Abstract
Transposon Tn558 integrated in the chromosomal radC gene was detected for the first time in Staphylococus pseudintermedius. It carried a novel fexA variant (fexAv) that confers only chloramphenicol resistance. The exporter FexAv exhibited two amino acid substitutions, Gly33Ala and Ala37Val, both of which seem to be important for substrate recognition. Site-directed mutagenesis that reverted the mutated base pairs to those present in the original fexA gene restored the chloramphenicol-plus-florfenicol resistance phenotype.
TEXT
In staphylococci, phenicol resistance is mediated either by the chloramphenicol acetyltransferase-encoding cat genes, which confer resistance to nonfluorinated phenicols (e.g., chloramphenicol), or either of the two genes fexA (coding for a phenicol-specific efflux pump) and cfr (coding for a rRNA methyltransferase), both of which mediate combined resistance to fluorinated phenicols (e.g., florfenicol) and nonfluorinated phenicols (1). Since the first description of the fexA gene in a bovine Staphylococcus lentus isolate (2), this gene has been detected—either as part of the small nonconjugative transposon Tn558 or in combination with the cfr gene in transposition-deficient Tn558 variants—in S. aureus and several coagulase-negative staphylococcal species from healthy and diseased cattle, swine, horses, or humans (3–9). The fexA gene has also been detected in a Bacillus isolate from swine feces and in environmental pollutants from swine feedlots in China (10, 11).
In a previous study that focused on the occurrence of methicillin-resistant coagulase-positive staphylococci in dogs in La Rioja, Spain, one methicillin-susceptible S. pseudintermedius isolate, named C2719, was identified (12). This isolate was recovered from a healthy dog admitted to a veterinary clinic for a routine checkup. No additional information on the living conditions of the dog and/or the possible contact with rural areas or livestock was recorded. Susceptibility testing by agar disk diffusion and/or broth microdilution (13) showed that isolate C2719 was resistant to penicillin (due to the blaZ gene) and to chloramphenicol (MIC 64 μg/ml) but exhibited a low MIC of 2 μg/ml for florfenicol. None of the three cat genes known to occur in staphylococci—catpC194, catpC221, and catpC223 (1)—was detected by PCR (14). The objective of this study was to identify the gene responsible for chloramphenicol resistance in this strain and to characterize its genetic environment.
PCR analysis for the chloramphenicol/florfenicol resistance genes fexA and cfr (5) showed that, despite its low florfenicol MIC, strain C2719 harbored the fexA gene. PCR mapping and sequencing revealed that this fexA variant, named fexAv, exhibited 99.7% nucleotide sequence identity (99.2% identity and 99.6% similarity at the protein level) to the prototype fexA gene of S. lentus (GenBank accession no. AJ549214) (2). Moreover, the fexAv gene was found to be part of the Tn558 transposon (3, 15) (Fig. 1). Tn558 is a member of the Tn554 family (15). Transposons of this family exhibit several features that distinguish them from most other transposable elements: (i) their ends are asymmetric, lacking either inverted or direct terminal repeats, (ii) they do not generate a duplication of the target sequence upon transposition, and (iii) they are extremely site specific, almost always inserting into the staphylococcal chromosome at the same location. This unique target site att, is located within the radC gene, which codes for a DNA repair protein (16–22). In addition to Tn558, first described in S. lentus (15), this group also includes at least another four members: Tn554 harboring the erm(A) gene for resistance to macrolides, lincosamides, and streptogramin B and the spc gene for spectinomycin resistance (19, 20); Tn5406 carrying the vga(A) gene for resistance to lincosamides, pleuromutilins, and streptogramin A (16); Tn559 harboring the dfrK gene for trimethoprim resistance (17); and Tn6133, which is composed of a Tn554-like element into which a segment with the novel gene vga(E) for resistance to lincosamides, pleuromutilins, and streptogramin A has been inserted (22). All these transposons have been found in S. aureus, with Tn559 and Tn6133 first detected in livestock-associated S. aureus of the lineage ST398.
