Abstract
Nontoxigenic Clostridium difficile (NTCD) has been shown to prevent fatal C. difficile infection in the hamster model when hamsters are challenged with standard toxigenic C. difficile strains. The purpose of this study was to determine if NTCD can prevent C. difficile infection in the hamster model when hamsters are challenged with restriction endonuclease analysis group BI C. difficile strains. Groups of 10 hamsters were given oral clindamycin, followed on day 2 by 106 CFU of spores of NTCD strain M3 or T7, and were challenged on day 5 with 100 CFU of spores of BI1 or BI6. To conserve animals, results for control hamsters challenged with BI1 or BI6 from the present study and controls from previous identical experiments were combined for statistical comparisons. NTCD strains M3 and T7 achieved 100% colonization and were 100% protective against challenge with BI1 (P ≤ 0.001). M3 colonized 9/10 hamsters and protected against BI6 challenge in the colonized hamsters (P = 0.0003). T7 colonized 10/10 hamsters, but following BI6 challenge, cocolonization occurred in 5 hamsters, 4 of which died, for protection of 6/10 animals (P = 0.02). NTCD colonization provides protection against challenge with toxigenic BI group strains. M3 is more effective than T7 in preventing C. difficile infection caused by the BI6 epidemic strain. Prevention of C. difficile infection caused by the epidemic BI6 strain may be more challenging than that of infections caused by historic BI1 and non-BI C. difficile strains.
INTRODUCTION
Clostridium difficile is a Gram-positive, spore-forming anaerobic bacterium known to be the leading infectious cause of nosocomial diarrhea and pseudomembranous colitis. Since the year 2000, the incidence of C. difficile infection (CDI) in many North American hospitals has shown a marked increase. The severity of CDI also appears to have increased and has been characterized by an increase in patient mortality and morbidity, an increase in the frequencies of admissions to the intensive care unit, and the need for a greater number of emergent interventions, such as colectomy (1–3). An outbreak strain of C. difficile identified as restriction endonuclease analysis (REA) group BI, PCR ribotype 027, and pulse-field gel electrophoresis type NAP1 (BI/NAP1/027) is in part responsible for the increased CDI incidence and severity (1, 2). It has been implicated in multiple hospital outbreaks, is more difficult to treat, and has higher recurrence rates than those for other C. difficile strains (4).
Previous studies have shown that asymptomatic C. difficile colonization by either toxigenic or nontoxigenic strains has been associated with a decreased risk of CDI in humans (5). An uncolonized patient who receives antibiotics is rendered susceptible to C. difficile colonization and infection if C. difficile spores are ingested. If the patient is colonized with nontoxigenic C. difficile (NTCD), there is a reduced incidence of CDI, presumably because the organism is unable to produce toxins and is able to prevent colonization by toxigenic C. difficile. Thus, it is hypothesized that after NTCD colonization, a patient exposed to toxigenic C. difficile will be protected. NTCD has been used to colonize and prevent CDI in two patients who were experiencing multiple relapses of CDI (6) and has been shown to be safe in phase 1 volunteer trials of healthy adults, and enrollment in a phase 2 dose-ranging and safety trial using strain M3, designated VP20621, for prevention of CDI recurrence in patients has recently been completed (7).
We have previously shown that colonization of hamsters with specific nontoxigenic strains of C. difficile, M3, M23, and T7, prevented fatal disease in hamsters when challenged with three epidemic toxigenic strains (J9, K14, and B1) of C. difficile typed by REA (8). We have also shown that the epidemic BI/NAP1/027 strains of C. difficile are highly virulent in the hamster model, causing fatal disease within approximately 48 h of inoculation (9). The aim of the current experiments was to determine whether colonization with nontoxigenic C. difficile would prevent infection due to BI/NAP1/027 in the hamster model. Two nontoxigenic strains, M3 and T7, were chosen for protective colonization, and both M3 and T7 have successfully been used in previous protection assays (8). Strain type BI6 was chosen as a representative of current BI epidemic strains in the United States. BI1 is a historic, nonepidemic BI strain and was used for comparison. Both have previously been shown to be 100% fatal in the hamster model following successful colonization (9).
(This study was presented in part at the Fifth International Meeting on the Molecular Biology and Pathogenesis of the Clostridia, 2006 [ClostPath 2006], Nottingham, United Kingdom [10].)
MATERIALS AND METHODS
Spore preparation.
