Abstract
Alanine substitutions and selected deletions have been used to localize amino acids in QnrB essential for its protective activity. Essential amino acids are found at positions i and i−2 in the pentapeptide repeat module and in the larger of two loops, where deletion of only a single amino acid compromises activity. Deletion of 10 amino acids at the N terminus is tolerated, but removal of 3 amino acids in the C-terminal dimerization unit destroys activity.
TEXT
Qnr proteins bind to and protect bacterial DNA gyrase and topoisomerase IV from inhibition by quinolones (1–3). Qnr belongs to the pentapeptide repeat protein (PRP) family (1, 4). These proteins are composed of tandemly repeated units of five amino acids that fold into a right-handed quadrilateral β-helix and link end to end to form rod-shaped dimers with size, shape, and surface charge resembling those of B-form DNA (5, 6). In Gram-negative organisms, each monomer has a smaller loop of 8 and a larger loop of 12 amino acids needed for activity (6, 7). The helix is stabilized by hydrogen bonding between backbone atoms of neighboring coils. By convention, the central amino acid in the pentapeptide unit is termed i, with neighboring amino acids i+1 and i−1 and i+2 and i−2 (5). The side chains of the i and i−2 residues pack in the interior of the β-helix, stabilizing its structure while the side chains of the other residues point outward and form the surface of the protein interacting with DNA gyrase and topoisomerase IV.
The aim of this work was to determine essential features of QnrB1 necessary for its protective activity. It was initiated in 2005 before the structural features of the molecule were completely known. To identify potential essential amino acids in Qnr proteins, the sequences of the then-known six alleles of QnrA, six of QnrB, and two of QnrS were aligned. Sites with a common amino acid were considered potentially essential. Site-directed mutagenesis was then used to replace these amino acids individually with alanine, which has a small, neutral, nonpolar side chain, and the protective activity of the resulting mutant for ciprofloxacin was measured by agar dilution MIC. In addition, the effects of deleting amino acids at each end of the molecule and from within the largest loop were evaluated.
The construction of IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible Escherichia coli M15(pREP4)(pQE-60-QnrB1) expressing QnrB1 with a C-terminal His6 tag and E. coli BL21(DE3)(pET28a:QnrB1) expressing QnrB1 with an N-terminal His6 tag has been described elsewhere (6, 8). Site-directed mutagenesis was performed with the QuikChange site-directed mutagenesis kit (Agilent, Santa Clara, CA) using E. coli XL1-Blue as recipient and primer pairs that matched the qnrB1 sequence except for the desired base substitutions or deletion centrally located in the primers. After sequencing to confirm mutagenesis, plasmids were transformed into M15(pREP4) or BL21(DE3) by electroporation. Ciprofloxacin MICs were determined in at least triplicate by agar dilution according to CLSI guidelines (9) using Mueller-Hinton agar, an inoculum of ∼104 CFU per spot, and incubation at 35°C for 16 to 20 h. MICs were determined with 0, 10, and 100 μM IPTG to evaluate basal and stimulated expression levels. Wild-type and mutant QnrB1-His6 proteins were purified as previously described (6). In brief, BL21(DE3) derivatives were grown in autoinduction medium (10), harvested by centrifugation, and treated with lysozyme and sonication. After centrifugation, the supernatant was applied to a nickel-nitrilotriacetic acid (Ni-NTA) column (HiTrap Chelating HP; GE Healthcare). Fractions were eluted with increasing concentrations of imidazole, evaluated for purity by SDS-PAGE, dialyzed, and stored in liquid nitrogen. Gyrase supercoiling assays were performed with recombinant E. coli DNA gyrase, gyrase substrate, and reaction buffer from NE Biolabs (Ipswich, MA). Supercoiled band intensity was evaluated with the Gel Doc EZ imager (Bio-Rad, Hercules, CA), which provides quantitative data from which half-protective concentration can be estimated.
Table 1 shows results with pQE-60-QnrB1 in E. coli M15(pREP4). The first column lists the site of alanine replacement or deletion numbered from the start of translation at the second potential ATG site (11), using standard single-letter amino acid designations. These sites were those conserved among the 2005 set of QnrA, QnrB, and QnrS alleles, excluding 5 native alanine residues. The second column shows the position of the mutated amino acid within the pentapeptide repeat sequence or loop (i, i−1, etc.) as determined from the structure of QnrB1 (6). Alanine replacements were constructed at 9 i−2, 5 i−1, 21 i, 4 i+1, and 7 i+2 sites. The third column shows the 28 amino acids that are invariant, and hence most likely to be essential, in a current collection of 64 Qnr sequences determined by chromosomal or plasmid genes in gammaproteobacteria (not including QnrB alleles) (12). The fourth column shows naturally occurring amino acid variants at these sites among the 72 currently known QnrB alleles (http://www.lahey.org/qnrstudies). The next three columns give the ciprofloxacin MICs for the alanine mutant at each site under various degrees of IPTG stimulation.
