Abstract
The broth microdilution method for fosfomycin and Pseudomonas aeruginosa was assessed and compared with the approved agar dilution method in 206 genetically unrelated P. aeruginosa clinical isolates. Essential agreement between the two methods was 84%, and categorical agreement was 89.3%. Additionally, Etest and disk diffusion assays were performed. Results validate broth microdilution as a reliable susceptibility testing method for fosfomycin against P. aeruginosa. Conversely, unacceptable concordance was established between Etest and disk diffusion results with agar dilution results.
TEXT
Multi-drug resistance (MDR) in clinical Pseudomonas aeruginosa isolates has been widely reported and is of particularly concern in severe infections for which few therapeutic options remain available (1). Fosfomycin shows no cross-resistance with other antimicrobials and has demonstrated safety and efficacy over a broad range of infections and organisms (2). This antibiotic may act synergistically with many antimicrobials, and the intravenous form could be administered in combination for the treatment of systemic infections caused by MDR P. aeruginosa (3, 4).
The approved in vitro antimicrobial susceptibility testing method for fosfomycin is agar dilution (5); however, broth microdilution is the basis of automated systems currently used in clinical microbiology laboratories. Neither the Clinical and Laboratory Standards Institute (CLSI) (5) nor the European Committee on Antimicrobial Susceptibility Testing (EUCAST) (6) includes fosfomycin breakpoints for P. aeruginosa, although EUCAST has defined an epidemiological cutoff (ECOFF) value (6).
Our study was designed to evaluate the in vitro fosfomycin activity against a collection of P. aeruginosa isolates by standard broth microdilution and to compare that method's performance with the performance of agar dilution, considered in this study as the reference method. In addition, agar dilution and broth microdilution were also assessed without glucose-6-phosphate (G6P) against 25 strains exhibiting different fosfomycin MIC values. Etest and disk diffusion were also assessed. A total of 206 genetically unrelated clinical P. aeruginosa isolates, 148 carbapenem susceptible and 58 non-carbapenem susceptible, were included (7). P. aeruginosa ATCC 27853 was used as the control.
(Part of this research was presented at the 22nd European Congress of Clinical Microbiology and Infectious Diseases, London, United Kingdom, 31 March to 3 April 2012.)
Susceptibility to fosfomycin (Laboratorios Ern, S.A., Barcelona, Spain) was determined concomitantly by agar dilution and broth microdilution using BBL Mueller-Hinton II cation-adjusted agar and broth, respectively (Becton, Dickinson [BD], Sparks, MD), both supplemented with 25 μg/ml of G6P (Sigma-Aldrich Chemical Co., St. Louis, MO). The MIC was defined as the lowest antibiotic concentration that inhibited visible growth of the organism. MIC interpretation was done using an ECOFF value of ≤128 μg/ml (6). Fosfomycin disk diffusion using disks of two different strengths (200 μg [BD] and 50 μg [Oxoid Ltd., Basingstoke, United Kingdom]), both supplemented with 50 μg of G6P (5, 8), and the Etest (containing 25 μg/ml of G6P; bioMérieux, Marcy-l'Étoile, France) were performed in duplicate on the same day. Isolates showing colonies inside both the inhibition ellipse of the Etest and the inhibition zone diameters of disks but excluding an objective measurement were recorded as “resistant.” Quantification, susceptibility status, and mutation rate of these colonies were not assessed, as this appraisal was beyond the scope of the present work.
Categorical error rates (categorical agreement [CA]) were calculated after comparing the MICs obtained by microdilution and Etest with those obtained by the agar dilution method. Essential agreement (EA) was defined as when the MIC results obtained by the two methods were identical or agreed within approximately one 2-fold dilution. Major and very major errors were recorded according to published guidelines (9). Error levels were computed as percentages along with their corresponding exact 95% confidence intervals (10). The log2-transformed MICs obtained by the different procedures were compared using a nonparametrical test (Wilcoxon rank sum test), and statistical significance was established at a P value of <0.05. Agreement between procedures was assessed by following the Bland and Altman method (11). Briefly, the mean MIC values obtained with the two methods were plotted (x axis) against the difference between such MIC values (y axis). A linear model, to test the presence of any linear trend between the two variables (mean and difference), was fitted. Values for the kappa coefficient, which gives a measure of the percentage of agreement between MICs obtained by the different methods beyond that expected by chance, were interpreted according to the classification described by Landis and Koch (12). Statistical analysis was performed using the Stata statistical software for Windows (Data Analysis and Statistical Software, version 11.0).
