Abstract
Doripenem-colistin exerts synergy against some, but not all, Klebsiella pneumoniae carbapenemase (KPC)-producing K. pneumoniae strains in vitro. We determined if doripenem MICs and/or ompK36 porin gene mutations impacted the responses of 23 sequence type 258 (ST258), KPC-2-producing strains to the combination of doripenem (8 μg/ml) and colistin (2 μg/ml) during time-kill assays. The median doripenem and colistin MICs were 32 and 4 μg/ml. Doripenem MICs did not correlate with KPC-2 expression levels. Five and 18 strains had wild-type and mutant ompK36, respectively. The most common mutations were IS5 promoter insertions (n = 7) and insertions encoding glycine and aspartic acid at amino acid (aa) positions 134 and 135 (ins aa134-135 GD; n = 8), which were associated with higher doripenem MICs than other mutations or wild-type ompK36 (all P values ≤ 0.04). Bactericidal activity (24 h) was achieved by doripenem-colistin against 12%, 43%, and 75% of ins aa134-135 GD, IS5, and wild-type/other mutants, respectively (P = 0.04). Doripenem-colistin was more active in time-kill studies than colistin at 12 and 24 h if the doripenem MIC was ≤8 μg/ml (P = 0.0007 and 0.09, respectively), but not if the MIC was >8 μg/ml (P = 0.10 and 0.16). Likewise, doripenem-colistin was more active at 12 and 24 h against the wild type/other mutants than ins aa134-135 GD or IS5 mutants (P = 0.007 and 0.0007). By multivariate analysis, the absence of ins aa134-135 GD or IS5 mutations was the only independent predictor of doripenem-colistin responses at 24 h (P = 0.002). In conclusion, ompK36 genotypes identified ST258 KPC-K. pneumoniae strains that were most likely to respond to doripenem-colistin.
INTRODUCTION
Carbapenem-resistant (CR) Klebsiella pneumoniae has emerged as a major pathogen (1–9), causing infections that are associated with crude mortality rates as high as 50% (1, 10–13). Observational studies indicate that outcomes may be improved with combination antimicrobial therapy compared to monotherapy (12, 14, 15), particularly if the regimens include a carbapenem (16). Nevertheless, optimal combinations are not defined, and treatment failure rates remain unacceptably high. Our group and others have reported bactericidal activity and synergy for doripenem-colistin against some, but not all, CR K. pneumoniae strains in vitro (17–19). Indeed, “one-size-fits-all” approaches to identifying optimal antimicrobial regimens against CR K. pneumoniae and other carbapenem-resistant Enterobacteriaceae are not likely to be effective. Carbapenem resistance is mediated through multiple mechanisms, including production of metallo-β-lactamases and nonmetallocarbapenemases (such as Klebsiella pneumoniae carbapenemase [KPC] and OXA-type carbapenemase). Alternatively, strains with defects in outer membrane proteins (OMPs) may express AmpC β-lactamase or extended-spectrum β-lactamases (ESBLs). Levels of carbapenem resistance in individual strains can be impacted by the presence of several mechanisms. We hypothesize that the specific resistance mechanisms manifested by CR K. pneumoniae strains dictate the efficacy of carbapenem-containing combinations.
We recently demonstrated that the vast majority of CR K. pneumoniae bacteremia among solid-organ transplant recipients at our center is caused by the international epidemic clone sequence type 258 (ST258) (20). We further showed that the majority of our ST258 strains are KPC-2 producers that express TEM-1 and SHV-12 β-lactamases and possess a mutant ompK35 porin gene (20). Within this clonal background, however, several ompK36 porin mutations were identified. Two ompK36 mutations in particular were associated with significantly higher carbapenem MICs than wild-type ompK36, but the impact of ompK36 mutations on responses to a carbapenem or the combination of a carbapenem and colistin during time-kill experiments is not known. The objective of this study was to determine if certain ompK36 mutations were associated with a decreased likelihood of bactericidal activity or synergy for doripenem-colistin. We have chosen to study doripenem among the carbapenems because it is the formulary agent at our center and it is less susceptible to hydrolysis by KPCs than other agents in the class (21).
MATERIALS AND METHODS
Strains.
Bloodstream isolates from 23 unique patients at the University of Pittsburgh Medical Center (UPMC) were selected for this study. They were stored at −80°C in the repository at the UPMC XDR Pathogen Laboratory and subcultured onto Mueller-Hinton agar twice before undergoing MIC determinations and time-kill assays. This study was performed under the protocol “Clinical database and biorepository of multi-drug resistant pathogens” that was reviewed and approved by the University of Pittsburgh Institutional Review Board.
MICs and time-kill assays.
MICs were determined by standard broth microdilution (22). In time-kill assays, fixed drug concentrations (doripenem, 8 μg/ml; colistin, 2 μg/ml) were tested against inocula of each strain (1 × 106 CFU/ml) suspended in 10 ml of cation-adjusted Mueller-Hinton broth, and bacterial colonies were enumerated following incubation at 37°C with shaking. Drug concentrations were chosen based on achievable serum drug levels in pharmacokinetic studies (23, 24). The level of killing by a drug or the doripenem-colistin combination was defined as follows: log10 CFU/ml at time of interest − log10 CFU/ml at time zero. The extent of synergy between doripenem and colistin was defined as follows: log10 CFU/ml of a strain in the presence of doripenem-colistin − log10 CFU/ml of the strain in the presence of colistin. Bactericidal activity was defined as a level of killing of ≥3 log10 CFU/ml. Synergy was deemed to be present if the extent of synergy was ≥2 log10 CFU/ml.
Molecular characterization of strains.
Strains from −80°C stock were express mailed on dry ice to the Public Health Research Institute for detection of resistance determinants. PCR and DNA sequencing were used as previously described to characterize blaKPC, blaIMP, blaNDM, blaVIM, blaOXA-48, blaTEM, blaSHV, blaCTX-M, blaAmpC, and porin genes (25–29). PCR primers for ompK35 and ompK36 are listed in Table 1. Nucleotide sequences were compared with data from GenBank (http://www.ncbi.nlm.nih.gov/blast/).
Table 1.
