Abstract
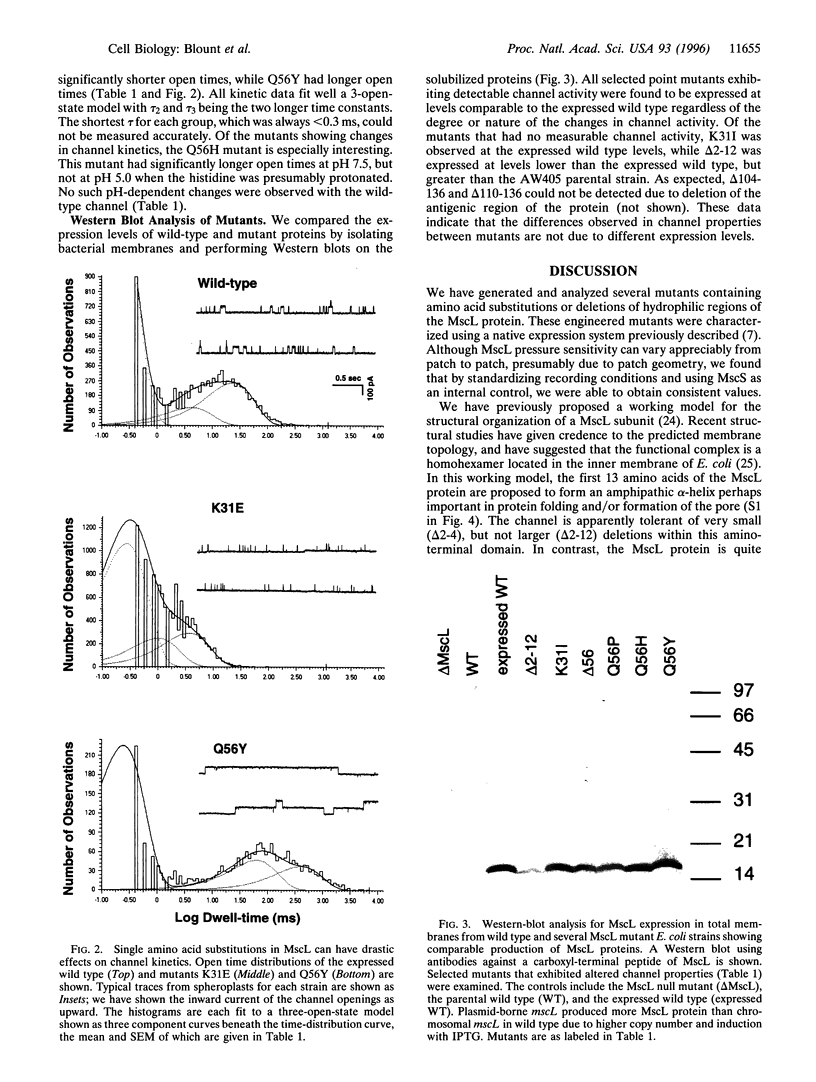
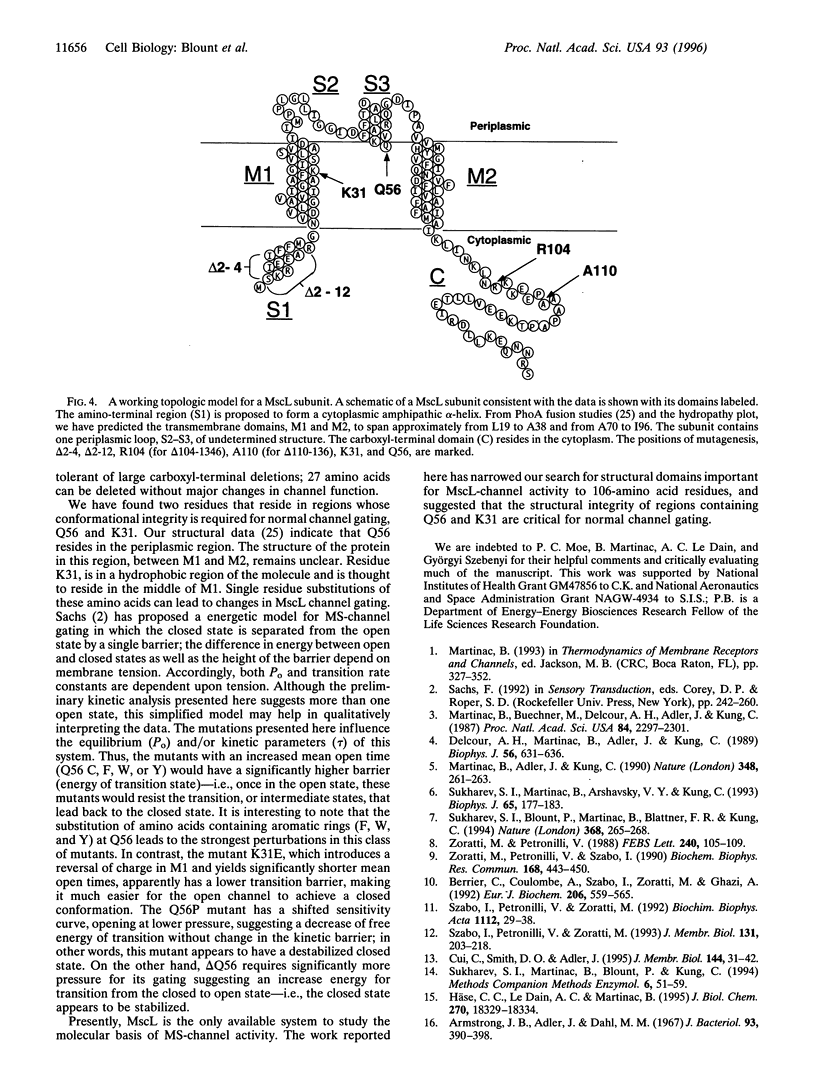
MscL is a channel that opens a large pore in the Escherichia coli cytoplasmic membrane in response to mechanical stress. Previously, we highly enriched the MscL protein by using patch clamp as a functional assay and cloned the corresponding gene. The predicted protein contains a largely hydrophobic core spanning two-thirds of the molecule and a more hydrophilic carboxyl terminal tail. Because MscL had no homology to characterized proteins, it was impossible to predict functional regions of the protein by simple inspection. Here, by mutagenesis, we have searched for functionally important regions of this molecule. We show that a short deletion from the amino terminus (3 amino acids), and a larger deletion of 27 amino acids from the carboxyl terminus of this protein, had little if any effect in channel properties. We have thus narrowed the search of the core mechanosensitive mechanism to 106 residues of this 136-amino acid protein. In contrast, single residue substitutions of a lysine in the putative first transmembrane domain or a glutamine in the periplasmic loop caused pronounced shifts in the mechano-sensitivity curves and/or large changes in the kinetics of channel gating, suggesting that the conformational structure in these regions is critical for normal mechanosensitive channel gating.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Armstrong J. B., Adler J., Dahl M. M. Nonchemotactic mutants of Escherichia coli. J Bacteriol. 1967 Jan;93(1):390–398. doi: 10.1128/jb.93.1.390-398.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrier C., Coulombe A., Szabo I., Zoratti M., Ghazi A. Gadolinium ion inhibits loss of metabolites induced by osmotic shock and large stretch-activated channels in bacteria. Eur J Biochem. 1992 Jun 1;206(2):559–565. doi: 10.1111/j.1432-1033.1992.tb16960.x. [DOI] [PubMed] [Google Scholar]