Confluent cultures of C. difficile were grown on blood-agar plates (Columbia base; BBL) for 4 to 6 days in a 36°C anaerobic incubator to encourage sporulation. They were harvested and washed in sterile phosphate-buffered saline (PBS) with no added calcium or magnesium and then heat shocked at 56°C for 10 min to kill surviving vegetative cells. Spores were centrifuged, resuspended in Dulbecco's modified Eagle medium, aliquoted, and frozen at −80°C. Tenfold serial dilutions of the frozen spores were grown on trifluoroacetic acid plates to determine the concentration of spores in the freezer culture.
Hamsters.
Male Syrian Golden hamsters (age, 6 to 8 weeks; weight, approximately 90 to 100 g) were purchased from Charles River Laboratories, Wilmington, MA. Hamsters were housed in individual polycarbonate cages with filter tops and disposable air filters to prevent cross-contamination. Food, bedding, water and water bottles, cages, wire tops, filters, and filter tops were autoclaved before usage. The hamsters were allowed to rest for a week in an isolation room before beginning the experiment. During the course of the experiment, bedding was changed once a day; the cage and water were changed once a week. The experimental protocol was approved by the Hines VA Institutional Animal Care and Use Committee.
Clindamycin treatment.
Hamsters received clindamycin (30 mg/kg of body weight) by oral gavage on day 0.
Colonization with nontoxigenic C. difficile.
On day 2, groups of 10 hamsters were orally inoculated with 106 CFU of spores of strain T7 or M3. Two control hamsters received sham inoculation as unprotected controls for each challenge toxigenic strain. The hamsters were inoculated using an oral gavage needle as previously described (8).
Challenge with toxigenic C. difficile.
On day 5, study hamsters were challenged with 100 CFU of toxigenic spores of either strain BI6 or BI1 using an oral gavage needle as previously described (8). Control hamsters were given the same inoculum of BI6 or BI1 spores to ensure that the inoculum was effective, but to conserve animals, data for historic controls from the study of Razaq et al. (9) were included for statistical comparisons. These strains caused 100% mortality in colonized hamsters (90% overall mortality).
Culturing and colonization.
Hamster fecal pellets were cultured once daily from days 5 to 9 and then twice weekly until the end of the study. For each hamster, three fecal pellets were collected into a 4-ml tube with 750 μl of sterile PBS solution. The collections were then plated onto a prereduced selective taurocholate-cycloserine-cefoxitin-fructose agar (TCCFA) plate using a sterile cotton swab and 4-quadrant streaking. Selective agar plates were made following the method of Wilson et al. (11). After 48 h of incubation, plates were assessed for the presence of characteristic C. difficile colonies. Hamsters that showed any positive C. difficile growth were noted as being colonized. The distinction between the toxigenic and nontoxigenic strains was made via culture on selective TCCFA medium containing erythromycin at 5 mg/ml for BI6 (which is resistant to erythromycin) and confirmed by REA typing. For strain BI1, which is not resistant to erythromycin, isolates from at least 3 colonized hamster groups were typed by REA to confirm their identities.
REA typing of C. difficile strains.
C. difficile isolates from hamsters were inoculated into 20 ml of Trypticase soy broth from a 48-h blood agar plate culture and grown at 36°C in an anaerobic chamber. DNA was isolated as previously described and digested using restriction endonuclease HindIII (12). The agarose gel patterns were then compared to those of previously identified isolates defined by letter groups (in which there is a ≤6-band difference) and numerical subtypes (identical banding patterns). In all four studies, at least three colonized hamsters were chosen for REA typing of the isolates to confirm the identity of the infecting or protecting organism.
Statistical methods.
The mean time interval between challenge with toxigenic C. difficile and colonization, the mean time interval between colonization and death, and the mean time interval between challenge with C. difficile and death were compared using Student's t test. For hamsters found dead before daily colonization was confirmed, colonization was presumed to have occurred on the same day as the death and the time between colonization detection and death was defined to be 0 days. The numbers of animals protected was compared using the two-tailed Fisher's exact test. The results for the 2 BI1-infected animals and the 2 BI6-infected control animals from this experiment were each combined with those for the 10 BI1-infected and 10 BI6-infected animals (90% mortality each) from a previous study that were given the same 100-CFU spore dose on the same schedule of clindamycin to create the control group (9).
