Abstract
We investigated the mechanisms of resistance to carbapenems, aminoglycosides, glycylcyclines, tetracyclines, and quinolones in 90 multiresistant clinical strains of Acinetobacter baumannii isolated from two genetically unrelated A. baumannii clones: clone PFGE-ROC-1 (53 strains producing the OXA-58 β-lactamase enzyme and 18 strains with the OXA-24 β-lactamase) and clone PFGE-HUI-1 (19 strains susceptible to carbapenems). We used real-time reverse transcriptase PCR to correlate antimicrobial resistance (MICs) with expression of genes encoding chromosomal β-lactamases (AmpC and OXA-51), porins (OmpA, CarO, Omp33, Dcap-like, OprB, Omp25, OprC, OprD, and OmpW), and proteins integral to six efflux systems (AdeABC, AdeIJK, AdeFGH, CraA, AbeM, and AmvA). Overexpression of the AdeABC system (level of expression relative to that by A. baumannii ATCC 17978, 30- to 45-fold) was significantly associated with resistance to tigecycline, minocycline, and gentamicin and other biological functions. However, hyperexpression of the AdeIJK efflux pump (level of expression relative to that by A. baumannii ATCC 17978, 8- to 10-fold) was significantly associated only with resistance to tigecycline and minocycline (to which the TetB efflux system also contributed). TetB and TetA(39) efflux pumps were detected in clinical strains and were associated with resistance to tetracyclines and doxycycline. The absence of the AdeABC system and the lack of expression of other mechanisms suggest that tigecycline-resistant strains of the PFGE-HUI-1 clone may be associated with a novel resistance-nodulation-cell efflux pump (decreased MICs in the presence of the inhibitor Phe-Arg β-naphthylamide dihydrochloride) and the TetA(39) system.
INTRODUCTION
Acinetobacter baumannii is an important pathogen that causes nosocomial infections associated with high morbidity and mortality (1). Multidrug-resistant (MDR) strains of A. baumannii have emerged in the last few decades as a result of the combination of two main factors: (i) a high level of genomic plasticity (2) and (ii) mutation of endogenous genes, alteration of which is associated with antimicrobial resistance, such as overexpression of the chromosomally encoded ADC β-lactamase (AmpC) (3) and the OXA-51-like β-lactamase (4), loss of expression of porins (CarO and Omp33) (5, 6), mutation in the gyrA and parC genes (7), and overexpression of efflux systems (8).
Overexpression of the OXA-51-like β-lactamase has been associated with resistance to carbapenems and decreased expression of CarO and Omp33 (5, 6, 9).
Efflux pumps have multifactorial roles. These mechanisms are important for detoxification of intracellular metabolites, bacterial virulence (in both animal and plant hosts), intercellular signaling and trafficking, and cell homeostasis (10). Three resistance-nodulation-cell division (RND) systems, AdeABC, AdeIJK, and AdeFGH, have been characterized and reported to cause MDR in A. baumannii (8). AdeABC is the RND system most frequently involved in MDR in clinical strains; it has been found in approximately 80% of clinical isolates (the rates reported vary from 53% to 97%) (11) but was not detected in 32 environmental isolates (12). AdeRS is a two-component system that regulates AdeABC expression (13). Mutations in this system and the presence of an ISAba1 insertion sequence in this system can lead to overexpression of the AdeABC operon (13–15). However, strains of A. baumannii that express AdeABC without mutations have been found in association with AdeRS (16, 17). Recently, the adeN gene has been found to be associated with the regulation of the AdeIJK system (18), and mutations in the adeL gene have been associated with overexpression of the AdeFGH pump (11). Three other types of efflux systems have been described in A. baumannii: CraA (a major facilitator superfamily [MFS] pump), which confers intrinsic chloramphenicol resistance (19); AbeM (a member of the multidrug and toxic compound extrusion [MATE] family of pumps), which extrudes several antimicrobials and biocides (20); and AmvA (an MFS pump), which confers resistance to detergents, disinfectants, dyes, and erythromycin. Overexpression of the AmvA efflux pump has been associated with increased drug resistance in A. baumannii clinical isolates (21). Finally, several tetracycline efflux pumps (systems acquired from the MFS superfamily) have been described in A. baumannii. The most prevalent of these are TetA, which is associated with resistance to tetracycline, and TetB, which is implicated in resistance to tetracycline, doxycycline, and minocycline (8). TetA(39) is an important tetracycline resistance mechanism in clinical strains (22).
Because of the complexity of clinical strains of A. baumannii, many researchers have used ATCC reference strains to investigate the mechanisms of resistance. However, very few studies have analyzed the combinations of mechanisms and their interrelation in clinical isolates of Acinetobacter baumannii. Here, we studied the interplay between the mechanisms of multidrug resistance in clinical A. baumannii strains, particularly those involving efflux pumps, the influx of antimicrobials, and chromosomally encoded β-lactamases.
MATERIALS AND METHODS
Bacterial isolates and molecular typing.
In 2010, 444 strains of A. baumannii were isolated (from 273 patients) in 42 participating hospitals and identified as part of the second multicenter study on this pathogen in Spain (GEIH-REIPI-2010-Ab) (23). The strains were identified by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry (24) and amplified rRNA gene restriction analysis (ARDRA) (25). Species identification was confirmed by detection of the OXA-51 gene by PCR (26) and also by detection of the bsp gene (a novel target) by real-time quantitative PCR (27, 28).
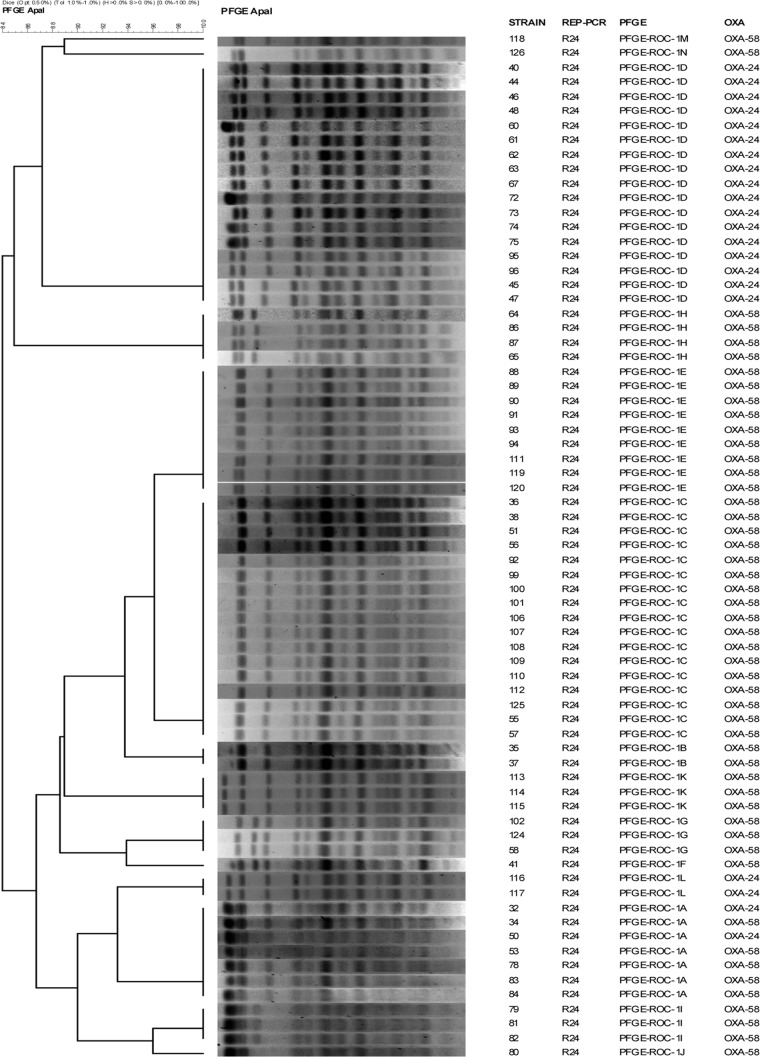
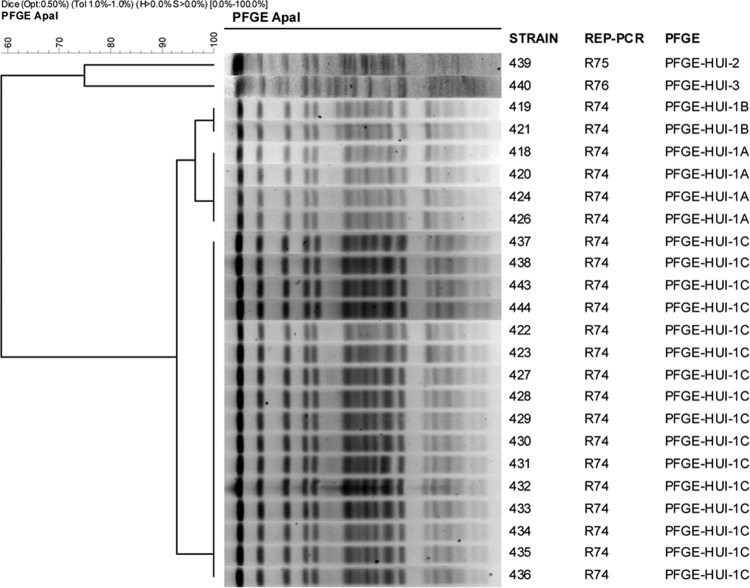
The clonal relationship between all strains displaying various levels of antibiotic susceptibility (n = 71) from a hospital in southern Spain and a hospital in the Canary Islands (n = 19) was determined by pulsed-field gel electrophoresis (PFGE) (29, 30) of samples of chromosomal DNA digested with ApaI (Roche, Mannheim, Germany) and embedded in low-melting point agarose. The restriction fragments thus obtained were separated in a CHEF DR-III system (Bio-Rad, Hercules, CA). FPQuest II software, version 4.5 (Bio-Rad), was used to analyze the band patterns in the agarose gel electrophoresis images (cutoff = 85%). Strains of both clones were analyzed by multilocus sequence typing, according to the system developed by Nemec and coworkers (31).