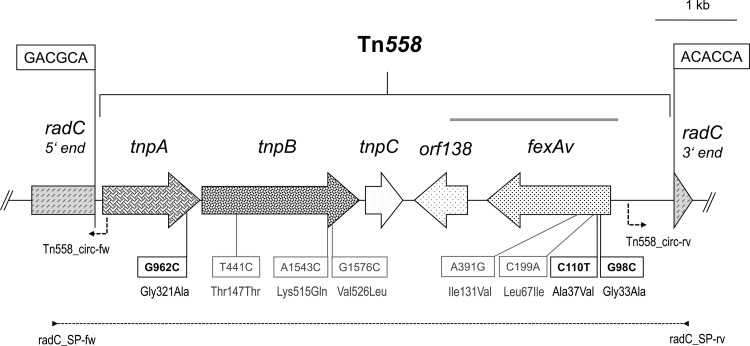
Fig 1.
Schematic presentation of the Tn558 structure carrying the fexAv gene detected in this study as well as its integration region (within the radC gene) in the chromosomal DNA of S. pseudintermedius C2719 (EMBL accession no. HF679552). The arrows indicate the extents and directions of transcription of chloramphenicol resistance (fexAv) and transposase (tnpA, tnpB, tnpC) genes as well as reading frame orf138. The 6-bp core nucleotide sequences at the transposon junctions are shown within boxes. The positions of primers used to amplify the region between the radC gene extremes are indicated as arrowheads, with a dashed line showing the extension length. The primers employed to detect circular intermediates, as well as the direction of amplification, are also shown. The gray bar over the fexAv gene indicates the region that has been used for cloning and mutagenesis experiments. A size scale in kilobases (kb) is displayed in the upper right-hand corner of the figure.
The chromosomal/plasmid location of Tn558, as well as its specific integration site, was determined by plasmid preparation, specific PCRs, and sequencing (3, 23). As no plasmids were detected in S. pseudintermedius C2719, a chromosomal location of the fexAv-carrying transposon appeared to be likely. Primers radC_SP-fw and radC_SP-rv (Table 1) were designed to determine whether Tn558 was integrated within the chromosomal radC gene. The target recognition sequence, att558, was identified on the basis of the nucleotide similarity of (i) the 6-bp “core” sequence (att, 5′-GATGTA-3′) of the radC gene of the original S. lentus isolate carrying Tn558 (accession no. AJ715531) (15) and that detected on the radC gene of S. pseudintermedius HKU10-03 and (ii) the sequences flanking this att site, which are also essential for transposition (19, 20).
Table 1.
Primers used to detect the chromosomal integration site of the fexA-carrying Tn558 of S. pseudintermedius isolate C2719, those to amplify the complete fexA gene, and those to perform the site-directed-mutagenesis
| Primer name | Primer sequence (5′→3′)a | Amplicon size (bp) | PCR conditions | Polymerase |
|---|---|---|---|---|
| radC_SP-fw | GTTTGTCGGAATAGGGCGTA | 437b/7,082 | 1 min at 94°C; 30 cycles of 10 s at 98°C, 15 min at 68°C and 10 min at 72°C | La Taq (TaKaRa) |
| radC_SP-rv | ACGATTCTTCCCCAATCACA | |||
| entire_fexA-1 | GATCCGTAAGCCCATCCATA | 2,081 | 3 min at 94°C; 30 cycles of 30 s at 94°C, 45 s at 55°C, and 2 min at 72°C; 5 min at 72°C | BioTaq (Bioline) |
| entire_fexA-2 | AGGCACCGGTTGTTAAACTG | |||
| fexA_33-inv1 | ATCTGTACTTGTAGGTGCAATTACGGTTG | 6,012c | 3 min at 98°C; 20 cycles of 1 min at 98°C, 1 min at 55°C, and 2.5 min at 72°C; 10 min at 72°C | Phusion (Finnzymes) |
| fexA_33-inv2 | CAACCGTAATTGCACCTACAAGTACAGAT | |||
| fexA_37-inv1 | TAGCTGCAATTACGGCTGATTTAGTCAATCC | |||
| fexA_37-inv2 | GGGATTGACTAAATCAGCCGTAATTGCAGCTA | |||
| fexA_33 + 37-inv1 | CTATCTGTACTTGTAGGTGCAATTACGGCTGATTTAGTCAATCCC | |||
| fexA_33 + 37-inv2 | GGGATTGACTAAATCAGCCGTAATTGCACCTACAAGTACAGATAG |
Nucleotides in bold type and underlined are those modified by the site-directed-mutagenesis PCR.
Amplicon size obtained when an intact copy of the radC gene was present.
