Abstract
To compare the in vitro antibacterial efficacies and resistance profiles of rifampin-based combinations against methicillin-resistant Staphylococcus aureus (MRSA) in a biofilm model, the antibacterial activities of vancomycin, teicoplanin, daptomycin, minocycline, linezolid, fusidic acid, fosfomycin, and tigecycline alone or in combination with rifampin against biofilm-embedded MRSA were measured. The rifampin-resistant mutation frequencies were evaluated. Of the rifampin-based combinations, rifampin enhances the antibacterial activities of and even synergizes with fusidic acid, tigecycline, and, to a lesser extent, linezolid, fosfomycin, and minocycline against biofilm-embedded MRSA. Such combinations with weaker rifampin resistance induction activities may provide a therapeutic advantage in MRSA biofilm-related infections.
TEXT
Methicillin-resistant Staphylococcus aureus (MRSA)-associated infections are difficult to cure, and treatment failure or recurrent diseases are common in patients who retain an infected graft. Notably, treatment failure may be related to biofilm formation (1, 2). Rifampin is a component of the active combinations against MRSA and biofilms formed by these organisms (3, 4). However, despite the in vitro susceptibility to rifampin of MRSA strains, rapid emergence of rifampin-resistant MRSA isolates, especially biofilm-embedded isolates, has been found (5). Therefore, the use of rifampin monotherapy for MRSA infections is commonly discouraged, and rifampin-based combinations are alternative options for treatment of biofilm-associated MRSA infections. In this study, eight MRSA strains with distinct genotypes identified by pulse-field gel electrophoresis from 33 clinical isolations were used (6, 7). The antimicrobial susceptibility of planktonic or biofilm-embedded MRSA to vancomycin, rifampin, minocycline, fosfomycin, linezolid, tigecycline, fusidic acid, teicoplanin, and daptomycin was tested. All eight isolates were susceptible to all the drugs but minocycline and fusidic acid in common planktonic form. Of note, all isolates had low rifampin MICs (<0.06 μg/ml). In general, the minimum biofilm eradication concentrations (MBECs) were higher than the MICs for all isolates (data not shown).
Biofilms of individual strains were prepared in 24-well plates according to a previously described method (8) and were treated with vancomycin, teicoplanin, daptomycin, minocycline, linezolid, fusidic acid, fosfomycin, tigecycline, or rifampin alone or with rifampin-based combinations. The concentrations of most drugs were adjusted to the susceptible breakpoint concentrations (SBCs) recommended by the CLSI (9). The SBCs of fusidic acid and tigecycline were adopted from the British Society for Antimicrobial Chemotherapy (10) and Food and Drug Administration (11) guidelines. Specifically, the following concentrations were used: for vancomycin, 2 μg/ml; for teicoplanin, 8 μg/ml; for daptomycin, 1 μg/ml; for minocycline, 4 μg/ml; for linezolid, 4 μg/ml; for fusidic acid, 1 μg/ml; for fosfomycin, 64 μg/ml; for tigecycline, 0.5 μg/ml; and for rifampin, 1 μg/ml. The drug-containing medium was gently aspirated after 1 day at 37°C, and the biofilm on the wells was incubated with fresh drug dilutions for 5 consecutive days, followed by counting at day 5. The bacterial loads determined for the rifampin-based combinations were compared to those seen with monotherapy. The presence of synergism was defined as a ≥100-fold reduction of the bacterial load after treatment with a combination regimen versus the load determined after treatment with the more active drug in the combination. The inhibition effects of each drug alone and in combination with rifampin are shown in Fig. 1 and Table 1.