RESULTS
Protection against challenge with historic toxigenic strain BI1 was similar in hamsters colonized with NTCD M3 or T7 (Fig. 1). By day 3, 9 of 10 M3 hamsters were colonized and 10 of 10 T7 hamsters were colonized. The two controls remained uncolonized to this point, but all the hamsters given NTCD were colonized with their specific strain, either M3 or T7, by day 5. On day 5, all hamsters were challenged with 100 CFU of BI1. On day 6, only the two controls were positive for colonization with BI1. By day 8, both controls died of BI1 toxicity, and all the nontoxigenic colonized hamsters survived until the end of the study at day 36.
Fig 1.
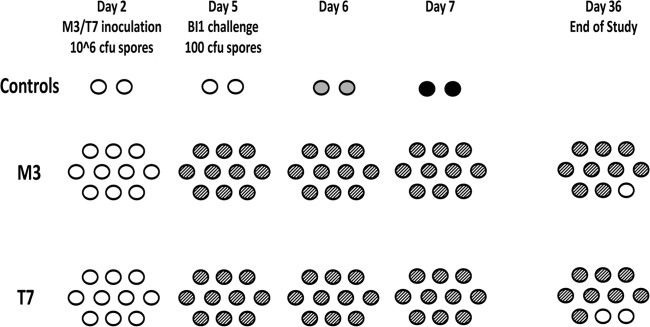
Hamsters (n = 10/group) challenged with nonepidemic historic toxigenic BI1 Clostridium difficile. Day 2, 2 days after clindamycin treatment; white ovals, uncolonized hamster; gray ovals, toxigenic colonized hamster; striped ovals, nontoxigenic colonized hamsters; black ovals, dead hamsters.
The results of challenge of hamsters colonized with M3 with toxigenic epidemic strain BI6 are shown in Fig. 2. By day 5, 9 of 10 hamsters demonstrated colonization with M3. On day 6, 1 day following challenge with BI6, the two controls and single uncolonized hamster were shown to be colonized with the epidemic BI6 strain. On day 7, all the hamsters colonized with BI6 died. The remaining 9 NTCD M3-colonized hamsters survived until the end of the study on day 36.
Fig 2.
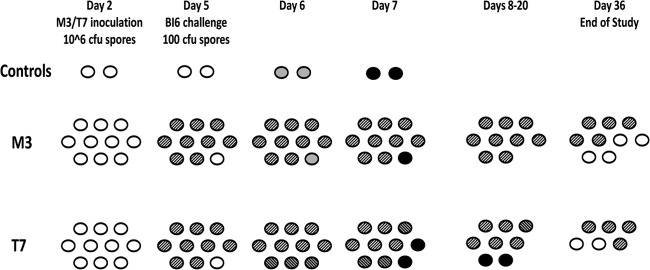
Hamsters (n = 10/group) challenged with epidemic toxigenic BI6 Clostridium difficile. Day 2, 2 days after clindamycin treatment; white ovals, uncolonized hamster; gray ovals, toxigenic colonized hamster; striped ovals, nontoxigenic colonized hamsters; gray striped ovals, cocolonized hamsters; black ovals, dead hamsters.
The results of BI6 challenge of hamsters given nontoxigenic strain T7 are also shown in Fig. 2. Ten hamsters were inoculated with T7 on day 2, and by day 4, 9 of 10 were culture positive for T7. On day 5, these hamsters were challenged with BI6. All 10 were culture positive for T7 by day 6, but 3 hamsters showed cocolonization with BI6 on that day. By day 7, a total of 5 hamsters were cocolonized with T7 and BI6. Two of the cocolonized hamsters died on day 7, one died on day 13, and one died on day 15. The last cocolonized hamster survived, losing BI6 colonization on day 21, and was euthanized with the remaining five of the T7-colonized hamsters at the end of the experiment (Fig. 2).
The administration of T7 followed by BI6 challenge resulted in the death of 4/10 hamsters. There was a statistically significant difference in both the time from challenge with BI6 to death (8.8 days versus 1.9 days, T7-colonized/BI6-challenged hamsters versus controls, P = 0.006) and the time from colonization with BI6 to death (7.3 days versus 0.8 days, T7-colonized/BI6-challenged hamsters versus controls, P = 0.002) in hamsters that were given T7 (Fig. 3).
Fig 3.