Size of the complete recombinant and mutated plasmid, composed of the PCR 2.1-TOPO vector (3,931 bp) and the insertion obtained with the entire_fexA-1 and entire_fexA-2 primers (2,081 bp).
An amplicon of 7,082 bp that comprised the complete Tn558 (6,645 bp) and part of the radC gene (437 bp) was obtained (Fig. 1). Complete analysis of the sequence of this transposon revealed 99.7% nucleotide identity to that of S. lentus (GenBank accession no. AJ715531). Sequence analysis identified three semiconserved amino acid substitutions in two reading frames: (i) Gly321Ala in the transposase protein TnpA (nucleotide G962C in the transposase gene tnpA) and (ii) Gly33Ala and Ala37Val in the FexAv protein (nucleotides G98C and C110T, respectively, within the fexAv gene) (Fig. 1). Four additional conserved amino acid substitutions were observed in Tn558: (i) Lys515Gln and Val526Leu in the TnpB transposase (nucleotides A1543C and G1576C, respectively) and (ii) Leu67Ile and Ile131Val within the FexAv protein (nucleotides C199A and A391G, respectively). No circular intermediates of Tn558, which are indicative for the mobility of the transposon, were detected by specific PCR (Fig. 1) (3) either when the bacteria were grown under normal growth conditions (13) or after growth under stress conditions (overnight cultures were exposed to ultraviolet light for 10 min or to a heat shock of 60°C for 2 h; growth of bacteria under conditions of anaerobiosis or at 45°C). Whether the observed amino acid changes in TnpA and/or TnpB or other factors account for the lack of mobility of Tn558 under the tested conditions remains to be determined.
To confirm that the fexAv gene is in fact responsible for chloramphenicol but not for florfenicol resistance, a PCR assay using primers entire_fexA-1 and entire_fexA-2 (Table 1) that amplified the complete fexAv gene (1,428 bp), including 201 bp and 452 bp of its upstream and downstream region, respectively, was conducted. This 2,081-bp amplicon was first cloned into the pCR 2.1-TOPO vector and transformed into the recipient Escherichia coli TOP10 strain using a TOPO TA cloning kit (Invitrogen, Groningen, The Netherlands). Subsequent transformation into E. coli HB101 was conducted to test the functionality of the fexAv gene in a phenicol-susceptible E. coli host (chloramphenicol MIC, 2 μg/ml; florfenicol MIC, 4 μg/ml). E. coli HB101 carrying the recombinant vector exhibited a 16-fold increase in the chloramphenicol MIC (32 μg/ml), while the florfenicol MIC value remained unchanged.
In a study on the FloR chloramphenicol/florfenicol efflux protein, Braibant et al. (24) determined by site-directed mutagenesis (SDM) that the Asp23 residue seems to participate directly in the affinity pocket involved in phenicol-derivative recognition. When Asp23 was mutated to Glu23, the corresponding FloR protein still conferred chloramphenicol resistance but lost its ability to export florfenicol. A model (http://www.ch.embnet.org/software/TMPRED_form.html) of the transmembrane segments (TMS) of the 12-TMS FloR protein and the 14-TMS FexA protein predicted that the Asp23 residue in FloR and the Gly33/Ala37 residues in the prototype FexA protein are all located in transmembrane segment 1 and hence may have similar functions. Based on these data, SDM was conducted on the fexAv nucleotide substitutions G98C and C110T separately and on G98C and C110T in combination to revert them to those of the fexA prototype from S. lentus. For this, inverse PCR assays with specific “mutagenesis primers” were performed (Table 1). A plasmid preparation (Qiagen Plasmid Midikit; Qiagen, Hilden, Germany) of the aforementioned recombinant pCR 2.1-TOPO vector served as the template for SDM. After SDM, digestion of the obtained SDM products with the DpnI restriction enzyme (Fermentas, St. Leon-Rot, Germany) was performed to eliminate the original methylated template plasmid DNA. An aliquot from the SDM approach was then transferred by electrotransformation into E. coli HB101. Correctly mutated recombinant vectors (TOPO/fexAvC98G, TOPO/fexAvT110C, and TOPO/fexAvC98G+T110C) were confirmed by PCR of the complete fexA gene and sequence analysis of the amplicon (Table 1). Macrodilution assays for chloramphenicol and florfenicol (13) showed a florfenicol MIC of 16 μg/ml for the E. coli HB101 transformants carrying TOPO/fexAvC98G+T110C that was 4-fold higher than that seen with the recipient strain alone (MIC 4 μg/ml) and clones carrying each of the reverted positions separately, while the chloramphenicol MIC remained unchanged in all mutants (MIC, 32 μg/ml). This increase in the florfenicol MIC was similar to that described for E. coli JM109 carrying the cloned fexA gene from S. lentus (2) and confirms that reversion of the naturally mutated positions restores the florfenicol resistance phenotype. It should be noted that the observed MIC changes might differ, at least slightly, in different bacterial hosts, such as in Gram-positive bacteria.