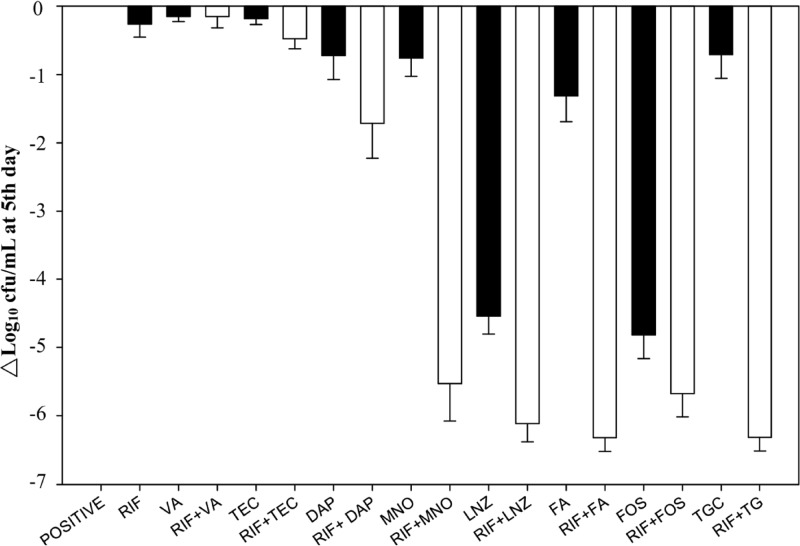
Fig 1.
Bacterial load reduction (log10 CFU/ml) of eight methicillin-resistant Staphylococcus aureus strains in biofilm after exposure to nine antibodies alone or in combination with rifampin for 5 days at the following susceptible breakpoint concentrations: vancomycin (VA) 2 μg/ml, teicoplanin (TEC) 8 μg/ml, daptomycin (DAP) 1 μg/ml, minocycline (MNO) 4 μg/ml, linezolid (LNZ) 4 μg/ml, fusidic acid (FA) 1 μg/ml, fosfomycin (FOS) 64 μg/ml, tigecycline (TGC) 0.5 μg/ml, and rifampin (RIF) 1 μg/ml. The black bar represents the single antibiotic, and the white bar represents the rifampin base combination with the left antibiotic.
Table 1.
Compared to the bacterial loads without antibiotic exposure, bacterial load changes of eight methicillin-resistant Staphylococcus aureus strains in biofilm after 5 days of exposure to nine antibiotics alone or in combination with rifampin at susceptible breakpoint concentrations
| Drug(s)a | Bacterial load change (log10 CFU/ml) for isolateb: |
|||||||
|---|---|---|---|---|---|---|---|---|
| 252 | 3315 | 3337 | 3509 | 3562 | 3626 | 3641 | 3643 | |
| No antibiotic | 7.78 ± 0.54 | 8.40 ± 0.32 | 8.45 ± 0.71 | 8.61 ± 0.23 | 8.40 ± 0.11 | 8.38 ± 0.36 | 7.88 ± 0.64 | 8.34 ± 0.54 |
| RIF | −0.25 ± 0.04 | −0.17 ± 0.08 | 0.13 ± 0.07 | 0.32 ± 0.07 | −0.44 ± 1.10 | −0.27 ± 0.54 | −0.37 ± 0.23 | −0.75 ± 0.69 |
| VA | −0.18 ± 0.18 | −0.22 ± 0.28 | −0.15 ± 0.14 | −0.17 ± 0.40 | −0.05 ± 0.30 | −0.12 ± 0.04 | 0.17 ± 0.04 | −0.26 ± 0.05 |
| RIF + VA | −0.21 ± 0.11 | −0.17 ± 0.25 | 0.00 ± 0.30 | −0.20 ± 0.11 | 0.49 ± 0.24 | −0.06 ± 0.07 | 0.23 ± 0.85 | −0.46 ± 0.07 |
| TEC | −0.12 ± 0.16 | −0.17 ± 0.17 | 0.00 ± 0.15 | −0.27 ± 0.06 | −0.22 ± 0.47 | −0.06 ± 0.07 | −0.02 ± 0.09 | −0.30 ± 0.01 |
| RIF + TEC | −0.22 ± 0.31 | −0.28 ± 0.11 | −0.54 ± 0.35 | −0.53 ± 0.12 | −0.65 ± 0.05 | −0.15 ± 0.42 | −0.33 ± 0.08 | −0.63 ± 0.19 |
| DAP | −0.21 ± 0.65 | −1.28 ± 0.33 | −0.17 ± 0.07 | −0.33 ± 0.07 | −0.14 ± 0.47 | −1.04 ± 0.13 | −0.44 ± 0.03 | −0.11 ± 0.08 |
| RIF + DAP | −0.74 ± 0.39 | −1.19 ± 0.18 | −0.19 ± 1.23 | −0.95 ± 0.06 | −1.36 ± 0.13 | −2.54 ± 0.61 | −0.36 ± 0.22 | −0.62 ± 0.29 |