Primers for PCR and qRT-PCR
| Primer | Sequence | Expected band size (bp) | Reference |
|---|---|---|---|
| PCR | |||
| ompK35-F1 | 5′GGATGGAAAGATGCCTTCAG3′ | 1,392 | This study |
| ompK35-R1 | 5′CATGACGAGGTTCCATTGTG3′ | ||
| ompK36-F1 | 5′GGGAAGAATCGCACGAAATA3′ | 1,744 | This study |
| ompK36-R1 | 5′TCTTACCAGGGCGACAAGAG3′ | ||
| qRT-PCR | |||
| RT-ompK35-F1 | 5′GCAATATTCTGGCAGTGGTGATC3′ | This study | |
| RT-ompK35-R1 | 5′ACCATTTTTCCATAGAAGTCCAGT3′ | ||
| RT-ompK36-F1 | 5′TTAAAGTACTGTCCCTCCTGG3′ | This study | |
| RT-ompK36-R1 | 5′TCAGAGAAGTAGTGCAGACCGTCA3′ | ||
| RT- rpoB-F1 | 5′AAGGCGAATCCAGCTTGTTCAGC3′ | 39 | |
| RT- rpoB-R1 | 5′TGACGTTGCATGTTCGCACCCATCA3′ | ||
| RT-KPC-F1 | 5′CGTGACGGAAAGCTTACAAA3′ | 30 | |
| RT-KPC-R1 | 5′ AGCCAATCAACAAACTGCTG3′ |
Expression of blaKPC and ompK36 porin genes was measured in the UPMC XDR Pathogen Laboratory. DNase-treated RNA was obtained from late-exponential-phase cultures using a RiboPure-Bacteria kit (Ambion), and cDNA was made using qScript cDNAMix (Quanta Biosciences). Quantitative reverse transcription (qRT)-PCR was performed in a TaqMan gene expression assay (ABI) using the one-step SYBR green kit (SYBR green FastMix ROX; Quanta Biosciences) with primers listed in Table 1. Expression levels were measured in threshold cycle (CT) units (the first cycle at which the signal is above a preset threshold). The relative mRNA level for each gene was determined by the comparative CT, with normalization to the housekeeping gene rpoB. Data are presented as expression relative to a control strain that expresses ompK35 and ompK36 (K. pneumoniae ATCC 13883) (29, 30). Experiments were performed in triplicate.
Statistical analysis.
Numbers of CFU and MICs were logarithmically transformed prior to statistical analyses. Comparisons between groups of antimicrobial agents were made by Fisher's exact test for categorical variables and Student's t test or analysis of variance (ANOVA) for continuous variables. Correlations between pairs of variables were assessed using the Spearman rank test. A multiple-linear-regression model was used to identify factors independently associated with time-kill results at 24 h; the factors included in the model were those identified by univariate analyses to be significant at a P value of <0.10 or those that could be performed in real time in clinical microbiology laboratories. Significance was defined as a P value of ≤0.05 (two tailed).
RESULTS
ompK36 porin mutations and gene expression.
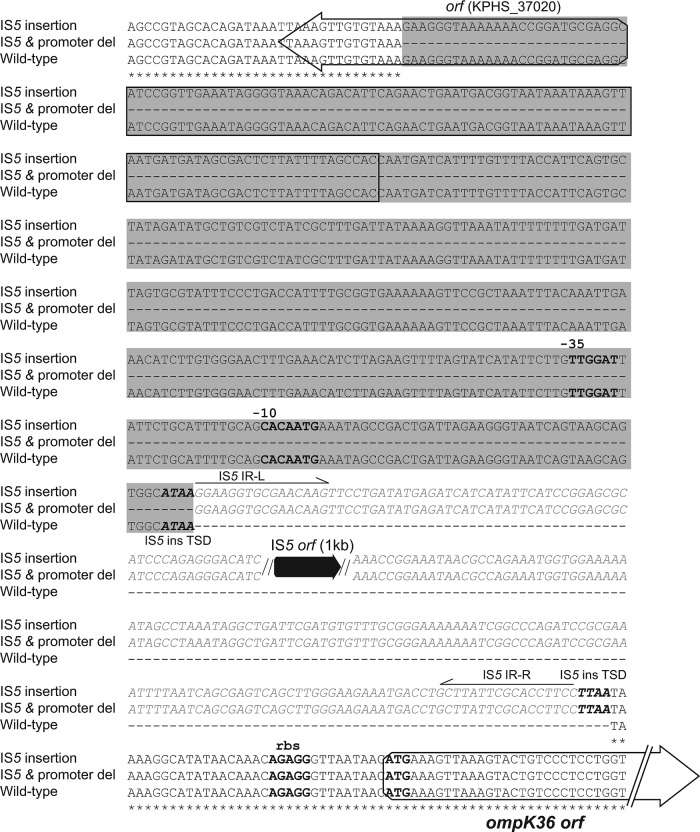
Each of the 23 strains carried blaKPC-2, blaTEM-1, blaSHV-12, and mutant ompK35 (guanine insertion at nucleotide [nt] 121, resulting in a premature stop codon at amino acid [aa] 89). Five and 18 strains had wild-type and mutant ompK36, respectively. The ompK36 mutations included a 6-bp insertion that encodes glycine and aspartic acid at amino acid 134 (ins aa134-135 GD; n = 8), an IS5 insertion sequence within the promoter with or without partial promoter deletion (IS5 mutants; n = 7) (Fig. 1), a guanine insertion at nt 382 causing a frameshift (n = 1), a mutation of alanine to valine at aa 11 (A11V; n = 1), and an asparagine-asparagine-threonine-glutamic acid (NNTE) deletion at amino acid positions 84 to 87 (del aa84-87NNTE; n = 1).
Fig 1.
Nucleotide sequences of the upstream regions of the ompK36 gene in strains with wild-type sequence, a full-length IS5 insertion, and an IS5 insertion along with a promoter deletion. Nucleotide positions shared by the three strains are marked with asterisks; missing bases are indicated by dashes. The ompK36 ribosome binding site (rbs), start codon, and −10 and −35 promoter sequences are shown in boldface, while the presumed IS5 element integration sites (target site duplications [TSD]) are shown in boldface italics. Open reading frames (ORFs) are portrayed by arrows, with the IS5 transposase ORF indicated in black and the upstream ORF (KPHS_37020) and ompK36 ORF in white. The inserted IS5 element, including the IS5 transposase ORF (black arrow) and its right and left inverted repeats (IR-R and IR-L), are shown in gray italic letters. Single-line arrows beneath the inverted repeats indicate the direction of the repeats. A 1,200-bp IS5 element is inserted 33 bp upstream of the ompK36 start code from the TSD site (TTAA), which qRT-PCR data indicate is disrupting the transcription of ompK36. The gray shading denotes a 395-bp deletion region, encompassing a partial sequence of an unknown ORF (KPHS_37020) and the −30 and −10 promoters. The IS5 insertion and promoter deletion also disrupt ompK36 expression.
We evaluated the impacts of the major mutations on ompK36 gene expression for representative KPC-2-producing K. pneumoniae (KPC-K. pneumoniae) strains by qRT-PCR (3 strains per ompK36 variant). Strains with IS5 promoter insertions exhibited significantly lower levels of ompK36 expression than strains with wild-type ompK36 or the ins aa134-135 GD insertion mutation (median relative expression [range]: 0.30 [0.09 to 8.7] versus 100.5 [87.4 to 113.6] and 78.0 [61.0 to 95.0], respectively; P = 0.003 and 0.002).
Activity of doripenem and colistin alone.
The median doripenem MIC was 32 μg/ml (Table 2). There was no association between doripenem MICs and the level of KPC-2 expression (P = 0.52). IS5 and ins aa134-135 GD mutants were associated with higher doripenem MICs (median log10 MIC, 7) than ompK36 wild-type strains (median log10 MIC, 3.5; P = 0.0002) or ins nt382 G, A11V, and del aa84-87NNTE mutants (median log10 MIC, 5; P = 0.04). There were no differences in doripenem MICs between the last group of 3 mutants and wild-type ompK36 strains (P = 0.65). For the remainder of this report, these 8 strains will be considered together and are referred to as other mutant/wild-type ompK36 strains.