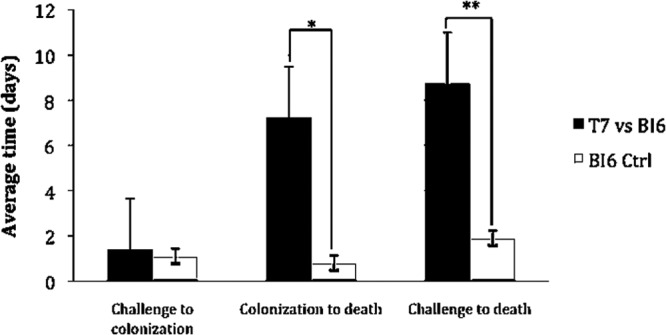
Effect of nontoxigenic strain T7 on time to death of hamsters challenged with epidemic toxigenic strain BI6. *, P = 0.002, time from colonization to death; **, P = 0.006, time from BI6 challenge to death.
Overall, the administration of M3 reduced the incidence of fatal CDI from 92% to 10% in hamsters challenged with BI6 (P < 0.0003) and from 92% to 0% in hamsters challenged with BI1 (P < 0.00003). The administration of T7 reduced the incidence of fatal disease from 92% to 40% in hamsters challenged with BI6 (P < 0.02) and 92% to 0% in hamsters challenged with BI1 (P < 0.00003) (Fig. 4).
Fig 4.
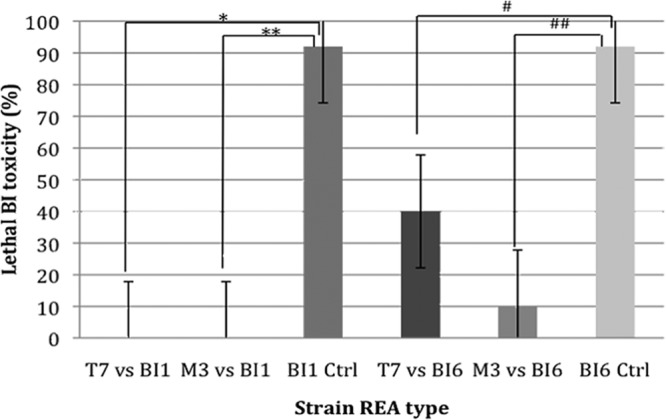
Protection of hamsters from death by nontoxigenic strains M3 and T7 when challenged with toxigenic historic strain BI1 and toxigenic epidemic strain BI6. For each toxigenic strain, a total of 12 control (Ctrl) animals were compared to 10 animals receiving M3 or T7. *, P = 0.00003, T7 versus BI1; **, P = 0.00003, M3 versus BI1; #, P = 0.02, T7 versus BI6; ##, P = 0.0003, M3 versus BI6.
DISCUSSION
Epidemic BI organisms have been found to produce 16 times more toxin A and 23 times more toxin B than nonepidemic C. difficile isolates from the same institutions (13). They contain a deletion and frameshift in the tcdC gene, a putative downregulator of toxin production, and carry a third toxin known as binary toxin. Of clinical importance is the fact that epidemic BI strains are highly resistant to newer fluoroquinolones, such as gatifloxacin and moxifloxacin (2, 14). There are 29 subtypes of BI strains recognized by REA typing. Most have been found in Canada and the United States but have also been cultured from multiple international sites in the United Kingdom, the European Union, and, more recently, Asia and Australia (15–18).
Historic strains of the BI group are BI1 through BI5; they are nonepidemic and were found only in individual patients from 1984 to 1994. The strains from only 8 patients in Minnesota (19), 1 in New York, and 4 in Arizona were typed as being of one of the five historic BI types (2). These CDI cases were sporadic and did not exhibit unique clinical severity. However, the historic BI isolates are toxinotype III and binary toxin positive and have the tcdC frameshift and deletion. They differ from the epidemic strains in that they are not resistant to the newer fluoroquinolones (2).
Given the above-described major similarities between historic and epidemic BI group isolates, it is somewhat surprising to note the difference in protection afforded by nontoxigenic strain T7 against challenge with the historic type BI1 strain and the epidemic type BI6 strain. However, genetic comparisons of historic and epidemic BI/NAP1/027 strains have demonstrated five unique genetic regions in the epidemic BI/NAP1/027 strain not present in historic BI/NAP1/027 strains or in non-027 strain 630 (20, 21). Nontoxigenic strain M3 was fully protective in colonized animals when they were challenged with toxigenic types BI1 and BI6 (92% protective against BI6 overall), but type T7 was fully protective only against challenge with the historic BI1 strain and was only 60% protective against challenge with the epidemic BI6 strain. The 60% protection afforded a statistically significant reduction in mortality compared with that for the controls (P = 0.02). It was not statistically significantly inferior to the 100% protection afforded by M3 against BI6 in colonized hamsters (P = 0.087). This trend toward a difference in protection between NTCD strains is the first that we have seen to date and suggests that all NTCD strains may not be equally protective or durable, as suggested by earlier studies by Wilson and Sheagren (22) and Borriello and Barclay (23).