| MNO | 0.12 ± 1.06 | −0.71 ± 0.14 | −0.78 ± 0.31 | −0.96 ± 0.16 | −0.82 ± 0.30 | −1.65 ± 0.16 | 0.18 ± 0.47 | −0.17 ± 0.09 |
| RIF + MNO | −4.27 ± 1.15* | −5.02 ± 0.12* | −2.74 ± 1.39 | −4.89 ± 1.25* | −1.62 ± 0.47 | −8.38 ± 0.00* | −0.82 ± 1.41 | −8.34 ± 0.00* |
| LNZ | −3.68 ± 0.83 | −4.92 ± 0.19 | −4.03 ± 0.11 | −4.90 ± 0.16 | −3.81 ± 0.18 | −4.23 ± 0.20 | −4.20 ± 0.11 | −4.34 ± 0.57 |
| RIF + LNZ | −5.48 ± 0.35 | −5.55 ± 0.20 | −8.45 ± 0.00* | −5.53 ± 0.24 | −6.22 ± 0.47* | −5.20 ± 0.09 | −7.88 ± 0.00* | −8.34 ± 0.00* |
| FA | −1.06 ± 0.16 | −1.22 ± 0.82 | −2.00 ± 0.81 | −0.91 ± 0.11 | −0.55 ± 0.02 | −1.66 ± 1.25 | −0.23 ± 0.35 | −1.20 ± 0.30 |
| RIF + FA | −7.78 ± 0.00* | −8.40 ± 0.00* | −8.45 ± 0.00* | −8.61 ± 0.00* | −8.40 ± 0.00* | −8.38 ± 0.00* | −7.88 ± 0.00* | −5.26 ± 0.24* |
| FOS | −5.18 ± 0.35 | −2.94 ± 0.24 | −2.79 ± 0.47 | −4.91 ± 0.28 | −1.19 ± 1.41 | −4.54 ± 0.61 | −4.36 ± 1.31 | −5.34 ± 0.28 |
| RIF + FOS | −5.60 ± 0.47 | −5.32 ± 0.24* | −4.94 ± 0.09* | −5.01 ± 1.38 | −4.81 ± 0.40* | −5.43 ± 0.47 | −7.88 ± 0.00* | −5.87 ± 0.47 |
| TGC | −0.05 ± 0.31 | −1.14 ± 0.31 | −0.24 ± 0.18 | −0.66 ± 1.10 | −0.22 ± 0.28 | −1.20 ± 0.47 | 0.10 ± 0.09 | 0.13 ± 0.14 |
| RIF + TGC | −5.00 ± 0.94* | −5.85 ± 1.01* | −8.45 ± 0.00* | −8.61 ± 0.00* | −8.40 ± 0.00* | −8.38 ± 0.00* | −7.88 ± 0.00* | −8.34 ± 0.00* |
RIF, rifampin; VA, vancomycin; TEC, teicoplanin; DAP, daptomycin; MNO, minocycline; LNZ, linezolid; FA, fusidic acid; FOS, fosfomycin; TGC, tigecycline.
Data are shown as means ± standard deviations. An asterisk (*) indicates the presence of synergism.
Rifampin alone showed a poor inhibitory effect, despite low MICs for MRSA. In contrast, linezolid alone exhibited a potent inhibitory effect, namely, a 3.68 to 4.92 log10 reduction of bacterial load. Fosfomycin alone also produced a 2.79 to 5.34 log10 reduction against all but one resistant isolate (no. 3562). Synergism was noted in the combinations rifampin plus linezolid (four MRSA isolates), rifampin plus fosfomycin (four), rifampin plus minocycline (five), rifampin plus fusidic acid (eight), and rifampin plus tigecycline (eight).
