Abstract
T-705 (favipiravir; 6-fluoro-3-hydroxy-2-pyrazinecarboxamide) selectively and strongly inhibits replication of the influenza virus in vitro and in vivo. T-705 has been shown to be converted to T-705-4-ribofuranosyl-5-triphosphate (T-705RTP) by intracellular enzymes and then functions as a nucleotide analog to selectively inhibit RNA-dependent RNA polymerase (RdRp) of the influenza virus. To elucidate these inhibitory mechanisms, we analyzed the enzyme kinetics of inhibition using Lineweaver-Burk plots of four natural nucleoside triphosphates and conducted polyacrylamide gel electrophoresis of the primer extension products initiated from 32P-radiolabeled 5′Cap1 RNA. Enzyme kinetic analysis demonstrated that T-705RTP inhibited the incorporation of ATP and GTP in a competitive manner, which suggests that T-705RTP is recognized as a purine nucleotide by influenza virus RdRp and inhibited the incorporation of UTP and CTP in noncompetitive and mixed-type manners, respectively. Primer extension analysis demonstrated that a single molecule of T-705RTP was incorporated into the nascent RNA strand of the influenza virus and inhibited the subsequent incorporation of nucleotides. These results suggest that a single molecule of T-705RTP is incorporated into the nascent RNA strand as a purine nucleotide analog and inhibits strand extension, even though the natural ribose of T-705RTP has a 3′-OH group, which is essential for forming a covalent bond with the phosphate group.
INTRODUCTION
Influenza viruses have claimed many lives since ancient times and remain a serious threat to mankind. Influenza viruses enter the body, bind to the sialic acid of glycoproteins on the cell surface, and are then endocytosed into the cell, where they release virus RNA and RNA-bound protein complexes (RNP) into the cytoplasm (uncoating). RNP released into the cytoplasm enter the nucleus and are transcribed and replicated by the RNA-dependent RNA polymerase (RdRp) of the influenza virus. Subsequently, translated virus proteins, i.e., hemagglutinin, neuraminidase, and M2 proteins, migrate with the nascent RNP to the cell surface. Virus particles are then constructed and released from the cell membrane by neuraminidase. Conventional anti-influenza virus drugs target any one of the above proliferation processes of influenza viruses. For example, M2 channel inhibitors (e.g., amantadine), which inhibit virus uncoating and neuraminidase inhibitors (e.g., oseltamivir and zanamivir), which inhibit virus release, are used as anti-influenza virus agents (1). However, resistant viruses have already been reported for neuraminidase inhibitors and amantadine (2, 3). Thus, novel anti-influenza virus agents with different mechanisms of action need to be developed. T-705 (favipiravir; 6-fluoro-3-hydroxy-2-pyrazinecarboxamide) is a novel anti-influenza virus agent developed by Toyama Chemical Co., Ltd. (Fig. 1) (4–6). T-705 was shown to have different characteristics from amantadine and neuraminidase inhibitors. T-705 strongly inhibits the replication of influenza virus types A, B, and C and has broad-spectrum activities against various RNA viruses, such as arenaviridae, bunyaviridae, flaviviridae, picornaviridae, and paramyxoviridae (4, 7–11). T-705 is currently undergoing phase II clinical trials for the treatment of influenza virus infections in the United States. In addition, T-705 is less likely to allow virus regrowth after agent removal than a neuraminidase inhibitor. The combination therapy with a neuraminidase inhibitor has synergistic effects (6, 12, 13). Furthermore, no resistant virus developed until 30 passages were completed in the presence of various concentrations of T-705, which indicates that T-705 is unlikely to cause resistant viruses (4; unpublished data). The above favorable properties specific to T-705 may result from its mechanism of action. We previously demonstrated that the in vitro anti-influenza virus activity of T-705 was decreased in the presence of purine nucleic acids, suggesting that T-705 may induce antivirus activity as a nucleobase analog (5). In addition, we demonstrated that T-705 was converted to T-705-4-ribofuranosyl-5-monophosphate (T-705RMP) and then to T-705-4-ribofuranosyl-5-triphosphate (T-705RTP; Fig. 1) by intracellular enzymes and then selectively inhibited the RdRp of the influenza virus (5, 14). Influenza virus RdRp, which consists of three subunits (PA, PB1, and PB2) transcribes and replicates virus RNA in the nuclei of infected cells. Its transcription includes a specific process called cap snatching. Specifically, 9- to 15-nucleotide (nt) fragments containing the 5′Cap1 structure of host mRNA are excised by the endonuclease function of influenza virus RdRp to serve as transcription primers. On the other hand, its replication requires no primer. Progeny vRNA is synthesized through cRNA (15). To elucidate the inhibitory mechanisms of T-705RTP, which is the active form of T-705, we herein analyzed enzyme kinetics and conducted primer extension analysis using an mRNA fragment with an artificial 5′Cap1 structure, instead of host mRNA.
Fig 1.
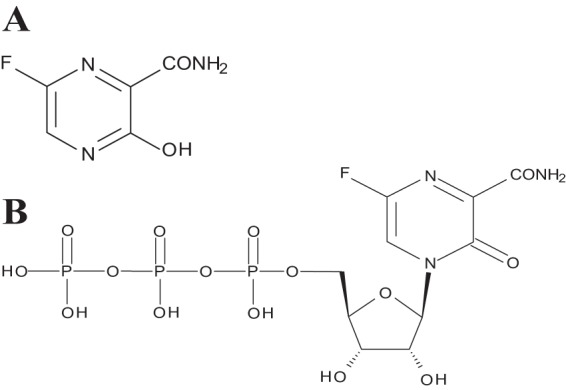
Chemical structures of T-705 (A) and T-705RTP (B). (A) Prodrug; (B) active form.