Susceptibility testing.
The antibiotic susceptibility profile was determined according to CLSI recommendations (23). In strains of the PFGE-HUI-1 clone, we determined MICs in the presence of Phe-Arg β-naphthylamide dihydrochloride (PAbetaN; a commonly assumed inhibitor of the RND efflux pump) (32).
DNA amplifications studies.
We used PCR to detect the genes coding for common aminoglycoside-modifying enzymes (AacA4, AacC1, AacC2, AadB, AadA1, AphA1, AphA6, and AadA2) (33); CHDL enzymes (OXA-23, -24, -51, -58, and -143) (34, 35); MBL enzymes (IMP, VIM, SPM-1, GIM-1, SIM-1, BIC, DIM, and NDM) (36); extended-spectrum β-lactamases (ESBLs), such as GES enzymes; and carbapenemases, such as KPC enzymes. We sequenced the gyrA and parC genes to study the presence of the mutations. Finally, we analyzed the tet genes most commonly detected in isolates of A. baumannii [tetA, tetB, tetA(34), and tetA(39)] (37, 38).
Real-time RT-PCR studies.
We used real-time reverse transcriptase PCR (RT-PCR) to examine all isolates for expression of adeB, adeJ, adeG, abeM, craA, and amvA (genes belonging to efflux pumps systems); oprC, oprD, ompW, ompA,carO, omp33, dcap-like, oprB, and omp25 (genes harboring porins or outer membrane protein); and finally, the OXA-51 and ampC β-lactamase genes. We obtained DNase-treated RNA from late-log-phase cultures (optical density = 0.4 to 0.6 absorbance units) by using a High Pure RNA isolation kit (Roche, Germany) and 50 ng of RNA. Analysis of controls without reverse transcriptase confirmed the absence of contaminating DNA in the samples. We used a LightCycler 480 RNA master hydrolysis probe (Roche, Germany) for the RT-PCR studies. The Universal Probe Library (UPL) TaqMan probes (Roche, Germany) and primers used are listed in Table 1. All were designed from conserved regions of DNA after the alignment of the genomes of the following strains of A. baumannii: AB 307-0294, AB 0057, ACICU, SDF, AYE, and ATCC 17978. We adjusted the concentrations of the samples to achieve efficiencies of 90% to 110% and performed all experiments in triplicate from three RNA extractions. For each strain, we normalized the levels of expression of all genes relative to those of the single-copy housekeeping genes rpoB and gyrB. We then calibrated the normalized expression of each gene of interest relative to its expression by A. baumannii ATCC 17978, which was assigned a value of 1.0.
Table 1.
Primers used in this study
| Analysis and gene | Orientation | Primer sequence (5′–3′) | UPL probea | Reference or source |
|---|---|---|---|---|
| RT-PCR analysis | ||||
| rpoB | Forward | CGTGTATCTGCGCTTGG | 131 | This study |
| Reverse | CGTACTTCGAAGCCTGCAC | |||
| gyrB | Forward | TGGTGGAACGTGGTCATATTT | 76 | This study |
| Reverse | TGCTCTTGCTTACCCTTTTTG | |||
| adeB | Forward | CGAGTGGCACAACTAGCATC | 61 | This study |
| Reverse | CCTTGTCTTGGCTGCACTCT | |||
| adeJ | Forward | CCTATTGCACAATATCCAACGA | 119 | This study |
| Reverse | AGGATAAGTCGCAGCAATCG | |||
| adeG | Forward | GTCCTGAAATGGTCGTTCGT | 43 | This study |
| Reverse | AGCTTCTGCTTGGCTAGATGA | |||
| craA | Forward | TTCATTGCTTGCGCCTTT | 125 | This study |
| Reverse | CCAGTGCCATGAAACATAATCA | |||
| abeM | Forward | AGGGACGTATTATGGCGAAA | 165 | This study |
| Reverse | CTGCTGTGCTTAGACCAATTTTT | |||
| amvA | Forward | GCAGAGAAATTTTGCACTTGG | 10 | This study |
| Reverse | CGACTAATGGACCAAAAGCTG | |||
| ompA | Forward | GGTATTCAGATAATTTTTCAGCAACTT | 129 | This study |
| Reverse | AACAAATCAAACATCAAAGACCAA | |||
| ompW | Forward | GCCTTATTTGCTCTGCCAAC | 60 | This study |
| Reverse | CGTTTGAAACCATCACCATCT | |||
| dcap-like | Forward | TGATCGACTTCTCGACAAACA | 77 | This study |
| Reverse | GTGTAGTTGGGCCTAGTTTGTAGTT | |||
| oprC | Forward | ACTCGATACAAAGCGGTGGA | 9 | This study |
| Reverse | TTTAATACGTGAACCAAACATACCTC | |||
| oprB | Forward | GCCCCACACTTCTTGAACAG | 67 | This study |
| Reverse | ATGGGCAATCGCTTTCTG | |||
| omp25 | Forward | CGAACGTGAAATCGACAACA | 128 | This study |
| Reverse | CGTAACCTTTAACACCTAGAGCAAG | |||
| omp33-omp36 | Forward | CAAGTGTTGCTAACCAATTCGCT | FAM-CCAAACTGCTGCTATCCAAAACGACCAA-BBQ | This study |
| Reverse | GTTTTCTTGACCGAATGCACC | |||
| carO | Forward | TGTTCATGACAGCTATGCATTCGATA | FAM-CGCTCGTGCTGAAGTAGGTACTACAGGTT-BBQ | This study |
| Reverse | CCCAATGCTAAACCTACATATGGGT | |||
| Sequencing analysis | ||||
| adeR | Forward | ACTACGATATTGGCGACATT | ||
| Reverse | GCGTCAGATTAAGCAAGATT | 13–15 | ||
| adeS | Forward | TTGGTTAGCCACTGTTATCT | ||
| Reverse | AGTGGACGTTAGGTCAAGTT | 13–15 | ||
| adeN | Forward | GCTGTTAGGTTGGGGTCGTA | ||
| Reverse | CGTGACCAAAAGTACGAATCA | 18 |
FAM, 6-carboxyfluorescein; BBQ, BlackBerry Quencher.
Sequencing of the genes regulating AdeABC and AdeIJK efflux pumps.
We sought mutations in the regulatory genes adeR-adeS and adeN, which have previously been associated with upregulation of the AdeABC and AdeIJK efflux systems, respectively. We amplified the genes by using the primers listed in Table 1. We used the Macrogen program (Macrogen Europe, Amsterdam, Netherlands) for DNA sequencing and the NCBI BLAST (www.ncbi.nlm.gov/BLAST) and CLUSTAL (www.ebi.ac.uk/Tools/msa/clustalw2/) programs for posterior analyses.
Statistical analysis.
We categorized the strains into two groups according to antimicrobial susceptibility (not following the CLSI or EUCAST clinical breakpoints). We worked with the Student's t test to compare differences in gene expression between groups and thus evaluate any associations with antibiotic resistance. Differences were considered significant at a P value of <0.05.
Nucleotide sequence accession numbers.
The nucleotide sequences of the adeR, adeS, and adeN genes from strains of the PFGE-ROC-1 clone were submitted to the GenBank database and have been assigned accession numbers KF147860, KF147861, and KF147862, respectively.
RESULTS
MICs, typing, and PCR detection of genes of the isolates.
To study the expression levels of efflux pump systems, porins, and chromosomal β-lactamases, we selected clonally related strains with different antibiotic susceptibilities (39). In Spain, OXA-type enzymes are prevalent in carbapenem-resistant strains of A. baumannii (40, 41). Isolates of A. baumannii from two hospitals in Spain that showed some clonal relation were designated clone PFGE-ROC-1 (sequence type 2 [ST2]) (n = 71; Fig. 1) and clone PFGE-HUI-1 (ST79) (n = 19; Fig. 2). Moreover, 53 strains of the PFGE-ROC-1 clone carried the OXA-58 β-lactamase gene (designated PFGE-ROC-1OXA-58; imipenem MICs, 8 to 64 mg/liter; meropenem MICs, 8 to 16 mg/liter) and 18 isolates carried the OXA-24 β-lactamase gene (designated PFGE-ROC-1OXA-24; imipenem MICs, ≥64 mg/liter; meropenem MICs, 32 to 64 mg/liter). The isolates of clone PFGE-HUI-1 (n = 19) were susceptible to carbapenems. We studied the variability in the MICs of glycylcyclines, aminoglycosides, tetracyclines, rifampin, and doripenem for all isolates, with the following results: (i) for PFGE-ROC-1OXA-58 (Table 2) tigecycline MICs were ≤0.25 to 2 mg/liter, gentamicin MICs were 1 to >64 mg/liter, amikacin MICs were <2 to 64 mg/liter, doxycycline MICs were 16 to >64 mg/liter, minocycline MICs were 1 to 8 mg/liter, tetracycline MICs were >64 mg/liter, netilmicin MICs were 1 to >64 mg/liter, rifampin MICs were 1 to 64 mg/liter, tobramycin MICs were <0.5 to 64, and doripenem MICs were 4 to 8 mg/liter. (ii) For PFGE-ROC-1OXA-24 (Table 3), tigecycline MICs were ≤0.25 to 1 mg/liter, gentamicin MICs were 2 to >64 mg/liter, amikacin MICs were <2 to 64 mg/liter, doxycycline MICs were 16 to 32 mg/liter, minocycline MICs were <0.5 to 4 mg/liter, tetracycline MICs were >64 mg/liter, netilmicin MICs were 64 to >64 mg/liter, rifampin MICs were <0.5 to 4 mg/liter, tobramycin MICs were 4 to 64 mg/liter, and doripenem MICs were 64 to >64 mg/liter. (iii) For PFGE-HUI-1 (Table 4), tigecycline MICs were 1 to 2 mg/liter, gentamicin MICs were 16- to >64 mg/liter, amikacin MICs were 4 to 64 mg/liter, doxycycline MICs were <0.5 to 8 mg/liter, minocycline MICs were <0.5 to 1 mg/liter, tetracycline MICs were 4 to >64 mg/liter, netilmicin MICs were 4 to >64 mg/liter, rifampin MICs were 2 to 32 mg/liter, tobramycin MICs were 8 to 64 mg/liter, and doripenem MICs were <0.5 to 2 mg/liter.