Table 2.
Killing activities of doripenem and colistin alone or in combination against 23 KPC-K. pneumoniae isolates
| Isolate no. | MIC (μg/ml)a |
KPC expressionb | ompK36 genotype | Level of killing (log10 CFU/ml) at (h): |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dori | Col | Dori |
Col |
Dori + Col |
||||||||||||
| 4 | 8 | 12 | 24 | 4 | 8 | 12 | 24 | 4 | 8 | 12 | 24 | |||||
| 83 | 32 | 128 | 4.88 | Wild type | −2.30 | −2.32 | −0.14 | 3.48 | −0.83 | −0.30 | −1.09 | −2.49 | −1.35 | −2.41 | −2.83 | −4.27 |
| 25 | 16 | 16 | 4.10 | Wild type | −0.90 | −1.59 | −2.14 | −3.17 | −0.59 | −0.91 | −1.01 | 1.85 | −1.19 | −1.95 | −2.65 | −3.47 |
| 216 | 4 | 8 | 5.02 | Wild type | −2.57 | −1.13 | 2.68 | 3.72 | 0.27 | 1.79 | 3.51 | 3.91 | −2.91 | −3.34 | −3.14 | 0.72 |
| 347 | 4 | 0.125 | 5.04 | Wild type | −3.14 | −1.00 | 2.21 | 3.90 | −3.81 | −2.08 | −3.16 | 2.07 | −3.41 | −4.27 | −6.05 | −4.57 |
| 383 | 8 | 0.125 | 4.11 | Wild type | 2.17 | 3.48 | 3.63 | 3.90 | −4.37 | −3.66 | −1.61 | 3.47 | −4.30 | −3.51 | −6.08 | −6.08 |
| 41 | 4 | 64 | 5.02 | Del aa84-87NNTE | −3.71 | −1.36 | 1.68 | 3.68 | −2.56 | −4.34 | −2.76 | 3.60 | −3.77 | −6.03 | −6.03 | −6.03 |
| 44 | 32 | 4 | 4.8 | A11V | −2.47 | −0.25 | 2.79 | 3.64 | −0.48 | −0.48 | −0.38 | −1.36 | −1.38 | −1.97 | −2.42 | −2.75 |
| 539 | 64 | 0.25 | 4.3 | Frameshift (ins nt382 G) | 2.47 | 3.42 | 3.78 | 3.80 | −3.00 | −1.47 | −2.29 | −1.10 | −3.80 | −3.62 | −3.42 | −4.69 |
| 175 | 64 | 2 | 4.75 | Ins aa134-135 GD | 3.19 | 3.95 | 4.11 | 4.106 | −0.82 | −1.08 | −1.32 | −0.71 | −2.27 | 0.10 | −0.02 | −1.88 |
| 89 | 128 | 64 | 5.02 | Ins aa134-135 GD | 2.48 | 3.22 | 3.19 | 3.19 | −1.14 | −1.57 | −1.44 | −2.29 | −2.02 | −3.55 | −3.08 | −4.40 |
| 168 | 128 | 32 | 3.26 | Ins aa134-135 GD | 2.71 | 3.26 | 3.12 | 3.51 | −0.35 | −0.72 | −0.26 | 3.81 | −2.15 | −3.25 | −1.32 | −0.77 |
| 115 | 32 | 0.125 | 4.30 | Ins aa134-135 GD | 2.40 | 3.66 | 4.42 | 3.84 | −4.53 | −3.92 | −2.85 | 2.20 | −3.70 | −3.55 | −2.00 | 3.28 |
| 184 | 32 | 8 | 3.88 | Ins aa134-135 GD | 2.63 | 3.57 | 3.81 | 3.83 | 0.27 | 0.27 | 1.95 | 3.96 | −0.84 | −1.02 | 0.74 | 3.80 |
| 155 | 32 | 2 | 4.60 | Ins aa134-135 GD | 2.47 | 3.43 | 3.80 | 3.70 | −6.10 | −2.08 | −1.36 | 1.68 | −4.10 | −2.39 | −2.01 | 2.77 |
| 484 | 128 | 0.25 | 4.75 | Ins aa134-135 GD | 2.24 | 3.51 | 3.89 | 3.95 | −4.33 | −2.09 | −1.76 | 1.48 | −3.64 | −3.97 | −2.58 | 2.15 |
| 436 | 32 | 0.125 | 3.67 | Ins aa134-135 GD | 2.30 | 3.47 | 3.58 | 3.82 | −4.03 | −2.44 | −0.83 | 3.79 | −4.00 | −2.59 | −0.85 | 3.78 |
| 178 | 256 | 16 | 4.89 | IS5 ins and promoter del | −1.63 | 0.61 | 3.43 | 3.79 | −0.48 | −0.48 | −0.37 | −1.11 | −0.27 | −0.24 | −0.33 | −1.23 |
| 220 | 256 | 16 | 5.15 | IS5 ins and promoter del | −2.07 | 0.85 | 2.66 | 3.27 | −0.81 | −0.85 | −1.44 | −0.39 | −0.67 | −1.13 | −2.38 | −2.62 |
| 94 | 128 | 4 | 4.457 | IS5 | 1.92 | 3.65 | 3.77 | 3.78 | −0.66 | −0.81 | −1.14 | −2.29 | −1.07 | −1.73 | −2.73 | −3.96 |
| 121 | 256 | 4 | 5.34 | IS5 | −0.77 | −1.29 | −2.21 | −1.32 | −0.82 | −0.99 | −2.05 | −1.46 | −1.05 | −1.32 | −2.35 | −3.24 |
| 1 | 128 | 8 | 4.80 | IS5 | −0.15 | 3.08 | 3.64 | 3.85 | 2.22 | 3.56 | 3.80 | 3.85 | 1.48 | 3.30 | 3.70 | 3.83 |
| 404 | 64 | 2 | 5.02 | IS5 | 1.84 | 3.46 | 3.57 | 1.48 | −1.32 | −0.10 | −1.72 | −1.42 | −1.78 | −3.11 | −3.36 | −3.99 |
| 584 | 32 | 32 | 5.23 | IS5 | 1.60 | 1.38 | 2.08 | 3.67 | 1.97 | 1.77 | 2.47 | 3.83 | −1.59 | −1.82 | −1.11 | 3.60 |
During the first 8 h of time-kill experiments, doripenem (8 μg/ml) was more active in killing KPC-K. pneumoniae strains for which the doripenem MIC was ≤8 μg/ml than strains for which the MIC was >8 μg/ml (Fig. 2A). Thereafter, strains that were originally inhibited by doripenem demonstrated regrowth, and there were no significant differences in median time kills based on doripenem MICs.
Fig 2.
