Abstract
Resistance to the recently approved nonnucleoside reverse transcriptase inhibitor (NNRTI) rilpivirine (RPV) commonly involves substitutions at positions E138K and K101E in HIV-1 reverse transcriptase (RT), together with an M184I substitution that is associated with resistance to coutilized emtricitabine (FTC). Previous biochemical and virological studies have shown that compensatory interactions between substitutions E138K and M184I can restore enzyme processivity and the viral replication capacity. Structural modeling studies have also shown that disruption of the salt bridge between K101 and E138 can affect RPV binding. The current study was designed to investigate the impact of K101E, alone or in combination with E138K and/or M184I, on drug susceptibility, viral replication capacity, and enzyme function. We show here that K101E can be selected in cell culture by the NNRTIs etravirine (ETR), efavirenz (EFV), and dapivirine (DPV) as well as by RPV. Recombinant RT enzymes and viruses containing K101E, but not E138K, were highly resistant to nevirapine (NVP) and delavirdine (DLV) as well as ETR and RPV, but not EFV. The addition of K101E to E138K slightly enhanced ETR and RPV resistance compared to that obtained with E138K alone but restored susceptibility to NVP and DLV. The K101E substitution can compensate for deficits in viral replication capacity and enzyme processivity associated with M184I, while M184I can compensate for the diminished efficiency of DNA polymerization associated with K101E. The coexistence of K101E and E138K does not impair either viral replication or enzyme fitness. We conclude that K101E can play a significant role in resistance to RPV.
INTRODUCTION
The reverse transcriptase (RT) of human immunodeficiency virus type 1 (HIV-1) is crucial for HIV-1 replication and has been an important target of antiretroviral (ARV) therapy (1). Both nucleos(t)ide reverse transcriptase inhibitors [N(t)RTIs] and nonnucleoside reverse transcriptase inhibitors (NNRTIs) are key components of ARV therapy (2), which has led to significant declines in HIV-associated morbidity and mortality (3, 4). N(t)RTIs that act as competitive inhibitors and cause chain termination of the growing viral DNA chain include zidovudine (AZT, ZDV), didanosine (ddI), stavudine (d4T), lamivudine (3TC), emtricitabine (FTC), abacavir (ABC), and tenofovir (TFV). In contrast, NNRTIs act allosterically by binding to the NNRTI binding pocket (BP) located 10 Å from the polymerase active site (3) and include earlier drugs, such as nevirapine (NVP), delavirdine (DLV), and efavirenz (EFV), and newer products, such as etravirine (ETR) and rilpivirine (RPV). However, the rapid replication rate of HIV-1 and the error-prone nature of its RT can drive the development of resistance to all ARVs currently in use (5).
The earlier NNRTIs, such as NVP and EFV, have a low genetic barrier for development of resistance, and cross-resistance among NNRTIs is common (6, 7). Recently, however, several newer NNRTIs that are diarylpyrimidine (DAPY) compounds, ETR (8, 9) and RPV (10), have been developed and are active against viruses containing mutations associated with resistance to NVP and EFV (11, 12). However, two phase III clinical trials, ECHO and THRIVE, showed that treatment failure in HIV-infected patients receiving coformulated RPV-FTC-tenofovir disoproxil fumarate (TDF) was most frequently associated with an E138K substitution or less often with a K101E substitution, both of which are known to cause NNRTI resistance, in most cases together with the M184I substitution, known to cause resistance to 3TC and FTC (12). Until now, biochemical and virological studies to explain resistance to RPV have focused on interactions between E138K and M184I/V (13–17). We and others have shown that the decreased viral replication capacity of M184I/V was restored in the presence of E138K (14). Furthermore, biochemical analyses showed that the addition of E138K to M184I in RT restored the processivity of DNA synthesis by enhancing deoxynucleoside triphosphate (dNTP) usage (14, 15). It was also reported that the addition of M184I to E138K enhanced the levels of resistance to both ETR and RPV compared to those obtained with E138K alone (12, 16, 17). These data provide mechanistic insights into the favored emergence of E138K and M184I in patients failing RPV-containing regimens. However, the role of K101E and its interactions with other substitutions, such as M184I and E138K, in resistance to RPV and other NNRTIs has not been determined.
K101E is known to cause resistance to NVP and EFV (18, 19) and has been observed, usually in combination with other NNRTI resistance substitutions, in patients failing therapy (20, 21). K101E can also be a transmitted minority variation in acute HIV-1 infection (22) and may compromise the treatment efficacy of NNRTI-containing regimens. The addition of K101E to other NNRTI substitutions, such as K103N or G190S, can increase levels of resistance to EFV (19, 20). K101E, together with M184I, resulted in a diminution in susceptibility to RPV (12). Although the salt bridge between amino acid K101 of the p66 subunit of RT and E138 in the p51 subunit is involved in NNRTI binding (15, 16, 23), it is not known how the K101E/E138K double substitution might impact NNRTI susceptibility. The study of interactions between K101E and other resistance substitutions in RT is the topic of this report.
MATERIALS AND METHODS
Chemicals, cells, and nucleic acids.
ETR and RPV were gifts of Janssen Pharmaceuticals (Titusville, NJ). FTC was kindly provided by Gilead Sciences (Foster City, CA). 3TC was a gift of GlaxoSmithKline (Greenford, United Kingdom).
Cord blood mononuclear cells (CBMCs) were obtained through the Department of Obstetrics, Jewish General Hospital, Montreal, QC, Canada. The HEK293T cell line was obtained from the American Type Culture Collection (ATCC). The following reagents and cells were obtained through the NIH AIDS Research and Reference Reagent Program: the infectious molecular clone pNL4-3 from Malcolm Martin and TZM-bl (JC53-bl) cells from John C. Kappes, Xiaoyun Wu, and Tranzyme Inc.
The pNL4.3PFB plasmid DNA was a generous gift from Tomozumi Imamichi, National Institutes of Health, Bethesda, MD. The plasmid pRT6H-PROT was a generous gift from Stuart F. J. Le Grice, National Institutes of Health, Bethesda, MD.
An HIV-1 RNA template of ∼500 nucleotides (nt) in size spanning the 5′ untranslated region (UTR) to the primer binding site (PBS) was transcribed in vitro from AccI-linearized pHIV-PBS DNA (24) by using an Ambion T7-MEGAshortscript kit (Invitrogen, Burlington, ON, Canada) as described previously (25). The oligonucleotides used in this study were synthesized by Integrated DNA Technologies Inc. (Coralville, IA) and purified by polyacrylamide-urea gel electrophoresis. For 5′-end labeling of oligonucleotides with [γ-32P]ATP, an Ambion KinaseMax kit was used, followed by purification through Ambion NucAway spin columns, according to protocols provided by the supplier (Invitrogen, Burlington, ON, Canada).
Site-directed mutagenesis and preparation of virus stocks.
To construct HIV-1 RT expression plasmids and recombinant HIV-1 clonal variants harboring the desired mutations in the RT gene, site-directed mutagenesis (SDM) reactions were first carried out using a QuikChange II XL site-directed mutagenesis kit (Stratagene, La Jolla, CA) on an HIV-1 RT expression plasmid, pRT6H-PROT. Then, fragments spanning RT amino acids (aa) 25 to 314 were generated by PCR and were used to substitute for corresponding RT fragments in pNL4.3PFB plasmid DNA (26), as described previously (25), to generate recombinant HIV-1 isolates containing the desired RT mutations. DNA sequencing was performed to verify the absence of spurious mutations and the presence of any desired mutation. Recombinant wild-type (WT) and mutant HIV-1 isolates were generated by transfection of the corresponding proviral plasmid DNAs into HEK293T cells using Lipofectamine 2000 (Invitrogen, Burlington, ON, Canada) according to the manufacturer's instructions. Viral supernatants were harvested at 48 h posttransfection, centrifuged for 5 min at 800 × g to remove the cellular debris, filtered through a 0.45-μm-pore-size filter, aliquoted, and stored at −80°C. The levels of p24 in the viral supernatant were measured by a Perkin-Elmer HIV-1 p24 antigen enzyme-linked immunosorbent assay (ELISA) kit. Virion-associated RT activity was verified by an in vitro recombinant RT assay, as described previously (27).
Selection of HIV-1 mutants in CBMCs under drug selection pressure.
