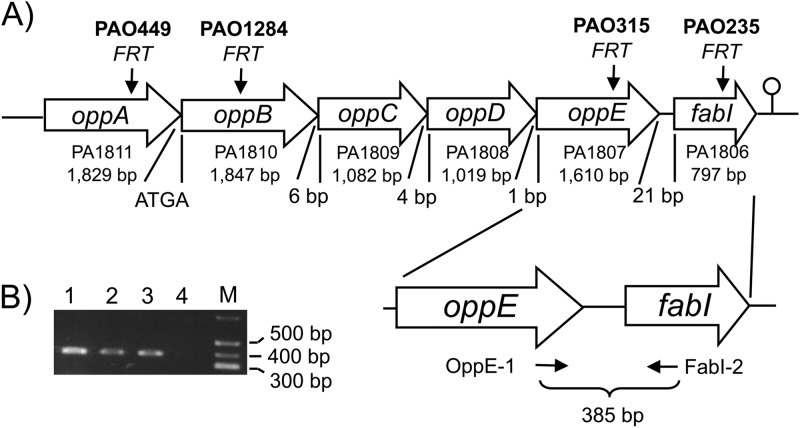
The two oppA transposon mutants PW4183 and PW4184 were obtained from the sequence-verified two-allele P. aeruginosa PAO1 transposon mutant library (13). The two mutants contain transposon ISlacZ/hah inserted either early (PA4184; nucleotide 85 of 1830) or late (PA4183; nucleotide 1528 of 1830) in oppA. In both mutants the transposon is oriented in the same orientation as oppA (or reverse with respect to the annotation of the chromosome), such that the outward-facing neomycin resistance gene promoter engineered into the transposon (25) should direct transcription of downstream genes, minimizing or eliminating polar effects on genes in the same transcriptional unit, in this instance oppB (Fig. 2). Transposon locations were verified by PCR amplification with the recommended primers, followed by sequencing of the PCR products.
Fig 2.
Gene organization in the P. aeruginosa opp-fabI operon. (A) The genes present in the opp-fabI operon are indicated by gene name, PAO1 gene annotation number, and size. Distances between genes are also shown. ATGA indicates overlap of the oppB ATG initiation codon with the oppA TGA stop codon. Arrows mark the relative positions of FRT insertions in site-specific mutants with mutant names shown above the arrows. The lollipop symbol indicates a putative transcriptional terminator. oppA and oppB encode periplasmic binding proteins; oppC and oppD encode membrane transport proteins; oppE encodes a traffic ATPase; fabI encodes an enoyl-acyl carrier protein reductase. (B) RT-PCR analysis of oppE and fabI coexpression. Amplification was achieved with primers OppE1 and FabI-2, which yielded the expected indicated 384-bp product. PCR fragments were analyzed by agarose gel electrophoresis. Lanes: 1, 2, and 3, PCR products from three separate RNA and cDNA preparations; 4, blank (same as lane 1 but without reverse transcriptase addition); M, Hi-Lo DNA size ladder from Minnesota Molecular (Minneapolis, MN).
Gene fusion constructs.
A fabI′-lacZ transcriptional fusion was constructed by PCR amplifying a 377-bp DNA fragment from PAO1 chromosomal DNA using the primer pair P244 and P245. The fabI fragment contained the last 68 codons of oppE, the 21-bp oppE-fabI intergenic region, and the first 45 codons of fabI. This fragment was then subcloned into the integrative lacZ fusion vector mini-CTX-lacZ (26) between the EcoRI and SmaI sites and integrated at the φCTX attachment site on the PAO1 chromosome using established procedures (27). Strain PAO306 (fabI′-lacZ) was obtained after removal of the tetracycline resistance selection marker using Flp recombinase (27). The presence of the correct fusion was verified by amplifying PCR fragments containing the 5′ lacZ sequences, the cloned chromosomal DNA segment, and portions of the mini-CTX-lacZ vector sequences with primers lacZ-148 and Pser-down. The resulting PCR fragment was sequenced with primer lacZ-148. The strains harboring the fabI′-lacZ fusion were grown in various media to an optical density at 600 nm of ∼1, and β-galactosidase activities were measured (28).
RT-PCR analysis.