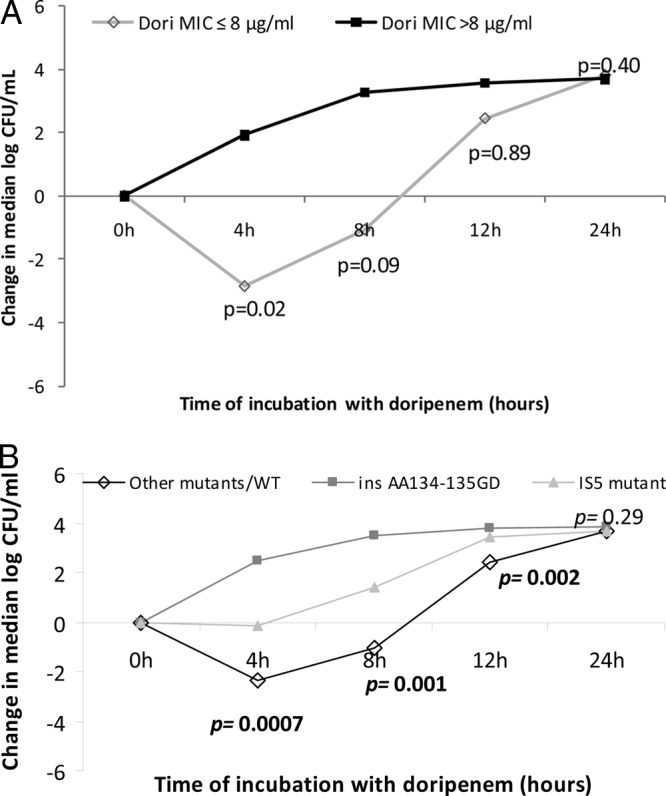
Killing activity of doripenem (Dori) against KPC-K. pneumoniae strains, stratified by doripenem MICs (A) and the presence of ompK36 mutations (B). The data represent the median log10 kills by doripenem. (A) P values indicate the differences in killing by doripenem against 4 strains with doripenem MICs of ≤8 μg/ml and 19 strains with MICs of >8 μg/ml. (B) P values indicate the differences in killing by doripenem against 8 ins aa134-135 GD mutants, 7 IS5 mutants, and 8 ompK36 other mutant/wild-type strains (ANOVA).
Likewise, median levels of killing differed at earlier time points between ins aa134-135 GD mutants, IS5 mutants, and other mutant/wild-type ompK36 strains (Fig. 2B). Responses to doripenem were hierarchical. The ins aa134-135 GD mutants were most resistant to killing, as doripenem failed to inhibit the growth of any strain at any time point. IS5 mutants exhibited intermediate responses; reductions from the starting inocula were evident for 57% (4/7) of strains at 4 h, but there was regrowth of all but one strain by 8 h. There was no difference in median levels of killing between wild-type ompK36 and other mutants. Other mutant/wild-type ompK36 strains were most sensitive at 4 h, as reductions from the starting inocula were demonstrated for 75% (6/8) of the strains; bactericidal activity (defined as ≥3 log10 kill) was achieved against 38% (3/8) of these strains for at least one time point compared to none of the ins aa134-135 GD or IS5 mutants. After 24 h of incubation with doripenem, there were no significant differences between the three groups of ompK36 variants.
The median colistin MIC was 4 μg/ml (range, 0.125 to 128 μg/ml), which did not differ based on the type of ompK36. Colistin MICs were >2 μg/ml against 61% (14/23) of the strains. During the first 12 h of time-kill experiments, colistin (2 μg/ml) was less active against strains for which the colistin MIC was >2 μg/ml (Fig. 3). Nevertheless, colistin at 2 μg/ml was able to inhibit the growth of the latter strains, consistent with bacteriostatic activity. By 24 h, differences based on colistin MICs were no longer apparent. The type of ompK36 did not impact time-kill responses to colistin at any time point (P = 0.12, 0.06, 0.66, and 0.56 at 4, 8, 12, and 24 h, respectively; ANOVA).
Fig 3.
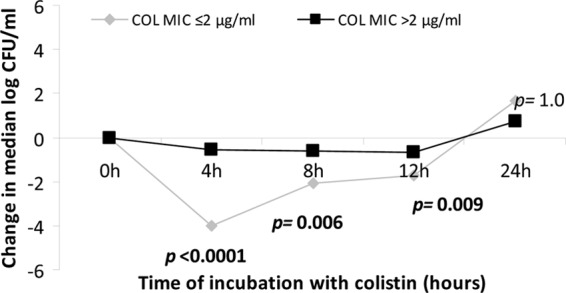
Killing activity of colistin (COL) against KPC-K. pneumoniae strains, stratified by colistin MICs. The data represent median log10 kills by colistin; the P values indicate the differences between KPC-K. pneumoniae strains for which colistin MICs were ≤2 μg/ml and >2 μg/ml.
Activity of the doripenem-colistin combination.
Doripenem-colistin was significantly more active than colistin alone at 12 and 24 h against strains with doripenem MICs of ≤8 μg/ml (P = 0.0007 and 0.09, respectively) (Fig. 4A). In contrast, median time kills were not significantly different for strains with doripenem MICs of >8 μg/ml (P = 0.10 and 0.16, respectively). Doripenem-colistin exerted bactericidal activity against 100% (4/4) and 16% (3/19) of strains with doripenem MICs of ≤8 μg/ml and >8 μg/ml, respectively, at 12 h (P = 0.0004). At 24 h, the corresponding rates were 75% (3/4) and 32% (6/19) (P = 0.15). Synergy (defined as ≥2 log10 more kill with doripenem-colistin than colistin alone) was achieved against 100% (4/4) and 11% (2/19) of strains with doripenem MICs of ≤8 μg/ml and >8 μg/ml, respectively, at 12 h (P = 0.002). At 24 h, the corresponding rates were 100% (4/4) and 32% (6/19) (P = 0.02). There was no difference in synergy between strains with colistin MICs of ≤2 μg/ml and >2 μg/ml at 12 h (22% [2/9] versus 29% [4/14]; P = 1.0) and 24 h (44% [4/9] versus 43% [6/14]; P = 1.0).
Fig 4.