CBMCs stimulated by phytohemagglutinin A (PHA) were isolated and cultured as described previously (28) in 10% RPMI 1640 medium supplemented with 10% qualified fetal bovine serum, 20 U of human interleukin-2 (IL-2)/ml, 5 μg of hydrocortisone/ml, 2 mM l-glutamine/ml, 100 U of penicillin/ml, and 100 μg of streptomycin/ml. Cells in 24-well tissue culture plates were infected with recombinant viral clones at a similar multiplicity of infection (MOI). Selection of viral resistance mutations was performed using increasing concentrations of RT inhibitors at starting concentrations below the 50% effective concentration (EC50), as described previously (28, 29). Virus-containing culture media were harvested and kept at −80°C for subsequent standard genotypic analysis by population sequencing. Selections for resistance were performed over a period of 19 weeks.
Analysis of phenotypic drug susceptibility using the Monogram PhenoSense HIV RT assay.
Resistance test vectors (RTVs) containing HIV-1 sequences with the protease sequence and the first 305 amino acids of RT were constructed and used to evaluate NRTI and NNRTI susceptibility as previously described (19). Pseudotyped viruses were produced by cotransfecting HEK293T cell cultures with RTV plasmid DNA, together with an expression plasmid encoding the Env protein of amphotropic murine leukemia virus. RTVs contain a luciferase reporter gene to monitor recombinant virus infection in cell culture following a single cycle of replication. Results are expressed as the fold change (FC) in the EC50 for the mutant variants compared to the EC50 for a wild-type reference virus (NL4-3). Assay techniques have been optimized to minimize variability, and assay performance has been extensively validated. Several modifications have further enhanced assay performance (30).
Measurements of HIV-1 replication kinetics in CBMCs.
CBMCs were isolated and cultured as previously described (28). Recombinant wild-type viruses and viruses containing the desired mutations were normalized by p24 in order to minimize interinoculum effects, as described previously (31). Briefly, 2 × 106 CBMCs were infected with viruses containing 8 × 106 pg of p24 for 2 h. The cells were washed with medium and resuspended in 4 ml complete medium after centrifugation, and each sample was split and placed into 2 wells of a 12-well plate. The replication kinetics of mutant and WT viral stocks were assessed on the basis of p24 levels in culture supernatants sampled at days 1, 3, 4, 7, and 8 postinfection, and viral growth kinetics were monitored by p24 levels, as measured by ELISA as described above.
Recombinant RT expression and purification.
Recombinant RTs in heterodimeric form were expressed from plasmid pbRT6H-PROT (25) and purified as described previously (32, 33) with minor modifications. In brief, RT expression in Escherichia coli M15(pREP4) (Qiagen, Mississauga, ON, Canada) was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at room temperature. Pelleted bacteria were lysed under native conditions with the BugBuster protein extraction reagent containing Benzonase (Novagen, Madison, WI) according to the manufacturer's instructions. After clarification by high-speed centrifugation, the clear supernatant was subjected to the batch method of Ni-nitrilotriacetic acid (NTA) metal affinity chromatography (QIAexpressionist; Qiagen, Mississauga, ON, Canada). All buffers contained complete protease inhibitor cocktail (Roche, Mississauga, ON, Canada). Hexahistidine-tagged RT was eluted using an imidazole gradient. RT-containing fractions were pooled, passed through DEAE-Sepharose (GE Healthcare, Mississauga, ON, Canada), and further purified using SP-Sepharose (GE Healthcare, Mississauga, ON, Canada). Fractions containing purified RT were pooled, dialyzed against storage buffer (50 mM Tris-HCl [pH 7.8], 50 mM NaCl, 50% glycerol), and concentrated to 4 to 8 mg/ml with a Centricon Plus-20 filter with a molecular mass cutoff of 30 kDa (Millipore, Etobicoke, ON, Canada). Aliquots of proteins were stored at −80°C. The protein concentration was measured by a Bradford protein assay kit (Bio-Rad Laboratories, Saint-Laurent, QC, Canada), and the purity of the recombinant RT preparations was verified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The RNA-dependent DNA polymerase activity of each recombinant RT preparation was evaluated as described previously (34) using various concentrations of RT and a synthetic homopolymeric poly(rA)/p(dT)12-18 template/primer (T/P; Midland Certified Reagent Company, Midland, TX).
Enzyme processivity assays.
The processivities of recombinant RT enzymes were analyzed as described previously using a heteropolymeric HIV-1 PBS RNA template in the presence of a heparin enzyme trap to ensure a single processive cycle, i.e., a single round of binding and of primer extension and dissociation (14). The T/P was prepared by annealing the HIV-1 PBS RNA with the 25-nt DNA primer D25 labeled with 32P at the 5′ end at a molar ratio of 1:1, denatured at 85°C for 5 min, and then slowly cooled to room temperature to allow specific annealing of the primer to the template. RT enzymes with equal amounts of activity and 40 nM T/P were preincubated for 5 min at 37°C in a buffer containing 50 mM Tris-HCl (pH 7.8), 50 mM NaCl, 6 mM MgCl2. Reactions were initiated by the addition of dNTPs and a heparin trap (final concentration, 3.2 mg/ml), and the reaction mixtures were incubated at 37°C for 30 min; then, 2 volumes of stop solution (90% formamide, 10 mM EDTA, 0.1% each xylene cyanol and bromophenol blue) were added to stop the reaction. Reaction products were denatured by heating at 95°C and analyzed using 6% denaturing polyacrylamide gel electrophoresis and phosphorimaging. The effectiveness of the heparin trap was verified in control reactions in which the trap was preincubated with substrate before the addition of RT enzymes and dNTP.
RNA-dependent DNA polymerase activity assay.
The same HIV-1 PBS RNA template and the D25 primer labeled with 32P at the 5′ end described above were used to assess primer extension efficiency in processive DNA synthesis by recombinant RT enzymes in gel-based time course experiments (13, 14). Final reaction mixtures contained 20 nM T/P, 400 nM RT enzyme, 50 mM Tris-HCl (pH 7.8), and 50 mM NaCl. Reactions were initiated by adding 6 mM MgCl2 and dNTP at 200 μM, and the reaction mixtures were mixed with 2 volumes of stop solution at various time points. Reaction products were separated by 6% denaturing polyacrylamide gel electrophoresis and analyzed by phosphorimaging.
RT-catalyzed RNase H activity.
RNase H activity was assayed using a 40-mer RNA template (kim40R) labeled with 32P at the 5′ end annealed to a complementary 32-mer DNA primer (kim32D) (35) at a 1:4 molar ratio. Reactions were conducted as described previously (13, 14, 36) at 37°C in mixtures containing an RNA-DNA hybrid duplex substrate with purified RT enzymes normalized by activity in assay buffer, 50 mM Tris-HCl, pH 7.8, 50 mM NaCl, 0.1 mM EDTA, pH 8.0, 6 mM MgCl2, in the absence or presence of the heparin trap at a final concentration of 2 mg/ml. RNase H cleavage was monitored in time course experiments, with aliquots of samples removed at different time points after initiation of the reactions and with the reactions quenched by adding 3 volumes of formamide loading buffer (96% formamide, 20 mM EDTA, 0.05% xylene cyanol FF, and 0.05% bromophenol blue). The samples were heated to 90°C for 3 min, cooled on ice, and resolved on 6% polyacrylamide–7 M urea gels. The dried gels were exposed to phosphoscreens and analyzed by a phosphorimager (GE Healthcare, Mississauga, ON, Canada) using ImageQuant software. The efficacy of the heparin trap was verified by preincubation experiments performed through a 10-min preincubation of RT enzymes with various concentrations of the heparin trap, followed by initiation with the 32P-labeled RNA-DNA hybrid duplex substrate and magnesium in the same assay buffer described above.
RESULTS
Selection of K101E by NNRTIs in CBMCs.