Of note, 5 of 10 animals were cocolonized with T7 and BI6, and 4 of these went on to succumb to BI6 infection. This indicates that despite the benefit of a much higher inoculum (106 versus 102) and a 3-day lead time, BI6 was able to overcome the protection offered by T7 in some of the hamsters. This is the highest frequency of cocolonization and dominance of a toxigenic strain over an NTCD strain that we have seen in hamster studies to date and suggests the possibility of enhanced virulence by epidemic BI strains compared to conventional strains. T7 offered some protection to these cocolonized animals, as evidenced by the marked increase in the time from colonization to death and challenge to death and the survival of one of the hamsters.
Both M3 and T7 have undergone extensive PCR testing for toxins A and B and binary toxin, and no PCR products have been obtained. They are also phenotypically toxin negative by cell cytotoxin testing, enzyme immunoassay, and ileal loop assay. Thus, their ability to cause clinical illness in hamsters or humans appears to be absent; however, the mechanism by which colonization is protective has not been determined.
Preliminary in vitro assays for adherence to Caco-2 human intestinal epithelial cells indicate that M3 and T7 have greater adherence to this cell line than many toxigenic strains of C. difficile, including BI1 and BI6, suggesting one possible mechanism of protection (24). However, it has also been shown that epidemic strains of C. difficile associated with increased human mortality, such as REA group BI (including BI6) and PCR ribotype 078 (REA group BK), possess a third toxin, binary toxin CDT, which has been shown in vitro on Caco-2 cells and in vivo in mice to increase C. difficile adherence through the formation of microtubule-based protrusions on the cell surface (25). This enhanced cellular adherence could be one possible explanation for how these binary toxin-positive strains are able to compete favorably with NTCD strains; however, it does not explain the differences found in this study between BI1 and BI6, both of which are binary toxin positive.
Colonization with nontoxigenic C. difficile, particularly REA type M3, is highly effective in preventing CDI in hamsters caused by historic and epidemic strains of BI/NAP1/027 C. difficile. Previous hamster studies have also shown it to be effective against epidemic C. difficile strains J9, B1, and K14, which lack binary toxin but which have been responsible for hospital outbreaks of CDI (8, 26). As with previous hamster studies using NTCD, this study utilized a high inoculum of 106 spores and a low inoculum of 102 spores of toxigenic C. difficile, the minimum found to consistently infect hamsters (8, 9, 26). Human trials to date have also used high doses of up to 108 spores twice a day. (7) This is a novel approach with the potential to prevent the development of CDI caused by both the newer epidemic C. difficile BI group strains and other C. difficile strains in humans. NTCD strain M3 has undergone phase 1 human safety testing and has completed successful phase 2 clinical evaluation for prevention of recurrence in patients treated for CDI, and preliminary results have been reported in a press release (7, 27). Efficacy in preventing primary and recurrent CDI in patients warrants continued clinical evaluation.
ACKNOWLEDGMENTS
K.J.N., S.T.P., A.K.C., S.P.S., and W.E.Z. have no conflicts of interest to report. S.J. has served as a consultant for Optimer, Pfizer, and Bio-K+. D.N.G. holds patents for the treatment and prevention of CDI licensed to ViroPharma, is a consultant for ViroPharma, Merck, Optimer, Cubist, GlaxoSmithKline, BioRelix, Novartis, Medicines Co., Cangene, and Actelion, and holds research grants from the U.S. Department of Veterans Affairs, the U.S. Centers for Disease Control and Prevention, GOJO, and Sanofi Pasteur.
This work was supported by grants from the U.S. Department of Veterans Affairs Research Service to D.N.G. and S.J.