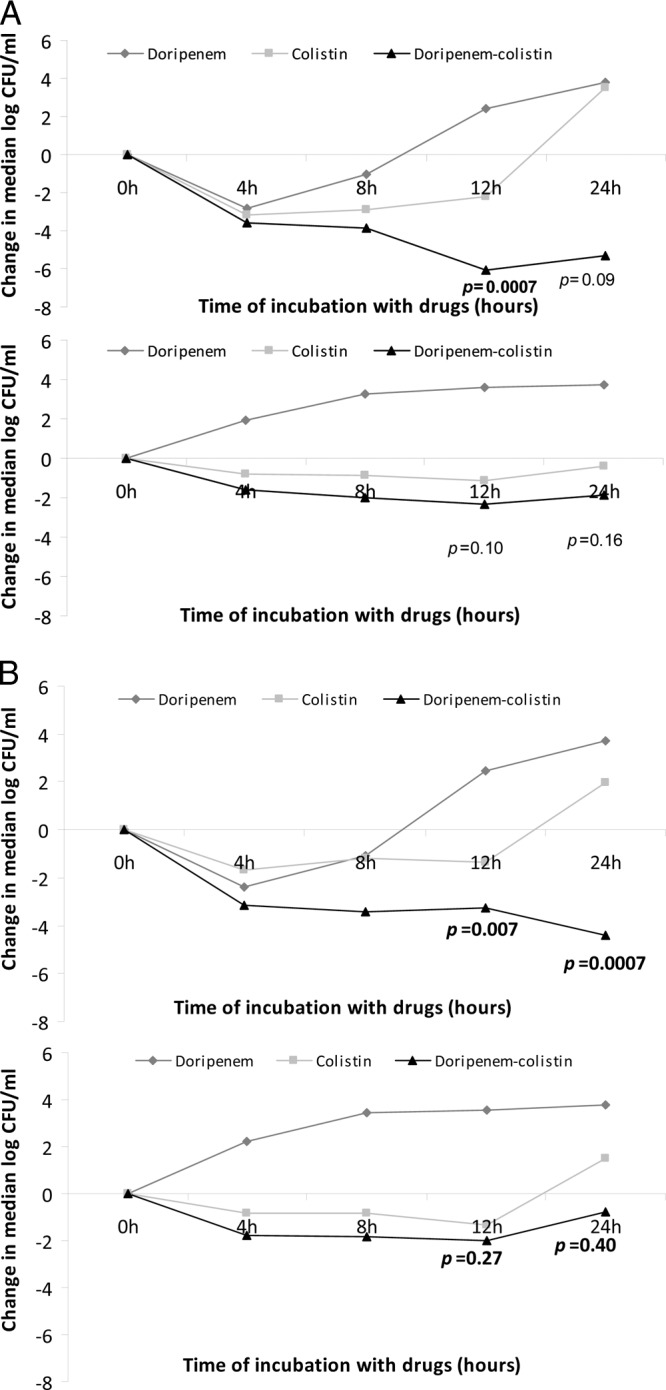
Killing activities of doripenem, colistin, and colistin-doripenem against KPC-K. pneumoniae strains, stratified by doripenem MICs (A) and the presence of ompK36 mutations (B). The P values indicate the differences in median time kills by colistin versus doripenem-colistin in combination. (A) Median log10 kills by doripenem, colistin, and doripenem-colistin against KPC-K. pneumoniae strains with MICs of ≤8 μg/ml (upper graph) and >8 μg/ml (lower graph). Note the lower level of kills for strains with MICs of >8 μg/ml at each time point. (B) Median log10 kills by doripenem, colistin, and doripenem-colistin against other mutant/wild-type ompK36 strains (upper graph) and strains with ins aa134-135 GD or IS5 mutations (lower graph). Note the lower level of kills for ins aa134-135 GD or IS5 mutants.
Bactericidal activity was achieved by colistin-doripenem against 62% (5/8) of other mutant/wild-type ompK36 strains, 14% (1/7) of IS5 mutants, and 12% (1/8) of ins aa134-135 GD mutants at 12 h (P = 0.05; ANOVA). At 24 h, the corresponding rates were 75% (6/8), 43% (3/7), and 12% (1/8) (P = 0.04; ANOVA). Synergy was achieved against 62% (5/8), 14% (1/7), and 0% (0/8) of other mutant/wild-type ompK36 strains, IS5 insertion mutants, and ins aa134-135 GD mutants, respectively, at 12 h (P = 0.01; ANOVA). At 24 h, the corresponding rates were 75% (6/8), 29% (2/7), and 25% (2/8) (P = 0.14; ANOVA). The performances of doripenem-colistin were comparable against IS5 insertion and ins aa134-135 GD mutants, whether measured by the achievement of bactericidal activity (P = 1.0 and 0.28 at 12 and 24 h, respectively) or synergy (P = 0.47 and 1.0, respectively).
Doripenem-colistin was significantly more active than colistin alone against wild-type ompK36 and other mutant strains, as measured by levels of killing during time-kill assays (P = 0.28, 0.035, 0.007, and 0.0007 at 4, 8, 12, and 24 h, respectively) (Fig. 4B). The activities of doripenem-colistin and colistin alone during time-kill assays were comparable against aa134-135 GD and IS5 mutants (P = 0.54, 0.15, 0.27, and 0.40 at 4, 8, 12, and 24 h, respectively).
Predictors of responses to doripenem-colistin.
By univariate analysis, the absence of aa134-135 GD or IS5 mutations was significantly associated with the level of killing by doripenem-colistin at 24 h (P = 0.006). The absence of aa134-135 GD or IS5 mutations and lower doripenem MICs were associated with the extent of synergy between doripenem and colistin at 24 h (P = 0.0004 and 0.004, respectively) (Table 3). By multivariate analysis, the absence of aa134-135 GD or IS5 mutations was the only independent factor associated with the level of killing by doripenem-colistin and the extent of synergy during time-kill assays at 24 h (P = 0.002 and 0.04, respectively) (Table 3).
Table 3.
Predictors of responses to doripenem-colistin during time-kill experiments in vitroa
| Factor |
P value for doripenem-colistin at 24 h |
|||||
|---|---|---|---|---|---|---|
| Level of killing |
Extent of synergy |
|||||
| Univariate analysis | Multivariate analyses |
Univariate analysis | Multivariate analyses |
|||
| log2 doripenem MIC | 0.52 | 0.06 | 0.057 | 0.004 | 0.44 | 0.42 |
| log2 colistin MIC | 0.53 | 0.81 | 0.68 | 0.89 | 0.64 | 0.64 |
| log10 KPC expression | 0.32 | 0.40 | Not included | 0.94 | 0.87 | Not included |
| Absence of ins aa134-135 GD or IS5 mutation | 0.006 | 0.002 | 0.002 | 0.0004 | 0.04 | 0.037 |
Univariate and multivariate analyses were performed using linear-regression and multiple-linear-regression models, respectively. Factors included in the multivariate analyses were those found significant by the univariate analyses or those that would be feasible as real-time tests in clinical microbiology laboratories. Multivariate analyses were performed including and excluding log10 KPC expression. Boldface numbers represent significant P values at the level of <0.05.
DISCUSSION
This is one of the first studies to evaluate the in vitro efficacy of a carbapenem and carbapenem-colistin combination against CR K. pneumoniae strains in the context of molecular mechanisms of resistance. We demonstrated that ST258, KPC-2-producing K. pneumoniae strains causing bacteremia at our center harbor a variety of mutant ompK36 genes, which impact responses to doripenem and doripenem-colistin in vitro. Our most striking finding is that ompK36 ins aa134-135 GD and ompK36 promoter IS5 mutations were associated with significantly higher doripenem MICs and diminished efficacy of doripenem-colistin during time-kill assays, as measured by median kills and the likelihood of achieving bactericidal activity or synergy. Indeed, the absence of these mutations in KPC-K. pneumoniae strains was an independent predictor of killing by doripenem-colistin and synergy between the agents. In linking molecular data for strains with responses to antimicrobials, this study provides a model for future laboratory and clinical investigations into the treatment of infections due to CR K. pneumoniae and other carbapenem-resistant Enterobacteriaceae.