K101E has been observed to be a minority NNRTI substitution, generally in combination with other known NNRTI resistance substitutions, in patients failing NVP- and EFV-containing regimens (20, 21). Recently, K101E was also reported to be the second most frequent NNRTI substitution, together with E138K (usually accompanied by M184I), in patients with virologic failure during treatment with RPV, although combinations of K101E/M184I and K101E/M184V/and other substitutions were also observed (12). To verify whether K101E can be independently selected by NNRTIs, we performed tissue culture selection experiments using CBMCs in the presence of two DAPY compounds, ETR and dapivirine (DPV), as well as EFV. Table 1 shows that K101E emerged following selection with all three compounds, alone or in combination with NRTIs. In some instances, K101E emerged after 13 weeks of ETR-FTC pressure but was eventually outgrown by viruses containing E138K by week 19, suggesting that these two substitutions might be exclusive or antagonistic. In the ECHO and THRIVE trials, K101E and 138K were observed together only in the presence of M184I/V and other substitutions. Since exclusion/antagonism between these two substitutions may be related to viral replication capacity and/or enhanced drug susceptibility, we performed biochemical and virological analyses to evaluate this relationship.
Table 1.
Summary of emergent substitutions in selection experiments using CBMCsa
| Virusb | Subtype | Background substitution(s) | Drug(s) | Selected substitution(s) |
|---|---|---|---|---|
| 8336 | B | A98S, G190A | EFV | K101E |
| NL4-3 WT | B | None | EFV | K101E, V108I |
| 10583 | C | Y181C | EFV | K101E/K, V106M, Y181C |
| 8116 | B | A98S, G190A | DPV | K101E, Y181C |
| NL4- -Y181C | B | Y181C | DPV | K101E, V179I, Y181C |
| NL4-3-Y181C | B | Y181C | DPV-TDF | K101K/E, V108I/V, V179I/V, Y181C, H221Y |
| 10583 | C | Y181C | DPV-TDF | K101E/K, V108I/V, V179I/V, Y181C, H221Y |
| Mole 03c | C | None | ETR-FTC | K101E/K, E138E/K |
| Mole 03d | C | None | ETR-FTC | E138K, M184I. |
| Mole 03 | C | None | ETR-3TC | K101E, M184I |
Selection experiments were performed over 19 weeks. Abbreviations: 3TC, lamivudine; DPV, dapivirine; EFV, efavirenz; ETR, etravirine; FTC, emtricitabine; TDF, tenofovir disoproxil fumarate.
Viruses other than NL4-3 clones are clinical isolates from treatment-naive patients undergoing acute HIV-1 infection.
Viruses were genotyped at week 13.
Viruses were genotyped at week 19.
Phenotypic drug susceptibilities determined by the Monogram PhenoSense HIV RT assay.
The drug susceptibilities of recombinant HIV-1 isolates containing specific mutations were determined and compared to those of a wild-type reference virus using the Monogram PhenoSense HIV RT assay (19, 30). Susceptibility to five NNRTIs and seven NRTIs was evaluated (Table 2). The K101E substitution conferred large reductions in susceptibility to NVP and DLV, while more modest reductions in susceptibility to EFV, ETR, or RPV were observed. In contrast, the E138K substitution did not confer large reductions in susceptibility to any of the NNRTIs and only small reductions in susceptibility to ETR and RPV.
Table 2.
Changes in drug susceptibilities for recombinant HIV-1 WT and site-directed mutants containing various substitutions compared to the sequence of WT NL4-3a
| Mutation(s) | FC in EC50 (nM) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RPV | ETR | EFV | NVP | DLV | ZDV | D4T | 3TC | FTC | DDI | ABC | TDF | |
| K101E | 3.0 | 4.9 | 2.1 | 170 | 50.8 | 0.4 | 1.0 | 0.7 | 0.7 | 0.8 | 0.8 | 0.7 |
| E138K | 2.3 | 2.5 | 1.3 | 0.9 | 1.6 | 1.8 | 1.4 | 1.3 | 1.1 | 1.2 | 1.4 | 1.2 |
| K101E/E138K | 3.2 | 3.1 | 2.1 | 3.3 | 3.0 | 1.2 | 1.4 | 1.2 | 1.1 | 1.2 | 1.4 | 1.1 |
| E138K/M184I | 3.2 | 3.3 | 1.6 | 0.8 | 1.8 | 0.75 | 1.6 | >103.8 | >75.0 | 2.4 | 3.2 | 0.8 |
| K101E/E138K/M184I | 3.3 | 3.2 | 1.9 | 2.0 | 2.6 | 0.5 | 1.4 | >103.8 | >75.0 | 2.2 | 2.8 | 0.6 |
The changes were assessed by the Monogram PhenoSense HIV RT assay. Shown are the FC in the EC50 values (nM) normalized to the value for the HIV-1 wild type.
The addition of E138K to K101E did not further reduce susceptibility to RPV, ETR, or EFV compared to the reductions conferred by K101E alone, yet it notably restored susceptibility to NVP and DLV. Combinations of E138K/M184I or K101E/E138K/M184I comparably conferred very modest and comparable reductions in NNRTI susceptibility. These findings suggest that the salt bridge between residues K101 and E138 (15, 16) can play a different role in regard to resistance to NVP-DLV and ETR-RPV.
In addition, we verified that the mutant with the E138K/M184I double substitution exhibited slightly larger reductions in ETR and RPV susceptibility than the mutant with the single E138K substitution, in agreement with previous findings that M184I enhances ETR-RPV resistance in the background of E138K (16, 17). It was recently reported that K101E in the presence of M184I conferred a further reduction in RPV susceptibility compared to that conferred by K101E alone (12), and in the present study, we found that the susceptibility to RPV, ETR, EFV, and all N(t)RTIs of the mutant with the K101E/E138K/M184I triple substitution was comparable to that of the mutant with the E138K/M184I double substitution.
The K101E substitution compensates for the impaired viral replication capacity of HIV-1 containing M184I.
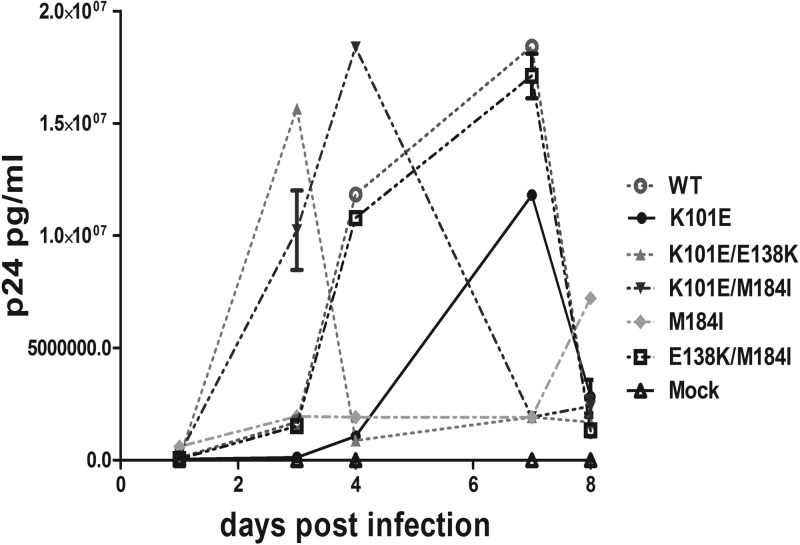
We previously showed that the E138K substitution can restore the replication capacity of HIV-1 that harbors the M184I substitution in a short-term viral replication assay in TZM-bl cells (14). Then, we wished to investigate the impact of interactions between M184I and K101E and between E138K and K101E on viral replication over multiple cycles in CBMCs that are more physiologically relevant in regard to HIV-1 infection than cell lines. Therefore, we infected CBMCs with viruses that were normalized in terms of inoculum on the basis of the amount of p24 (37). Quantification of virus production at various time points was carried out by measurement of p24 antigen, and some samples were also tested for RT activity (37, 38). As shown in Fig. 1, the relative replication ability of viruses containing M184I alone was severely impaired compared to that of the WT, while the replication capacity of the mutant virus with the E138K/M184I double substitution was restored to WT levels, in agreement with previously published data (14), (17). An even more pronounced compensatory effect on viral replication capacity was observed with viruses with the K101E/M184I double substitution, indicating that K101E can function together with M184I to restore viral fitness. The mutant virus with the K101E/E138K double substitution replicated faster than either WT virus or virus with the K101E/M184I substitutions. In contrast, viruses containing K101E alone replicated less efficiently than WT virus, as assessed by p24 antigen measurement, but attained a peak of replication at the same time as did WT virus, in agreement with the observation that K101E impaired replication fitness (39). These findings are also in agreement with the enzymatic data described below which show that K101E, like E138K, can function as a compensatory substitution for M184I in regard to RT enzymatic fitness.