- Blount P., Merlie J. P. Mutational analysis of muscle nicotinic acetylcholine receptor subunit assembly. J Cell Biol. 1990 Dec;111(6 Pt 1):2613–2622. doi: 10.1083/jcb.111.6.2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke D. T., Olson M. V. Oligodeoxynucleotide-directed mutagenesis of Escherichia coli and yeast by simple cotransformation of the primer and template. DNA. 1986 Aug;5(4):325–332. doi: 10.1089/dna.1986.5.325. [DOI] [PubMed] [Google Scholar]
- Cui C., Smith D. O., Adler J. Characterization of mechanosensitive channels in Escherichia coli cytoplasmic membrane by whole-cell patch clamp recording. J Membr Biol. 1995 Mar;144(1):31–42. doi: 10.1007/BF00238414. [DOI] [PubMed] [Google Scholar]
- Delcour A. H., Martinac B., Adler J., Kung C. Modified reconstitution method used in patch-clamp studies of Escherichia coli ion channels. Biophys J. 1989 Sep;56(3):631–636. doi: 10.1016/S0006-3495(89)82710-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häse C. C., Le Dain A. C., Martinac B. Purification and functional reconstitution of the recombinant large mechanosensitive ion channel (MscL) of Escherichia coli. J Biol Chem. 1995 Aug 4;270(31):18329–18334. doi: 10.1074/jbc.270.31.18329. [DOI] [PubMed] [Google Scholar]
- Ingham C., Buechner M., Adler J. Effect of outer membrane permeability on chemotaxis in Escherichia coli. J Bacteriol. 1990 Jul;172(7):3577–3583. doi: 10.1128/jb.172.7.3577-3583.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinac B., Adler J., Kung C. Mechanosensitive ion channels of E. coli activated by amphipaths. Nature. 1990 Nov 15;348(6298):261–263. doi: 10.1038/348261a0. [DOI] [PubMed] [Google Scholar]