To study the emergence of rifampin-resistant mutants from biofilm-embedded MRSA isolates incubated with rifampin alone or in combination with other agents, surviving colonies were collected and the highest MICs of rifampin were evaluated. Rifampin-resistant isolates (those with the highest MIC > 64 μg/ml) emerged readily in the eight MRSA isolates incubated with SBCs of rifampin alone or in combination with vancomycin, teicoplanin, or daptomycin (Table 2). In contrast, the rifampin MICs of the surviving colonies from eight MRSA isolates treated with the combinations of rifampin plus linezolid, rifampin plus fusidic acid, rifampin plus fosfomycin, or rifampin plus tigecycline were not higher than 0.06 μg/ml. For the combination of rifampin and minocycline, the MIC results were divergent. Among five MRSA isolates in which rifampin plus minocycline showed synergistic antibacterial activity, rifampin-resistant isolates (>64 μg/ml) could not be found, but they were easily found among the MRSA isolates without rifampin-minocycline synergism.
Table 2.
The highest rifampin MICs of the viable isolates from eight methicillin-resistant Staphylococcus aureus strains in biofilm after 5 days of exposure to rifampin alone or in combination at susceptible breakpoint concentrations
| Drug(s)a | Highest rifampin MIC (μg/ml) for isolate: |
|||||||
|---|---|---|---|---|---|---|---|---|
| 252 | 3315 | 3337 | 3509 | 3562 | 3626 | 3641 | 3643 | |
| RIF | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 |
| VA + RIF | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 |
| TEC + RIF | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 |
| DAP + RIF | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 |
| MNO + RIF | <0.06 | <0.06 | >64 | <0.06 | >64 | <0.06 | >64 | <0.06 |
| LNZ + RIF | <0.06 | <0.06 | <0.06 | <0.06 | <0.06 | <0.06 | <0.06 | <0.06 |
| FA + RIF | <0.06 | <0.06 | <0.06 | <0.06 | <0.06 | <0.06 | <0.06 | <0.06 |
| FOS + RIF | <0.06 | <0.06 | <0.06 | <0.06 | <0.06 | <0.06 | <0.06 | <0.06 |
| TGC + RIF | <0.06 | <0.06 | <0.06 | <0.06 | <0.06 | <0.06 | <0.06 | <0.06 |
RIF, rifampin; VA, vancomycin; TEC, teicoplanin; DAP, daptomycin; MNO, minocycline; LNZ, linezolid; FA, fusidic acid; FOS, fosfomycin; TGC, tigecycline.
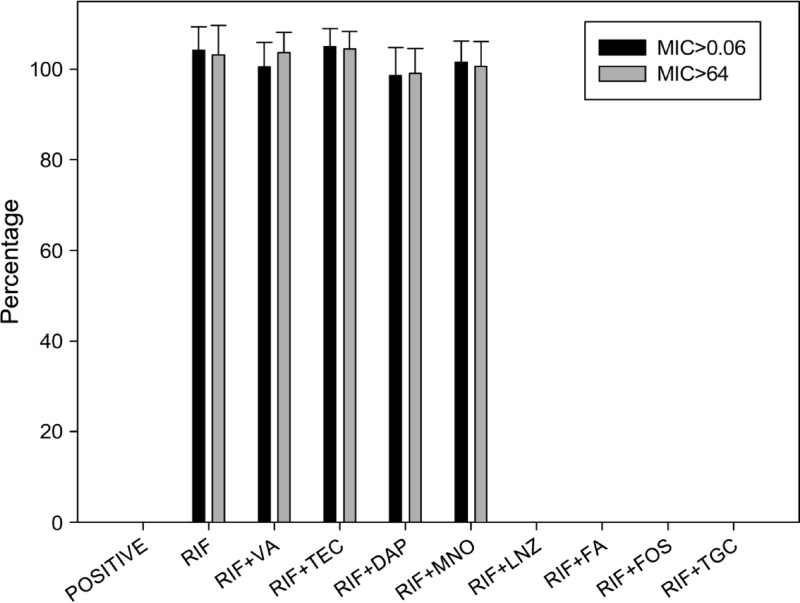
In order to understand how many survival strains of biofilm-embedded MRSA have higher rifampin MICs after antibiotic treatment, resistant strains from surviving colonies were further plated with or without 0.06 or 64 μg/ml rifampin to measure the mutation and high-level-resistance rates. The mutation and high-level-resistance rates were 104.2% and 103.2% for rifampin alone, 100.5% and 103.6% for rifampin plus vancomycin, 105.0% and 104.5% for rifampin plus teicoplanin, 98.6% and 99.1% for rifampin plus daptomycin, and 101.5% and 100.6% for the nonsynergistic rifampin-plus-minocycline combination. In contrast, the mutation and high-level-resistance rates were 0% and 0% for the combinations of rifampin plus linezolid, rifampin plus fusidic acid, rifampin plus fosfomycin, rifampin plus tigecycline, and synergistic rifampin plus minocycline. Nearly all surviving bacteria from the plates treated with rifampin alone or in combination with vancomycin, teicoplanin, daptomycin, or minocycline were highly resistant to rifampin, with MICs > 64 μg/ml (Fig. 2).