Fig 1.
Replication kinetics in CBMCs of WT and mutant viruses. Viruses were harvested from transfection of HEK293T cells and were used to infect CBMCs through normalization of the p24 antigen level of the inoculum. Virus production was monitored at the indicated time points by measuring p24 levels in cell-free supernatants by ELISA. Each experiment was performed in triplicate. The results are representative of those from two independent experiments.
Activity of recombinant HIV-1 RT enzymes.
Amino acid residues K101 in the p66 subunit of HIV-1 RT and E138K in the p51 subunit constitute the floor of the NNRTI binding pocket (40–42) at the p66/p51 interface. It was previously shown that E138K alone and E138K/M184I in tandem did not interfere with either heterodimer formation or enzyme purification (14, 43, 44). However, it is possible that the addition of K101E to E138 alone or with M184I in tandem might have negative effects. Therefore, WT enzymes as well as four recombinant heterodimeric (p66/p51) RT enzymes containing K101E substitutions in both RT subunits, i.e., K101E, K101E/M184I, K101E/E138K, and K101E/E138K/M184I, and E138K/M184I, were purified to >95% homogeneity, as demonstrated by Coomassie blue staining in SDS-polyacrylamide gels (data not shown). We also purified enzymes that contained these substitutions together with M184I or E138K/M184I. The RT p66 and p51 subunits were processed to similar molar ratios in each case, verifying that the substitutions did not affect proteolytic cleavage, p66/p51 heterodimer formation, or RT enzyme purification.
The RT preparations were titrated by standard RNA-dependent DNA polymerase activity assay (25, 34), and all of the mutant RTs showed activities similar to the activity of WT RT (data not shown). These results validate the quality of the recombinant RT preparations used for biochemical analyses.
The K101E substitution restores the enzyme processivity of RT containing M184I.
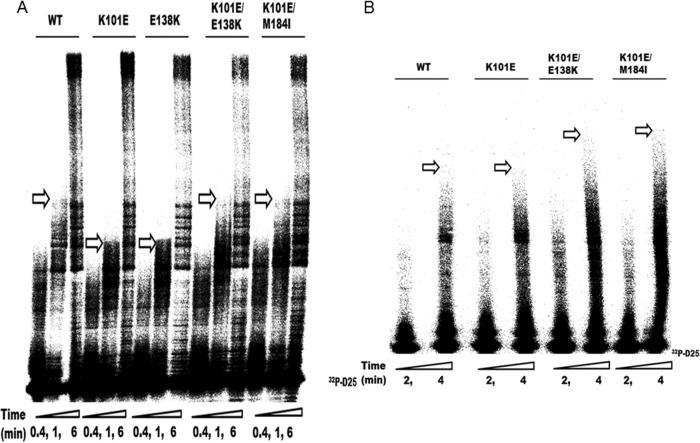
HIV-1 RT drug resistance substitutions, especially those conferring resistance to NRTIs, can often affect RT enzymatic fitness by decreasing enzyme processivity (45–47) (i.e., the number of nucleotides incorporated in a single round of enzyme binding, elongation, and dissociation). Earlier studies showed that diminished HIV-1 RT processivity is a major determinant of the impaired viral replication capacity that is associated with the M184I/V substitutions, especially at low dNTP concentrations (45, 48). Previous reports also showed that E138K can compensate for the replication capacity deficit of viruses containing either M184I or M184V by restoring RT enzyme processivity (13, 14) but that the Y181C substitution in RT can diminish the processivity of E138K-containing RT. Indeed, interactions among different drug resistance substitutions, whether compensatory or antagonistic, may constitute the molecular basis for the occurrence of certain patterns of drug resistance. Then, we wished to determine how the K101E substitution, alone or in combination with M184I and/or E138K, might affect RT processivity. Therefore, we performed single-cycle processivity assays using recombinant RT enzymes at both low (0.5 μM) and high (200 μM) dNTP concentrations as previously described (13, 14). The results of these assays at low dNTP concentration showed that K101E-containing mutant enzymes, in combination with M184I or E138K/M184I, had higher processivity than enzymes with the M184I substitution alone, indicating that K101E, like E138K, can compensate for the diminished enzyme processivity associated with M184I (Fig. 2). Furthermore, the K101E substitution, alone or in combination with E138K, did not display diminished processivity compared to that of the WT. At high dNTP concentrations, all enzymes displayed similar processivities, on the basis of the production of full-length DNA products (Fig. 2). This is in agreement with the findings of previous studies that the processivity defect of the M184I RT mutant is minimized at high dNTP concentrations (14, 45, 49). These results imply that K101E is able to function as a compensatory substitution for M184I and are confirmed in the cell-based replication capacity assays presented below.
Fig 2.
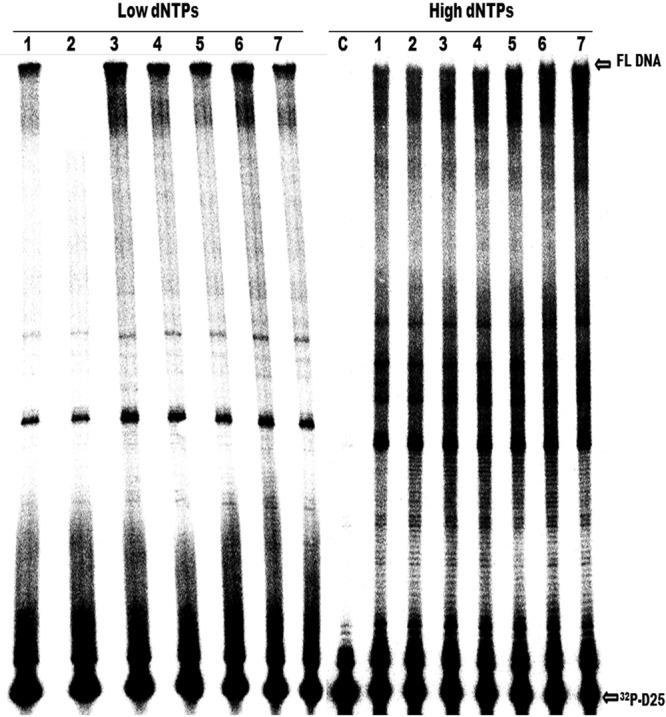
Comparative analysis of enzyme processivity of WT RT and RT enzymes containing the indicated substitutions. The processivity of purified recombinant RT enzymes was analyzed using a 5′-end-labeled DNA primer (D25) annealed to a 471-nt HIV-1 PBS RNA template as the substrate; the resulting full-length DNA is 471 nt in length. Processivities were determined by the size distribution of DNA products in fixed-time experiments at low (0.5 μM) and high (200 μM) concentrations of dNTPs in the presence of a heparin trap. All reaction products were resolved by denaturing 6% polyacrylamide gel electrophoresis and visualized by phosphorimaging. The positions of the 32P-labeled D25 primer (32P-D25) and the 471-nt full-length (FL) extension DNA product are indicated on the right. Lanes: 1, WT; 2, M184I mutant; 3, K101E/M184I mutant; 4, E138K/M184I mutant; 5, K101E/E138K/M184I mutant; 6, K101E mutant; 7, K101E/E138K mutant. The results of a control reaction to verify the efficiency of the heparin trap by preincubation with substrate prior to addition of WT RT are also shown (lane C). Each experiment was repeated at least twice and yielded similar results on each occasion. The figure shows a gel from a representative experiment.
Impact of K101E on the efficiency of processive DNA synthesis.
Cell culture passage experiments have shown that the K101E and E138K substitutions are antagonistic, unless other substitutions, such as M184I, are simultaneously present, and in the ECHO and THRIVE clinical trials, K101E and M184I were often observed together. Previously, it was shown that E138K can impair the efficiency of DNA polymerization in a dNTP concentration-dependent manner (14). At high dNTP concentrations, E138K-containing RT possessed a diminished efficiency of DNA polymerization, while M184I was able to compensate for the E138K deficit in polymerization. At low dNTP concentrations, E138K was able to restore the efficiency of polymerization of M184I. These mutually compensatory effects constitute the molecular basis for competent replication of doubly mutated E138K/M184I viruses at variable dNTP concentrations, i.e., in cells with both large and low dNTP pools. Then, we wished to determine how the K101E substitution, in a background of WT, E138K, and/or M184I, might impact the efficiency of processive DNA synthesis.