MATERIALS AND METHODS
Compound.
T-705RTP (6-fluoro-3,4-dihydro-4-[5-O-(hydroxy{[hydroxy(phosphonooxy)phosphinyl]oxy}phosphinyl)-β-d-ribofuranosyl]-3-oxo-2-pyrazinecarboxamide) was synthesized at Toyama Chemical Co., Ltd. (Toyama, Japan) (Fig. 1). 2FdGTP (2-deoxy-2-fluoroguanosine-5-triphosphate) was purchased from TriLink Biotechnologies (San Diego, CA).
Virus.
Influenza virus strain A/PR/8/34 (H1N1) was kindly provided by Kimiyasu Shiraki, Department of Virology, Graduate School of Medicine and Pharmaceutical Sciences, University of Toyama, Toyama, Japan.
Cells.
Madin-Darby canine kidney (MDCK) cells were purchased by Dainippon Sumitomo Pharma Co., Ltd. (Osaka, Japan), maintained in Eagle minimum essential medium (EMEM; Sigma Chemical Co., St. Louis, MO) supplemented with 10% fetal bovine serum and 60 μg of kanamycin/ml, and then cultured at 37°C in 5% CO2.
Preparation of the crude influenza virus RdRp.
MDCK cells were seeded into 100-mm plates at 3 × 106 cells per dish and incubated overnight at 37°C in 5% CO2. Cells were infected with influenza virus (multiplicity of infection = 0.001) in 7 ml/dish of medium containing EMEM supplemented with 1% bovine serum albumin, 3% vitamin (Life Technologies, Carlsbad, CA), and 3 μg of TPCK (tolylsulfonyl phenylalanyl chloromethyl ketone) trypsin/ml and were cultured at 35°C in 5% CO2 for 2 days. Culture supernatant (∼300 ml) was harvested by ultracentrifugation (50,860 × g at 4°C for 2.5 h) and stored in 1 to 2 ml of the storage buffer containing 10 mM Tris-HCl (pH 7.5), 1 mM EDTA, and 100 mM NaCl at −80°C until use. Isolated virions were disrupted by exposure to the detergent (200 mM Tris-HCl [pH 8.0], 200 mM KCl, 10 mM MgCl2, 3 mM dithiothreitol [DTT], 10% [wt/vol] glycerol, 3% [vol/vol] Triton N-101, and 2% [wt/vol] LPC) (5) and were used as the crude influenza virus RdRp containing the viral RNA genome.
Measurement of ApG-primed RNA polymerase activity.
In all experiments, 2 μl (0.1 μg) of the crude influenza virus RdRp was incubated at 30°C for 1 h in 30 μl of the transcription buffer containing 100 mM Tris-HCl (pH 8.0), 100 mM KCl, 5 mM MgCl2, 1 mM DTT, 4 μg of tRNA/ml, and 0.25 (vol/vol)% Triton N-101. The crude influenza virus RdRp was added with 0.25 mM dinucleotide ApG, 100 μM ATP, 50 μM CTP, 50 μM UTP, 1 μM GTP, 0.028 μM (2.5 μCi) [α-32P]GTP (3,000 Ci/mmol; Perkin-Elmer, Inc., Waltham, MA), and T-705RTP in the GTP incorporation assay (5). In the UTP incorporation assay, the crude influenza virus RdRp was added with 0.25 mM dinucleotide ApG, 100 μM ATP, 50 μM CTP, 50 μM GTP, 0.75 μM UTP, and 0.25 μM [5,6-3H]UTP (35 Ci/mmol; Perkin-Elmer). The reaction mixture was filtrated on a DE81 filter (Whatman Japan, Ltd., Tokyo, Japan) soaked in 500 mM EDTA. The filters were dried, washed three times for 10 min with 5% Na2HPO4 (pH 10.4), and rinsed twice for 5 min with distilled water and once with methanol. The filters were air dried and measured for radioactivity in 10 ml of Ultima Gold (Perkin-Elmer) with a Tri-Carb 3110TR liquid scintillation counter (Perkin-Elmer). The 50% inhibitory concentration (IC50) was calculated by the logistic curve fitting using SAS analytical software (release 8.2; SAS Institute Japan, Ltd., Tokyo, Japan). Kinetic analyses of ATP, CTP, GTP, and UTP were performed under the same conditions as described above. The concentrations of the nucleotides were shown in Table 1. The data were fitted to Lineweaver-Burk plots for kinetic analyses. All results were expressed as means ± the standard deviations (SD) of triplicate experiments.
Table 1.
Kinetic analysis of NTP concentrations
| NTP | NTP concn(s) (μM)a |
|||
|---|---|---|---|---|
| ATP | CTP | GTP | UTP | |
| ATP | 800, 400, 200, 100, 50 | 50 | 50 | 1† |
| CTP | 100 | 100, 50, 25, 12.5, 6.25 | 50 | 1† |
| GTP | 100 | 50 | 100, 50, 25, 12.5, 6.25 | 1† |
| UTP | 100 | 50 | 1* | 100, 50, 25, 12.5, 6.25 |
*, 1 μM GTP and 2.5 μCi of [α-32P]GTP; †, 0.75 μM UTP and 0.25 μM [5,6-3H]UTP.
Preparation of RNA substrates by in vitro transcription.