For RNA isolation, cells of PAO1 were grown overnight at 37°C in 1% proteose peptone broth 3 (Difco). The cells were diluted 1:100 into the same medium and incubated at 37°C until the cell density reached an optical density (540 nm) of 0.5 to 0.7. Total RNA was isolated from these cells using the RNeasy kit (Qiagen, Valencia, CA). The reverse transcriptase (RT) reaction mixture contained 1 μg of total RNA, 10 pmol of primer FabI-2, and 5% DMSO. This mixture was heated at 70°C for 10 min before addition of RNasin, RT buffer, dithiothreitol (DTT), and deoxynucleoside triphosphates (dNTPs). After 2 min at 48°C, 10 U reverse AMV transcriptase was added, and the mixture was incubated at 48°C for 50 min before the RT was inactivated by heating to 70°C for 15 min. Five-μl aliquots of the PCR mixes were used as the templates in PCR mixtures containing 0.2 mM dNTPs, 10 pmol of FabI-2, 10 pmol of OppE-1, 5% DMSO, 1× PCR buffer, and 5 U Taq polymerase. Cycle conditions were 35 cycles of 95°C for 45 s, 65°C for 45 s, 72°C for 1 min, and a final extension step for 10 min at 72°C. The sizes of the amplicons were analyzed by electrophoresis on a 2% agarose gel in Tris-acetate-EDTA (TAE) buffer (19), and their nucleotide sequences were determined.
RESULTS AND DISCUSSION
Identification of spontaneous pacidamycin-resistant mutants.
Exposure of PAO1 (MIC, 4 to 16 μg/ml) resulted in ready emergence of pacidamycin-resistant mutants of two types (Table 3). Type 1 mutants were obtained with a frequency of 2 × 10−6. These so-called pac-1 mutants, exemplified by PAM1135, were highly resistant to pacidamycin (MIC, >500 μg/ml) but not cross-resistant to other antibiotics. Type 2 mutants emerged less frequently (10−7 to 10−8), showed intermediate resistance to pacidamycin (64 μg/ml), and exhibited cross-resistance with other antibiotics. Antibiotic resistance profiles of two type 2 mutants analyzed in more detail, PAM1043 and PAM1044, were similar to MexAB-OprM- and MexCD-OprJ-expressing control strains (Table 3). RT-PCR analysis confirmed overexpression of mexB and mexD in PAM1043 and PAM1044, respectively (data not shown). This indicated that the intermediate pacidamycin resistance levels observed in PAM1043 and PAM1044 most likely were due to upregulation of MexAB-OprM and MexCD-OprJ, respectively. Pacidamycin was not a substrate for MexEF-OprN, since the MICs of PAO1 and a MexEF-OprN-overexpressing strain were the same.
Table 3.
Properties of spontaneous pacidamycin-resistant P. aeruginosa mutants
| Strain | Description | MIC (μg/ml) |
||||
|---|---|---|---|---|---|---|
| Pacidamycin | Levofloxacin | Carbenicillin | Erythromycin | Imipenem | ||
| PAO1 | Wild type | 4–16 | 0.25 | 32 | 128 | 1 |
| Type 1 | High-level resistance | |||||
| PAM1135 | pac-1 | >512 | 0.25 | 32 | 128 | 1 |
| Type 2 | Low-level resistance | |||||
| PAM1043 | MexAB-OprM up | 64 | 1 | 256 | 256 | 1 |
| PAM1044 | MexCD-OprJ up | 64 | 2 | 16 | >512 | 0.125 |
| Controls | ||||||
| PAM1032 | nalB (MexAB-OprM up) | 64 | 1 | 256 | 256 | 1 |
| PAM1033 | nfxB (MexCD-OprJ up) | 64 | 2 | 16 | >512 | 0.25 |
| PAM1034 | nfxC (MexEF-OprN up) | 16 | 4 | 16 | 128 | 4 |
Because it has been well established that MexAB-OprM plays a major role in the intrinsic antibacterial resistance of P. aeruginosa (29–31), we sought to further characterize the impact of this efflux pump on pacidamycin susceptibility (Table 4). Cells expressing an inactive MexAB-OprM system (PAM1154) were more susceptible to pacidamycin than wild-type PAO1, indicating that this efflux pump plays a role in intrinsic pacidamycin resistance. However, inactivation of MexAB-OprM or the presence of the efflux pump inhibitor PAβN (32 and data not shown) did not reduce resistance to pacidamycin in type 1 mutants, corroborating the notion that high-level resistance was not affected by this efflux system. As expected, in control experiments, inactivation of MexAB-OprM or the presence of PAβN did reduce the levofloxacin MIC for PAM1135 from 0.25 to 0.015 μg/ml (data not shown). It should be noted that inactivation of OprM also impairs the function of the MexXY efflux pump, resulting in hypersusceptibility to antimicrobials not extruded by MexAB-OprM, e.g., aminoglycosides, but the role of MexXY in pacidamycin resistance, if any, remains to be assessed.