Taken together, our data suggest that doripenem MICs and ompK36 genotyping of KPC-K. pneumoniae strains can be used to rapidly identify strains that are most likely to respond to doripenem-colistin in vitro (Fig. 5). In our paradigm, strains are stratified based on doripenem MICs. The presence of lower-level doripenem resistance, as reflected by a MIC of ≤8 μg/ml in our data set, indicates that doripenem-colistin is likely to be effective against a given strain. It is anticipated that the vast majority of these strains, if genotyped, would be shown to carry wild-type ompK36 or mutations other than ins aa134-135 GD or IS5 insertions. Molecular typing is most useful for strains with higher-level doripenem resistance, defined here as a MIC of >8 μg/ml. The data indicate that strains carrying IS5 or ins aa134-135 GD mutations in ompK36 are not likely to respond any more robustly to doripenem-colistin than to colistin alone. In contrast, our findings suggest that the combination is likely to retain activity against other mutant/wild-type ompK36 strains, even if doripenem MICs are 16 to 64 μg/ml. If validated, our findings have two important clinical implications. First, they provide an argument for the measurement of precise doripenem MICs, rather than relying on breakpoint values to simply classify strains as susceptible or resistant. Second, they suggest that our paradigm could be useful for guiding the selection of antimicrobial regimens against KPC-K. pneumoniae infections.
Fig 5.
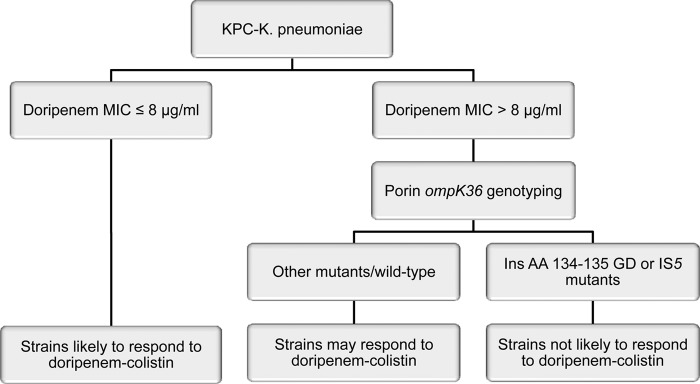
Model for predicting responses of KPC-K. pneumoniae strains to doripenem-colistin. The model is based on data demonstrating that lower doripenem MICs were associated with the extent of synergy at 24 h and that the absence of ins aa134-135 GD and IS5 mutations was independently associated with levels of killing and extent of synergy at 24 h.
Our data also have important implications for understanding the mechanisms by which susceptibility or resistance to carbapenems and carbapenem-colistin combinations are mediated. Porins are outer membrane channels that allow β-lactams and other molecules to diffuse into periplasmic active sites (31), and porin modifications are well recognized in CR K. pneumoniae strains. ompK36 ins aa134-135 GD and IS5 mutations have been linked to elevated carbapenem MICs (30, 32). The site between OmpK36 amino acids 134 and 135 falls within a transmembrane β-strand loop 3 (L3) that constitutes the porin channel eyelet (30, 32, 33). L3 mutations can alter the porin channel and reduce carbapenem uptake (33). Our aa134-135 GD insertion mutants did not exhibit diminished ompK36 expression, implying that elevated carbapenem MICs stemmed from porin structural changes rather than porin loss. IS5-like insertions (∼1.2 kb) were identified previously within the ompK36 open reading frame and promoter (34–36). To our knowledge, however, the IS5 insertions with or without a promoter deletion found in this study have not been described. Moreover, the promoter site of insertion is novel, suggesting that the mutations arose de novo at our center. The IS5 insertions were associated with impaired ompK36 expression, which implies that the high-level carbapenem resistance exhibited by mutant strains resulted from diminished levels of OmpK36.
The kinetics by which strains manifested their resistance to doripenem during time-kill assays were also impacted by the OmpK36 status. The ins aa134-135 GD mutants did not respond to doripenem at any time point, indicating that changes to the porin channel severely restricted access of the drug to penicillin binding protein targets. IS5 mutants, on the other hand, were often inhibited 4 h before they exhibited regrowth, indicating that doripenem had some early access to target sites via limited porins that were then exhausted. In keeping with a hypothesis that the time-kill data were shaped by access to drug targets via porins, strains with wild-type ompK36 or weak ompK36 mutants were most strongly inhibited by doripenem prior to regrowth. Regardless of differences in responses to doripenem at earlier time points, the bacterial concentrations achieved by ins aa134-135 GD mutants, IS5 mutants, and other mutants or wild-type OmpK36 strains at 24 h in the presence of doripenem (8 μg/ml) were indistinguishable (Table 2).
Synergy between colistin and doripenem is consistent with a model in which membrane permeabilization by the former agent facilitates increased access of the latter to target sites, allowing it to overcome hydrolysis by KPC (37, 38). Our data demonstrate that intact OmpK36 porins are also necessary for synergy in this model. We cannot assess the impact of ompK35 mutations on responses to either doripenem or doripenem-colistin, since all ST258 strains carried a TGA nonsense mutation at codon 89. Loss or mutation of ompK35 or analogous porins is common in ESBL-producing Enterobacteriaceae (39). The ompK35 mutation in our strains is described in previous studies of ST258 strains, suggesting that it is widespread (30, 32, 40). The lower doripenem MICs against strains with wild-type ompK36 support the general belief that ompK35 mutations are not linked to high-level carbapenem resistance in the absence of coexisting ompK36 mutations (32, 41–43). It is also notable that doripenem-colistin responses were not impacted by colistin MICs. On one hand, we showed that colistin alone (2 μg/ml) was significantly more active during the first 12 h of time-kill experiments against KPC-K. pneumoniae strains for which colistin MICs were ≤2 μg/ml. However, these differences were no longer observed at 24 h. Moreover, colistin exerted bacteriostatic activity throughout time-kill experiments against strains for which MICs were >2 μg/ml. Therefore, colistin retained activity against strains that were putatively resistant to the drug as determined by the MIC.
In conclusion, our data clearly demonstrate that ST258 KPC-K. pneumoniae strains bear genetic heterogeneity that impacts in vitro responses to a widely used antimicrobial combination. Based on these findings, the detailed molecular characterization of strains should be considered an essential component of future studies of antimicrobial regimens against KPC-K. pneumoniae. Along these lines, we acknowledge that the genotypes of strains from our patients may differ from those at other centers. Therefore, studies comparable to ours must be conducted using strains with various genetic backgrounds from different geographic locations, and the results for particular mutations must be corroborated. The mechanistic explanations that we propose for our findings also will need to be investigated in follow-up studies. Nevertheless, the current study is an important early step in the development of methods to rapidly identify antimicrobial regimens that are best suited to treat patients infected with CR K. pneumoniae or other highly resistant bacteria. In the future, the molecular signature of infecting strains should be one of the factors routinely considered by clinicians as they make treatment decisions, along with the type of infection being treated, underlying diseases, host factors, and pharmacokinetic/pharmacodynamic parameters.