Fosfomycin MICs demonstrated almost identical distributions irrespective of carbapenem susceptibility status, and MIC distributions (agar dilution and broth microdilution) were virtually superimposable over those recorded by EUCAST (6). According to the ECOFF value, 86.4% and 80.6% of isolates were susceptible and 13.6% and 19.4% were resistant to fosfomycin by the agar dilution and broth microdilution methods, respectively. A modal value of 64 μg/ml was observed in both cases. The MIC50 value by both methods was also 64 μg/ml, while the MIC90 values were 256 μg/ml and 512 μg/ml for agar dilution and broth microdilution, respectively. The MIC results obtained for P. aeruginosa strain ATCC 27853 by the two methods were identical (4 μg/ml). When broth microdilution was compared with agar dilution, the EA was 84% and the CA was 89.3%, with a kappa value of 0.65, which indicates “substantial agreement.” Rates of very major and major errors were 17.9% and 9.6%, respectively (Table 1). Using the Wilcoxon test, significant differences were not found between the MIC values obtained with the two methods (P = 0.1327). By using the Bland-Altman analysis (Fig. 1), good agreement between the two methods was corroborated, as the majority of values were distributed between the ±1 range. Linear fit showed a significant negative slope (P = 0.002). At low fosfomycin concentrations, agar dilution tended to give slightly higher MICs, and at high fosfomycin concentrations, microdilution was the method that showed slightly higher MICs.
Table 1.
Correlation of broth microdilution and Etest results with agar dilution results for 206 P. aeruginosa isolatesa
| Performance value | Agar dilution vs microdilution |
Agar dilution vs Etest |
||
|---|---|---|---|---|
| % correlation (no. of isolates/total no.) | 95% CI | % correlation (no. of isolates/total no.) | 95% CI | |
| Essential agreement | 84.0 (173/206) | 78.2–88.7 | 65.5 (135/206) | 58.6–72.0 |
| Categorical agreement | 89.3 (184/206) | 84.3–93.2 | 73.3 (151/206) | 66.7–79.2 |
| Major error | 9.6 (17/178)a | 6.1–36.9 | 26.4 (47/178)b | 17.3–29.2 |
| Very major error | 17.9 (5/28)b | 5.7–14.9 | 28.6 (8/28)c | 13.2–48.7 |
A EUCAST ECOFF value of ≤128 mg/liter was considered for MIC categorization.
Considering the number of susceptible isolates.
Considering the number of resistant isolates.
Fig 1.
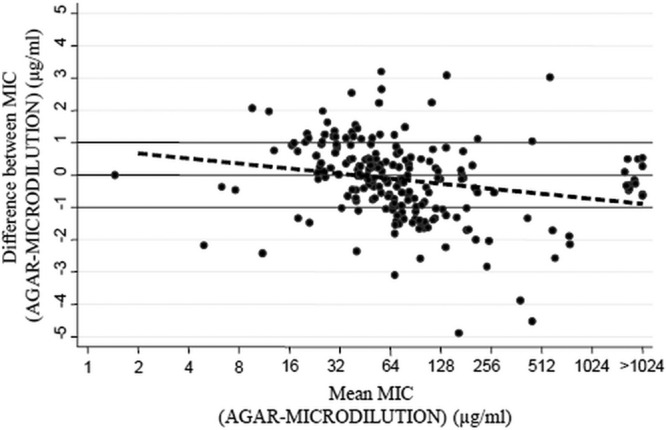
Agreement observed by plotting the difference between the broth microdilution and agar dilution methods against their mean MIC values. The broken line represents the linear fit between mean MIC values and the difference between MICs (solid circles) obtained with the two methods (P = 0.002).
Identical MICs were observed by the agar dilution and microdilution methods for the 25 selected isolates and the control strain without supplementation with G6P (data not shown). This fact corroborates that, as previously stated, P. aeruginosa seems to lack a specific G6P transporter (13).
The presence of colonies inside the Etest inhibition ellipses led to high rates of major and very major errors, 26.4% and 28.6%, respectively (Table 1). The occurrence of such colonies inside disk inhibition zones (irrespective of disk strength) was high enough to preclude the measurement of zone diameters, yielding unacceptable error levels with this method (data not shown). Unlike for the Etest, in which the presence of a dominantly inhibited population (except in those cases of complete resistance) could be more easily differentiated from the scattered colonies of the resistant subpopulation, the colonies of resistant mutants within disk inhibition zones led to unreliable measurements, preventing the use of this method.
Validating the broth microdilution method as an alternative to the approved agar dilution test for fosfomycin and P. aeruginosa was the main goal of this work. By taking into account the fact that automated systems used worldwide for antimicrobial susceptibility testing are microdilution-adapted devices, the correlation observed between the agar dilution results and those obtained with broth microdilution may validate the latter as an adequate susceptibility testing method. Even though a rate of 17.9% (5/28) of very major errors with broth microdilution is rather high, it should be taken into consideration that the total number of resistant isolates (n = 28) obtained with the agar dilution method is lower than the desirable value established by guidelines (n > 30) (9). Moreover, three out of the five isolates contributing to the EA of 84% differed only in approximately one 2-fold dilution from the reference MIC values.
According to this study, fosfomycin MICs against P. aeruginosa appear to be scarcely dependent on the method used when agar dilution is compared with broth microdilution. In contrast, Etest and disk diffusion are not reliable alternatives for routine laboratory susceptibility testing. The same conclusion was obtained in studies conducted with other microorganisms (14, 15). We also note that supplementation with G6P when determining fosfomycin MICs against P. aeruginosa was demonstrated to be unnecessary.