The synthetic capped mRNA described by Hagen et al. (16) and Chung et al. (17) was used as a primer for studies on the inhibition of initiation and elongation (see Fig. S1 in the supplemental material). RNA transcripts were prepared from SmaI-digested pGEM-7zf(+) DNA (Promega Corp., Madison, WI) by in vitro runoff transcription with SP6 RNA polymerase. After gel purification, the 67-nt transcript was converted to a 32P-radiolabeled cap-0 structure (m7G32pppGAAUACUCAAGCUAUN52) by concurrent capping and methylation reactions in 20 μl of the reaction buffer using ScriptCap m7G Capping System (CellScript, Inc., Madison, WI). The reaction buffer contained ∼15 pmol of the transcript, 0.1 mM SAM (S-adenosylmethionine), 20 U of RNase inhibitor, 10 U of capping enzyme mix (mRNA triphosphatase, guanylyltransferase, and guanine-7-methyltransferase), and 50 μCi of [α-32P]GTP. After incubation for 1 h at 37°C, the RNAs were extracted with phenol-chloroform, spin chromatographed (Sephadex G-25 column) and precipitated by ethanol. The cap-0 transcript was finally converted to a 32P-radiolabeled cap-1 structure (m7G32pppGmAAUACUCAAGCUAUN52) by a methylation reaction using ScriptCap 2′-O-methyltransferase (CellScript). After incubation for 1 h at 37°C, the RNAs were extracted with phenol-chloroform and precipitated by ethanol.
Inhibition of the initiation and elongation of the influenza virus RNA strand.
First, 10 μl of the crude influenza virus RdRp was incubated with the 32P-radiolabeled cap-1 RNA (67 nt) in 60 μl of the transcription buffer containing100 mM Tris-HCl (pH 8.0) 100 mM KCl, 5 mM MgCl2, 1 mM DTT, 4 μg of tRNA/ml, and 0.25% Triton N-101 in the absence or presence of nucleoside triphosphates (NTPs) and T-705RTP. After incubation for 1 h at 30°C, the reaction products were extracted with phenol-chloroform, precipitated by ethanol, and analyzed on 25% polyacrylamide–7 M urea gels, followed by image analysis with a FujiFilm BAS-5000 Bio-Image Analyzer (Fujifilm Corporation, Tokyo, Japan).
RESULTS
Effects of GTP and UTP concentrations on the inhibitory activity of T-705RTP against influenza virus RdRp.
We previously demonstrated that purine bases (including guanine), but not pyrimidine bases (including uracil), inhibited the anti-influenza virus activity of T-705 (5). We investigated here the effects of GTP and UTP concentrations on the inhibitory activity of T-705RTP, the intracellular active compound of T-705 on influenza virus RdRp. All of the experiments were performed in triplicate. In the labeled GTP incorporation assay, the IC50 of T-705RTP was 0.360 μM (95% confidence interval [CI] = 0.274 to 0.472) for the ∼1 μM GTP-containing solution. In the labeled UTP incorporation assay, the IC50 of T-705RTP was 10.5 μM (95% CI = 8.60 to 12.9) for the 50 μM GTP-containing solution. Thus, the inhibitory activity of T-705RTP against influenza virus RdRp was influenced by the type of competing nucleotide, with inhibition versus GTP being much stronger than that seen with UTP (Fig. 2).
Fig 2.
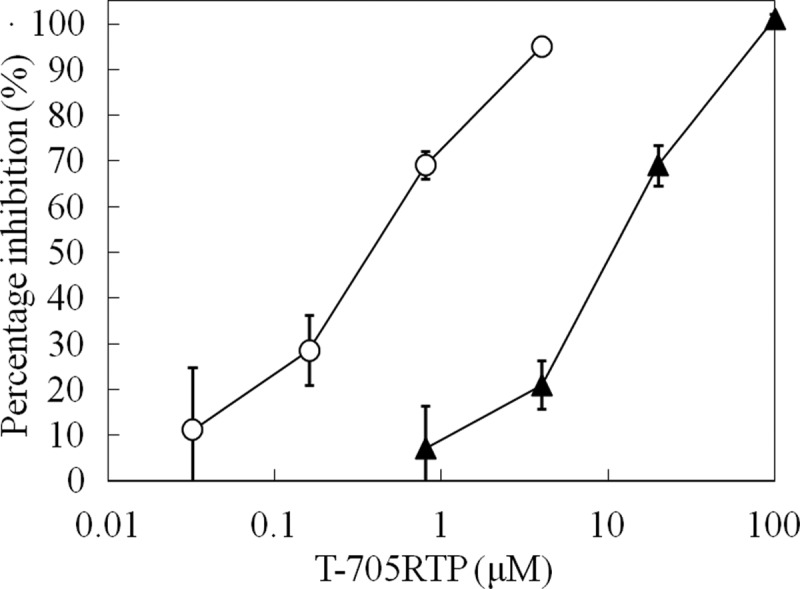
Inhibition of influenza virus RNA polymerase by T-705RTP. The percentage inhibition of influenza virus RNA polymerase at different concentrations of T-705RTP was determined as described in Materials and Methods. All samples were in triplicate, and means ± the SD values are shown. All samples were incubated at 30°C for 1 h and counted in each sample with [α-32P]GTP (○) or [5,6-3H] UTP (▲) as labeled precursors.
Enzyme kinetics of the inhibition of T-705RTP on influenza virus RdRp.