Fig 2.
The percentage of rifampin-resistant mutants (MIC > 0.06 or > 64 mg/liter) of MRSA isolates from biofilm at day 5 after exposure to rifampin alone or in combination with eight antibiotics at the following susceptible breakpoint concentrations: vancomycin (VA) 2 μg/ml, teicoplanin (TEC) 8 μg/ml, daptomycin (DAP) 1 μg/ml, minocycline (MNO) 4 μg/ml, linezolid (LNZ) 4 μg/ml, fusidic acid (FA) 1 μg/ml, fosfomycin (FOS) 64 μg/ml, tigecycline (TGC) 0.5 μg/ml, and rifampin (RIF) 1 μg/ml.
In the present study, the rifampin combinations with fusidic acid, tigecycline, and, to a lesser extent, linezolid, minocycline, and fosfomycin exhibited synergistic activity against eight MRSA isolates. Linezolid monotherapy, as we previously reported (8), exhibited excellent inhibitory effects against biofilm-embedded MRSA. Rifampin augmented the antibacterial effect of linezolid in biofilm, as we previously found (8, 12). Vancomycin plus rifampin has exhibited an in vitro synergistic effect against MRSA isolates with a high vancomycin MIC (2 μg/ml) (13).However, the emergence and spread of rifampin-resistant MRSA isolates during vancomycin-rifampin combination therapy in an intensive care unit has been reported (14). Such a combination could easily induce high-level rifampin resistance (MIC > 64 μg/ml) in biofilm-embedded MRSA isolates, according to our in vitro study (8).
Although tigecycline has been considered for treatment of biofilm-embedded MRSA infection (5), our results question the therapeutic value of tigecycline monotherapy. However, at the susceptible breakpoint concentrations used in our experiments, which are feasible and compatible with achievable serum levels with currently recommended dosages, rifampin plus tigecycline could have synergistic antibacterial activity against MRSA in biofilm, with rare rifampin-resistant mutants. On the other hand, the combination of minocycline and rifampin, as used in our previous study (12), possesses a synergistic effect and leads to fewer mutants in MRSA in biofilm. That combination has not been mentioned before.
In vitro, daptomycin is considered effective and the fastest in eradicating MRSA in biofilm (5). However, in vivo studies have reported different results. Miró et al. found that the addition of gentamicin or rifampin did not enhance the effectiveness of daptomycin in the treatment of experimental endocarditis due to MRSA (15). Furthermore, our study found a poor effect of daptomycin monotherapy, and even of daptomycin combined with rifampin, against MRSA in biofilm. Certainly, rifampin-resistant mutants, especially isolates with high-level resistance, could easily be selected during such combination therapy. Such diversity might also be related to the concentrations used during the experiments, as we previously mentioned (8).
The combination of fusidic acid and rifampin is effective against MRSA infections (4, 16–18). Occasionally, such a combination can be a treatment option for heterogeneous vancomycin-intermediate S. aureus (hVISA) or VISA infections (19), as supported by our finding that fusidic acid plus rifampin is effective in decreasing bacterial loads in the biofilm model. In addition, the frequency of rifampin-resistant mutations in the presence of fusidic acid and rifampin in S. aureus is lower than that seen with rifampin monotherapy (10−11 versus <10−8) (20). Therefore, the rifampin-fusidic acid combination with synergism and low rifampin mutation-inducing activity could be useful as a long-term maintenance therapy for biofilm-associated MRSA infections.