Fig 1.
Pulsed-field electrophoresis of strains of the PFGE-ROC-1 clone. REP-PCR, repetitive element palindromic PCR.
Fig 2.
Pulsed-field electrophoresis of strains of the PFGE-HUI-1 clone.
Table 2.
MICs and RE of genes harboring efflux pumpsc
| Straina | MICd (mg/liter) |
REb |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TIG | GEN | AMK | DOX | MIN | NET | TET | RIF | TOB | DOR | adeB | adeJ | adeG | craA | abeM | amvA | |
| 65 | ≤0.25 | 8 | <2 | 16 | 2 | 1 | >64 | 2 | 1 | 4 | 1.13 | 3.10 | 1.00 | 0.43 | 0.21 | 0.002 |
| 34 | 0.5 | 2 | 32 | 32 | 2 | >64 | >64 | 64 | 32 | 8 | 27.73 | 0.55 | 0.70 | 1.31 | 0.60 | 0.003 |
| 35 | 0.5 | 4 | 16 | 32 | 2 | >64 | >64 | 64 | 64 | 8 | 31.97 | 0.40 | 0.82 | 1.12 | 0.66 | 0.003 |
| 37 | 0.5 | 4 | 16 | 32 | 2 | >64 | >64 | 64 | 32 | 8 | 28.10 | 0.42 | 0.70 | 1.25 | 0.78 | 0.003 |
| 41 | 0.5 | 4 | 32 | 16 | 2 | >64 | >64 | 64 | 64 | 4 | 26.89 | 0.57 | 0.66 | 1.34 | 0.50 | 0.003 |
| 51 | 0.5 | 4 | 16 | 32 | 4 | >64 | >64 | 64 | 64 | 8 | 16.21 | 1.62 | 0.41 | 0.65 | 0.70 | 0.004 |
| 53 | 0.5 | 4 | 16 | 32 | 4 | >64 | >64 | 64 | 32 | 8 | 17.03 | 1.06 | 0.49 | 0.70 | 0.48 | 0.003 |
| 55 | 0.5 | 4 | 16 | 32 | 4 | >64 | >64 | 64 | 64 | 8 | 18.25 | 1.38 | 0.85 | 0.64 | 0.37 | 0.004 |
| 57 | 0.5 | 4 | 16 | 32 | 4 | >64 | >64 | 64 | 64 | 8 | 10.43 | 4.69 | 0.37 | 0.70 | 0.78 | 0.003 |
| 78 | 0.5 | 32 | <2 | 16 | 2 | 4 | >64 | 64 | <0.5 | 4 | 23.88 | 5.44 | 0.96 | 1.14 | 0.40 | 0.007 |
| 88 | 0.5 | 4 | 32 | 16 | 2 | >64 | >64 | 32 | 32 | 4 | 11.77 | 6.60 | 0.92 | 0.73 | 0.24 | 0.003 |
| 118 | 0.5 | 32 | 16 | 32 | 2 | >64 | >64 | 32 | 64 | 4 | 35.08 | 0.61 | 1.31 | 0.64 | 0.27 | 0.003 |
| 126 | 0.5 | 16 | <2 | 2 | 4 | 4 | >64 | 32 | <0.5 | 4 | 1.77 | 1.06 | 1.27 | 0.64 | 0.32 | 0.004 |
| 36 | 1 | 4 | 16 | 32 | 2 | >64 | >64 | 64 | 64 | 4 | 32.25 | 0.61 | 0.80 | 1.03 | 0.69 | 0.004 |
| 38 | 1 | 4 | 32 | 32 | 2 | >64 | >64 | 64 | 64 | 8 | 30.51 | 1.34 | 0.78 | 1.14 | 0.51 | 0.002 |
| 58 | 1 | 4 | 32 | 32 | 4 | >64 | >64 | 64 | 64 | 8 | 26.93 | 2.20 | 0.44 | 0.85 | 0.45 | 0.005 |
| 83 | 1 | 2 | 32 | 32 | 4 | >64 | >64 | 64 | 32 | 4 | 33.76 | 7.98 | 1.37 | 0.80 | 0.19 | 0.002 |
| 84 | 1 | 2 | 32 | 32 | 4 | >64 | >64 | 64 | 64 | 4 | 13.45 | 10.02 | 1.08 | 0.85 | 0.24 | 0.003 |
| 86 | 1 | 64 | 8 | 32 | 4 | 32 | >64 | 2 | 2 | 4 | 22.39 | 6.85 | 0.97 | 0.69 | 0.19 | 0.004 |
| 87 | 1 | 64 | 4 | 64 | 4 | 16 | >64 | 2 | 2 | 4 | 43.48 | 6.82 | 0.92 | 0.67 | 0.08 | 0.003 |
| 89 | 1 | >64 | 4 | 64 | 4 | 32 | >64 | 64 | 64 | 4 | 7.25 | 5.61 | 1.17 | 0.89 | 0.21 | 0.003 |
| 90 | 1 | 2 | 16 | 64 | 4 | >64 | >64 | 64 | 32 | 8 | 20.86 | 10.33 | 1.00 | 0.72 | 0.23 | 0.004 |
| 110 | 1 | 2 | 16 | 16 | 4 | >64 | >64 | 32 | 64 | 4 | 30.22 | 5.18 | 1.25 | 0.31 | 0.30 | 0.003 |
| 111 | 1 | 2 | 16 | 16 | 2 | >64 | >64 | 32 | 32 | 4 | 9.86 | 5.39 | 1.55 | 0.31 | 0.21 | 0.003 |
| 112 | 1 | 2 | 8 | 16 | 2 | >64 | >64 | 32 | 32 | 8 | 11.45 | 3.06 | 1.23 | 0.21 | 0.18 | 0.36 |
| 113 | 1 | 2 | <2 | 32 | 2 | 4 | >64 | 32 | <0.5 | 4 | 24.40 | 4.27 | 0.89 | 0.31 | 0.25 | 0.006 |
| 114 | 1 | 1 | <2 | 32 | 2 | 4 | >64 | 32 | 1 | 4 | 7.01 | 3.87 | 0.77 | 0.27 | 0.26 | 0.004 |
| 115 | 1 | 1 | <2 | 16 | 2 | 4 | >64 | 16 | <0.5 | 4 | 30.32 | 3.13 | 0.82 | 0.75 | 0.19 | 0.003 |
| 119 | 1 | 32 | 16 | 32 | 2 | >64 | >64 | 32 | 32 | 4 | 43.01 | 0.60 | 1.23 | 0.54 | 0.33 | 0.003 |
| 120 | 1 | 32 | 16 | 16 | 2 | >64 | >64 | 32 | 32 | 4 | 35.43 | 0.66 | 0.75 | 0.65 | 0.19 | 0.003 |
| 124 | 1 | 4 | 64 | 64 | 4 | >64 | >64 | 32 | 64 | 4 | 46.58 | 0.72 | 0.79 | 0.63 | 0.30 | 0.004 |
| 125 | 1 | 4 | 16 | 32 | 8 | >64 | >64 | 64 | 32 | 4 | 32.77 | 8.97 | 0.45 | 0.70 | 0.30 | 0.005 |
| 56 | 2 | 2 | <2 | 32 | 4 | 8 | >64 | 64 | 1 | 8 | 21.05 | 5.82 | 0.48 | 0.87 | 0.38 | 0.003 |
| 64 | 2 | 64 | 4 | 32 | 2 | 16 | >64 | 2 | 4 | 4 | 68.98 | 2.94 | 0.36 | 0.62 | 0.21 | 0.004 |
| 79 | 2 | >64 | 4 | 16 | 1 | 16 | >64 | 2 | 2 | 8 | 37.66 | 12.58 | 1.57 | 0.59 | 0.22 | 0.004 |
| 80 | 2 | >64 | 8 | 32 | 4 | 32 | >64 | 1 | 2 | 8 | 32.70 | 10.77 | 0.97 | 0.63 | 0.19 | 0.002 |
| 81 | 2 | 64 | 4 | 32 | 4 | 16 | >64 | 2 | 2 | 4 | 46.54 | 11.16 | 0.92 | 0.86 | 0.18 | 0.002 |
| 82 | 2 | 64 | <2 | 32 | 2 | 16 | >64 | 1 | 1 | 4 | 27.63 | 10.04 | 1.00 | 0.97 | 0.15 | 0.002 |
| 101 | 2 | 2 | 16 | 32 | 4 | >64 | >64 | 32 | 32 | 4 | 12.44 | 4.13 | 1.25 | 0.24 | 0.13 | 0.003 |
| 102 | 2 | 2 | 16 | 32 | 4 | >64 | >64 | 32 | 32 | 4 | 8.91 | 7.26 | 1.17 | 0.36 | 0.28 | 0.005 |
| 106 | 2 | 2 | 8 | 64 | 8 | >64 | >64 | 64 | 32 | 4 | 12.06 | 10.55 | 1.38 | 0.76 | 0.31 | 0.006 |
| 107 | 2 | 2 | 32 | 64 | 8 | >64 | >64 | 64 | 32 | 4 | 16.38 | 10.80 | 0.91 | 0.25 | 0.32 | 0.004 |
| 108 | 2 | 4 | 32 | 64 | 8 | >64 | >64 | 64 | 32 | 8 | 23.92 | 10.94 | 1.07 | 0.21 | 0.29 | 0.002 |
| 109 | 2 | 4 | 32 | 64 | 8 | >64 | >64 | 64 | 32 | 8 | 23.37 | 6.32 | 0.68 | 0.32 | 0.22 | 0.003 |
Strains are ranked according to the MIC of tigecycline.