Table 4.
Impact of MexAB-OprM on P. aeruginosa susceptibility to pacidamycin
| Strain | Description | MIC (μg/ml) |
||
|---|---|---|---|---|
| Pacidamycin | Levofloxacin | Carbenicillin | ||
| PAO1 | Wild type | 16 | 0.25 | 32 |
| PAM1154 | PAO1 (PAM1020) oprM::Hgr | 4 | 0.015 | 0.5 |
| PAM1135 | PAO1 (PAM1020) pac-1 | >512 | 0.25 | 32 |
| PAM1348 | PAM1135 oprM::Hgr | >512 | 0.015 | 0.5 |
A transposon mutant exhibiting high-level pacidamycin resistance maps to an oligopeptide permease operon.
To assess the molecular mechanism(s) underlying high-level pacidamycin resistance in type 1 mutants, wild-type PAO1 was subjected to mini-D171 transposon mutagenesis, and mutants exhibiting high-level (>200 μg/ml) pacidamycin resistance were selected. One of the resulting mutants, PAM1194 (pac::mini-D171), was further characterized (Table 5). The resistance profile of this mutant was the same as that of the spontaneous pac-1 mutant PAM1135, and inactivation of MexAB-OprM had no effect on the pacidamycin resistance. Transduction of mini-D171 from PAM1194 into the wild-type PAO1 resulted in high-level resistance (PAM2386), demonstrating that mini-D171 insertion was linked to a pacidamycin resistance locus.
Table 5.
Characterization of P. aeruginosa transposon and defined opp mutants exhibiting high-level pacidamycin resistance
| Strain | Description | MIC (μg/ml) |
|||
|---|---|---|---|---|---|
| Pacidamycin | Levofloxacin | Carbenicillin | Tetracycline | ||
| PAO1 | Wild type | 16 | 0.25 | 32 | 4 |
| PAM1194 | PAO1 pac::mini-D171b | >512 | 0.25 | 32 | 512a |
| PAM1135 | PAO1 pac-1 | >512 | 0.25 | 32 | ND |
| PAM1316 | PAO1194 oprM::Hgr | >512 | 0.015 | 0.5 | 32 |
| PAM2386 | PAO1 pac::mini-D171b,c | >512 | 1 | 256 | >512a |
| PAO235 | PAO1 fabI::FRT | 32 | 0.25 | ND | ND |
| PAO315 | PAO1 oppE::FRT | >512 | 0.25 | ND | ND |
| PAO1284 | PAO1 ΔoppB::FRT | >512 | 0.25 | ND | ND |
| PAO449 | PAO1 oppA::FRT | 256 | 0.25 | ND | ND |
| PW4183 | PAO1 oppA::ISlacZ/hah | >512 | ND | ND | ND |
| PW4184 | PAO1 oppA::ISlacZ/hah | >512 | ND | ND | ND |
The mini-D171 element contains a tetracycline resistance marker.
Insertion mapping revealed that in this strain, pac is identical to oppB.
The tetracycline resistance marker was transferred from PAM1194 to PAO1 by F116L-mediated transduction.
By inverse PCR, determination of the DNA sequence adjacent to mini-D171, and BLAST searches of the published P. aeruginosa chromosomal sequences, including PAO1 (11), the transposon insertion in PAM1194 was localized to the oppB gene, encoding a protein with 63% similarity to a periplasmic binding protein encoded by the oppA gene of Helicobacter pylori (33).