ACKNOWLEDGMENTS
This project was supported by funding provided to the XDR Pathogen Laboratory by the University of Pittsburgh Medical Center and in part by grants (to B.N.K. and Y.D.) from the National Institutes of Health (1R01AI090155 and R21AI107302, respectively).
Footnotes
Published ahead of print 12 August 2013
REFERENCES
- 1.Bratu S, Landman D, Haag R, Recco R, Eramo A, Alam M, Quale J. 2005. Rapid spread of carbapenem-resistant Klebsiella pneumoniae in New York City: a new threat to our antibiotic armamentarium. Arch. Intern. Med. 165:1430–1435 [DOI] [PubMed] [Google Scholar]
- 2.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. 2009. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 48:1–12 [DOI] [PubMed] [Google Scholar]
- 3.Nordmann P, Cuzon G, Naas T. 2009. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect. Dis. 9:228–236 [DOI] [PubMed] [Google Scholar]
- 4.Maltezou HC, Giakkoupi P, Maragos A, Bolikas M, Raftopoulos V, Paphatzaki H, Vrouhos G, Liakou V, Vatopouos AC. 2009. Outbreak of infections due to KPC-2-producing Klebsiella pneumoniae in a hospital in Crete (Greece). J. Infect. 58:213–219 [DOI] [PubMed] [Google Scholar]
- 5.Rhee JY, Park YK, Shin JY, Choi JY, Lee MY, Peck KR, Song HG, Ko KS. 2010. KPC-producing extreme drug-resistant Klebsiella pneumoniae isolate from a patient with diabetes mellitus and chronic renal failure on hemodialysis in South Korea. Antimicrob. Agents Chemother. 54:2278–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cuzon G, Naas T, Truong H, Villegas MV, Wisell KT, Carmeli Y, Gales AC, Venezia SN, Quinn JP, Nordmann P. 2010. Worldwide diversity of Klebsiella pneumoniae that produce beta-lactamase blaKPC-2 gene. Emerg. Infect. Dis. 16:1349–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Souli M, Galani I, Antoniadou A, Papadomichelakis E, Poulakou G, Panagea T, Vourli S, Zerva L, Armaganidis A, Kanellakopoulou K, Giamarellou H. 2010. An outbreak of infection due to beta-lactamase Klebsiella pneumoniae carbapenemase 2-producing K. pneumoniae in a Greek university hospital: molecular characterization, epidemiology, and outcomes. Clin. Infect. Dis. 50:364–373 [DOI] [PubMed] [Google Scholar]
- 8.Nordmann P, Naas T, Poirel L. 2011. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 17:1791–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta N, Limbago BM, Patel JB, Kallen AJ. 2011. Carbapenem-resistant Enterobacteriaceae: epidemiology and prevention. Clin. Infect. Dis. 53:60–67 [DOI] [PubMed] [Google Scholar]
- 10.Marchaim D, Navon-Venezia S, Schwaber MJ, Carmeli Y. 2008. Isolation of imipenem-resistant Enterobacter species: emergence of KPC-2 carbapenemase, molecular characterization, epidemiology, and outcomes. Antimicrob. Agents Chemother. 52:1413–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gasink LB, Edelstein PH, Lautenbach E, Synnestvedt M, Fishman NO. 2009. Risk factors and clinical impact of Klebsiella pneumoniae carbapenemase-producing K. pneumoniae. Infect. Control Hosp. Epidemiol. 30:1180–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zarkotou O, Pournaras S, Tselioti P, Dragoumanos V, Pitiriga V, Ranellou K, Prekates A, Themeli-Digalaki K, Tsakris A. 2011. Predictors of mortality in patients with bloodstream infections caused by KPC-producing Klebsiella pneumoniae and impact of appropriate antimicrobial treatment. Clin. Microbiol. Infect. 17:1798–1803 [DOI] [PubMed] [Google Scholar]
- 13.Borer A, Saidel-Odes L, Riesenberg K, Eskira S, Peled N, Nativ R, Schlaeffer F, Sherf M. 2009. Attributable mortality rate for carbapenem-resistant Klebsiella pneumoniae bacteremia. Infect. Control Hosp. Epidemiol. 30:972–976 [DOI] [PubMed] [Google Scholar]
- 14.Qureshi ZA, Paterson DL, Potoski BA, Kilayko MC, Sandovsky G, Sordillo E, Polsky B, Adams-Haduch JM, Doi Y. 2012. Treatment outcome of bacteremia due to KPC-producing Klebsiella pneumoniae: superiority of combination antimicrobial regimens. Antimicrob. Agents Chemother. 56:2108–2113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirsch EB, Tam VH. 2010. Detection and treatment options for Klebsiella pneumoniae carbapenemases (KPCs): an emerging cause of multidrug-resistant infection. J. Antimicrob. Chemother. 65:1119–1125 [DOI] [PubMed] [Google Scholar]
- 16.Tumbarello M, Viale P, Viscoli C, Trecarichi EM, Tumietto F, Marchese A, Spanu T, Ambretti S, Ginocchio F, Cristini F, Losito AR, Tedeschi S, Cauda R, Bassetti M. 2012. Predictors of mortality in bloodstream infections caused by Klebsiella pneumoniae carbapenemase-producing K. pneumoniae: importance of combination therapy. Clin. Infect. Dis. 55:943–950 [DOI] [PubMed] [Google Scholar]
- 17.Deris ZZ, Yu HH, Davis K, Soon RL, Jacob J, Ku CK, Poudyal A, Bergen PJ, Tsji BT, Bulitta JB, Forrest A, Paterson DL, Velkov T, Li J, Nation RL. 2012. The combination of colistin and doripenem is synergistic against Klebsiella pneumoniae at multiple inocula and suppresses colistin resistance in an in vitro pharmacokinetic/pharmacodynamic model. Antimicrob. Agents Chemother. 56:5103–5112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jernigan MG, Press EG, Nguyen MH, Clancy CJ, Shields RK. 2012. The combination of doripenem and colistin is bactericidal and synergistic against colistin-resistant, carbapenemase-producing Klebsiella pneumoniae. Antimicrob. Agents Chemother. 56:3395–3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pankey GA, Ashcraft DS. 2011. Detection of synergy using the combination of polymyxin B with either meropenem or rifampin against carbapenemase-producing Klebsiella pneumoniae. Diagn. Microbiol. Infect. Dis. 70:561–564 [DOI] [PubMed] [Google Scholar]
- 20.Clancy CJ, Chen L, Shields RK, Zhao Y, Cheng S, Chavda KD, Hao B, Hong JH, Doi Y, Kwak EJ, Silveira FP, Abdel-Massih R, Bogdanovich T, Humar A, Perlin DS, Kreiswirth BN, Nguyen MH. Epidemiology and molecular characterization of bacteremia due to carbapenem-resistant Klebsiella pneumoniae in transplant recipients. Am. J. Transplant., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Queenan AM, Shang W, Flamm R, Bush K. 2010. Hydrolysis and inhibition profiles of beta-lactamases from molecular classes A to D with doripenem, imipenem, and meropenem. Antimicrob. Agents Chemother. 54:565–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.CLSI 2012. Performance standards for antimicrobial susceptibility testing; 22nd informational supplement, M100-S22. CLSI, Wayne, PA [Google Scholar]