We first performed RNA-dependent DNA polymerase reactions using all of the enzymes with single mutations and combinations of mutations in time course experiments at high dNTP concentrations (200 μM) and compared the results with those obtained with WT RT (Fig. 3A). RT molecules were used at a ∼20-fold excess over the amount of substrate, so that any RTs that dissociated from the primer terminus during synthesis would be rapidly replaced. In this case, the rate-limiting step would be nucleotide addition (49). The efficiency of DNA polymerization was assessed and compared in terms of the length of the longest extension products (indicated by arrows in Fig. 3A) that were synthesized after 1 min. E138K RT was included as a control in regard to diminished efficiency of DNA polymerization. The data in Fig. 3A show that K101E exhibited a diminished efficiency of DNA polymerization, as did E138K, at high dNTP concentrations, while the RT enzymes with the K101E/E138K and K101E/M184I double substitutions showed the same efficiency of DNA polymerization as the WT enzyme. These results indicate that the diminished efficiency of DNA polymerization associated with K101E contributes to the impaired replication capacity of mutated HIV-1 in CBMCs and that E138K, like M184I, can rescue the efficiency of DNA polymerization.
Fig 3.
Analysis of efficiency of processive DNA synthesis. (A) Comparative analysis of the enzyme efficiency of processive DNA synthesis of HIV-1 WT RT and mutant RT at high dNTP concentrations (200 μM). The D25 primer labeled with 32P at the 5′ end (32P-D25) was annealed to the 471-nt HIV-1 PBS RNA template, and primer extension assays were performed at an excess of recombinant RT enzymes at high dNTP concentrations (200 μM). Reactions were stopped at 0.4 min, 1 min, and 6 min. The longest extension products generated at 1 min are identified by arrows and indicate differences in the efficiency of polymerization. Each experiment was repeated at least twice and yielded similar results on each occasion. The figure shows a gel from a representative experiment. (B) Comparative analysis of enzyme efficiency of processive DNA synthesis of HIV-1 WT RT and mutant RT at low dNTP concentrations (0.5 μM). Reactions were stopped at 2 min and 4 min. The longest extension products generated at 4 min are identified by arrows and indicate differences in the efficiency of polymerization. Each experiment was repeated at least twice and yielded similar results on each occasion. The figure shows a gel from a representative experiment.
In addition, at low dNTP concentrations, E138K can compensate for the diminished polymerization efficiency of the M184I RT (25), which is defective in regard to dNTP usage (45, 49), even though the E138K mutant, on its own, possesses a diminished catalytic efficiency compared to WT at high dNTP concentrations. Then, we evaluated how K101E, alone or in combination with M184I and/or E138K, might impact the efficiency of DNA polymerization at low dNTP concentrations. The data in Fig. 4B show that the K101E RT displayed a similar efficiency of DNA polymerization under these conditions as the WT enzyme and that the addition of E138K to K101E or E138K/M184I to K101E enhanced DNA polymerization, as demonstrated by the presence of longer extension products after 4 min (Fig. 3B).
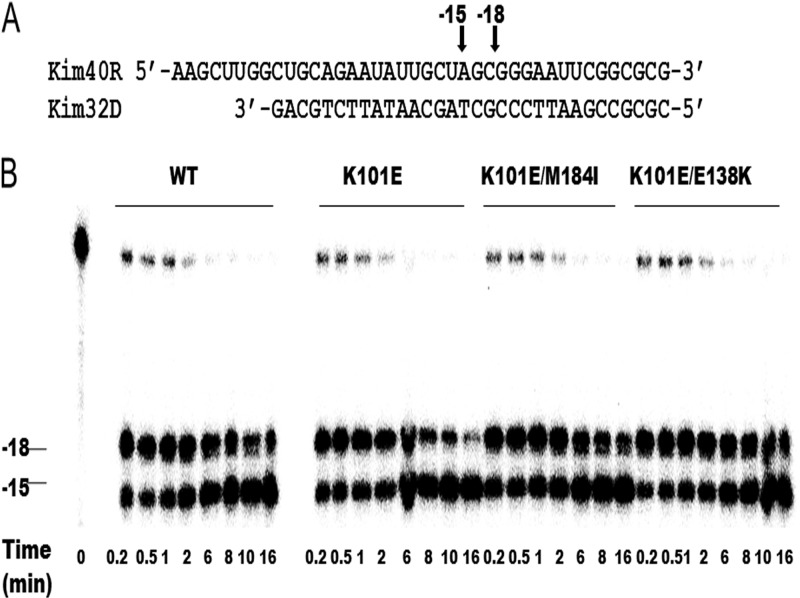
Fig 4.
RNase H activity of WT and mutant recombinant RT enzymes. (A) Graphic representation of the RNA-DNA (kim40R-kim32D) substrate duplex used to monitor the cleavage efficiency of mutant and WT RTs. The 40-mer RNA kim40R was labeled at its 5′ terminus with 32P and annealed to 32-mer DNA oligonucleotide kim32D. The positions of RNase H cleavage relative to the 3′ terminus of the DNA primer are indicated by arrows. (B) RNase H activity was analyzed by monitoring substrate cleavage in time course experiments. The time points were 0.2, 0.5, 1, 2, 6, 8, 10, and 16 min. The lane labeled time point 0 shows the uncleaved substrate in a control reaction without RT enzyme. The positions of cleaved products relative to the 3′ terminus of the DNA primer are indicated on the left. All reactions were resolved by denaturing 6% polyacrylamide gel electrophoresis. Each experiment was repeated at least twice and yielded similar results on each occasion. The figure shows a gel from a representative experiment.
RNase H activity.
As stated earlier, E138K in HIV RT impairs RNase H activity and, in combination with a diminished efficiency in processive DNA synthesis, contributes to reductions in viral replication capacity (13, 14). We then wished to test the impact of the K101E substitution alone or in combination with M184I on RNase H activity. Therefore, we performed an RNase H time course study in the presence of a heparin trap by using a recessed 32-mer DNA primer hybridized to a 5′-end-labeled 40-mer RNA to monitor 3′-DNA-directed RNase H activity (Fig. 4). The presence of the heparin trap permitted the analysis of cleaved products from a single event of RT binding to the substrate. The results show that the RNase H activity of the K101E mutant RT was comparable to that of WT RT and that the addition of the M184I or E138K substitution did not impair the RNase H activity of the K101E RT (Fig. 4). Thus, the K101E substitution does not impair RNase H activity, despite the fact that it results in a deficit in the efficiency of polymerization, as shown above.
DISCUSSION
RPV and ETR are compounds that can bind efficiently to the NNRTI BP and possess activity against HIV-1 variants that are resistant to earlier NNRTIs, such as NVP, DLV, and EFV (10, 50, 51). The once-daily fixed-dose combination of RPV-FTC-TFV has provided an additional option for management of HIV-1 infection in treatment-naive patients (52, 53). Most data sets regarding RPV resistance are based on the ECHO and THRIVE clinical studies, and recent work has focused on the RT enzymatic properties and replication capacity of viruses resistant to RPV (12, 50), as well as interactions between the E138K and M184I/V substitutions (13–17).
The ECHO and THRIVE clinical trials also showed that K101E was the second most frequent substitution in patients who failed RPV-FTC-TDF treatment (12). Here, we have shown that K101E can be selected in cell culture by each of ETR, DPV, and EFV and that K101E can restore the enzyme processivity of RT and the viral replication capacity of viruses containing the M184I substitution. Thus, doubly mutated K101E/M184I viruses are competent in regard to replication capacity while displaying resistance to each of FTC, 3TC, RPV, and ETR.
Both K101E and E138K emerged in cell culture selection experiments with RPV, and, interestingly, K101E was frequently observed together with Y181C (50). In contrast, antagonism seems to exist between E138K and Y181C, and the mechanisms involved have been reported (54, 55). In the present study, we have compared the effects of the K101E and E138K substitutions on drug susceptibility using the Monogram PhenoSense HIV RT assay. Our results confirm that K101E and E138K possess different NNRTI resistance profiles; K101E confers high-level resistance to the earlier NNRTIs NVP and DLV but only low-level resistance to EFV; K101E also confers higher-level resistance to ETR and RPV than EFV. In contrast, E138K-containing virus is susceptible to NVP and modestly resistant to DLV and EFV, and E138K confers resistance to the newer NNRTIs ETR and RPV. We have shown that the K101E/E138K double substitution can restore susceptibility to NVP and DLV, to which mutants with the K101E substitution alone are resistant, without affecting resistance to RPV and ETR. This is the first demonstration that resistance to an NNRTI can be reversed by the presence of a different NNRTI resistance mutation.