- Martinac B., Buechner M., Delcour A. H., Adler J., Kung C. Pressure-sensitive ion channel in Escherichia coli. Proc Natl Acad Sci U S A. 1987 Apr;84(8):2297–2301. doi: 10.1073/pnas.84.8.2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthe H. J., Adler J. Fusion of bacterial spheroplasts by electric fields. Biochim Biophys Acta. 1985 Sep 25;819(1):105–113. doi: 10.1016/0005-2736(85)90200-7. [DOI] [PubMed] [Google Scholar]
- Sukharev S. I., Blount P., Martinac B., Blattner F. R., Kung C. A large-conductance mechanosensitive channel in E. coli encoded by mscL alone. Nature. 1994 Mar 17;368(6468):265–268. doi: 10.1038/368265a0. [DOI] [PubMed] [Google Scholar]
- Sukharev S. I., Martinac B., Arshavsky V. Y., Kung C. Two types of mechanosensitive channels in the Escherichia coli cell envelope: solubilization and functional reconstitution. Biophys J. 1993 Jul;65(1):177–183. doi: 10.1016/S0006-3495(93)81044-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabó I., Petronilli V., Zoratti M. A patch-clamp investigation of the Streptococcus faecalis cell membrane. J Membr Biol. 1993 Feb;131(3):203–218. doi: 10.1007/BF02260109. [DOI] [PubMed] [Google Scholar]
- Szabó I., Petronilli V., Zoratti M. A patch-clamp study of Bacillus subtilis. Biochim Biophys Acta. 1992 Nov 23;1112(1):29–38. doi: 10.1016/0005-2736(92)90250-p. [DOI] [PubMed] [Google Scholar]
- Zoratti M., Petronilli V. Ion-conducting channels in a gram-positive bacterium. FEBS Lett. 1988 Nov 21;240(1-2):105–109. doi: 10.1016/0014-5793(88)80348-x. [DOI] [PubMed] [Google Scholar]
- Zoratti M., Petronilli V., Szabo I. Stretch-activated composite ion channels in Bacillus subtilis. Biochem Biophys Res Commun. 1990 Apr 30;168(2):443–450. doi: 10.1016/0006-291x(90)92341-v. [DOI] [PubMed] [Google Scholar]