- 23.Garonzik SM, Li J, Thamlikitkul V, Paterson DL, Shoham S, Jacob J, Silveira FP, Forrest A, Nation RL. 2011. Population pharmacokinetics of colistin methanesulfonate and formed colistin in critically ill patients from a multicenter study provide dosing suggestions for various categories of patients. Antimicrob. Agents Chemother. 55:3284–3294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Wart SA, Andes DR, Ambrose PG, Bhavnani SM. 2009. Pharmacokinetic-pharmacodynamic modeling to support doripenem dose regimen optimization for critically ill patients. Diagn. Microbiol. Infect. Dis. 63:409–414 [DOI] [PubMed] [Google Scholar]
- 25.Chen L, Mediavilla JR, Endimiani A, Rosenthal ME, Zhao Y, Bonomo RA, Kreiswirth BN. 2011. Multiplex real-time PCR assay for detection and classification of Klebsiella pneumoniae carbapenemase gene (bla KPC) variants. J. Clin. Microbiol. 49:579–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dallenne C, Da Costa A, Decre D, Favier C, Arlet G. 2010. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 65:490–495 [DOI] [PubMed] [Google Scholar]
- 27.Essack SY, Hall LM, Pillay DG, McFadyen ML, Livermore DM. 2001. Complexity and diversity of Klebsiella pneumoniae strains with extended-spectrum beta-lactamases isolated in 1994 and 1996 at a teaching hospital in Durban, South Africa. Antimicrob. Agents Chemother. 45:88–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poirel L, Walsh TR, Cuvillier V, Nordmann P. 2011. Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 70:119–123 [DOI] [PubMed] [Google Scholar]
- 29.Hong JH, Clancy CJ, Cheng S, Shields RK, Chen L, Doi Y, Zhao Y, Perkin DS, Kreiswirth BN, Nguyen MH. 2013. Characterization of porin expression in Klebsiella pneumoniae carbapenemase (KPC)-producing Klebsiella pneumoniae identifies isolates most susceptible to the combination of colistin and carbapenems. Antimicrob. Agents Chemother. 57:2147–2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Landman D, Bratu S, Quale J. 2009. Contribution of OmpK36 to carbapenem susceptibility in KPC-producing Klebsiella pneumoniae. J. Med. Microbiol. 58:1303–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsai YK, Fung CP, Lin JC, Chen JH, Chang FY, Chen TL, Sui LK. 2011. Klebsiella pneumoniae outer membrane porins OmpK35 and OmpK36 play roles in both antimicrobial resistance and virulence. Antimicrob. Agents Chemother. 55:1485–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garcia-Fernandez A, Villa L, Carta C, Venditti C, Giordano A, Venditti M, Mancini C, Carattoli A. 2012. Klebsiella pneumoniae ST258 producing KPC-3 identified in Italy carries novel plasmids and OmpK36/OmpK35 porin variants. Antimicrob. Agents Chemother. 56:2143–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alberti S, Rodriquez-Quinones F, Schirmer T, Rummel G, Tomas JM, Rosenbusch J, Benedi VJ. 1995. A porin from Klebsiella pneumoniae: sequence homology, three-dimensional model, and complement binding. Infect. Immun. 63:903–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hernandez-Alles S, Benedi VJ, Martinez-Martinez L, Pascual A, Aguilar A, Tmas JM, Alberti S. 1999. Development of resistance during antimicrobial therapy caused by insertion sequence interruption of porin genes. Antimicrob. Agents Chemother. 43:937–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song W, Suh B, Choi JY, Jeong SH, Jeon EH, Lee YK, Hong SG, Lee K. 2009. In vivo selection of carbapenem-resistant Klebsiella pneumoniae by OmpK36 loss during meropenem treatment. Diagn. Microbiol. Infect. Dis. 65:447–449 [DOI] [PubMed] [Google Scholar]
- 36.Lee CH, Chu C, Liu JW, Chen YS, Chiu CJ, Su LH. 2007. Collateral damage of flomoxef therapy: in vivo development of porin deficiency and acquisition of blaDHA-1 leading to ertapenem resistance in a clinical isolate of Klebsiella pneumoniae producing CTX-M-3 and SHV-5 beta-lactamases. J. Antimicrob. Chemother. 60:410–413 [DOI] [PubMed] [Google Scholar]
- 37.Bergen PJ, Tsuji BT, Bulitta JB, Forrest A, Jacob J, Sidjabat HE, Paterson DL, Natino RL, Li J. 2011. Synergistic killing of multidrug-resistant Pseudomonas aeruginosa at multiple inocula by colistin combined with doripenem in an in vitro pharmacokinetic/pharmacodynamic model. Antimicrob. Agents Chemother. 55:5685–5695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoon J, Urban C, Terzian C, Mariano N, Rahal JJ. 2004. In vitro double and triple synergistic activities of polymyxin B, imipenem, and rifampin against multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 48:753–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Doumith M, Ellington MJ, Livermore DM, Woodford N. 2009. Molecular mechanisms disrupting porin expression in ertapenem-resistant Klebsiella and Enterobacter spp. clinical isolates from the UK. J. Antimicrob. Chemother. 63:659–667 [DOI] [PubMed] [Google Scholar]
- 40.Kitchel B, Rasheed JK, Endimiani A, Hujer AM, Anderson KF, Bonomo RA, Patel JB. 2010. Genetic factors associated with elevated carbapenem resistance in KPC-producing Klebsiella pneumoniae. Antimicrob. Agents Chemother. 54:4201–4207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garcia-Fernandez A, Miriagou V, Papagiannitsis CC, Giordano A, Venditti M, Mancini C, Carattoli A. 2010. An ertapenem-resistant extended-spectrum-beta-lactamase-producing Klebsiella pneumoniae clone carries a novel OmpK36 porin variant. Antimicrob. Agents Chemother. 54:4178–4184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Domenech-Sanchez A, Martinez-Martinez L, Hernandez-Alles S, del Carmen Conejo M, Pasual A, Tomas JM, Alberti S, Benedi VJ. 2003. Role of Klebsiella pneumoniae OmpK35 porin in antimicrobial resistance. Antimicrob. Agents Chemother. 47:3332–3335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu JJ, Wang LR, Liu YF, Chen HM, Yan JJ. 2011. Prevalence and characteristics of ertapenem-resistant Klebsiella pneumoniae isolates in a Taiwanese university hospital. Microb. Drug Resist. 17:259–266 [DOI] [PubMed] [Google Scholar]