Nucleotide sequence accession number.
The 7,698-bp nucleotide sequence enclosing the complete Tn558 and the radC gene of S. pseudintermedius strain C2719 has been deposited on the EMBL database under accession number HF679552.
(Part of these data was presented at the 2nd Conference on Methicillin-resistant Staphylococci in Animals [ASM-ESCMID], Washington, DC, 8 to 11 September 2011, and at the 15th International Symposium on Staphylococci and Staphylococcal Infections [ISSSI], Lyon, France, 26 to 30 August 2012.)
ACKNOWLEDGMENTS
The work conducted at the University of La Rioja was financially supported by Project SAF2012-35474 from the Ministerio de Economía y Competitividad of Spain and FEDER. The work conducted at the Friedrich-Loeffler-Institut was financially supported by the German Federal Ministry of Education and Research (BMBF) through the German Aerospace Center (DLR), grant number 01KI1014D (MedVet-Staph). E.G.-S. is supported by a fellowship from the Gobierno de La Rioja of Spain.
We thank Kerstin Meyer for excellent technical assistance.
Footnotes
Published ahead of print 26 August 2013
REFERENCES
- 1.Schwarz S, Kehrenberg C, Doublet B, Cloeckaert A. 2004. Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol. Rev. 28:519–542 [DOI] [PubMed] [Google Scholar]
- 2.Kehrenberg C, Schwarz S. 2004. fexA, a novel Staphylococcus lentus gene encoding resistance to florfenicol and chloramphenicol. Antimicrob. Agents Chemother. 48:615–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kehrenberg C, Schwarz S. 2006. Distribution of florfenicol resistance genes fexA and cfr among chloramphenicol-resistant Staphylococcus isolates. Antimicrob. Agents Chemother. 50:1156–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kadlec K, Ehricht R, Monecke S, Steinacker U, Kaspar H, Mankertz J, Schwarz S. 2009. Diversity of antimicrobial resistance pheno- and genotypes of methicillin-resistant Staphylococcus aureus ST398 from diseased swine. J. Antimicrob. Chemother. 64:1156–1164 [DOI] [PubMed] [Google Scholar]
- 5.Shore AC, Brennan OM, Ehricht R, Monecke S, Schwarz S, Slickers P, Coleman DC. 2010. Identification and characterization of the multidrug resistance gene cfr in a Panton-Valentine leukocidin-positive sequence type 8 methicillin-resistant Staphylococcus aureus IVa (USA300) isolate. Antimicrob. Agents Chemother. 54:4978–4984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feßler A, Scott C, Kadlec K, Ehricht R, Monecke S, Schwarz S. 2010. Characterization of methicillin-resistant Staphylococcus aureus ST398 from cases of bovine mastitis. J. Antimicrob. Chemother. 65:619–625 [DOI] [PubMed] [Google Scholar]
- 7.Argudín MA, Tenhagen BA, Fetsch A, Sachsenröder J, Käsbohrer A, Schroeter A, Hammerl JA, Hertwig S, Helmuth R, Bräunig J, Mendoza MC, Appel B, Rodicio MR, Guerra B. 2011. Virulence and resistance determinants of German Staphylococcus aureus ST398 isolates from nonhuman sources. Appl. Environ. Microbiol. 77:3052–3060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, Zhang W, Wang J, Wu C, Shen Z, Fu X, Yan Y, Zhang Q, Schwarz S, Shen J. 2012. Distribution of the multidrug resistance gene cfr in Staphylococcus species isolates from swine farms in China. Antimicrob. Agents Chemother. 56:1485–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lozano C, Ruiz-García M, Gómez-Sanz E, López-García P, Royo-García G, Zarazaga M, Torres C. 2012. Characterization of a cfr-positive methicillin-resistant Staphylococcus epidermidis strain of the lineage ST22 implicated in a life-threatening human infection. Diagn. Microbiol. Infect. Dis. 73:380–382 [DOI] [PubMed] [Google Scholar]