Increased gene RE values of ≥2 are indicated in bold.
The reference strain used was A. baumannii ATCC 17978. RNA (50 μg/ml) was from strains of the PFGE-ROC-1OXA58 clone.
TIG, tigecycline; GEN, gentamicin; AMK, amikacin; DOX, doxycycline; MIN, minocycline; NET, netilmicin; TET, tetracycline; RIF, rifampin; TOB, tobramycin; DOR, doripenem.
Table 3.
MICs and RE of genes harboring efflux pumpsc
| Straina | MICd (mg/liter) |
REb |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TIG | GEN | AMK | DOX | MIN | TET | NET | RIF | TOB | DOR | adeBb | adeJ | adeG | craA | abeM | amvA | |
| 44 | ≤0.25 | 2 | 32 | 16 | 1 | >64 | >64 | 2 | 32 | 64 | 26.60 | 1.69 | 0.68 | 1.47 | 0.39 | 0.002 |
| 45 | ≤0.25 | 2 | 32 | 16 | 1 | >64 | >64 | 1 | 64 | >64 | 15.00 | 1.21 | 0.67 | 0.71 | 0.43 | 0.001 |
| 47 | ≤0.25 | 4 | 16 | 16 | 1 | >64 | >64 | 1 | 64 | 64 | 19.50 | 1.29 | 0.80 | 0.53 | 0.45 | 0.002 |
| 61 | ≤0.25 | 4 | 16 | 16 | 4 | >64 | >64 | 2 | 64 | 64 | 23.25 | 1.24 | 0.34 | 0.99 | 0.15 | 0.005 |
| 62 | ≤0.25 | 2 | 32 | 16 | 1 | >64 | >64 | 1 | 64 | >64 | 20.19 | 1.67 | 0.49 | 0.49 | 0.17 | 0.002 |
| 95 | ≤0.25 | 4 | 32 | 16 | 1 | >64 | >64 | <0.5 | 32 | >64 | 33.95 | 1.33 | 1.06 | 0.32 | 0.22 | 0.002 |
| 40 | 0.5 | 4 | 32 | 16 | 1 | >64 | >64 | 1 | 64 | 64 | 12.88 | 0.25 | 0.59 | 0.91 | 0.36 | 0.001 |
| 46 | 0.5 | 2 | 32 | 16 | 1 | >64 | >64 | 1 | 64 | >64 | 10.82 | 1.70 | 0.84 | 1.18 | 0.38 | 0.002 |
| 48 | 0.5 | 8 | 32 | 16 | 1 | >64 | >64 | 1 | 64 | 64 | 10.28 | 1.13 | 1.04 | 0.81 | 0.33 | 0.002 |
| 50 | 0.5 | 2 | 64 | 16 | <0.5 | >64 | >64 | 2 | 64 | 64 | 11.91 | 1.25 | 0.44 | 0.70 | 0.41 | 0.002 |
| 60 | 0.5 | 4 | 64 | 16 | 1 | >64 | >64 | 1 | 64 | 64 | 29.20 | 1.52 | 0.36 | 1.18 | 0.41 | 0.003 |
| 72 | 0.5 | 2 | 32 | 16 | <0.5 | >64 | >64 | 1 | 32 | >64 | 24.94 | 1.19 | 0.87 | 0.73 | 0.23 | 0.004 |
| 73 | 0.5 | 2 | 32 | 16 | <0.5 | >64 | >64 | 2 | 64 | >64 | 7.53 | 1.21 | 0.60 | 0.72 | 0.17 | 0.004 |
| 74 | 0.5 | >64 | <2 | 32 | 4 | >64 | 64 | 4 | 4 | >64 | 22.19 | 1.08 | 0.51 | 0.68 | 0.17 | 0.003 |
| 75 | 0.5 | >64 | 4 | 16 | 4 | >64 | 64 | 4 | 4 | >64 | 13.95 | 0.84 | 0.49 | 0.52 | 0.17 | 0.002 |
| 96 | 0.5 | 4 | 32 | 16 | 1 | >64 | >64 | 1 | 64 | 64 | 8.55 | 1.46 | 0.97 | 0.31 | 0.22 | 0.002 |
| 63 | 1 | 4 | 16 | 16 | 1 | >64 | >64 | 1 | 32 | 64 | 43.11 | 1.87 | 0.41 | 0.59 | 0.23 | 0.005 |
| 67 | 1 | 64 | 32 | 16 | <0.5 | >64 | >64 | 1 | 64 | >64 | 44.95 | 0.81 | 0.63 | 0.45 | 0.19 | 0.002 |
Strains are ranked according to the MIC of tigecycline.
Increased gene RE values of ≥2 are indicated in bold.
The reference strain used was A. baumannii ATCC 17978. RNA (50 μg/ml) was from strains of the PFGE-ROC-1OXA24 clone.
TIG, tigecycline; GEN, gentamicin; AMK, amikacin; DOX, doxycycline; MIN, minocycline; TET, tetracycline; NET, netilmicin; RIF, rifampin; TOB, tobramycin; DOR, doripenem.
Table 4.
MICs and RE of efflux pumpsc
| Straina | MICd (mg/liter) |
REb |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TIG | GEN | AMK | DOX | MIN | TET | NET | RIF | TOB | DOR | adeB | adeJ | adeG | craA | abeM | amvA | |
| 421 | 1 | >64 | 64 | <0.5 | <0.5 | 4 | 64 | 2 | 64 | 1 | NA | 1.44 | 1.23 | 1.20 | 0.04 | 0.01 |
| 422 | 1 | 16 | 16 | <0.5 | <0.5 | 8 | 8 | 2 | 8 | <0.5 | NA | 1.37 | 2.11 | 0.82 | 0.05 | 0.02 |
| 423 | 1 | 16 | 16 | <0.5 | <0.5 | 8 | 8 | 2 | 16 | 1 | NA | 1.21 | 1.16 | 0.63 | 0.07 | 0.02 |
| 424 | 1 | 16 | 64 | 1 | 1 | 4 | 8 | 16 | 32 | <0.5 | NA | 1.41 | 0.89 | 0.68 | 0.04 | 0.02 |
| 426 | 1 | 64 | 32 | 1 | 1 | 8 | 64 | 16 | 32 | <0.5 | NA | 1.69 | 1.00 | 0.45 | 0.04 | 0.02 |
| 427 | 2 | 16 | 16 | 8 | <0.5 | >64 | 8 | 4 | 8 | 1 | NA | 1.46 | 0.83 | 0.42 | 0.04 | 0.02 |
| 428 | 2 | 16 | 16 | 4 | <0.5 | >64 | 4 | 2 | 8 | 1 | NA | 1.64 | 0.89 | 0.49 | 0.05 | 0.02 |
| 429 | 2 | 64 | 4 | 4 | <0.5 | >64 | 8 | 4 | 8 | 1 | NA | 1.75 | 0.91 | 0.48 | 0.04 | 0.01 |
| 430 | 2 | 32 | 16 | 8 | <0.5 | >64 | 16 | 4 | 16 | 1 | NA | 1.22 | 0.76 | 0.48 | 0.04 | 0.02 |
| 431 | 2 | 64 | 4 | 8 | 1 | >64 | 32 | 32 | 32 | 1 | NA | 1.28 | 1.00 | 0.44 | 0.04 | 0.02 |
| 432 | 2 | 16 | 16 | 4 | <0.5 | >64 | 4 | 4 | 16 | 2 | NA | 1.61 | 0.90 | 0.53 | 0.04 | 0.02 |
| 433 | 2 | 16 | 16 | 8 | <0.5 | >64 | 8 | 4 | 16 | 2 | NA | 1.51 | 0.89 | 0.54 | 0.04 | 0.02 |
| 434 | 2 | 16 | 16 | 4 | <0.5 | >64 | 8 | 4 | 8 | 2 | NA | 1.82 | 1.09 | 0.50 | 0.04 | 0.02 |
| 435 | 2 | 32 | 32 | 4 | <0.5 | >64 | 8 | 2 | 32 | 1 | NA | 1.27 | 0.90 | 0.34 | 0.05 | 0.01 |
| 436 | 2 | 16 | 16 | 8 | 1 | >64 | 16 | 4 | 8 | 1 | NA | 1.32 | 0.79 | 0.43 | 0.04 | 0.01 |
| 437 | 2 | 16 | 16 | 4 | 1 | >64 | 4 | 4 | 16 | <0.5 | NA | 0.72 | 1.18 | 0.78 | 0.03 | 0.03 |
| 438 | 2 | 32 | 16 | 8 | 1 | >64 | 8 | 4 | 16 | <0.5 | NA | 1.25 | 1.51 | 0.60 | 0.03 | 0.02 |
| 443 | 2 | 16 | 16 | 4 | <0.5 | >64 | 8 | 4 | 8 | 1 | NA | 0.91 | 0.80 | 0.52 | 0.04 | 0.01 |
Strains are ranked according to the MIC of tigecycline.