The P. aeruginosa PAO1 opp operon (Fig. 2) contains five genes that encode proteins of a probable oligopeptide transport system. The first two genes, oppA and oppB, whose coding sequences overlap, contain type I signal peptides; thus, they are probably periplasmic binding proteins. The next three genes of the operon, oppC, oppD, and oppE, are separated from oppB by 6 bp of untranslated sequence. Based on similarity with amino acid motifs found in proteins from other binding protein-dependent systems, oppC and oppD encode membrane transporter components and oppE encodes an ATPase component. Interestingly, there is no transcriptional terminator downstream of oppE. Rather, this gene is separated from another gene, fabI, by only 21 bp. A transcriptional terminator is found downstream of fabI (34). The previously identified fabI gene encodes the triclosan-sensitive enoyl-acyl carrier protein (ACP) reductase gene of the de novo fatty acid biosynthetic pathway (35).
To determine whether the opp genes and fabI are located in the same operon, reverse transcription-PCR (RT-PCR) was performed. Preliminary analyses using a fabI′-lacZ transcriptional fusion had shown that the opp operon was transcribed at low but detectable levels in most rich (e.g., LB, PTSB, SOB, or proteose peptone broth) and minimal (e.g., M9 plus 1% Casitone) media. The β-galactosidase activity levels in PAO1 containing fabI′-lacZ integrated at the mini-CTX attachment site on the chromosome (26, 27) ranged between 24 and 171 Miller units (28) in cells grown in either LB, PTSB, SOB, proteose peptone broth, or M9 plus 1% Casitone. Therefore, RNA was isolated from cells grown in proteose peptone broth and reverse transcribed using a primer (FabI-2) that anneals to sequences early in fabI (Fig. 2A depicts approximate locations of primer binding sites), and the resulting cDNA was PCR amplified using primers OppE-1 and FabI-2. RT-PCR yielded a single amplicon of the expected size (Fig. 2B), and its identity was verified by nucleotide sequencing. This finding demonstrates that the opp genes and fabI are cotranscribed as part of the same operon. The RT-PCR product was also produced in P. aeruginosa cells grown under various growth conditions; for example, no significant differences were observed in rich or defined minimal media (data not shown).
Opp proteins but not FabI are responsible for high-level pacidamycin resistance.
To determine the requirement of various opp operon genes and their products for pacidamycin transport, several defined insertion or deletion mutants were isolated and characterized. Inactivation of oppA, oppB, and oppE resulted in high-level pacidamycin resistance (256 to >512 μg/ml), but the susceptibility of the fabI mutant PAO235 was similar to that of wild-type PAO1 (Table 5). These results are consistent with the notion that the Opp system is required for uptake of pacidamycin across the inner membrane and that the activity of this transport system is responsible for the observed pacidamycin susceptibility of wild-type P. aeruginosa. This mutant analysis also indicated that both OppA and OppB periplasmic binding proteins were required for pacidamycin uptake and that they cannot compensate for each other. OppA (609 amino acids) and OppB (615 amino acids) are closely related (42.5% identity in CLUSTALW alignments), but neither P. aeruginosa OppA nor OppB shows significant homology to the 543-amino-acid E. coli OppA (identity, 16.4 and 15.7%, respectively). Despite expression of OppB in the nonpolar deletion mutant PAO449 or the nonpolar transposon mutants PW4182 and PW4184 (downstream genes are transcribed from the strong neomycin resistance gene promoter engineered into the transposon; see Materials and Methods), these three mutants still exhibit high-level pacidamycin resistance. Similarly, despite expression of OppA, the ΔoppB mutant PAO1284 still exhibits high-level resistance.
Pacidamycin-resistant mutants are affected in peptide transport.