Fosfomycin has long been used in combination therapies against MRSA, especially in biofilm (8, 13). When used in a combination against biofilm-embedded MRSA, it possesses less mutation-inducing potential than rifampin (8). However, the combination of fosfomycin with rifampin, which had never been used before, seemed to enhance the inhibitory activity and even synergism when used against some of the tested MRSA isolates. More evidence is needed to determine whether this combination also has low rifampin mutation-inducing activity.
In conclusion, we found that rifampin enhances the antibacterial activities of and even synergizes with fusidic acid, tigecycline, and, to a lesser extent, linezolid, fosfomycin, and (in part) minocycline against biofilm-embedded MRSA. Such combinations also possess weaker rifampin resistance induction activities. Animal experiments and clinical studies are essential to validate the usefulness of these rifampin-based combination regimens in clinical management of biofilm-related MRSA infections.
ACKNOWLEDGMENTS
We acknowledge Mei-Feng Lee for his critical review of this article and the members of the Research Laboratory of Infectious Diseases of the Chi-Mei Medical Center for their assistance in the statistical analyses of these data.
We declare that we have no conflicts of interest.
The study was supported by a grant from the National Science Council (NSC102-2314-13-384-009-MY2) and by the Chi-Mei Medical Center Research Foundation (CMFHT10201).
Footnotes
Published ahead of print 19 August 2013
REFERENCES
- 1.Prince AS. 2002. Biofilms, antimicrobial resistance, and airway infection. N. Engl. J. Med. 347:1110–1111 [DOI] [PubMed] [Google Scholar]
- 2.Edmiston CE, Jr, Goheen MP, Seabrook GR, Johnson CP, Lewis BD, Brown KR, Towne JB. 2006. Impact of selective antimicrobial agents on staphylococcal adherence to biomedical devices. Am. J. Surg. 192:344–354 [DOI] [PubMed] [Google Scholar]
- 3.Saginur R, Stdenis M, Ferris W, Aaron SD, Chan F, Lee C, Ramotar K. 2006. Multiple combination bactericidal testing of staphylococcal biofilms from implant-associated infections. Antimicrob. Agents Chemother. 50:55–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu C, Bayer A, Cosgrove SE, Daum RS, Fridkin SK, Gorwitz RJ, Kaplan SL, Karchmer AW, Levine DP, Murray BE, J Rybak M, Talan DA, Chambers HF; Infectious Diseases Society of America 2011. Clinical practice guidelines by the Infectious Diseases Society of America for the treatment of methicillin-resistant Staphylococcus aureus infections in adults and children. Clin. Infect. Dis. 52:e18–e55. 10.1093/cid/ciq146 [DOI] [PubMed] [Google Scholar]
- 5.Raad I, Hanna H, Jiang Y, Dvorak T, Reitzel R, Chaiban G, Sherertz R, Hachem R. 2007. Comparative activities of daptomycin, linezolid, and tigecycline against catheter-related methicillin-resistant Staphylococcus bacteremic isolates embedded in biofilm. Antimicrob. Agents Chemother. 51:1656–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chung M, de Lencastre H, Matthews P, Tomasz A, Adamsson I, Aires de Sousa M, Camou T, Cocuzza C, Corso A, Couto I, Dominguez A, Gniadkowski M, Goering R, Gomes A, Kikuchi K, Marchese A, Mato R, Melter O, Oliveira D, Palacio R, Sa-Leao R, Santos Sanches I, Song JH, Tassios PT, Villari P. 2000. Molecular typing of methicillin-resistant Staphylococcus aureus by pulsed-field gel electrophoresis: comparison of results obtained in a multilaboratory effort using identical protocols and MRSA strains. Microb. Drug Resist. 6:189–198 [DOI] [PubMed] [Google Scholar]