Primers for adeA and adeC genes were also used. NA, not applicable.
The reference strain used was A. baumannii ATCC 17978. RNA (50 μg/ml) was from strains of the PFGE-HUI-1 clone (susceptible to carbapenems).
TIG, tigecycline; GEN, gentamicin; AMK, amikacin; DOX, doxycycline; MIN, minocycline; TET, tetracycline; NET, netilmicin; RIF, rifampin; TOB, tobramycin; DOR, doripenem.
We detected tet genes in both clones: the tetB gene in all strains of PFGE-ROC-1 and the tetA(39) gene in strains of the PFGE-HUI-1 clone (except for strains 421, 422, 423, 424, and 426).
In both A. baumannii clones, we detected the AacC1/AphA1/AadB combination of acetylases in strains displaying some resistance to aminoglycosides. We also detected mutations in the gyrA (Ser83 → Leu) and parC (Ser80 → Leu) genes in strains showing resistance to quinolones.
Relative gene expression.
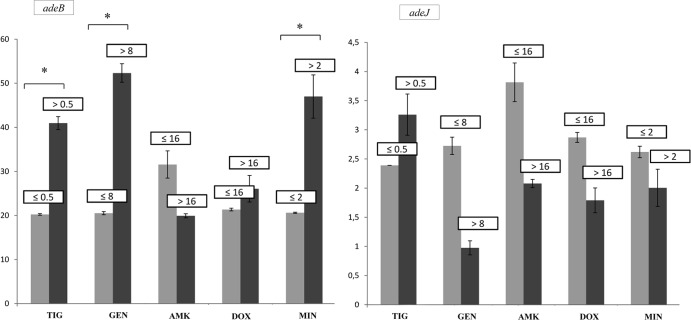
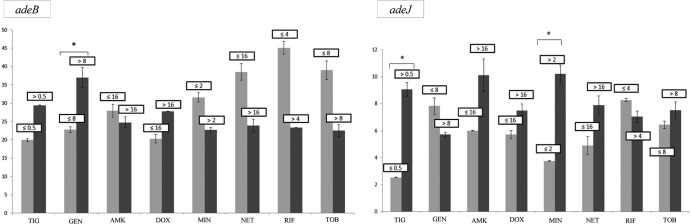
The levels of expression of the efflux pump genes in the isolates relative to that by A. baumannii ATCC 17978 (relative expression ([RE] values) are shown in Tables 2 to 4. For clone PFGE-ROC-1, we applied statistical analysis to genes with RE values higher than 8 (i.e., genes adeB and adeJ) to determine how gene expression was related to the antibiotic MICs (for strains carrying the OXA-58 β-lactamase gene, see Fig. 3; for strains carrying the OXA-24 β-lactamase gene, see Fig. 4). However, we were not able to analyze the adeB gene in strains of clone PFGE-HUI-1, because the internal and external primers used did not amplify the genes in the AdeABC operon of these strains. Moreover, the relative expression of adeJ in this clone was not higher than 2.
Fig 3.
Relative expression of the adeB and adeJ genes from strains of the PFGE-ROC-1OXA-58 clone in relation to the MICs of different antibiotics.*, P < 0.05 (Student's t test). Light gray bars, strains susceptible to several antibiotics (grouped by MIC [in mg/liter]); dark gray bars, strains resistant to several antibiotics (grouped by MIC [in mg/liter]); TIG, tigecycline; GEN, gentamicin; AMK, amikacin; DOX, doxycycline; MIN, minocycline; NET, netilmicin; RIF, rifampin; TOB, tobramycin.
Fig 4.
Relative expression of the adeB and adeJ genes from strains of the PFGE-ROC-1OXA-24 clone in relation to the MICs of different antibiotics.*, P < 0.05 (Student's t test). Light gray bars, strains susceptible to several antibiotics (grouped by MIC [in mg/liter]); dark gray bars, strains resistant to several antibiotics (grouped by MIC [in mg/liter]); TIG, tigecycline; GEN, gentamicin; AMK, amikacin; DOX, doxycycline; MIN, minocycline.
The RE values of adeG, craA, abeM, and amvA in all strains ranged from 0.003 to 1.
Porin expression was not significantly related to antibiotic resistance in strains of clone PFGE-ROC-1 or PFGE-HUI-1. However, in strains of clone PFGE-ROC-1OXA-24, the RE values of the carO and omp25 genes were lower than those in strains of clone PFGE-ROC-1OXA-58 (Fig. 5). However, the RE values of the OXA-51 and ampC β-lactamase genes were similar among the isolates.
Fig 5.
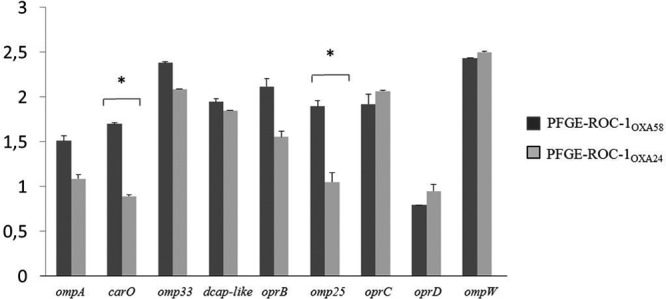
Relative expression of porin genes by strains of the PFGE-ROC-1OXA-58 and PFGE-ROC-1OXA-24 clones. *, P < 0.05 (Student's t test).
Polymorphisms of the regulatory genes of the AdeABC and AdeIJK efflux pumps.
Strains of the PFGE-ROC-1 clone that overexpressed the AdeABC efflux pump had three mutations in the adeS gene (Ala94 → Val, Gly186 → Val, and Phe214 → Leu) and one mutation in the adeR gene (Ala136 → Val). Only two strains of this clone had mutations in the adeN regulatory gene in the AdeIJK efflux pump (His111 → Pro, Ile112 → Phe). The adeS and adeR genes in strains of clone PFGE-HUI-1 were not successfully amplified. In all strains of this clone, the adeN gene had one mutation (Pro16 → Lys).
Relation between relative gene expression and MICs.
Possible interactions between the mechanisms of resistance of the clinical strains of the two clones are summarized in Table 5.
Table 5.
Interplay of mechanisms of resistance to several antibiotics of the strains of clones PFGE-ROC-1 and PFGE-HUI-1 under study
| Antibiotic(s) | Mechanism of resistance |
||
|---|---|---|---|
| PFGE-ROC-1OXA-58 | PFGE-ROC-1OXA-24 | PFGE-HUI-1 | |
| Tigecycline | Overexpression of AdeIJK | Overexpression of AdeABC | New RND efflux system/TetA(39) efflux pumpc |
| Gentamicin | Overexpression of AdeABC/acetylases (aacC1, aphA1, aadB) | Overexpression of AdeABC/acetylases (aacC1, aphA1, aadB) | Acetylases (aacC1, aphA1, aadB) |
| Minocycline | Overexpression of AdeIJK/TetB efflux pumps | Overexpression of AdeABCb | |
| Netilmicin, tobramycin, and amikacin | Acetylases (aacC1, aphA1, aadB) | Acetylases (aacC1, aphA1, aadB) | Acetylases (aacC1, aphA1, aadB) |
| Imipenem, meropenem, and doripenem | OXA-58 β-lactamase | OXA-24 β-lactamase | |
| Ciprofloxacin | Mutations in gyrA and parC | Mutations in gyrA and parC | Mutations in gyrA and parC |
| Doxycycline | Overexpression of AdeIJKa/TetB efflux pumps | TetB efflux pump | TetA(39) efflux pumpc |
| Tetracyclines | TetB efflux pump | TetB efflux pump | TetA(39) efflux pumpc |
Nonsignificantly increased expression relative to strains with doxycycline resistance.
Only three isolates, 61, 74, and 75.
Except for strains 421, 422, 423, 424, and 426.
(i) Carbapenems.
For the carbapenems (imipenem, meropenem, and doripenem), resistance was associated with the presence of the OXA-type enzymes (OXA-24 and OXA-58 β-lactamases) in strains of clone PFGE-ROC-1.
(ii) Aminoglycosides.
In gentamicin-resistant strains (MICs > 8 mg/liter) of clone PFGE-ROC-1, the AdeABC system was overexpressed and/or acetylases (AacC1/AphA1/AadB) were present. Moreover, in strains of both clones (PFGE-ROC-1 and PFGE-HUI-1), the presence of acetylases (AacC1/AphA1/AadB) was associated with resistance to netilmicin (MICs > 16 mg/liter), tobramycin (MICs > 8 mg/liter), and amikacin (MICs > 16 mg/liter).
(iii) Glycylcyclines.