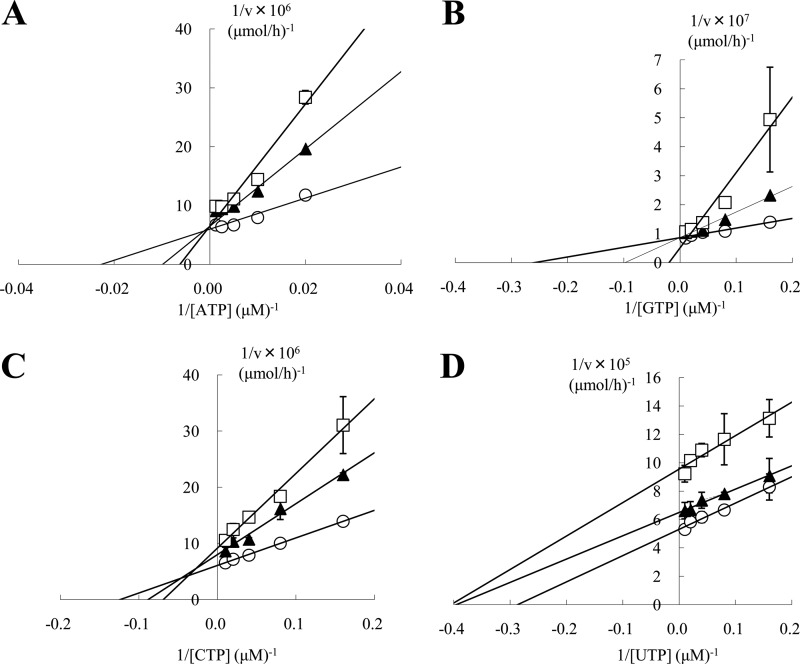
We investigated the inhibitory mechanism of T-705RTP on the activity of influenza virus RdRp. These experiments were conducted by measuring the extension activity of influenza virus RdRp, initiated from an ApG primer. The reaction time was set to 1 h to give significant counts of reaction products, and this period fell within the linear range. The concentrations of the NTP used here were shown in Table 1 and that of T-705RTP were determined on the basis of the value of IC50 calculated from results in Fig. 2. Lineweaver-Burk plots demonstrated that T-705RTP inhibited the incorporation of ATP and GTP in a competitive manner with Ki values of 7.72 and 1.56 μM, respectively (Fig. 3A and B), inhibited the incorporation of CTP in a mixed-type manner with a Ki value of 11.3 μM, and also inhibited the incorporation of UTP in a noncompetitive manner (Fig. 3C and D).
Fig 3.
Inhibitory activity of T-705RTP versus the incorporation of ATP, GTP, CTP, or UTP. The activity of influenza virus RNA polymerase in the presence of T-705RTP was determined at different concentrations of NTPs. The incorporation of [α-32P]GTP was measured when the UTP concentration was varied, and the incorporation of [5,6-3H] UTP was determined when the concentration of ATP, GTP, and CTP was varied. The results are presented as Lineweaver-Burk plots. All samples were incubated at 30°C for 1 h, and the nucleotide concentrations were indicated in Table 1. (A) Incorporation at different ATP concentrations with 0 (○), 5 (▲), and 10 (□) μM T-705RTP. (B) Incorporation at different GTP concentrations with 0 (○), 2.5 (▲), and 5 (□) μM T-705RTP. (C) Incorporation at different CTP concentrations with 0 (○), 5 (▲), and 10 (□) μM T-705RTP. (D) Incorporation at different UTP concentrations with 0 (○), 0.1 (▲), and 0.3 (□) μM T-705RTP. The results represent the means ± the SD of triplicate determinations.
Primer extension analysis of the inhibition of T-705RTP on influenza virus RdRp.
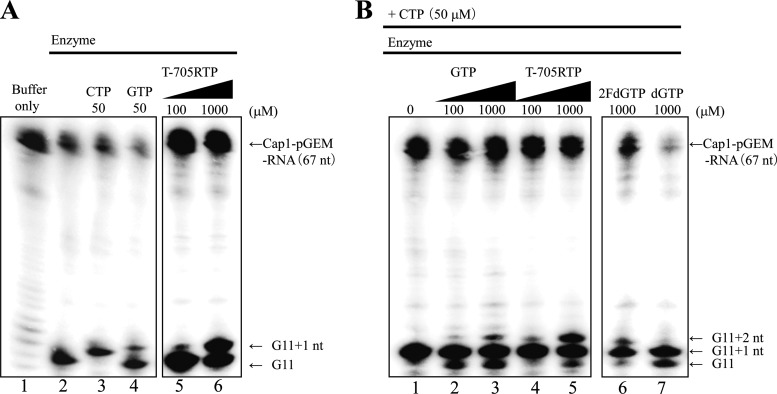
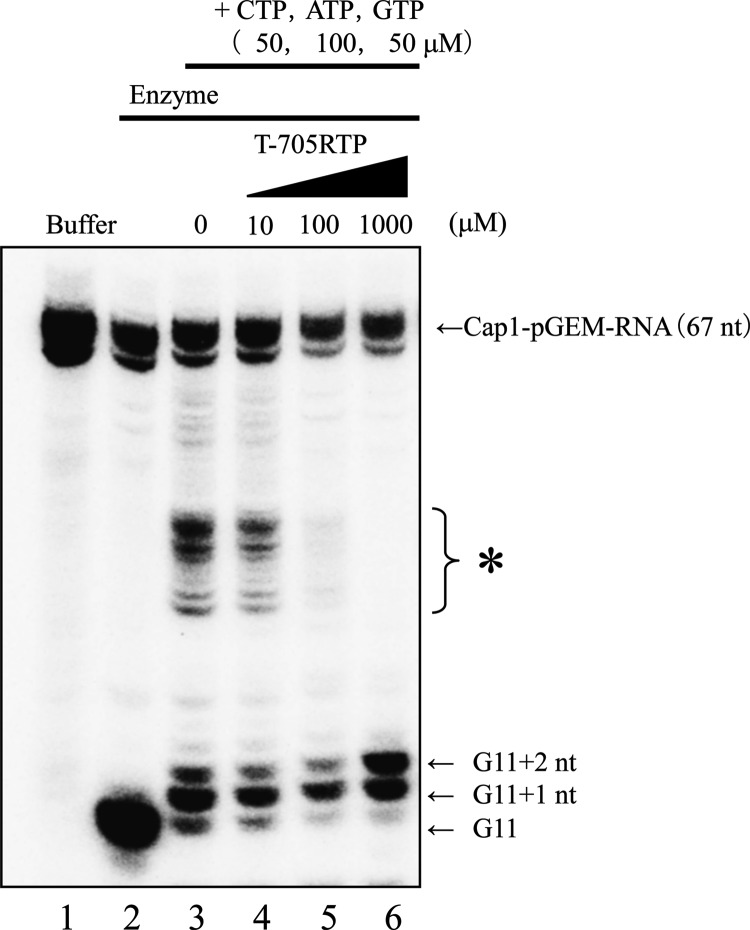
To further characterize the inhibitory mechanism of T-705RTP on influenza virus RdRp, the incorporation of T-705RTP into a nascent RNA strand was investigated using the primer extension method (17). RNA transcripts were prepared by in vitro runoff transcription (see Fig. S1 in the supplemental material). After incubating the runoff cap-1 transcript with the crude influenza virus RdRp, an 11-mer primer (G11 primer) with a 5′cap1 structure was produced (Fig. 4A, lane 2). All primer extension experiments were performed at least three times, and the results of a representative experiment are shown. NTP or T-705RTP was added to the reaction mixture, followed by incubation, to generate primer extension products using the viral RNA genome contained in the crude influenza virus RdRp. Sequences of transcribed RNA fragments from the 3′ end of the 8-segmented influenza virus genome are shown in Table 2. The products were analyzed by using polyacrylamide gel electrophoresis (Fig. 4A and B, lane 6). In the presence of only CTP, a band of G11+1 nt was detected (Fig. 4A, lane 3). In the presence of only GTP, a weak band of G11+1 nt was detected (Fig. 4A, lane 4). In the presence of only T-705RTP, a band of G11+1 nt was