Footnotes
Published ahead of print 12 August 2013
REFERENCES
- 1.Loo VG, Poirier L, Miller MA, Oughton M, Libman MD, Michaud S, Bourgault AM, Nguyen T, Frenette C, Kelly M, Vibien A, Brassard P, Fenn S, Dewar K, Hudson TJ, Horn R, René P, Monczak Y, Dascal A. 2005. A predominantly clonal multi-institutional outbreak of Clostridium difficile-associated diarrhea with high morbidity and mortality. N. Engl. J. Med. 353:2442–2449 [DOI] [PubMed] [Google Scholar]
- 2.McDonald LC, Killgore GE, Thompson A, Owens RC, Kazakova SV, Sambol SP, Johnson S, Gerding DN. 2005. An epidemic, toxin gene-variant strain of Clostridium difficile. N. Engl. J. Med. 353:2433–2441 [DOI] [PubMed] [Google Scholar]
- 3.Pépin J, Valiquette L, Cossette B. 2005. Mortality attributable to nosocomial Clostridium difficile-associated disease during an epidemic caused by a hypervirulent strain in Quebec. Can. Med. Assoc. J. 173:1037–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petrella LA, Sambol SP, Cheknis A, Nagaro K, Kean Y, Sears PS, Babakhani F, Johnson S, Gerding DN. 2012. Decreased cure and increased recurrence rate for Clostridium difficile infection caused by the epidemic C. difficile BI strain. Clin. Infect. Dis. 55:351–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shim JK, Johnson S, Samore MH, Bliss DZ, Gerding DN. 1998. Primary symptomless colonisation by Clostridium difficile and decreased risk of subsequent diarrhoea. Lancet 351:633–636 [DOI] [PubMed] [Google Scholar]
- 6.Seal D, Borriello SP, Barclay FE, Welch A, Piper M, Bonnycastle M. 1987. Treatment of relapsing Clostridium difficile diarrhoea by administration of a non-toxigenic strain. Eur. J. Clin. Microbiol. 6:51–53 [DOI] [PubMed] [Google Scholar]
- 7.Villano SA, Seiberling M, Tatarowicz W, Monnot-Chase E, Gerding DN. 2012. Evaluation of an oral suspension of VP20621, spores of nontoxigenic Clostridium difficile strain M3, in healthy subjects. Antimicrob. Agents Chemother. 56:5224–5229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sambol SP, Merrigan MM, Tang JK, Johnson S, Gerding DN. 2002. Colonization for the prevention of Clostridium difficile disease in hamsters. J. Infect. Dis. 186:1781–1789 [DOI] [PubMed] [Google Scholar]
- 9.Razaq N, Sambol S, Nagaro K, Zukowski W, Cheknis A, Johnson S, Gerding DN. 2007. Infection of hamsters with historical and epidemic BI types of Clostridium difficile. J. Infect. Dis. 196:1813–1819 [DOI] [PubMed] [Google Scholar]
- 10.Nagaro KJ, Cheknis AK, Sambol SP, Zukowski WE, Gerding DN. 2006. Non-toxigenic Clostridium difficile (CD) protects hamsters against historic and epidemic toxigenic BI strains. Abstr. Fifth Int. Meet. Mol. Biol. Pathogenesis Clostridia, Nottingham, United Kingdom [Google Scholar]
- 11.Wilson KJ, Kennedy MJ, Fekety FR. 1982. Use of sodium taurocholate to enhance spore recovery on a medium selective for Clostridium difficile. J. Clin. Microbiol. 15:443–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clabots CR, Johnson S, Bettin K, Mathie PA, Mulligan ME, Schaberg DR, Peterson LR, Gerding DN. 1993. Development of a rapid and efficient restriction endonuclease analysis typing system for Clostridium difficile and correlation with other typing systems. J. Clin. Microbiol. 31:1870–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Warny M, Pépin J, Fang A, Killgore G, Thompson A, Brazier J, Frost E, McDonald LC. 2005. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet 366:1079–1084 [DOI] [PubMed] [Google Scholar]