Resistance to tigecycline (MICs > 0.5 mg/liter) was associated with overexpression of the RND systems (AdeIJK efflux pump in PFGE-ROC-1OXA58 [Fig. 3] and AdeABC efflux pump in PFGE-ROC-1OXA24 [Fig. 4]) and the TetA(39) effux pump. The presence of PAbetaN (an inhibitor of the RND system) in strains of clone PFGE-HUI-1 was associated with decreased resistance to tigecycline. In the strains of this clone with no TetA efflux pump (strains 421, 422, 423, 424, and 426), the tigecycline MIC decreased from 1 to ≤0.25 mg/liter (with PAbetaN). In those strains of clone PFGE-HUI-1 with a TetA system, the tigecycline MIC decreased from 2 to 1 mg/liter in the presence of PAbetaN. The AdeIJK and AdeABC efflux pumps (in strains of clone PFGE-ROC-1) were associated with resistance to minocycline (MICs > 2 mg/liter). However, PFGE-ROCOXA-58 displayed resistance to this antibiotic, possibly because of overexpression of the AdeIJK and TetB efflux pumps.
(iv) Tetracyclines.
Resistance to tetracyclines was associated with TetB and TetA(39) efflux pumps. In doxycycline-resistant isolates (strains of PFGE-ROC-1OXA-58 with MICs of >16 mg/liter), AdeIJK was overexpressed together with Tet systems.
(v) Quinolones.
Finally, mutations of the gyrA and parC genes conferred resistance to ciprofloxacin without any variations in MICs.
DISCUSSION
The impact of the interplay between different mechanisms of antimicrobial resistance in the susceptibility or resistance to antibiotics has been addressed in previous studies. Here, we focused on two of these studies in relation to the present study. One of these studies involved clinical strains of P. aeruginosa isolated from cystic fibrosis patients (representative of the Liverpool epidemic strain) (39), and the other involved strains isolated from bloodstream infections (40). In the present study, we attempted to determine if similar conclusions can be applied to clinical strains of A. baumannii in which resistance is associated with a multifactorial mechanism. We analyzed strains of two different clones, PFGE-ROC-1 and PFGE-HUI-1. The PFGE-ROC-1 clone included 53 strains carrying the OXA-58 β-lactamase gene (PFGE-ROC-1OXA-58) and 18 strains carrying the OXA-24 carbapenemase gene (PFGE-ROC-1OXA-24). The enzymes encoded by both of these genes are highly prevalent in isolates of A. baumannii in the Iberian Peninsula (41, 42). Only strains of the PFGE-HUI-1 clone (n = 19 strains) have previously shown susceptibility to carbapenems (23).
Overexpression of the AdeABC system (RE, 30- to 45-fold) was significantly associated with resistance to gentamicin (MICs > 8 mg/liter) in strains of PFGE-ROC-1 (which produce the OXA-58 and OXA-24 β-lactamases) (8). Moreover, in strains of PFGE-ROC-1OXA-24, resistance to tigecycline (MICs > 0.5 mg/liter) and minocycline (MICs > 2 mg/liter) was also significantly associated with expression of this efflux pump, as previously reported (8). All strains of the PFGE-ROC-1 clone had mutations in the adeR (Ala136 → Val) and adeS (Ala94 → Val, Gly186 → Val, and Phe214 → Leu) genes. Hornsey et al. associated the Ala94 → Val substitution with overexpression of the AdeABC efflux pump in A. baumannii strains representative of the prevalent United Kingdom lineage, OXA-23 clone 1 (16, 43). However, the other mutations have not previously been described. Peleg et al. (17) reported that increased (40- to 54-fold) expression of the adeB gene was associated with tigecycline MICs of 4 to 16 mg/liter in A. baumannii. We found that tigecycline-susceptible strains (MIC = 0.5 mg/liter) were associated with increased expression of the adeB gene (about 20- to 30-fold), which could indicate the role of the AdeABC efflux pump in others functions necessary for the pathogenesis of clinical strains of A. baumannii, such as colonization, infection, and the persistence of microorganisms in the host (10). We did not detect the AdeABC efflux pump or regulator genes in clinical strains of A. baumannii clone PFGE-HUI-1 (susceptible to carbapenems). As mentioned above, this efflux pump is present in 80% (range, 53% to 97%) of clinical isolates studied so far (8).
Increased expression of the AdeIJK efflux pump (RE, 8- to 10-fold) was significantly associated with tigecycline resistance (MICs > 0.5 mg/liter) and minocycline resistance (MICs > 2 mg/liter) in strains of PFGE-ROC-1OXA-58. However, this system was not significantly associated with resistance to netilmicin or tobramycin (aminoglycosides). These results are consistent with those obtained by Coyne et al. (44). These authors also noted that overexpression of this pump is always lower than that of the AdeABC system. These results could confirm the theory that high-level expression of the AdeIJK efflux pump is toxic to the host cell (45). The adeJ gene was not overexpressed in strains of the PFGE-ROC-1OXA-24 and PFGE-HUI-1 clones. Only two strains of the PFGE-ROC-1OXA-58 clone had two new mutations in a gene regulating the AdeIJK pump (adeN; His111 → Pro, Ile112 → Phe), and all strains of PFGE-HUI-1 had a Pro16 → Lys substitution in the adeN gene. None of these mutations have been associated with overexpression of AdeIJK, although other possible mechanisms of regulation cannot be ruled out (18).
Expression of adeG (AdeFGH), craA, abeM, and amvA was not increased (RE, 0.003 to 1) in strains of the PFGE-ROC-1 or PFGE-HUI-1 clone.
Gram-positive bacteria are the origin of tet genes detected in Gram-negative bacteria, such as A. baumannii (22, 38). Here, we detected the tetB gene in strains of the PFGE-ROC-1 clone, all of which were resistant to tetracycline (MICs = 16 to 64 mg/liter) and doxycycline (MICs = 16 to 64 mg/liter). Moreover, in some strains (PFGE-ROC-1OXA-58), overexpression of AdeIJK together with the presence of this acquired efflux pump was possibly associated with resistance to minocycline (MICs = 2 to 4 mg/liter). In other pathogens, such as Escherichia coli, the combination of AcrAB-TolC and TetA has been associated with a high degree of resistance to tetracycline (46). Our results showed that detection of the tetA(39) gene in all strains of this clone was associated with resistance to tetracycline and doxycycline. Agersø and Guardabassi (47) analyzed the presence of this gene in A. baumannii strains. These authors located the gene in both environmental and clinical strains, and they found the tetA(39) gene in 33 tigecycline-resistant strains (MICs ≥ 16 mg/liter). We noted that in strains of PFGE-HUI-1 harboring the tetA(39) gene, the tigecycline MIC was lower (2 to 1 mg/liter) in the presence of PAbetaN (an RND efflux pump inhibitor), and the MIC decreased from 1 to 0.25 mg/liter in the A. baumannii strains without this gene. This suggests the involvement of a new RND efflux pump, together with the TetA(39) system, in the resistance to tigecycline.
In relation to porins and unlike in other pathogens, such as Pseudomonas aeruginosa, in which OprD expression plays an important role in resistance to carbapenem antibiotics (39), we found that decreased expression of a porin was significantly associated with antimicrobial resistance. We observed decreased RE only of the carO and omp25 genes, on comparing strains of PFGE clone-ROC-1OXA-58 and PFGE clone-ROC-1OXA-24. This decrease was not associated with resistance to carbapenems, which is known to be associated with the presence of the β-lactamases (9). Moreover, the carbapenem resistance was not associated with expression of the OXA-51 and AmpC chromosomal β-lactamases. Overall, our data revealed that the presence of OXA-type enzymes (OXA-24 and/or OXA-58) is sufficient to confer resistance to carbapenem in the A. baumannii strains under study, as previously found (41, 42). Moreover, resistance to doripenem was also associated with the presence of the β-lactamases OXA-58 (MICs = 4 to 8 mg/liter) and OXA-24 (MICs = 64 to >64 mg/liter) (compared with MICs for strains of the PFGE-HUI-1 clone of 0.5 to 1 mg/liter). Marti and colleagues (48) analyzed the activity of doripenem against clinical isolates of A. baumannii and concluded that doripenem was more active than imipenem and meropenem in strains carrying the OXA-58 β-lactamase gene. However, in the present study, doripenem, imipenem, and meropenem MICs were high for the clinical strains producing the OXA-24 enzyme.
Quinolone resistance did not vary between the strains under study and was associated with previously reported mutations in gyrA and parC (7). Aminoglycoside-resistant isolates of clones PFGE-ROC-1 and PFGE-HUI-1 showed acetylases known to be common in A. baumannii strains (AacC1, AphA1, and AadB) (49).
In conclusion, (i) the clinical strains of Acinetobacter baumannii under study possess efflux systems and other mechanisms (possibly connected) that enable them to develop resistance to various antimicrobials and that also have other functions necessary in bacterial pathogenesis. (ii) Overexpression of the AdeABC system was found to be associated with resistance to glycylcycline (tigecycline and minocycline) and aminoglycosides (gentamicin), and possibly other biological functions, in the clinical strains under study. (iii) Hyperexpression of the AdeIJK efflux pump was significantly associated with resistance to tigecycline and minocycline but did not appear to be involved in other functions related to the pathogenesis of the bacterium. This efflux pump may be related to the TetB system and, thus, to minocycline resistance. (iv) Porins, AmpC β-lactamases, and OXA-51 were not involved in the antimicrobial resistance observed in the present study in the presence of OXA-type enzymes (OXA-24 and OXA-58). (v) The OXA-24 and OXA-58 β-lactamases were associated with resistance to meropenem, doripenem, and imipenem (especially the OXA-24 β-lactamase). (vi) The presence of the Tet efflux pumps in A. baumannii isolates was associated with resistance to tetracyclines and doxycycline. (vii) Finally, a new RND efflux pump may act in combination with the TetA(39) system to confer resistance to tigecycline in the absence of the AdeABC efflux pump and overexpression of the other systems in A. baumannii clinical strains susceptible to carbapenems.