also detected, depending on T-705RTP concentrations, whereas a band of G11+2 nt was not detected (Fig. 4A, lanes 5 and 6), indicating that a single molecule of T-705RTP, instead of CTP, was incorporated into the nascent RNA strand. T-705RTP had no effect on G11 primer synthesis, suggesting that T-705RTP does not inhibit the endonuclease activity of influenza virus RNA polymerase (Fig. 4A, lanes 5 and 6). When CTP was added to the reaction mixture in the absence or presence of GTP, bands of G11+1 nt and G11+2 nt were detected, respectively. A band of G11+2 nt was detected depending on the concentration of GTP. These results indicated that GTP was incorporated into the strand following CTP incorporation (Fig. 4B, lanes 1, 2, and 3). When T-705RTP, instead of GTP, was added to the mixture in the presence of CTP, a band of G11+2 nt was detected depending on the concentrations (Fig. 4B, lanes 4 and 5). 2FdGTP, which was previously shown to be a nonobligate chain terminator for influenza virus RdRp, was detected in the same position (Fig. 4B, lane 6) (19). dGTP, which is a substrate of DNA polymerase, was not incorporated (Fig. 4B, lane 7). These results suggested that T-705RTP was incorporated into the strand following the incorporation of CTP, similar to the results in the presence of GTP. When CTP, ATP, and GTP were added to the mixture in the absence of T-705RTP, bands of products, which were complementary to the 3′ ends of eight influenza virus segments and elongated from the G11 primer up to the position prior to the first UTP position of each segment, were detected (Fig. 5, lane 3) (17). When T-705RTP was added to the mixture, the bands of these elongated RNA faded and a band of G11+2 nt accumulated in a high concentration of T-705RTP (Fig. 5 lane 6), similar to the results in the absence of GTP and ATP (Fig. 4B lane 4, 5). A band of G11+2 nt, but not of G11+3 nt, was detected in the presence of T-705RTP (Fig. 5, lane 6). These results suggested that influenza virus RNA strand extension was inhibited by the incorporation of a single molecule of T-705RTP.
Fig 4.
Incorporation of T-705RTP or various nucleotides into a nascent RNA strand. (A) Incorporation of CTP, GTP, and T-705RTP at the position of G11+1. The 32P-labeled pGEM-7zf(+) DNA runoff transcript with a 5′Cap1 structure (Cap1-pGEM-mRNA), crude influenza virus RdRp containing a viral genome, and nucleotides including T-705RTP were incubated. Reaction products were then electrophoresed. Lane 1, Cap1-pGEM-mRNA; lanes 2 to 6, Cap1-pGEM-mRNA + crude enzyme solution; lane 3, conditions of lane 2 + 50 μM CTP; lane 4, conditions of lane 2 + 50 μM GTP; lanes 8 and 9, conditions of lane 2 + 100 or 1,000 μM T-705RTP. (B) Incorporation of GTP, T-705RTP, 2FdGTP, and dGTP at the position of G11+2. The 32P-labeled pGEM-7zf(+) DNA runoff transcript with a 5′Cap1 structure (Cap1-pGEM-mRNA), crude influenza virus RdRp containing a viral genome, and nucleotides including T-705RTP were incubated. Reaction products were then electrophoresed. Lanes 1 to 7, Cap1-pGEM-mRNA + crude enzyme solution + 50 μM CTP; lanes 2 and 3, conditions of lane 1 + 100 or 1,000 μM GTP; lanes 4 and 5, conditions of lane 2 + 100 or 1,000 μM T-705RTP; lane 6, conditions of lane 1 + 1,000 μM 2FdGTP; lane 7, conditions of lane 1 + 1,000 μM dGTP.