- 14.MacCannell DR, Louie TJ, Gregson DB, Laverdiere M, Labbe AC, Laing F, Henwick S. 2006. Molecular analysis of Clostridium difficile PCR ribotype 027 isolates from eastern and western Canada. J. Clin. Microbiol. 44:2147–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lim PL, Ling ML, Lee HY, Koh TH, Tan AL, Kuijper EJ, Goh SS, Low BS, Ang LP, Harmanus C, Lin RT, Krishnan P, James L, Lee CE, Lim PL, Ling ML, Lee HY, Koh TH, Tan AL, Kuijper EJ, Goh SS, Low BS, Ang LP, Harmanus C, Lin RT, Krishnan P, James L, Lee CE. 2011. Isolation of the first three cases of Clostridium difficile polymerase chain reaction ribotype 027 in Singapore. Singapore Med. J. 52:361–364 [PubMed] [Google Scholar]
- 16.Kuijper EJ, Barbut F, Brazier JS, Kleinkauf N, Eckmanns T, Lambert ML, Drudy D, Fitzpatrick F, Wiuff C, Brown DJ, Coia JE, Pituch H, Reichert P, Even J, Mossong J, Widmer AF, Olsen KE, Allerberger F, Notermans DW, Delmée M, Coignard B, Wilcox M, Patel B, Frei R, Nagy E, Bouza E, Marin M, Akerlund T, Virolainen-Julkunen A, Lyytikäinen O, Kotila S, Ingebretsen A, Smyth B, Rooney P, Poxton IR, Monnet DL. 2008. Update of Clostridium difficile infection due to PCR ribotype 027 in Europe, 2008. Euro Surveill. 13(31):pii=18942 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=18942 [PubMed] [Google Scholar]
- 17.Richards M, Knox J, Elliott B, Mackin K, Lyras D, Waring LJ, Riley TV. 2011. Severe infection with Clostridium difficile PCR ribotype 027 acquired in Melbourne, Australia. Med. J. Aust. 194:369–371 [DOI] [PubMed] [Google Scholar]
- 18.Clements AC, Magalhães RJ, Tatem AJ, Paterson DL, Riley TV. 2010. Clostridium difficile PCR ribotype 027: assessing the risks of further worldwide spread. Lancet Infect. Dis. 10:395–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Belmares J, Johnson S, Parada JP, Olson MM, Clabots CR, Bettin KM, Peterson LR, Gerding DN. 2009. Molecular epidemiology of Clostridium difficile over the course of 10 years in a tertiary care hospital. Clin. Infect. Dis. 49:1141–1147 [DOI] [PubMed] [Google Scholar]
- 20.Stabler RA, Gerding DN, Songer JG, Drudy D, Brazier JS, Trinh HT, Witney AA, Hinds J, Wren BW. 2006. Comparative phylogenomics of Clostridium difficile reveals clade specificity and microevolution of hypervirulent strains. J. Bacteriol. 188:7297–7305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stabler RA, He M, Dawson L, Martin M, Valiente E, Corton C, Lawley TD, Sebaihia M, Quail MA, Rose G, Gerding DN, Gibert M, Popoff MR, Parkhill J, Dougan G, Wren BW. 2009. Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. Genome Biol. 10:R102. 10.1186/gb-2009-10-9-r102 Epub 2009 Sep 25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson KH, Sheagren JN. 1983. Antagonism of toxigenic Clostridium difficile by nontoxigenic C. difficile. J. Infect. Dis. 147:733–736 [DOI] [PubMed] [Google Scholar]
- 23.Borriello SP, Barclay FE. 1985. Protection of hamsters against Clostridium difficile ileocaecitis by prior colonisation with non-pathogenic strains. J. Med. Microbiol. 19:339–350 [DOI] [PubMed] [Google Scholar]
- 24.Vedantam G, Merrigan MM, Gerding DN. 2006. Hypervirulent epidemic strains of Clostridium difficile have altered host cell adherence and protein expression. Abstr. Fifth Int. Meet. Mol. Biol. Pathogenesis Clostridia, Nottingham, United Kingdom [Google Scholar]
- 25.Schwan C, Stecher B, Tzivelekidis T, van Ham M, Rohde M, Hardt W-D, Wehland J, Aktories K. 2009. Clostridium difficile toxin CDT induces formation of microtubule-based protrusions and increases adherence of bacteria. PLoS Pathog. 5:e1000626. 10.1371/journal.ppat.1000626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambol SP, Tang JK, Merrigan MM, Johnson S, Gerding DN. 2001. Infection of hamsters with epidemiologically important strains of Clostridium difficile. J. Infect. Dis. 183:1760–1766 [DOI] [PubMed] [Google Scholar]
- 27.ViroPharma 2013. Treatment with VP20621 (non-toxigenic Clostridium difficile; NTCD) in a phase 2 study resulted in high rates of colonization and statistically significant reductions in recurrence of C. difficile infection. Press release. ViroPharma, Exton, PA [Google Scholar]