- 10.Dai L, Wu CM, Wang MG, Wang Y, Wang Y, Huang SY, Xia LN, Li BB, Shen JZ. 2010. First report of the multidrug resistance gene cfr and the phenicol resistance gene fexA in a Bacillus strain from swine feces. Antimicrob. Agents Chemother. 54:3953–3955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J, Shao B, Shen J, Wang S, Wu Y. 2013. Occurrence of chloramphenicol-resistance genes as environmental pollutants from swine feedlots. Environ. Sci. Technol. 47:2892–2897 [DOI] [PubMed] [Google Scholar]
- 12.Gómez-Sanz E, Torres C, Lozano C, Sáenz Y, Zarazaga M. 2011. Detection and characterization of methicillin-resistant Staphylococcus pseudintermedius in healthy dogs in La Rioja, Spain. Comp. Immunol. Microbiol. Infect. Dis. 34:447–453 [DOI] [PubMed] [Google Scholar]
- 13.Clinical and Laboratory Standards Institute 2013. Performance standards for antimicrobial susceptibility testing; Twenty-third informational supplement. CLSI Document M100-S23 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 14.Schnellmann C, Gerber V, Rossano A, Jaquier V, Panchaud Y, Doherr MG, Thomann A, Straub R, Perreten V. 2006. Presence of new mecA and mph(C) variants conferring antibiotic resistance in Staphylococcus spp. isolated from the skin of horses before and after clinic admission. J. Clin. Microbiol. 44:4444–4454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kehrenberg C, Schwarz S. 2005. Florfenicol-chloramphenicol exporter gene fexA is part of the novel transposon Tn558. Antimicrob. Agents Chemother. 49:813–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haroche J, Allignet J, El Solh N. 2002. Tn5406, a new staphylococcal transposon conferring resistance to streptogramin and related compounds including dalfopristin. Antimicrob. Agents Chemother. 46:2337–2343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kadlec K, Schwarz S. 2010. Identification of the novel dfrK-carrying transposon Tn559 in a porcine methicillin-susceptible Staphylococcus aureus ST398 strain. Antimicrob. Agents Chemother. 54:3475–3477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.López M, Kadlec K, Schwarz S, Torres C. 2012. First detection of the staphylococcal trimethoprim resistance gene dfrK and the dfrK-carrying transposon Tn559 in enterococci. Microb. Drug Resist. 18:13–18 [DOI] [PubMed] [Google Scholar]
- 19.Murphy E. 1989. Transposable elements in gram-positive bacteria, p 269–288 In Berg DE, Howe MM. (ed), Mobile DNA. ASM Press, Washington, DC [Google Scholar]
- 20.Murphy E. 1990. Properties of the site-specific transposable element Tn554, p 123–135 In Novick RP. (ed), Molecular biology of the staphylococci. VCH Publishers, New York, NY [Google Scholar]
- 21.Schwarz S, Feßler AT, Hauschild T, Kehrenberg C, Kadlec K. 2011. Plasmid-mediated resistance to protein biosynthesis inhibitors in staphylococci. Ann. N. Y. Acad. Sci. 1241:82–103 [DOI] [PubMed] [Google Scholar]
- 22.Schwendener S, Perreten V. 2011. New transposon Tn6133 in methicillin-resistant Staphylococcus aureus ST398 contains vga(E), a novel streptogramin A, pleuromutilin, and lincosamide resistance gene. Antimicrob. Agents Chemother. 55:4900–4904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kehrenberg C, Aarestrup FM, Schwarz S. 2007. IS21-558 insertion sequences are involved in the mobility of the multiresistance gene cfr. Antimicrob. Agents Chemother. 51:483–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Braibant M, Chevalier J, Chaslus-Dancla E, Pagès JM, Cloeckaert A. 2005. Structural and functional study of the phenicol-specific efflux pump FloR belonging to the major facilitator superfamily. Antimicrob. Agents Chemother. 49:2965–2971 [DOI] [PMC free article] [PubMed] [Google Scholar]