Table 2.
Sequences of nascent RNA transcripts transcribed from the 3′ ends of genome segments
| Segment no. | Segment name | Primer | Incorporated nucleotide at position: |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | |||
| 1 | PB2 | G11+ | C | G | A | A | A | G | C | A | G | G | U | |||||||
| 2 | PB1 | G11+ | C | G | A | A | A | G | C | A | G | G | C | A | A | A | C | C | A | U |
| 3 | PA | G11+ | C | G | A | A | A | G | C | A | G | G | U | |||||||
| 4 | HA | G11+ | C | A | A | A | A | G | C | A | G | G | G | G | A | A | A | A | U | |
| 5 | NP | G11+ | C | A | A | A | A | G | C | A | G | G | G | U | ||||||
| 6 | NA | G11+ | C | A | A | A | A | G | C | A | G | G | G | G | U | |||||
| 7 | M | G11+ | C | A | A | A | A | G | C | A | G | G | U | |||||||
| 8 | NS | G11+ | C | A | A | A | A | G | C | A | G | G | G | U | ||||||
Fig 5.
Inhibition of T-705RTP against influenza virus RdRp. The 32P-labeled pGEM-7zf(+) DNA runoff transcript with a 5′Cap1 structure (Cap1-pGEM-mRNA), crude influenza virus RdRp containing a viral genome, and nucleotides including T-705RTP were incubated. Reaction products were then electrophoresed. Lane 1, Cap1-pGEM-mRNA; lanes 2 to 6, Cap1-pGEM-mRNA + crude enzyme solution; lanes 3 to 6, conditions of lane 2 + 50 μM CTP, 100 μM ATP, or 50 μM GTP; lanes 4 to 6, conditions of lane 3 + 10, 100, or 1,000 μM T-705RTP. *, Elongated RNA was detected when GTP, ATP, and CTP were added to the reaction mixture.
DISCUSSION
In a previous study, T-705 was shown to be converted to T-705RTP by intracellular enzymes and then selectively inhibited influenza virus RdRp. However, the detailed inhibitory mechanisms of T-705RTP against influenza virus RdRp remain unclear. In the present study, we investigated the inhibitory mechanism of T-705RTP, an active form of T-705, on influenza virus RdRp. We first examined how natural nucleotides influenced the effects of T-705RTP on influenza virus RdRp. The inhibitory effects of T-705RTP on influenza virus RdRp were influenced by natural nucleotides in a concentration-dependent manner. Furthermore, enzyme kinetic analysis demonstrated that T-705RTP inhibited the incorporation of ATP and GTP in a competitive manner (Fig. 2 and Fig. 3A and B), suggesting that T-705RTP is recognized as a purine analog by influenza virus RdRp. These results were consistent with a previous report that the in vitro anti-influenza virus activity of T-705 was inhibited by purine bases and nucleosides (5). On the other hand, enzyme kinetic analysis demonstrated that T-705RTP inhibited the incorporation of UTP and CTP in noncompetitive and mixed-type manners, respectively (Fig. 3C and D). It was reported that GTP, instead of CTP, was incorporated into the first base during the RNA strand extension of the influenza virus (17, 18). This result may explain the mixed type of inhibition of T-705RTP for CTP incorporation and the incorporation of T-705RTP into the position of G11+1 nt instead of CTP.
Nucleoside analogs have been shown to competitively inhibit the extension reaction of virus polymerase by three inhibitory mechanisms: (i) a nucleoside analog is not incorporated into a nascent nucleic acid but shows competitive inhibition for natural substrates at enzyme active sites, (ii) a nucleoside analog is incorporated into a nascent nucleic acid to inhibit the extension reaction (e.g., acyclovir) (20), and (iii) nucleoside analogs and subsequent natural nucleotides are incorporated into a nascent nucleic acid to gradually inhibit the extension reaction (e.g., ribavirin) (21). We conducted primer-extension analysis to investigate the incorporation mechanism of T-705RTP into influenza virus RNA. The sequence of the vRNA template indicates that CTP and then ATP/GTP following the G11 primer should be incorporated in the presence of nucleotides (see Fig. S1 in the supplemental material). Although T-705RTP is a purine analog, it was incorporated instead of CTP in the absence of CTP and other NTPs. Two bands were detected in the presence of CTP and T-705RTP, suggesting that the band of G11+2 nt was a band of T-705RTP following CTP and the band of G11+1 nt had two capabilities, a band of CTP or T-705RTP incorporation. These results may reflect T-705RTP being a mixed inhibitor for the incorporation of CTP against influenza virus RdRp. The sequence of the vRNA template indicates that up to four molecules of T-705RTP, as a purine analog, should be competitively incorporated for ATP/GTP incorporation after CTP incorporation (see Fig. S1 in the supplemental material). However, two and more molecules of T-705RTP were not incorporated in the absence of NTP. This observation is similar to 2FdGTP, which is the analog of GTP and a well-known nonobligate chain terminator for influenza virus RdRp. Furthermore, only two bands were detected even in the presence of ATP, GTP, CTP, and T-705RTP (Fig. 5). This result was the same as that in the presence of CTP and T-705RTP (Fig. 4B, lanes 5 and 6), suggesting that a single molecule of T-705RTP was incorporated into influenza virus RNA for the incorporation of ATP/GTP and inhibited the incorporation of subsequent nucleotides.