The main limitation of the study was that we were not able to study the complex mechanisms of resistance to carbapenems in strains that did not produce OXA-type enzymes.
ACKNOWLEDGMENTS
We thank Neil Woodford for supplying strains AB210 and AB211 and Juan Vallejo for the elaboration of the figures.
Research in our laboratories is financially supported by grants from the Instituto de Salud Carlos III, the Spanish Network for Research in Infectious Diseases (REIPI RD 06/0008/0025), the Fondo de Investigaciones Sanitarias (PI08/1368 and PS09/00687 to G.B. and PI10/00056 to M.T.), SERGAS (PS07/90), and the Xunta de Galicia (07CSA050916PR). C.R. is funded by a doctoral grant (PFIS) from the Instituto de Salud Carlos III, and M.T. is supported by the Instituto de Salud Carlos III (program Miguel Servet), both from the Ministerio de Economia y Competitividad.
We are grateful to the following organizations and researchers who participated in the study: Hospital Virgen del Rocío (José Garnacho, Antonio Gutierrez Pizarraya, Juan Antonio Márquez Vácaro), Hospital Marqués de Valdecilla (María Eliecer Cano, M. Carmen Fariñas), Hospital SAS la Línea (Antonio Sánchez Porto, Gloria Esteban Meruendano, Luis Barbeyto Vales, Javier Casas Ciria, Luis Vallejo),Complejo Hospitalario de Ourense (Begona Fernández Pérez, José Carlos Villar Chao), Hospital Gregorio Maranón (Belén Padilla Ortega, Emilia Cercenado Mansilla), Hospital de Navarra (José Javier García Irure), Hospital Costa del Sol-Marbella (Alfonso del Arco Jiménez), Hospital General de Valencia (Concepción Gimeno Cardona, Juan Carlos Valía, Núria Tormo Palop, Vicente Abril, Josefina Rifa, Maria Jesus, Martinez Garcia), Consorci Hospitalari de Vic (Joseph Vilaró Pujals, Marian Navarro Aguirre, Ana Vilamala), Policlínica Guipúzkoa (José Antonio Jiménez Alfaro, Carlos Reviejo Jaca), Hospital Puerta del Mar (Pilar Marín Casanova, Francisca Guerreo, Evelyn Shaw, Virginia Plasencia,), Complejo Hospitalario de Soria (Teresa Nebreda Mayoral, María José Fernández Calavia, Susana García de Cruz, Carmen Aldea Mansilla), Hospital Universitario de Alicante (Esperanza Merino de Lucas, Alfredo Zorraquino, Sergio Reus Bañuls), Hospital Infanta Cristina (Eugenio Garduno Eseverri, Luis López Sánchez), Hospital Universitario Central de Asturias (Ana Fleites Gutiérrez, Azucena Rodríguez Guardado, Alfonso Moreno), Hospital Donostia (José María García-Arenzana Anguera),Complejo Hospitalario Torrecárdenas (Serafín López Palmero, Manuel Rodríguez Maresca), Complejo Hospitalario Xeral-Calde Lugo (Fernando García Garrote, José Varela Otero, María del Pilar Alonso), Hospital Universitario Reina Sofía de Córdoba (Elisa Vidal Verdú, Fernando Rodríguez López), Hospital Universitario Santiago Compostela (Fernanda Pardo Sánchez, E. Ferrer Vizoso, B. Regueiro Garcia), Hospital Sant Pau (Mercé Gurgui, Roser Pericas, Virginia Pomar), Hospital Galdakao-Usansolo (Pedro María Olaechea Astigarraga, Rafael Ayarza Igartua), Hospital Son Dureta (María Dolores Maciá Romero, Enrique Ruiz de Gopegui Bordes), Hospital Puerta de Hierro (María Isabel Sánchez Romero), Hospital Juan Grande (Jesús García Mata, María José Goyanes, Cristina Morales Mateos), Hospital San Cecilio (José Hernández Quero, Trinidad Escobar Lara), Hospital Sant Joan de Reus (Frederic Ballester Bastardie, Simona Iftimie, Isabel Pujol Bajador), Hospital de Motril (María Isabel Galán Navarro, María Luz Cádiz Gurrea), Hospital San Agustín (Carmen Amores Antequera, Montserrat Gómez, Purificación Cantudo), Hospital de Granollers (Carmina Martí Salas, Jordi Cuquet Peragosa, Antonio Moreno Flores, Luis Anibarro García), Hospital de Segovia (Susana Hernando Real, Pablo A. Carrero González), Complejo Hospitalario de Pontevedra (María Angeles Pallarés González, Sergio Rodríguez Fernández), Hospital de Bellvitge (Miquel Pujol Rojo, Fe Tubau), Hospital Virgen de la Victoria de Málaga (Enrique Nuno Alvarez, María Ortega Torres), Hospital Doctor Moliner (Salvador Giner Almaraz, María Rosa Roca Castelló, Manuela Castillo, Elena Hortelano), Hospital 12 de Octubre (Fernando Chaves Sánchez, Ana García Reyne), Hospital del Mar (Juan Pablo Horcajada Gallego, Concha Segura), Hospital San Agustín de Avilés (Gema Sierra Dorado, Raquel Yano Escudero), and Complejo Hospitalario Materno Insular de Gran Canaria (María Elena Dorta Hung, Cristóbal del Rosario Q).
Footnotes
Published ahead of print 12 August 2013
REFERENCES
- 1.Peleg AY, Seifert H, Paterson DL. 2008. Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21:538–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perez F, Hujer AM, Hujer KM, Decker BK, Rather PN, Bonomo RA. 2007. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 51:3471–3484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bou G, Martínez-Beltrán J. 2000. Cloning, nucleotide sequencing, and analysis of the gene encoding an AmpC beta-lactamase in Acinetobacter baumannii. Antimicrob. Agents Chemother. 44:428–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown S, Young HK, Amyes SG. 2005. Characterisation of OXA-51, a novel class D carbapenemase found in genetically unrelated clinical strains of Acinetobacter baumannii from Argentina. Clin. Microbiol. Infect. 11:15–23 [DOI] [PubMed] [Google Scholar]
- 5.del Mar Tomás M, Beceiro A, Pérez A, Velasco D, Moure R, Villanueva R, Martínez-Beltrán J, Bou G. 2005. Cloning and functional analysis of the gene encoding the 33- to 36-kilodalton outer membrane protein associated with carbapenem resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother. 49:5172–5175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Limansky AS, Mussi MA, Viale AM. 2002. Loss of a 29-kilodalton outer membrane protein in Acinetobacter baumannii is associated with imipenem resistance. J. Clin. Microbiol. 40:4776–4778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hujer KM, Hujer AM, Endimiani A, Thomson JM, Adams MD, Goglin K, Rather PN, Pennella TT, Massire C, Eshoo MW, Sampath R, Blyn LB, Ecker DJ, Bonomo RA. 2009. Rapid determination of quinolone resistance in Acinetobacter spp. J. Clin. Microbiol. 47:1436–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coyne S, Courvalin P, Perichon B. 2011. Efflux-mediated antibiotic resistance in Acinetobacter spp. Antimicrob. Agents Chemother. 55:947–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poirel L, Nordmann P. 2006. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin. Microbiol. Infect. 12:826–836 [DOI] [PubMed] [Google Scholar]
- 10.Martinez JL, Sanchez MB, Martinez-Solano L, Hernandez A, Garmendia L, Fajardo A, Alvarez-Ortega C. 2009. Functional role of bacterial multidrug efflux pumps in microbial natural ecosystems. FEMS Microbiol. Rev. 33:430–449 [DOI] [PubMed] [Google Scholar]
- 11.Coyne S, Rosenfeld N, Lambert T, Courvalin P, Périchon B. 2010. Overexpression of resistance-nodulation-cell division pump AdeFGH confers multidrug resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother. 54:4389–4393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huys G, Cnockaert M, Nemec A, Swings J. 2005. Sequence-based typing of ade B as a potential tool to identify intraspecific groups among clinical strains of multidrug-resistant Acinetobacter baumannii. J. Clin. Microbiol. 43:5327–5331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Magnet S, Courvalin P, Lambert T. 2001. Resistance-nodulation-cell division-type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii strain BM4454. Antimicrob. Agents Chemother. 45:3375–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun JR, Chan MC, Chang TY, Wang WY, Chiueh TS. 2010. Overexpression of the adeB gene in clinical isolates of tigecycline-nonsusceptible Acinetobacter baumannii without insertion mutations in adeRS. Antimicrob. Agents Chemother. 54:4934–4938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun JR, Perng CL, Chan MC, Morita Y, Lin JC, Su CM, Wang WY, Chang TY, Chiueh TS. 2012. A truncated AdeS kinase protein generated by ISAba1 insertion correlates with tigecycline resistance in Acinetobacter baumannii. PLoS One 7:e49534. 10.1371/journal.pone.0049534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hornsey M, Ellington MJ, Doumith M, Thomas CP, Gordon NC, Wareham DW, Quinn J, Lolans K, Livermore DM, Woodford N. 2010. AdeABC-mediated efflux and tigecycline MICs for epidemic clones of Acinetobacter baumannii. J. Antimicrob. Chemother. 65:1589–1593 [DOI] [PubMed] [Google Scholar]