Table 1.
Susceptibility of deletions and alanine substitutions in QnrB at conserved amino acids of the plasmid and Gram-negative chromosomal Qnr group
| Strain/mutant | PRP positiona | Conservedb | QnrB variantc | Ciprofloxacin MIC (μg/ml) at μM IPTG: |
||
|---|---|---|---|---|---|---|
| 0 | 10 | 100 | ||||
| M15(pREP4)(pQE-60) | 0.032 | 0.032 | 0.032 | |||
| M15(pREP4)(pQE-60-QnrB1) | 0.125 | 0.25 | 0.25 | |||
| F25A | i | Y | 0.032 | 0.032 | 0.25 | |
| F30A | i | Y | 0.032 | 0.064 | 0.25 | |
| L35A | i | M | 0.032 | 0.064 | 0.25 | |
| F40A | i | Y | 0.032 | 0.064 | 0.25 | |
| C43A | i−2 | Y | 0.032 | 0.064 | 0.25 | |
| F45A | i | Y | 0.032 | 0.032 | 0.125 | |
| G53A | Smaller loop | 0.032 | 0.032 | 0.125 | ||
| C54A | i−2 | 0.125 | 0.25 | 0.25 | ||
| F56A | i | 0.032 | 0.032 | 0.064 | ||
| L61A | i | Y | 0.125 | 0.25 | 0.25 | |
| F66A | i | Y | 0.032 | 0.032 | 0.064 | |
| C69A | i−2 | Y | S | 0.032 | 0.064 | 0.25 |
| L71A | i | 0.125 | 0.25 | 0.25 | ||
| F76A | i | 0.032 | 0.064 | 0.25 | ||
| G83A | i+2 | Y | 0.064 | 0.125 | 0.25 | |
| E85A | i−1 | 0.125 | 0.125 | 0.25 | ||
| C89A | i−2 | 0.064 | 0.125 | 0.25 | ||
| G93A | i+2 | Y | 0.064 | 0.125 | 0.25 | |
| F96A | i | Y | 0.032 | 0.032 | 0.125 | |
| F101A | i | Y | 0.032 | 0.032 | 0.25 | |
| F111A | Larger loop | Y | 0.032 | 0.032 | 0.032 | |
| C112A | Larger loop | 0.064 | 0.064 | 0.25 | ||
| I116A | i | 0.032 | 0.032 | 0.032 | ||
| N120A | i−1 | Y | 0.032 | 0.032 | 0.25 | |
| L121A | i | 0.032 | 0.032 | 0.25 | ||
| Y123A | i+2 | Y | 0.032 | 0.064 | 0.125 | |
| N125A | i−1 | 0.032 | 0.032 | 0.25 | ||
| E132A | i+1 | Y | 0.032 | 0.064 | 0.125 | |
| K133A | i+2 | 0.125 | 0.25 | 0.5 | ||
| C134A | i−2 | Y | 0.064 | 0.064 | 0.25 | |
| L136A | i | Y | 0.032 | 0.032 | 0.125 | |
| E138A | i+2 | 0.125 | 0.25 | 0.25 | ||
| N139A | i−2 | Y | 0.032 | 0.032 | 0.032 | |
| W141A | i | Y | 0.125 | 0.25 | 0.25 | |
| G148A | i+2 | 0.032 | 0.125 | 0.5 | ||
| S154A | i−2 | 0.032 | 0.125 | 0.5 | ||
| D155A | i−1 | 0.032 | 0.032 | 0.25 | ||
| L156A | i | Y | 0.032 | 0.032 | 0.125 | |
| S157A | i+1 | Y | A | 0.032 | 0.125 | 0.25 |
| G159A | i−2 | 0.032 | 0.032 | 0.25 | ||
| F161A | i | Y | 0.032 | 0.032 | 0.032 | |
| S162A | i+1 | T | 0.125 | 0.25 | 0.25 | |
| W166A | i | Y | 0.032 | 0.032 | 0.032 | |
| L176A | i | 0.032 | 0.032 | 0.064 | ||
| L181A | i | Y | 0.032 | 0.032 | 0.125 | |
| L184A | i−2 | 0.032 | 0.032 | 0.125 | ||
| R187A | i+1 | Y | 0.064 | 0.125 | 0.25 | |
| G193A | i+2 | 0.032 | 0.032 | 0.25 | ||
| K195A | i−1 | 0.064 | 0.125 | 0.25 | ||
| Q200A | dm | Y | 0.064 | 0.125 | 0.25 | |
| E206A | dm | 0.064 | 0.125 | 0.064 | ||
| L208A | dm | 0.064 | 0.064 | 0.064 | ||
| G209A | dm | Y | 0.032 | 0.032 | 0.125 | |
| ΔN5 | 0.125 | 0.25 | 0.25 | |||
| ΔN10 | 0.064 | 0.125 | 0.064 | |||
| ΔN15 | 0.032 | 0.032 | 0.032 | |||
| ΔN20 | 0.032 | 0.032 | 0.032 | |||
| ΔC3 | 0.032 | 0.032 | 0.032 | |||
| ΔC5 | 0.032 | 0.032 | 0.032 | |||
| ΔC10 | 0.032 | 0.032 | 0.032 | |||
In a pentapeptide repeat protein (PRP), the first amino acid is indicated by i−2, the next by i−1. the central residue by i, and the following two by i+1 and i+2. In QnrB1, the smaller loop comprises residues 46 to 53 and the larger loop comprises amino acids 102 to 113. dm indicates localization in the dimerization module. Δ signifies deletion of the indicated number of amino acids at the N or C terminus of the molecule.