The inhibitory mechanism of T-705RTP belongs to the class of mechanism ii above and is generally called “chain termination.” Three types of chain terminators have been reported for antivirus or anticancer nucleoside analogs: (i) an obligated-chain terminator, where a 3′-deoxy-nucleoside has an H instead of a 3′ OH group on ribose in NTP; (ii) a nonobligated-chain terminator, where the OH at the 3′ C of ribose remains, while its neighboring side chain is modified to sterically hinder the coordination of a subsequent base; and (iii) a delayed-chain terminator, where a nucleoside analog has an unnatural sugar that allows sugar chain extension but inhibits the extension at a few bases. All of these nucleoside analogs have artificially modified sugar structures and natural bases (22). T-705RTP, a ribose phosphate intracellularly converted from T-705, has an unnatural base moiety and natural ribose phosphate. The nucleic acid chain generally extends through a covalent bond between 3′-OH in ribose and the phosphate group in NTP. It is still unknown why T-705RTP is incorporated into virus RNA strands and inhibits its chain extension in spite of having a 3′ OH group in natural ribose in T-705RTP. As one possibility, the unnatural base of T-705RTP may alter the hydrogen bond of virus RNA bases and inhibit virus RNA extension. These inhibitory mechanisms should be investigated further based on influenza virus RdRp structural information.
Ribavirin is known to be an antivirus agent with an unnatural base and natural ribose similar to T-705 and T-705RTP. Ribavirin is intracellularly phosphorylated and exerts anti-influenza virus activity. Many reports have been published on its mechanisms (23), for example, (i) inhibition of host inosine-monophosphate dehydrogenase (IMPDH), (ii) inhibition of the capping mechanism, and (iii) inhibition of virus RNA transcription and extension by RdRp. The detailed inhibitory mechanisms of influenza virus RdRp remain unknown; however, ribavirin triphosphate was shown to be incorporated into a nascent RNA strand as a mutagen to provoke error catastrophe and inhibit poliovirus RdRp and HCV RdRp (21). T-705 has higher selective in vitro anti-influenza virus activity than ribavirin (4) and T-705RMP, a compound intracellularly converted from T-705, has few effects on host IMPDH (5). More recently, influenza A virus passage under the presence of T-705 lost its specific infectivity and increases the frequency of mutations with nucleotide transitions. Baranovich et al. concluded that the predominant mechanism of inhibition of T-705 against the influenza virus was lethal mutagenesis (24). Our results suggest that the inhibitory mechanism of T-705 against the influenza virus is predominantly chain termination against RNA extension by influenza virus RdRp. This finding suggests that mutagenesis does not occur as frequently as ribavirin on the polio virus. The structural difference in the moiety of the base between T-705 and ribavirin may contribute to the difference in the mechanism of action and their characteristics. In the future, the detailed inhibitory mechanisms of ribavirin on influenza virus RdRp will be investigated to examine these different mechanisms. In addition, T-705 has a high genetic barrier against resistance. It is similar to the results of anti-HCV agents which are nucleoside analogs (25), that is, when mutation is induced in the active site of influenza virus RdRp, its activity is significantly attenuated and mutant viruses are impaired for replication.
In the present study, we demonstrated that T-705RTP competitively incorporated ATP/GTP as purine analogs and that a single molecule of T-705RTP was incorporated into a nascent RNA strand, which then inhibited subsequent virus RNA extension. T-705 has potent antiviral activities against not only influenza virus but also various RNA viruses. One of the reasons for the wide spectrum of T-705 against various RNA viruses may be that T-705RTP, intracellularly converted from T-705, may inhibit RNA polymerase, the essential enzyme in various RNA viruses. A further investigation of the mechanism of action of T-705RTP on virus RdRp will contribute to the development of new drugs against not only the influenza virus but also various RNA viruses.
Supplementary Material
Footnotes
Published ahead of print 5 August 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00649-13.