In conclusion, the overall concordant distribution of fosfomycin MICs against P. aeruginosa with both agar dilution and broth microdilution methods indicates that the latter is a reliable procedure with a reliable performance that might validate those results obtained with routinely used, automated broth microdilution-based systems. The paucity of therapeutic options against P. aeruginosa reinforces the usefulness of routinely determining fosfomycin susceptibility, particularly to support the intravenous use of this compound in combination therapy when MIC values are below or equal to the ECOFF value of 128 μg/ml.
ACKNOWLEDGMENTS
R.D.C. has a contract from the Instituto de Salud Carlos III-FIS (CB05/137). This work was supported in part by the Ministerio de Economía y Competitividad, Instituto de Salud Carlos III, cofinanced by the European Development Regional Fund (ERDF) “A Way To Achieve Europe,” by the Spanish Network for the Research in Infectious Diseases (REIPI RD12/0015), and by the Microbial Sciences Foundation (Madrid, Spain).
We have no conflicts of interest.
Footnotes
Published ahead of print 12 August 2013
REFERENCES
- 1.Strateva T, Yordanov D. 2009. Pseudomonas aeruginosa—phenomenon of bacterial resistance. J. Med. Microbiol. 58:1133–1148 [DOI] [PubMed] [Google Scholar]
- 2.Michalopoulos AS, Livaditis IG, Gougoutas V. 2011. The revival of fosfomycin. Int. J. Infect. Dis. 15:e732–3739. 10.1016/j.ijid.2011.07.007 [DOI] [PubMed] [Google Scholar]
- 3.Dinh A, Salomon J, Bru JP, Bernard L. 2012. Fosfomycin: efficacy against infections caused by multidrug-resistant bacteria. Scand. J. Infect. Dis. 44:182–189 [DOI] [PubMed] [Google Scholar]
- 4.Falagas ME, Kastoris AC, Karageorgopoulos DE, Rafailidis PI. 2009. Fosfomycin for the treatment of infections caused by multidrug-resistant non-fermenting Gram-negative bacilli: a systematic review of microbiological, animal and clinical studies. Int. J. Antimicrob. Agents 34:111–120 [DOI] [PubMed] [Google Scholar]
- 5.Clinical and Laboratory Standards Institute 2012. Performance standards for antimicrobial susceptibility testing: 22nd informational supplement. M100-S22 CLSI, Wayne, PA [Google Scholar]
- 6.European Committee on Antimicrobial Susceptibility Testing 2012. Breakpoint tables for interpretation of MICs and zone diameter, version 2.0. http://www.eucast.org/
- 7.García-Castillo M, del Campo R, Morosini MI, Riera E, Cabot G, Willems R, van Mansfeld R, Oliver A, Cantón R. 2011. Wide dispersion of ST175 clone despite high genetic diversity of carbapenem-nonsusceptible Pseudomonas aeruginosa clinical strains in 16 Spanish hospitals. J. Clin. Microbiol. 49:2905–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu CL, Liu CY, Huang YT, Teng LJ, Turnidge JD, Hsueh PR. 2011. Antimicrobial susceptibilities of commonly encountered bacterial isolates to fosfomycin determined by agar dilution and disk diffusion methods. Antimicrob. Agents Chemother. 55:4295–4301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark RB, Lewinski MA, Loeffelholz MJ, Tibetts RJ. 2009. Cumitech 31A, Verification and validation of procedures in the clinical microbiology laboratory. Coordinating ed, Sharp SE ASM Press, Washington, DC [Google Scholar]
- 10.Leemis LM, Trivedi KS. 1996. A comparison of approximate interval estimators for the Bernoulli parameter. Am. Stat. 50:63–68 [Google Scholar]
- 11.Bland JM, Altman DG. 1986. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet 327:307–310 [PubMed] [Google Scholar]
- 12.Landis JR, Koch GG. 1977. The measurement of observer agreement for categorical data. Biometrics 33:159–174 [PubMed] [Google Scholar]
- 13.Castañeda-García A, Rodríguez-Rojas A, Guelfo JR, Blázquez J. 2009. The glycerol-3-phosphate permease GlpT is the only fosfomycin transporter in Pseudomonas aeruginosa. J. Bacteriol. 191:6968–6974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Cueto M, López L, Hernández JR, Morillo C, Pascual A. 2006. In vitro activity of fosfomycin against extended-spectrum-beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae: comparison of susceptibility testing procedures. Antimicrob. Agents Chemother. 50:368–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.López-Cerero L, de Cueto M, Díaz-Guerrero MA, Pascual A, Morillo C. 2007. Evaluation of the Etest method for fosfomycin susceptibility of ESBL-producing Klebsiella pneumoniae. J. Antimicrob. Chemother. 59:810–812 [DOI] [PubMed] [Google Scholar]