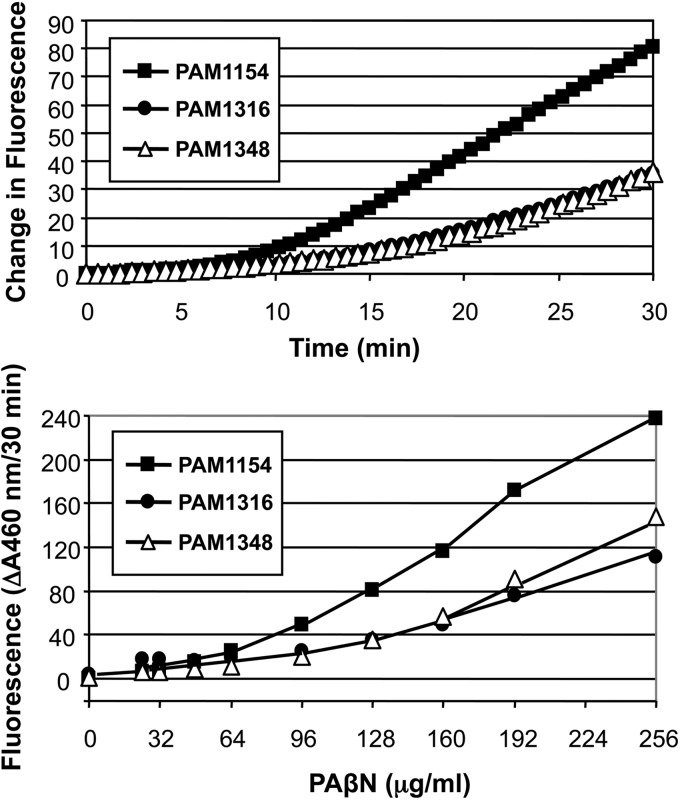
As mentioned above, using a single-copy, chromosomally integrated fabI′-lacZ transcriptional fusion, we demonstrated that the opp operon was expressed at low but detectable levels in cells grown in various media, and no significant differences were observed in rich or defined minimal media. Therefore, transport activity was assessed in LB broth-grown cells, utilizing PAβN as the substrate. Decreased uptake of PAβN was observed in the spontaneous or transposon-induced pacidamycin-resistant mutants (Fig. 3). The residual uptake of this dipeptide is probably due to the expression of a separate dipeptide transport system.
Fig 3.
P. aeruginosa opp operon encodes a functional peptide transport system. Cells of the indicated strains were grown in LB broth to an optical density (at 600 nm) of ∼1, washed, and resuspended in 50 mM K2HPO4 (pH 7.0), 1 mM MgSO4, and 0.4% glucose. Assays were initiated by addition of PAβN to suspensions of intact cells, and fluorescence was measured. Strains assayed were PAM1154 (PAO1 oprM::Hgr), PAM1316 (oppB::mini-D171 oprM::Hgr), and PAM1348 (pac-1 oprM::Hgr).
Concluding remarks.
In summary, we have identified the mechanisms associated with P. aeruginosa's resistance to the uridyl peptide antibiotic pacidamycin. High-level resistance emerges with high frequency through mutations in the Opp transporter, a binding protein-dependent ABC transporter used for oligopeptide import. Mutations in the oppA, oppB, and oppE genes, encoding two periplasmic binding proteins and an ATPase component, respectively, resulted in high-level pacidamycin resistance. These did not display cross-resistance to other antibiotics, consistent with high-level resistance being due to decreased uptake of pacidamycin rather than upregulation of multisubstrate efflux pumps. The Opp permease contains two putative periplasmic binding proteins, OppA and OppB. We observed that lack of OppA cannot be compensated for by OppB and vice versa, indicating that neither binding protein alone can mediate pacidamycin transport, which is indicative of an oligomeric binding protein requiring both OppA and OppB subunits. The Opp system was shown to be a bona fide oligopeptide transport system by demonstrating that transport of PAβN is impaired in opp mutants. Oligopeptide permease systems are often involved in uptake of toxic peptide compounds. For instance, the E. coli and Salmonella enterica serovar Typhimurium Opp permease is required for uptake of the phaseolotoxin ([Nδ,N′-sulfodiaminophosphinyl] l-ornithyl-alanyl-homoarginine) from Pseudomonas syringae pv. phaseolica, and opp mutants were resistant to phaseolotoxin (36). Spontaneous Listeria monocytogenes resistance to the tripeptide herbicide bialaphos (consisting of l-phosphinotricin, an analog of l-glutamate, and two l-alanine residues) occurs primarily via mutations affecting oligopeptide permease-mediated uptake (37). Disk diffusion susceptibility assays showed that in contrast to PAO1, which is susceptible to bialaphos, ΔoppB and oppE insertion mutants are bialaphos resistant, indicating that the opp system is also required for import of this tripeptide herbicide (data not shown). Isolates with low-level resistance of P. aeruginosa to pacidamycin were found at a lower frequency, and this resistance was mediated by the MexAB-OprM and MexCD-OprJ efflux pumps. Consistent with multidrug efflux pump expression, these isolates were cross-resistant to other antibiotics known to be pump substrates. In summary, development of pacidamycins as anti-Pseudomonas drugs faces a dual threat of resistance and reduced uptake and efflux.