The salt bridge between K101 of the p66 subunit and E138 of the p51 subunit of RT has been shown to be important for NNRTI binding by structural (23) and molecular modeling studies (15, 16). Recently, it was shown that E138K, which disrupts the salt bridge between p51E138 and p66K101, can change the dynamics of the entrance of the RPV binding pocket due to the proximity of these two positive residues. Quantum mechanics and biochemical molecular modeling analysis have shown that E138K in p51 affects access to the RPV binding site by disrupting this salt bridge (15).
This raises the question whether the salt bridge has the potential to re-form within RT when both the K101E and E138K substitutions are present, even though such doubly mutated viruses are still drug resistant, as shown here.
Ideally, this issue could be further explored through molecular modeling analysis, but such an analysis was beyond the scope of this study. However, it is worth pointing out that two groups have published different versions of the RT-RPV complex (56, 57), which may make it difficult to probe the impact of changes within the salt bridge on RPV binding affinity. Transit kinetic analysis has demonstrated that the mechanism of RPV resistance is largely due to an increase in the dissociation rate of RPV and RT rather than a direct decrease in binding affinity (15). The use of such a methodology may provide further insights into the mechanisms of resistance associated with the K101E and E138K substitutions.
Mechanisms of resistance to NNRTIs are often not as clear as those to NRTIs (58–63). The current data show that the disrupted salt bridge involving K101E and E138K might differentially impact drug binding, on the basis of the NNRTI involved. Further detailed structural and biochemical analysis of this salt bridge in NNRTI binding and resistance is warranted.
However, our biochemical and virological data do not explain why K101E occurs less often than E138K in patients who have failed RPV-TDF-FTC therapy, especially since the replication capacity of viruses with the K101E/M184I double substitution was higher than that of viruses with the E138K/M184I substitutions and compensation in regard to enzyme processivity and the efficiency of DNA polymerization was observed in both cases. The drug susceptibilities of these two doubly mutated viruses were also similar. One possibility might be that K101E derives from an A-to-G substitution (AAA to GAA), while E138K results from the G-to-A hypermutation (GAG to AAG), which also explains the prevalence of K138 over other substitutions at position E138 (64). Recently, deep sequencing analysis of patients who suffered from RPV virologic failure in the phase III studies ECHO and THRIVE showed that K101E and E138K rarely occurred on the same genome (65). However, no natural antagonism seems to exist between K101E and E138K; thus, the occurrence of other mutational pairs associated with NNRTI resistance, such as Y181C and E138K, may be in part a matter of chance in the context of RT enzymes that are functional and commensurate with viral replication capacity (55). In conclusion, enzymatic and cell-based assays show that K101E can play a significant role in resistance to RPV.
ACKNOWLEDGMENTS
This work was supported by research grants from the Canadian Institutes of Health Research (CIHR).
We thank Stuart Le Grice for providing pRT6H-PROT DNA and Tomozumi Imamichi for pNL4.3PFB plasmid DNA.
Footnotes
Published ahead of print 3 September 2013
REFERENCES
- 1.Arts EJ, Hazuda DJ. 2012. HIV-1 antiretroviral drug therapy. Cold Spring Harb. Perspect. Med. 2:a007161. 10.1101/cshperspect.a007161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thompson MA, Aberg JA, Hoy JF, Telenti A, Benson C, Cahn P, Eron JJ, Gunthard HF, Hammer SM, Reiss P, Richman DD, Rizzardini G, Thomas DL, Jacobsen DM, Volberding PA. 2012. Antiretroviral treatment of adult HIV infection: 2012 recommendations of the International Antiviral Society—USA Panel. JAMA 308:387–402 [DOI] [PubMed] [Google Scholar]
- 3.Mocroft A, Ledergerber B, Katlama C, Kirk O, Reiss P, d'Arminio Monforte A, Knysz B, Dietrich M, Phillips AN, Lundgren JD. 2003. Decline in the AIDS and death rates in the EuroSIDA study: an observational study. Lancet 362:22–29 [DOI] [PubMed] [Google Scholar]
- 4.Torres RA, Barr M. 1997. Impact of combination therapy for HIV infection on inpatient census. N. Engl. J. Med. 336:1531–1532 [DOI] [PubMed] [Google Scholar]
- 5.Wainberg MA. 2003. HIV resistance to nevirapine and other non-nucleoside reverse transcriptase inhibitors. J. Acquir. Immune Defic. Syndr. 34(Suppl 1):S2–S7 [DOI] [PubMed] [Google Scholar]
- 6.Delaugerre C, Rohban R, Simon A, Mouroux M, Tricot C, Agher R, Huraux JM, Katlama C, Calvez V. 2001. Resistance profile and cross-resistance of HIV-1 among patients failing a non-nucleoside reverse transcriptase inhibitor-containing regimen. J. Med. Virol. 65:445–448 [PubMed] [Google Scholar]
- 7.Hirsch MS, Brun-Vezinet F, D'Aquila RT, Hammer SM, Johnson VA, Kuritzkes DR, Loveday C, Mellors JW, Clotet B, Conway B, Demeter LM, Vella S, Jacobsen DM, Richman DD. 2000. Antiretroviral drug resistance testing in adult HIV-1 infection: recommendations of an International AIDS Society—USA Panel. JAMA 283:2417–2426 [DOI] [PubMed] [Google Scholar]
- 8.Andries K, Azijn H, Thielemans T, Ludovici D, Kukla M, Heeres J, Janssen P, De Corte B, Vingerhoets J, Pauwels R, de Bethune MP. 2004. TMC125, a novel next-generation nonnucleoside reverse transcriptase inhibitor active against nonnucleoside reverse transcriptase inhibitor-resistant human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 48:4680–4686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vingerhoets J, Azijn H, Fransen E, De Baere I, Smeulders L, Jochmans D, Andries K, Pauwels R, de Bethune MP. 2005. TMC125 displays a high genetic barrier to the development of resistance: evidence from in vitro selection experiments. J. Virol. 79:12773–12782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janssen PA, Lewi PJ, Arnold E, Daeyaert F, de Jonge M, Heeres J, Koymans L, Vinkers M, Guillemont J, Pasquier E, Kukla M, Ludovici D, Andries K, de Bethune MP, Pauwels R, Das K, Clark AD, Jr, Frenkel YV, Hughes SH, Medaer B, De Knaep F, Bohets H, De Clerck F, Lampo A, Williams P, Stoffels P. 2005. In search of a novel anti-HIV drug: multidisciplinary coordination in the discovery of 4-[[4-[[4-[(1E)-2-cyanoethenyl]-2,6-dimethylphenyl]amino]-2-pyrimidinyl]amino]benzonitrile (R278474, rilpivirine). J. Med. Chem. 48:1901–1909 [DOI] [PubMed] [Google Scholar]
- 11.Croxtall JD. 2012. Etravirine: a review of its use in the management of treatment-experienced patients with HIV-1 infection. Drugs 72:847–869 [DOI] [PubMed] [Google Scholar]
- 12.Rimsky L, Vingerhoets J, Van Eygen V, Eron J, Clotet B, Hoogstoel A, Boven K, Picchio G. 2012. Genotypic and phenotypic characterization of HIV-1 isolates obtained from patients on rilpivirine therapy experiencing virologic failure in the phase 3 ECHO and THRIVE studies: 48-week analysis. J. Acquir. Immune Defic. Syndr. 59:39–46 [DOI] [PubMed] [Google Scholar]