Y indicates amino acids conserved among 6 transmissible and 58 chromosomally determined Qnr proteins in a recent survey of Gram-negative organisms (12).
Amino acid substitutions in naturally occurring variants of QnrB (http://www.lahey.org/qnrstudies).
Without qnrB, M15(pREP4) had a basal ciprofloxacin MIC of 0.032 μg/ml. With pQE-60-QnrB1, the MIC increased 4-fold in the absence of IPTG and 8-fold in its presence. Some alanine replacements had no deleterious effect on MIC: C54A, L61A, L71A, E85A, K133A, E138A, W141A, and S162A. A few reduced the MIC to the basal level in the absence of QnrB1—F111A, I116A, N139A, F161A, and W166A—or showed minimal activity except with maximal IPTG stimulation—F56A, F66A, and L176A. Several of these sites were previously identified as important via site-directed substitution of amino acids other than alanine (13). One of these mutants was in the larger loop, and seven were in the structurally important i (n = 6) or i−2 (n = 1) positions of the pentapeptide repeat. Since the i and i−2 sites interact to maintain the β-helix, the alanine substitutions presumably destroy protective activity by disrupting the helical structure of the molecule. The remainder of the alanine substitutions had a reduced basal MIC but reached the same MIC as did the wild type with maximal IPTG stimulation. The lack of an effect of single alanine substitutions of externally oriented amino acids is thus consistent with a model in which the docking of the core Qnr backbone with gyrase is not dependent on the binding affinity of any single amino acid. Table 1 also shows the effect of N- or C-terminal deletions on QnrB1 activity. Five or 10 amino acids could be removed from the N terminus without total loss of activity, but removal of 15 or 20 amino acids from the N terminus and deletion of even 3 amino acids from the C-terminal dimerization module destroyed the protective effect, as reported previously for terminal deletions in QnrC (14). The formation of C-terminal end-to-end dimers is thus also critical for protective activity.
Table 2 shows the result of alanine substitutions in the essential loops of QnrB1. The amino acid composition of both loops varies among Qnr proteins. The first column shows the amino acid present in QnrB1. The second column shows the number of amino acids found at each site in the loops of naturally occurring Qnr proteins and varies from 1 (invariant F111) to 10 (12). The host BL21(DE3) had a ciprofloxacin MIC of 0.0005 μg/ml. With pET28a:QnrB1, the basal MIC was 0.008, increasing to 0.032 μg/ml with maximal IPTG stimulation. Alanine substitutions in the smaller loop (amino acids 46 to 53) had a relatively minor effect on activity. The effect of replacements in the larger loop (amino acids 102 to 113) was more pronounced. Activity was markedly reduced in N103A, W110A, and S113A and considerably diminished in the F111A and C112A mutants. Little or no loss of protective activity was seen in T106A, T107A, and R108A, but deletion of any of these amino acids individually reduced activity, while removal of two or three residues at these sites severely compromised protective activity. In general, the fewer the naturally occurring amino acids at a particular site (shown in Table 2, column 2), the greater was the effect of alanine substitution on protective activity, thus correlating protective function with conservation of amino acids at these positions.
Table 2.