ACKNOWLEDGMENTS
This work was supported by NIH grant GM56685 to H.P.S.
We thank Holly Settles, Branden Rhodes, and Nawarat Somprasong for their contributions to this work.
Footnotes
Published ahead of print 26 August 2013
REFERENCES
- 1.Chen RH, Buko AM, Whittern DN, McAlpine JB. 1989. Pacidamycins, a novel series of antibiotics with anti-Pseudomonas aeruginosa activity. II. Isolation and structural elucidation. J. Antibiot. 42:512–520 [DOI] [PubMed] [Google Scholar]
- 2.Fernandes PB, Swanson RN, Hardy DJ, Hanson CW, Coen L, Rasmussen RR, Chen RH. 1989. Pacidamycins, a novel series of antibiotics with anti-Pseudomonas aeruginosa activity. III. Microbiologic profile. J. Antibiot. 42:521–526 [DOI] [PubMed] [Google Scholar]
- 3.Karwowski JP, Jackson M, Theriault RJ, Chen RH, Barlow GJ, Maus ML. 1989. Pacidamycins, a novel series of antibiotics with anti-Pseudomonas aeruginosa activity. I. Taxonomy of the producing organism and fermentation. J. Antibiot. 42:506–511 [DOI] [PubMed] [Google Scholar]
- 4.Zhang W, Ostash B, Walsh CT. 2010. Identification of the biosynthetic gene cluster for the pacidamycin group of peptidyl nucleoside antibiotics. Proc. Natl. Acad. Sci. U. S. A. 107:16828–16833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rackham EJ, Gruschow S, Goss RJ. 2011. Revealing the first uridyl peptide antibiotic biosynthetic gene cluster and probing pacidamycin biosynthesis. Bioeng. Bugs 2:218–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brandish PE, Kimura KI, Inukai M, Southgate R, Lonsdale JT, Bugg TD. 1996. Modes of action of tunicamycin, liposidomycin B, and mureidomycin A: inhibition of phospho-N-acetylmuramyl-pentapeptide translocase from Escherichia coli. Antimicrob. Agents Chemother. 40:1640–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mao W, Warren M, Malouin F, Galazzo J, Lee A, Lomovskaya O. 2001. AcrAB-TolC efflux pump contributes to the intrinsic resistance to pacidomycin in Escherichia coli, poster A-35. Abstr. 101st Gen. Meet. Am. Soc. Microbiol. American Society for Microbiology, Washington, DC [Google Scholar]
- 8.Gotoh N, Murata T, Ozaki T, Kimura T, Kondo A, Nishino T. 2003. Intrinsic resistance of Escherichia coli to mureidomycin A and C due to expression of the multidrug efflux system AcrAB-TolC: comparison with the efflux systems of mureidomycin-susceptible Pseudomonas aeruginosa. J. Infect. Chemother. 9:101–103 [DOI] [PubMed] [Google Scholar]
- 9.Lomovskaya O, Mistry A, Warren M, Lee A. 2001. Characterization of pacidomycin resistant mutants in Pseudomonas aeruginosa, poster A-34 Abstr. Gen. Meet. Am. Soc. Microbiol. American Society for Microbiology, Washington, DC [Google Scholar]
- 10.Watson JM, Holloway BW. 1978. Chromosome mapping in Pseudomonas aeruginosa. J. Bacteriol. 133:1113–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stover CK, Pham X-Q, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FSL, Hufnagle WO, Kowalik DJ, Lagrou M, Garber RL, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody LL, Coulter SN, Folger KR, Kas A, Larbig K, Lim R, Spencer D, Wong GK-S, Wu Z, Paulsen IT, Reizer J, Saier MH, Hancock REW, Lory S, Olson MV. 2000. Complete genome sequence of Pseudomonas aeruginosa, an opportunistic pathogen. Nature 406:959–964 [DOI] [PubMed] [Google Scholar]
- 12.Lomovskaya O, Lee A, Hoshino K, Ishida H, Mistry A, Warren MS, Boyer E, Chamberland S, Lee VJ. 1999. Use of a genetic approach to evaluate the consequence of inactivation of efflux pumps in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43:1340–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Held K, Ramage E, Jacobs M, Gallagher L, Manoil C. 2012. Sequence-verified two-allele transposon mutant library for Pseudomonas aeruginosa PAO1. J. Bacteriol. 194:6387–6389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86 [DOI] [PubMed] [Google Scholar]
- 15.Liss L. 1987. New M13 host: DH5αF′ competent cells. Focus 9:13 [Google Scholar]
- 16.Schweizer HP. 1994. A method for construction of bacterial hosts for lac-based cloning and expression vectors: α complementation and regulated expression. Biotechniques 17:452–456 [PubMed] [Google Scholar]