- 13.Xu HT, Oliveira M, Quashie PK, McCallum M, Han Y, Quan Y, Brenner BG, Wainberg MA. 2012. Subunit-selective mutational analysis and tissue culture evaluations of the interactions of the E138K and M184I mutations in HIV-1 reverse transcriptase. J. Virol. 86:8422–8431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu HT, Asahchop EL, Oliveira M, Quashie PK, Quan Y, Brenner BG, Wainberg MA. 2011. Compensation by the E138K mutation in HIV-1 reverse transcriptase for deficits in viral replication capacity and enzyme processivity associated with the M184I/V mutations. J. Virol. 85:11300–11308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh K, Marchand B, Rai DK, Sharma B, Michailidis E, Ryan EM, Matzek KB, Leslie MD, Hagedorn AN, Li Z, Norden PR, Hachiya A, Parniak MA, Xu HT, Wainberg MA, Sarafianos SG. 2012. Biochemical mechanism of HIV-1 resistance to rilpivirine. J. Biol. Chem. 287:38110–38123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kulkarni R, Babaoglu K, Lansdon EB, Rimsky L, Van Eygen V, Picchio G, Svarovskaia E, Miller MD, White KL. 2012. The HIV-1 reverse transcriptase M184I mutation enhances the E138K-associated resistance to rilpivirine and decreases viral fitness. J. Acquir. Immune Defic. Syndr. 59:47–54 [DOI] [PubMed] [Google Scholar]
- 17.Hu Z, Kuritzkes DR. 2011. Interaction of reverse transcriptase (RT) mutations conferring resistance to lamivudine and etravirine: effects on fitness and RT activity of human immunodeficiency virus type 1. J. Virol. 85:11309–11314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Byrnes VW, Emini EA, Schleif WA, Condra JH, Schneider CL, Long WJ, Wolfgang JA, Graham DJ, Gotlib L, Schlabach AJ. 1994. Susceptibilities of human immunodeficiency virus type 1 enzyme and viral variants expressing multiple resistance-engendering amino acid substitutions to reserve transcriptase inhibitors. Antimicrob. Agents Chemother. 38:1404–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petropoulos CJ, Parkin NT, Limoli KL, Lie YS, Wrin T, Huang W, Tian H, Smith D, Winslow GA, Capon DJ, Whitcomb JM. 2000. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 44:920–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bacheler L, Jeffrey S, Hanna G, D'Aquila R, Wallace L, Logue K, Cordova B, Hertogs K, Larder B, Buckery R, Baker D, Gallagher K, Scarnati H, Tritch R, Rizzo C. 2001. Genotypic correlates of phenotypic resistance to efavirenz in virus isolates from patients failing nonnucleoside reverse transcriptase inhibitor therapy. J. Virol. 75:4999–5008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bacheler LT, Anton ED, Kudish P, Baker D, Bunville J, Krakowski K, Bolling L, Aujay M, Wang XV, Ellis D, Becker MF, Lasut AL, George HJ, Spalding DR, Hollis G, Abremski K. 2000. Human immunodeficiency virus type 1 mutations selected in patients failing efavirenz combination therapy. Antimicrob. Agents Chemother. 44:2475–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nicot F, Saliou A, Raymond S, Saune K, Dubois M, Massip P, Marchou B, Delobel P, Izopet J. 2012. Minority variants associated with resistance to HIV-1 nonnucleoside reverse transcriptase inhibitors during primary infection. J. Clin. Virol. 55:107–113 [DOI] [PubMed] [Google Scholar]
- 23.Ren J, Nichols CE, Stamp A, Chamberlain PP, Ferris R, Weaver KL, Short SA, Stammers DK. 2006. Structural insights into mechanisms of non-nucleoside drug resistance for HIV-1 reverse transcriptases mutated at codons 101 or 138. FEBS J. 273:3850–3860 [DOI] [PubMed] [Google Scholar]
- 24.Arts EJ, Li X, Gu Z, Kleiman L, Parniak MA, Wainberg MA. 1994. Comparison of deoxyoligonucleotide and tRNA(Lys-3) as primers in an endogenous human immunodeficiency virus-1 in vitro reverse transcription/template-switching reaction. J. Biol. Chem. 269:14672–14680 [PubMed] [Google Scholar]
- 25.Xu HT, Quan Y, Asahchop E, Oliveira M, Moisi D, Wainberg MA. 2010. Comparative biochemical analysis of recombinant reverse transcriptase enzymes of HIV-1 subtype B and subtype C. Retrovirology 7:80. 10.1186/1742-4690-7-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Imamichi T, Berg SC, Imamichi H, Lopez JC, Metcalf JA, Falloon J, Lane HC. 2000. Relative replication fitness of a high-level 3′-azido-3′-deoxythymidine-resistant variant of human immunodeficiency virus type 1 possessing an amino acid deletion at codon 67 and a novel substitution (Thr → Gly) at codon 69. J. Virol. 74:10958–10964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goff SP. 1990. Retroviral reverse transcriptase: synthesis, structure, and function. J. Acquir. Immune Defic. Syndr. 3:817–831 [PubMed] [Google Scholar]
- 28.Oliveira M, Brenner BG, Wainberg MA. 2009. Isolation of drug-resistant mutant HIV variants using tissue culture drug selection. Methods Mol. Biol. 485:427–433 [DOI] [PubMed] [Google Scholar]
- 29.Asahchop EL, Oliveira M, Wainberg MA, Brenner BG, Moisi D, Toni T, Tremblay CL. 2011. Characterization of the E138K resistance mutation in HIV-1 reverse transcriptase conferring susceptibility to etravirine in B and non-B HIV-1 subtypes. Antimicrob. Agents Chemother. 55:600–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parkin NT, Hellmann NS, Whitcomb JM, Kiss L, Chappey C, Petropoulos CJ. 2004. Natural variation of drug susceptibility in wild-type human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 48:437–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loemba H, Brenner B, Parniak MA, Ma'ayan S, Spira B, Moisi D, Oliveira M, Detorio M, Wainberg MA. 2002. Genetic divergence of human immunodeficiency virus type 1 Ethiopian clade C reverse transcriptase (RT) and rapid development of resistance against nonnucleoside inhibitors of RT. Antimicrob. Agents Chemother. 46:2087–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Le Grice SF, Cameron CE, Benkovic SJ. 1995. Purification and characterization of human immunodeficiency virus type 1 reverse transcriptase. Methods Enzymol. 262:130–144 [DOI] [PubMed] [Google Scholar]
- 33.Le Grice SF, Gruninger-Leitch F. 1990. Rapid purification of homodimer and heterodimer HIV-1 reverse transcriptase by metal chelate affinity chromatography. Eur. J. Biochem. 187:307–314 [DOI] [PubMed] [Google Scholar]
- 34.Quan Y, Brenner BG, Marlink RG, Essex M, Kurimura T, Wainberg MA. 2003. Drug resistance profiles of recombinant reverse transcriptases from human immunodeficiency virus type 1 subtypes A/E, B, and C. AIDS Res. Hum. Retroviruses 19:743–753 [DOI] [PubMed] [Google Scholar]
- 35.Operario DJ, Balakrishnan M, Bambara RA, Kim B. 2006. Reduced dNTP interaction of human immunodeficiency virus type 1 reverse transcriptase promotes strand transfer. J. Biol. Chem. 281:32113–32121 [DOI] [PubMed] [Google Scholar]
- 36.Gopalakrishnan V, Peliska JA, Benkovic SJ. 1992. Human immunodeficiency virus type 1 reverse transcriptase: spatial and temporal relationship between the polymerase and RNase H activities. Proc. Natl. Acad. Sci. U. S. A. 89:10763–10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quinones-Mateu ME, Arts EJ. 2002. Fitness of drug resistant HIV-1: methodology and clinical implications. Drug Resist. Updat. 5:224–233 [DOI] [PubMed] [Google Scholar]
- 38.Resch W, Ziermann R, Parkin N, Gamarnik A, Swanstrom R. 2002. Nelfinavir-resistant, amprenavir-hypersusceptible strains of human immunodeficiency virus type 1 carrying an N88S mutation in protease have reduced infectivity, reduced replication capacity, and reduced fitness and process the Gag polyprotein precursor aberrantly. J. Virol. 76:8659–8666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J, Liang H, Bacheler L, Wu H, Deriziotis K, Demeter LM, Dykes C. 2010. The non-nucleoside reverse transcriptase inhibitor efavirenz stimulates replication of human immunodeficiency virus type 1 harboring certain non-nucleoside resistance mutations. Virology 402:228–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pandey PK, Kaushik N, Talele TT, Yadav PN, Pandey VN. 2001. The beta7-beta8 loop of the p51 subunit in the heterodimeric (p66/p51) human immunodeficiency virus type 1 reverse transcriptase is essential for the catalytic function of the p66 subunit. Biochemistry 40:9505–9512 [DOI] [PubMed] [Google Scholar]