Susceptibility of alanine substitutions and deletions in QnrB1 loops
| Strain/mutant | No. of variantsa | Ciprofloxacin MIC (μg/ml) at μM IPTG: |
||
|---|---|---|---|---|
| 0 | 10 | 100 | ||
| BL21(DE3) | 0.0005 | 0.0005 | 0.0005 | |
| BL21(DE3)(pET28a:QnrB1) | 0.008 | 0.016 | 0.032 | |
| Y46Ab | 6 | 0.001 | 0.001 | 0.032 |
| D47A | 3 | 0.004 | 0.008 | 0.032 |
| R48A | 9 | 0.008 | 0.008 | 0.032 |
| E49A | 8 | 0.004 | 0.004 | 0.032 |
| S50A | 7 | 0.002 | 0.002 | 0.008 |
| Q51A | 10 | 0.008 | 0.016 | 0.032 |
| K52A | 10 | 0.008 | 0.016 | 0.032 |
| G53A | 4 | 0.004 | 0.008 | 0.032 |
| M102Ac | 6 | 0.004 | 0.004 | 0.016 |
| N103A | 2 | 0.001 | 0.001 | 0.002 |
| M104A | 6 | 0.002 | 0.002 | 0.016 |
| I105A | 2 | 0.002 | 0.002 | 0.008 |
| T106A | 4 | 0.008 | 0.016 | 0.064 |
| T107A | 10 | 0.004 | 0.008 | 0.032 |
| R108A | 6 | 0.004 | 0.004 | 0.016 |
| T109A | 7 | 0.004 | 0.004 | 0.008 |
| W110A | 5 | 0.001 | 0.002 | 0.002 |
| F111A | 1 | 0.001 | 0.001 | 0.004 |
| C112A | 2 | 0.002 | 0.004 | 0.004 |
| S113A | 5 | 0.0005 | 0.001 | 0.001 |
| Δ106T | 0.001 | 0.001 | 0.001 | |
| Δ107T | 0.001 | 0.001 | 0.004 | |
| Δ108R | 0.001 | 0.001 | 0.002 | |
| Δ106T-107T | 0.001 | 0.001 | 0.002 | |
| Δ107T-108R | 0.001 | 0.001 | 0.002 | |
| Δ106T-107T-108R | 0.0005 | 0.0005 | 0.0005 | |
Number of amino acids found at this position in naturally occurring Qnr proteins from Gram-negative bacteria (12).
The smaller external loop encompasses positions 46 to 53.
The larger external loop encompasses positions 102 to 113.
To validate that the loss of protective activity occurred at the level of mutant protein, the T106A and Δ106 QnrB1-His6 proteins were purified and compared with wild-type QnrB1-His6 for in vitro ability to protect DNA gyrase from inhibition by ciprofloxacin and inhibitory effect on gyrase at high concentration (Fig. 1). The protective activity of both mutant proteins was compromised. With 2.5 μM ciprofloxacin, the concentration of wild-type QnrB-His6 required for half-protection was ∼27 nM, but for the T106A mutant, it was ∼160 nM, 6-fold higher despite the absence of an effect on MIC (Table 2). The Δ106 mutant protein never achieved half-protective activity but, interestingly, was at least as active as the wild type in inhibiting gyrase, while the T106A mutant protein was less effective in both inhibition and protection than the wild type. Alanine substitution at position 106 thus decreases the ability of QnrB to interact with gyrase, while shrinkage of the larger loop by the single 106-amino-acid deletion compromises protective activity with little interference in the ability of the protein to interact with gyrase as an inhibitor.
Fig 1.
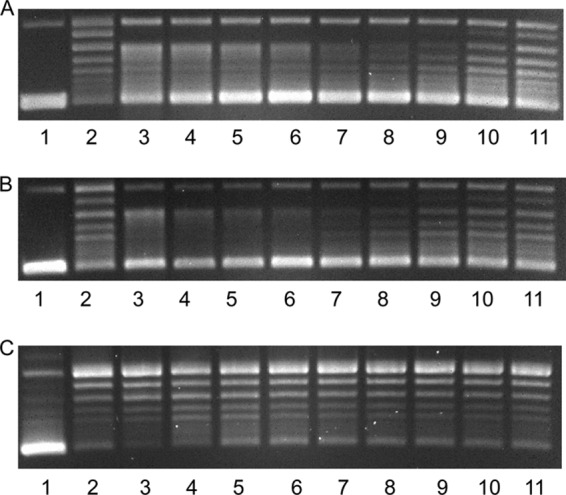
Protection of DNA gyrase from ciprofloxacin inhibition of supercoiling by wild-type QnrB1-His6 (A), the T106A mutant (B), and the ΔT106 deletion mutant (C). Reaction mixtures of 30 μl were analyzed by agarose gel electrophoresis. Each contained 0.5 μg relaxed pUC19 DNA, reaction buffer, and 1 unit gyrase (lanes 1 to 11), 2.5 μM ciprofloxacin (lanes 2 to 11), and QnrB1-His6 fusion protein at 7 μM (lane 3), 3.5 μM (lane 4), 1.75 μM (lane 5), 880 nM (lane 6), 440 nM (lane 7), 220 nM (lane 8), 110 nM (lane 9), 55 nM (lane 10), and 27 nM (lane 11).