- 17.Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557–580 [DOI] [PubMed] [Google Scholar]
- 18.Ohman DE, Cryz SJ, Iglewski BH. 1980. Isolation and characterization of a Pseudomonas aeruginosa PAO mutant that produces altered elastase. J. Bacteriol. 142:836–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook J, Russell DW. 2001. Molecular cloning, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 20.Clinical and Laboratory Standards Institute 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; M07-A9. Approved standard, 9th ed. CLSI, Wayne, PA [Google Scholar]
- 21.Darzins A, Casadaban MJ. 1989. Mini-D3112 bacteriophage transposable elements for genetic analysis of Pseudomonas aeruginosa. J. Bacteriol. 171:3909–3916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krishnapillai V. 1972. A novel transducing phage. Mol. Gen. Genet. 114:134–143 [DOI] [PubMed] [Google Scholar]
- 23.Goldbarg JA, Rutenburg AM. 1958. The colorimetric determination of leucine aminopeptidase in urine and serum of normal subjects and patients with cancer and other diseases. Cancer 11:283–291 [DOI] [PubMed] [Google Scholar]
- 24.de Lorenzo V, Timmis KN. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235:386–405 [DOI] [PubMed] [Google Scholar]
- 25.Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, Will O, Kaul R, Raymond C, Levy R, Chun-Rong L, Guenthner D, Bovee D, Olson MV, Manoil C. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 100:14339–14344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Becher A, Schweizer HP. 2000. Integration-proficient Pseudomonas aeruginosa vectors for isolation of single copy chromosomal lacZ and lux gene fusions. Biotechniques 29:948–954 [DOI] [PubMed] [Google Scholar]
- 27.Hoang TT, Kutchma AJ, Becher A, Schweizer HP. 2000. Integration proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid 43:59–72 [DOI] [PubMed] [Google Scholar]
- 28.Miller JH. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 29.Poole K, Krebes K, McNally C, Neshat S. 1993. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J. Bacteriol. 175:7363–7372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li XZ, Nikaido H, Poole K. 1995. Role of mexA-mexB-oprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:1948–1953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poole K, Srikumar R. 2001. Multidrug efflux in Pseudomonas aeruginosa: components, mechanisms and clinical significance. Curr. Topics Med. Chem. 1:59–71 [DOI] [PubMed] [Google Scholar]
- 32.Lomovskaya O, Warren MS, Lee A, Galazzo J, Fronko R, Lee M, Blais J, Cho D, Chamberland S, Renau T, Leger R, Hecker S, Watkins W, Hoshino K, Ishida H, Lee VJ. 2001. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination therapy. Antimicrob. Agents Chemother. 45:105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weinberg MV, Maier RJ. 2007. Peptide transport in Helicobacter pylori: Rokles of Dpp and Opp systems and evidence for additional peptide transporters. J. Bacteriol. 189:3392–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winsor GL, Lam DK, Fleming L, Lo R, Whiteside MD, Yu NY, Hancock RE, Brinkman FS. 2011. Pseudomonas Genome Database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res. 39:D596–D600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoang TT, Schweizer HP. 1999. Characterization of the Pseudomonas aeruginosa enoyl-acyl carrier protein reductase: a target for triclosan and its role in acylated homoserine lactone synthesis. J. Bacteriol. 181:5489–5497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Staskawicz B, Panopoulos N. 1980. Phaseolotoxin transport in Escherichia coli and Salmonella typhimurium via the oligopeptide permease. J. Bacteriol. 142:474–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borezee E, Pellegrini E, Berche P. 2000. OppA of Listeria monocytogenes, an oligopeptide-binding protein required for bacterial growth at low temperature and involved in intracellular survival. Infect. Immun. 68:7069–7077 [DOI] [PMC free article] [PubMed] [Google Scholar]