- 41.Pandey PK, Kaushik N, Singh K, Sharma B, Upadhyay AK, Kumar S, Harris D, Pandey VN. 2002. Insertion of a small peptide of six amino acids into the beta7-beta8 loop of the p51 subunit of HIV-1 reverse transcriptase perturbs the heterodimer and affects its activities. BMC Biochem. 3:18. 10.1186/1471-2091-3-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Esnouf RM, Ren J, Hopkins AL, Ross CK, Jones EY, Stammers DK, Stuart DI. 1997. Unique features in the structure of the complex between HIV-1 reverse transcriptase and the bis(heteroaryl)piperazine (BHAP) U-90152 explain resistance mutations for this nonnucleoside inhibitor. Proc. Natl. Acad. Sci. U. S. A. 94:3984–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ambrose Z, Herman BD, Sheen CW, Zelina S, Moore KL, Tachedjian G, Nissley DV, Sluis-Cremer N. 2009. The human immunodeficiency virus type 1 nonnucleoside reverse transcriptase inhibitor resistance mutation I132M confers hypersensitivity to nucleoside analogs. J. Virol. 83:3826–3833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nissley DV, Radzio J, Ambrose Z, Sheen CW, Hamamouch N, Moore KL, Tachedjian G, Sluis-Cremer N. 2007. Characterization of novel non-nucleoside reverse transcriptase (RT) inhibitor resistance mutations at residues 132 and 135 in the 51 kDa subunit of HIV-1 RT. Biochem. J. 404:151–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Back NK, Nijhuis M, Keulen W, Boucher CA, Oude Essink BO, van Kuilenburg AB, van Gennip AH, Berkhout B. 1996. Reduced replication of 3TC-resistant HIV-1 variants in primary cells due to a processivity defect of the reverse transcriptase enzyme. EMBO J. 15:4040–4049 [PMC free article] [PubMed] [Google Scholar]
- 46.Sharma PL, Crumpacker CS. 1999. Decreased processivity of human immunodeficiency virus type 1 reverse transcriptase (RT) containing didanosine-selected mutation Leu74Val: a comparative analysis of RT variants Leu74Val and lamivudine-selected Met184Val. J. Virol. 73:8448–8456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.White KL, Margot NA, Wrin T, Petropoulos CJ, Miller MD, Naeger LK. 2002. Molecular mechanisms of resistance to human immunodeficiency virus type 1 with reverse transcriptase mutations K65R and K65R + M184V and their effects on enzyme function and viral replication capacity. Antimicrob. Agents Chemother. 46:3437–3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Naeger LK, Margot NA, Miller MD. 2001. Increased drug susceptibility of HIV-1 reverse transcriptase mutants containing M184V and zidovudine-associated mutations: analysis of enzyme processivity, chain-terminator removal and viral replication. Antivir. Ther. 6:115–126 [PubMed] [Google Scholar]
- 49.Gao L, Hanson MN, Balakrishnan M, Boyer PL, Roques BP, Hughes SH, Kim B, Bambara RA. 2008. Apparent defects in processive DNA synthesis, strand transfer, and primer elongation of Met-184 mutants of HIV-1 reverse transcriptase derive solely from a dNTP utilization defect. J. Biol. Chem. 283:9196–9205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Azijn H, Tirry I, Vingerhoets J, de Bethune MP, Kraus G, Boven K, Jochmans D, Van Craenenbroeck E, Picchio G, Rimsky LT. 2010. TMC278, a next-generation nonnucleoside reverse transcriptase inhibitor (NNRTI), active against wild-type and NNRTI-resistant HIV-1. Antimicrob. Agents Chemother. 54:718–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Das K, Clark AD, Jr, Lewi PJ, Heeres J, De Jonge MR, Koymans LM, Vinkers HM, Daeyaert F, Ludovici DW, Kukla MJ, De Corte B, Kavash RW, Ho CY, Ye H, Lichtenstein MA, Andries K, Pauwels R, De Bethune MP, Boyer PL, Clark P, Hughes SH, Janssen PA, Arnold E. 2004. Roles of conformational and positional adaptability in structure-based design of TMC125-R165335 (etravirine) and related non-nucleoside reverse transcriptase inhibitors that are highly potent and effective against wild-type and drug-resistant HIV-1 variants. J. Med. Chem. 47:2550–2560 [DOI] [PubMed] [Google Scholar]
- 52.Patel N, Miller CD. 2012. New option for management of HIV-1 infection in treatment-naive patients: once-daily, fixed-dose combination of rilpivirine-emtricitabine-tenofovir. HIV AIDS (Auckl.) 4:61–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wainberg MA. 2013. Combination therapies, effectiveness, and adherence in patients with HIV infection: clinical utility of a single tablet of emtricitabine, rilpivirine, and tenofovir. HIV AIDS (Auckl.) 5:41–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Asahchop EL, Wainberg MA, Oliveira M, Xu H, Brenner BG, Moisi D, Ibanescu IR, Tremblay C. 2013. Distinct resistance patterns to etravirine and rilpivirine in viruses containing NNRTI mutations at baseline. AIDS 27:879–887 [DOI] [PubMed] [Google Scholar]
- 55.Xu HT, Oliveira M, Asahchop EL, McCallum M, Quashie PK, Han Y, Quan Y, Wainberg MA. 2012. Molecular mechanism of antagonism between the Y181C and E138K mutations in HIV-1 reverse transcriptase. J. Virol. 86:12983–12990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Das K, Bauman JD, Clark AD, Jr, Frenkel YV, Lewi PJ, Shatkin AJ, Hughes SH, Arnold E. 2008. High-resolution structures of HIV-1 reverse transcriptase/TMC278 complexes: strategic flexibility explains potency against resistance mutations. Proc. Natl. Acad. Sci. U. S. A. 105:1466–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lansdon EB, Brendza KM, Hung M, Wang R, Mukund S, Jin D, Birkus G, Kutty N, Liu X. 2010. Crystal structures of HIV-1 reverse transcriptase with etravirine (TMC125) and rilpivirine (TMC278): implications for drug design. J. Med. Chem. 53:4295–4299 [DOI] [PubMed] [Google Scholar]
- 58.Menendez-Arias L. 2008. Mechanisms of resistance to nucleoside analogue inhibitors of HIV-1 reverse transcriptase. Virus Res. 134:124–146 [DOI] [PubMed] [Google Scholar]
- 59.Menendez-Arias L. 2010. Molecular basis of human immunodeficiency virus drug resistance: an update. Antiviral Res. 85:210–231 [DOI] [PubMed] [Google Scholar]
- 60.Sarafianos SG, Marchand B, Das K, Himmel DM, Parniak MA, Hughes SH, Arnold E. 2009. Structure and function of HIV-1 reverse transcriptase: molecular mechanisms of polymerization and inhibition. J. Mol. Biol. 385:693–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Singh K, Marchand B, Kirby KA, Michailidis E, Sarafianos SG. 2010. Structural aspects of drug resistance and inhibition of HIV-1 reverse transcriptase. Viruses 2:606–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sluis-Cremer N, Arion D, Parniak MA. 2000. Molecular mechanisms of HIV-1 resistance to nucleoside reverse transcriptase inhibitors (NRTIs). Cell. Mol. Life Sci. 57:1408–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sluis-Cremer N, Tachedjian G. 2008. Mechanisms of inhibition of HIV replication by non-nucleoside reverse transcriptase inhibitors. Virus Res. 134:147–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu HT, Colby-Germinario SP, Asahchop EL, Oliveira M, McCallum M, Schader SM, Han Y, Quan Y, Sarafianos SG, Wainberg MA. 2013. Effect of mutations at position E138 in HIV-1 reverse transcriptase and their interactions with the M184I mutation on defining patterns of resistance to the nonnucleoside reverse transcriptase inhibitors rilpivirine and etravirine. Antimicrob. Agents Chemother. 57:3100–3109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Van Eygen V, Thys K, Vingerhoets J, Van Hove C, Rimsky L, Aerssens J, Picchio G. 2012. Deep sequencing analysis of baseline samples from patients treated with rilpivirine in the phase III studies ECHO and THRIVE shows no association between the presence of minority resistance-associated variants and virological failure. Antivir. Ther. 17:A35 [Google Scholar]