- 7.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang HJ, Chen CC, Cheng KC, Toh HS, Su BA, Chiang SR, Ko WC, Chuang YC. 2012. In vitro efficacy of fosfomycin-containing regimens against methicillin-resistant Staphylococcus aureus in biofilms. J. Antimicrob. Chemother. 67:944–950 [DOI] [PubMed] [Google Scholar]
- 9.Clinical and Laboratory Standards Institute 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A7 CLSI, Wayne, PA [Google Scholar]
- 10.Andrews JM. 2013. BSAC methods for antimicrobial susceptibility testing. (Version 12). British Society for Antimicrobial Chemotherapy, Birmingham, United Kingdom: http://www.bsac.org.uk/Susceptibility+Testing/Breakpoints [Google Scholar]
- 11.Brown SD, Traczewski MM. 2007. Comparative in vitro antimicrobial activity of tigecycline, a new glycylcycline compound, in freshly prepared medium and quality control. J. Clin. Microbiol. 45:2173–2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu WS. 2013. Efficacy of combination oral antimicrobial agnents against biofilm-embedded methicillin-resistant Staphyococcus aureus. J. Microbiol. Immunol. Infect. 46:89–95 [DOI] [PubMed] [Google Scholar]
- 13.Tang HJ, Chen CC, Ko WC, Yu WL, Chiang SR, Chuang YC. 2011. In vitro efficacy of antimicrobial agents against high-inoculum or biofilm-embedded meticillin-resistant Staphylococcus aureus with vancomycin minimal inhibitory concentrations equal to 2 mug/mL (VA2-MRSA). Int. J. Antimicrob. Agents 38:46–51 [DOI] [PubMed] [Google Scholar]
- 14.Ju O, Woolley M, Gordon D. 2006. Emergence and spread of rifampicin-resistant, methicillin-resistant Staphylococcus aureus during vancomycin-rifampicin combination therapy in an intensive care unit. Eur. J. Clin. Microbiol. Infect. Dis. 25:61–62 [DOI] [PubMed] [Google Scholar]
- 15.Miró JM, García-de-la-Mària C, Armero Y, Soy D, Moreno A, del Río A, Almela M, Sarasa M, Mestres CA, Gatell JM, Jiménez de Anta MT, Marco F; Hospital Clinic Experimental Endocarditis Study Group 2009. Addition of gentamicin or rifampin does not enhance the effectiveness of daptomycin in treatment of experimental endocarditis due to methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 53:4172–4177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howden BP, Grayson ML. 2006. Dumb and dumber—the potential waste of a useful antistaphylococcal agent: emerging fusidic acid resistance in Staphylococcus aureus. Clin. Infect. Dis. 42:394–400 [DOI] [PubMed] [Google Scholar]
- 17.Ferry T, Uckay I, Vaudaux P, Francois P, Schrenzel J, Harbarth S, Laurent F, Bernard L, Vandenesch F, Etienne J, Hoffmeyer P, Lew D. 2010. Risk factors for treatment failure in orthopedic device-related methicillin-resistant Staphylococcus aureus infection. Eur. J. Clin. Microbiol. Infect. Dis. 29:171–180 [DOI] [PubMed] [Google Scholar]
- 18.Matthews PC, Berendt AR, McNally MA, Byren I. 2009. Diagnosis and management of prosthetic joint infection. BMJ 338:b1773. 10.1136/bmj.b1773 [DOI] [PubMed] [Google Scholar]
- 19.Howden BP, Ward PB, Charles PG, Korman TM, Fuller A, du Cros P, Grabsch EA, Roberts SA, Robson J, Read K, Bak N, Hurley J, Johnson PD, Morris AJ, Mayall BC, Grayson ML. 2004. Treatment outcomes for serious infections caused by methicillin-resistant Staphylococcus aureus with reduced vancomycin susceptibility. Clin. Infect. Dis. 38:521–528 [DOI] [PubMed] [Google Scholar]
- 20.O'Neill AJ, Cove JH, Chopra I. 2001. Mutation frequencies for resistance to fusidic acid and rifampicin in Staphylococcus aureus. J. Antimicrob. Chemother. 47:647–650 [DOI] [PubMed] [Google Scholar]