These studies highlight conserved amino acid residues in Qnr proteins where substitution of alanine for the native amino acid predictably disrupts the overall β-helical structure of the molecule, the importance of C-terminal amino acids involved in linkage to create the active dimer, and the sensitivity of the larger nonhelical loop to alanine substitutions and especially the dramatic loss of protective but not inhibitory activity with deletion of a single loop amino acid. The latter finding implies that the larger external loop, and particularly its size, is key for determining the nature of the interaction of QnrB with gyrase.
ACKNOWLEDGMENT
This work was supported by grant R01 AI057576 (to D.C.H. and G.A.J.) from the National Institutes of Health, U.S. Public Health Service.
Footnotes
Published ahead of print 26 August 2013
REFERENCES
- 1.Tran JH, Jacoby GA. 2002. Mechanism of plasmid-mediated quinolone resistance. Proc. Natl. Acad. Sci. U. S. A. 99:5638–5642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tran JH, Jacoby GA, Hooper DC. 2005. Interaction of the plasmid-encoded quinolone resistance protein Qnr with Escherichia coli DNA gyrase. Antimicrob. Agents Chemother. 49:118–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tran JH, Jacoby GA, Hooper DC. 2005. Interaction of the plasmid-encoded quinolone resistance protein QnrA with Escherichia coli topoisomerase IV. Antimicrob. Agents Chemother. 49:3050–3052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vetting MW, Hegde SS, Fajardo JE, Fiser A, Roderick SL, Takiff HE, Blanchard JS. 2006. Pentapeptide repeat proteins. Biochemistry 45:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hegde SS, Vetting MW, Roderick SL, Mitchenall LA, Maxwell A, Takiff HE, Blanchard JS. 2005. A fluoroquinolone resistance protein from Mycobacterium tuberculosis that mimics DNA. Science 308:1480–1483 [DOI] [PubMed] [Google Scholar]
- 6.Vetting MW, Hegde SS, Wang M, Jacoby GA, Hooper DC, Blanchard JS. 2011. Structure of QnrB1, a plasmid-mediated fluoroquinolone resistance factor. J. Biol. Chem. 286:25265–25273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiong X, Bromley EH, Oelschlaeger P, Woolfson DN, Spencer J. 2011. Structural insights into quinolone antibiotic resistance mediated by pentapeptide repeat proteins: conserved surface loops direct the activity of a Qnr protein from a gram-negative bacterium. Nucleic Acids Res. 39:3917–3927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jacoby GA, Walsh KE, Mills DM, Walker VJ, Oh H, Robicsek A, Hooper DC. 2006. qnrB, another plasmid-mediated gene for quinolone resistance. Antimicrob. Agents Chemother. 50:1178–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.CLSI 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 9th ed. CLSI document M07-A9 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 10.Fox BG, Blommel PG. 2009. Autoinduction of protein expression. Curr. Protoc. Protein Sci. Chapter 5:Unit 5.23. 10.1002/0471140864.ps0523s56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacoby G, Cattoir V, Hooper D, Martínez-Martínez L, Nordmann P, Pascual A, Poirel L, Wang M. 2008. qnr gene nomenclature. Antimicrob. Agents Chemother. 52:2297–2299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacoby GA, Hooper DC. 2013. Phylogenetic analysis of chromosomally determined Qnr and related proteins. Antimicrob. Agents Chemother. 57:1930–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodríguez-Martínez JM, Briales A, Velasco C, Conejo MC, Martínez-Martínez L, Pascual A. 2009. Mutational analysis of quinolone resistance in the plasmid-encoded pentapeptide repeat proteins QnrA, QnrB and QnrS. J. Antimicrob. Chemother. 63:1128–1134 [DOI] [PubMed] [Google Scholar]
- 14.Guo Q, Weng J, Xu X, Wang M, Wang X, Ye X, Wang W. 2010. A mutational analysis and molecular dynamics simulation of quinolone resistance proteins QnrA1 and QnrC from Proteus mirabilis. BMC Struct. Biol. 10:33. 10.1186/1472-6807-10-33 [DOI] [PMC free article] [PubMed] [Google Scholar]