REFERENCES
- 1.von Itzstein M. 2007. The war against influenza: discovery and development of sialidase inhibitors. Nat. Rev. Drug Discov. 6:967–974 [DOI] [PubMed] [Google Scholar]
- 2.Bright RA, Shay DK, Shu B, Cox NJ, Klimov AI. 2006. Adamantane resistance among influenza A viruses isolated early during the 2005–2006 influenza season in the United States. JAMA 295:891–894 [DOI] [PubMed] [Google Scholar]
- 3.Shiraishi K, Mitamura K, Sakai-Tagawa Y, Goto H, Sugaya N, Kawaoka Y. 2003. High frequency of resistant viruses harboring different mutations in amantadine-treated children with influenza. J. Infect. Dis. 188:57–61 [DOI] [PubMed] [Google Scholar]
- 4.Furuta Y, Takahashi K, Fukuda Y, Kuno M, Kamiyama T, Kozaki K, Nomura N, Egawa H, Minami S, Watanabe Y, Narita H, Shiraki K. 2002. In vitro and in vivo activities of anti-influenza virus compound T-705. Antimicrob. Agents Chemother. 46:977–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Furuta Y, Takahashi K, Kuno-Maekawa M, Sangawa H, Uehara S, Kozaki K, Nomura N, Egawa H, Shiraki K. 2005. Mechanism of action of T-705 against influenza virus. Antimicrob. Agents Chemother. 49:981–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takahashi K, Furuta Y, Fukuda Y, Kuno M, Kamiyama T, Kozaki K, Nomura N, Egawa H, Minami S, Shiraki K. 2003. In vitro and in vivo activities of T-705 and oseltamivir against influenza virus. Antivir. Chem. Chemother. 14:235–241 [DOI] [PubMed] [Google Scholar]
- 7.Furuta Y, Takahashi K, Shiraki K, Sakamoto K, Smee DF, Barnard DL, Gowen BB, Julander JG, Morrey JD. 2009. T-705 (favipiravir) and related compounds: novel broad-spectrum inhibitors of RNA viral infections. Antivir. Res. 82:95–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gowen BB, Wong MH, Jung KH, Sanders AB, Mendenhall M, Bailey KW, Furuta Y, Sidwell RW. 2007. In vitro and in vivo activities of T-705 against arenavirus and bunyavirus infections. Antimicrob. Agents Chemother. 51:3168–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Julander JG, Shafer K, Smee DF, Morrey JD, Furuta Y. 2009. Activity of T-705 in a hamster model of yellow fever virus infection in comparison with that of a chemically related compound, T-1106. Antimicrob. Agents Chemother. 53:202–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mendenhall M, Russell A, Juelich T, Messina EL, Smee DF, Freiberg AN, Holbrook MR, Furuta Y, de la Torre JC, Nunberg JH, Gowen BB. 2010. T-705 (favipiravir) inhibition of arenavirus replication in cell culture. Antimicrob. Agents Chemother. 55:782–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morrey JD, Taro BS, Siddharthan V, Wang H, Smee DF, Christensen AJ, Furuta Y. 2008. Efficacy of orally administered T-705 pyrazine analog on lethal West Nile virus infection in rodents. Antivir. Res. 80:377–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smee DF, Hurst BL, Wong MH, Bailey KW, Tarbet EB, Morrey JD, Furuta Y. 2010. Effects of the combination of favipiravir (T-705) and oseltamivir on influenza A virus infections in mice. Antimicrob. Agents Chemother. 54:126–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tarbet EB, Maekawa M, Furuta Y, Babu YS, Morrey JD, Smee DF. 2012. Combinations of favipiravir and peramivir for the treatment of pandemic influenza A/California/04/2009 (H1N1) virus infections in mice. Antivir. Res. 94:103–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kiso M, Takahashi K, Sakai-Tagawa Y, Shinya K, Sakabe S, Le QM, Ozawa M, Furuta Y, Kawaoka Y. 2010. T-705 (favipiravir) activity against lethal H5N1 influenza A viruses. Proc. Natl. Acad. Sci. U. S. A. 107:882–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hay AJ, Skehel JJ, McCauley J. 1982. Characterization of influenza virus RNA complete transcripts. Virology 116:517–522 [DOI] [PubMed] [Google Scholar]
- 16.Hagen M, Chung TD, Butcher JA, Krystal M. 1994. Recombinant influenza virus polymerase: requirement of both 5′ and 3′ viral ends for endonuclease activity. J. Virol. 68:1509–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chung TD, Cianci C, Hagen M, Terry B, Matthews JT, Krystal M, Colonno RJ. 1994. Biochemical studies on capped RNA primers identify a class of oligonucleotide inhibitors of the influenza virus RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 91:2372–2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishihama A, Mizumoto K, Kawakami K, Kato A, Honda A. 1986. Proofreading function associated with the RNA-dependent RNA polymerase from influenza virus. J. Biol. Chem. 261:10417–10421 [PubMed] [Google Scholar]
- 19.Tisdale M, Ellis M, Klumpp K, Court S, Ford M. 1995. Inhibition of influenza virus transcription by 2′-deoxy-2′-fluoroguanosine. Antimicrob. Agents Chemother. 39:2454–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ilsley DD, Lee SH, Miller WH, Kuchta RD. 1995. Acyclic guanosine analogs inhibit DNA polymerases α, δ, and ε with very different potencies and have unique mechanisms of action. Biochemistry 34:2504–2510 [DOI] [PubMed] [Google Scholar]
- 21.Crotty S, Maag D, Arnold JJ, Zhong W, Lau JY, Hong Z, Andino R, Cameron CE. 2000. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat. Med. 6:1375–1379 [DOI] [PubMed] [Google Scholar]
- 22.Deval J. 2009. Antimicrobial strategies: inhibition of viral polymerases by 3′-hydroxyl nucleosides. Drugs 69:151–166 [DOI] [PubMed] [Google Scholar]
- 23.Crotty S, Cameron C, Andino R. 2002. Ribavirin's antiviral mechanism of action: lethal mutagenesis? J. Mol. Med. 80:86–95 [DOI] [PubMed] [Google Scholar]
- 24.Baranovich T, Wong SS, Armstrong J, Marjuki H, Webby RJ, Webster RG, Govorkova EA. 2013. T-705 (favipiravir) induces lethal mutagenesis in influenza A H1N1 viruses in vitro. J. Virol. 87:3741–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCown MF, Rajyaguru S, Le Pogam S, Ali S, Jiang WR, Kang H, Symons J, Cammack N, Najera I. 2008. The hepatitis C virus replicon presents a higher barrier to resistance to nucleoside analogs than to nonnucleoside polymerase or protease inhibitors. Antimicrob. Agents Chemother. 52:1604–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.