- 17.Peleg AY, Adams J, Paterson DL. 2007. Tigecycline efflux as a mechanism for nonsusceptibility in Acinetobacter baumannii. Antimicrob. Agents Chemother. 51:2065–2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosenfeld N, Bouchier C, Courvalin P, Périchon B. 2012. Expression of the resistance-nodulation-cell division pump AdeIJK in Acinetobacter baumannii is regulated by AdeN, a TetR-type regulator. Antimicrob. Agents Chemother. 56:2504–2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roca I, Marti S, Espinal P, Martinez P, Gibert I, Vila J. 2009. CraA, a major facilitator superfamily efflux pump associated with chloramphenicol resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother. 53:4013–4014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su XZ, Chen J, Mizushima T, Kuroda T, Tsuchiya T. 2005. AbeM, an H+-coupled Acinetobacter baumannii multidrug efflux pump belonging to the MATE family of transporters. Antimicrob. Agents Chemother. 49:4362–4364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rajamohan G, Srinivasan VB, Gebreyes WA. 2010. Molecular and functional characterization of a novel efflux pump, AmvA, mediating antimicrobial and disinfectant resistance in Acinetobacter baumannii. J. Antimicrob. Chemother. 65:1919–1925 [DOI] [PubMed] [Google Scholar]
- 22.Akers KS, Mende K, Yun HC, Hospenthal DR, Beckius ML, Yu X, Murray CK. 2009. Tetracycline susceptibility testing and resistance genes in isolates of Acinetobacter baumannii-Acinetobacter calcoaceticus complex from a U.S. military hospital. Antimicrob. Agents Chemother. 53:2693–2695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fernández-Cuenca F, Tomás-Carmona M, Caballero-Moyano F, Bou G, Martínez-Martínez L, Vila J, Pachón J, Cisneros JM, Rodríguez-Baño J, Pascual A, Grupo del Proyecto GEIH-REIPI-Ab 2010 2013. In vitro activity of 18 antimicrobial agents against clinical isolates of Acinetobacter spp.: multicenter national study GEIH-REIPI-Ab 2010. Enferm. Infecc. Microbiol. Clin. 31:4–9 (In Spanish.) [DOI] [PubMed] [Google Scholar]
- 24.Espinal P, Seifert H, Dijkshoorn L, Vila J, Roca I. 2012. Rapid and accurate identification of genomic species from the Acinetobacter baumannii (Ab) group by MALDI-TOF MS. Clin. Microbiol. Infect. 18:1097–1103 [DOI] [PubMed] [Google Scholar]
- 25.Vaneechoutte M, Dijkshoorn L, Tjernberg I, Elaichouni A, de Vos P, Claeys G, Verschraegen G. 1995. Identification of Acinetobacter genomic species by amplified ribosomal DNA restriction analysis. J. Clin. Microbiol. 33:11–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turton JF, Woodford N, Glover J, Yarde S, Kaufmann ME, Pitt TL. 2006. Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. J. Clin. Microbiol. 44:2974–2976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zander E, Higgins PG, Fernández-González A, Seifert H. 2013. Detection of intrinsic bla(OXA-51-like) by multiplex PCR on its own is not reliable for the identification of Acinetobacter baumannii. Int. J. Med. Microbiol. 303:88–89 [DOI] [PubMed] [Google Scholar]
- 28.Zhang L, Ding G, Wei L, Pan X, Mei L, Zhang Y, Lu Y. 2011. Establishment of a novel target-based real-time quantitative PCR method for Acinetobacter baumannii detection. Diagn. Mol. Pathol. 20:242–248 [DOI] [PubMed] [Google Scholar]
- 29.Seifert H, Dolzani L, Bressan R, van der Reijden T, van Strijen B, Stefanik D, Heersma H, Dijkshoorn L. 2005. Standardization and interlaboratory reproducibility assessment of pulsed-field gel electrophoresis-generated fingerprints of Acinetobacter baumannii. J. Clin. Microbiol. 43:4328–4335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nemec A, Krízová L, Maixnerová M, Diancourt L, van der Reijden TJ, Brisse S, van den Broek P, Dijkshoorn L. 2008. Emergence of carbapenem resistance in Acinetobacter baumannii in the Czech Republic is associated with the spread of multidrug-resistant strains of European clone II. J. Antimicrob. Chemother. 62:484–489 [DOI] [PubMed] [Google Scholar]
- 32.Pannek S, Higgins PG, Steinke P, Jonas D, Akova M, Bohnert JA, Seifert H, Kern WV. 2006. Multidrug efflux inhibition in Acinetobacter baumannii: comparison between 1-(1-naphthylmethyl)-piperazine and phenyl-arginine-beta-naphthylamide. J. Antimicrob. Chemother. 57:970–974 [DOI] [PubMed] [Google Scholar]
- 33.Akers KS, Chaney C, Barsoumian A, Beckius M, Zera W, Yu X, Guymon C, Keen EF, III, Robinson BJ, Mende K, Murray CK. 2010. Aminoglycoside resistance and susceptibility testing errors in Acinetobacter baumannii-calcoaceticus complex. J. Clin. Microbiol. 48:1132–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walther-Rasmussen J, Høiby N. 2006. OXA-type carbapenemases. J. Antimicrob. Chemother. 57:373–383 [DOI] [PubMed] [Google Scholar]
- 35.Higgins PG, Poirel L, Lehmann M, Nordmann P, Seifert H. 2009. OXA-143, a novel carbapenem-hydrolyzing class D beta-lactamase in Acinetobacter baumannii. Antimicrob. Agents Chemother. 53:5035–5038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ellington MJ, Kistler J, Livermore DM, Woodford N. 2007. Multiplex PCR for rapid detection of genes encoding acquired metallo-beta-lactamases. J. Antimicrob. Chemother. 59:321–322 [DOI] [PubMed] [Google Scholar]
- 37.Miranda CD, Kehrenberg C, Ulep C, Schwarz S, Roberts MC. 2003. Diversity of tetracycline resistance genes in bacteria from Chilean salmon farms. Antimicrob. Agents Chemother. 47:883–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ribera A, Ruiz J, Vila J. 2003. Presence of the Tet M determinant in a clinical isolate of Acinetobacter baumannii. Antimicrob. Agents Chemother. 47:2310–2312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tomás M, Doumith M, Warner M, Turton JF, Beceiro A, Bou G, Livermore DM, Woodford N. 2010. Efflux pumps, OprD porin, AmpC beta-lactamase, and multiresistance in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Antimicrob. Agents Chemother. 54:2219–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cabot G, Ocampo-Sosa AA, Tubau F, Macia MD, Rodríguez C, Moya B, Zamorano L, Suárez C, Peña C, Martínez-Martínez L, Oliver A, Spanish Network for Research in Infectious Diseases (REIPI) 2011. Overexpression of AmpC and efflux pumps in Pseudomonas aeruginosa isolates from bloodstream infections: prevalence and impact on resistance in a Spanish multicenter study. Antimicrob. Agents Chemother. 55:1906–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Merino M, Acosta J, Poza M, Sanz F, Beceiro A, Chaves F, Bou G. 2010. OXA-24 carbapenemase gene flanked by XerC/XerD-like recombination sites in different plasmids from different Acinetobacter species isolated during a nosocomial outbreak. Antimicrob. Agents Chemother. 54:2724–2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruiz M, Marti S, Fernandez-Cuenca F, Pascual A, Vila J. 2007. High prevalence of carbapenem-hydrolysing oxacillinases in epidemiologically related and unrelated Acinetobacter baumannii clinical isolates in Spain. Clin. Microbiol. Infect. 13:1192–1198 [DOI] [PubMed] [Google Scholar]
- 43.Hornsey M, Loman N, Wareham DW, Ellington MJ, Pallen MJ, Turton JF, Underwood A, Gaulton T, Thomas CP, Doumith M, Livermore DM, Woodford N. 2011. Whole-genome comparison of two Acinetobacter baumannii isolates from a single patient, where resistance developed during tigecycline therapy. J. Antimicrob. Chemother. 66:1499–1503 [DOI] [PubMed] [Google Scholar]
- 44.Coyne S, Guigon G, Courvalin P, Périchon B. 2010. Screening and quantification of the expression of antibiotic resistance genes in Acinetobacter baumannii with a microarray. Antimicrob. Agents Chemother. 54:333–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Damier-Piolle L, Magnet S, Bremont S, Lambert T, Courvalin P. 2008. AdeIJK, a resistance-nodulation-cell division pump effluxing multiple antibiotics in Acinetobacter baumannii. Antimicrob. Agents Chemother. 52:557–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Cristóbal RE, Vincent PA, Salomón RA. 2006. Multidrug resistance pump AcrAB-TolC is required for high-level, Tet(A)-mediated tetracycline resistance in Escherichia coli. J. Antimicrob. Chemother. 58:31–36 [DOI] [PubMed] [Google Scholar]
- 47.Agersø Y, Guardabassi L. 2005. Identification of Tet 39, a novel class of tetracycline resistance determinant in Acinetobacter spp. of environmental and clinical origin. J. Antimicrob. Chemother. 55:566–569 [DOI] [PubMed] [Google Scholar]
- 48.Marti S, Sánchez-Céspedes J, Alba V, Vila J. 2009. In vitro activity of doripenem against Acinetobacter baumannii clinical isolates. Int. J. Antimicrob. Agents 33:181–182 [DOI] [PubMed] [Google Scholar]
- 49.Asadollahi K, Taherikalani M, Maleki A, Alizadeh E, Valadbaigi H, Soroush S, Maleki H, Asadollahi P, Emaneini M. 2011. Diversity of aminoglycoside modifying enzyme genes among multidrug resistant Acinetobacter baumannii genotypes isolated from nosocomial infections in Tehran hospitals and their association with class 1 integrons. Acta Microbiol. Immunol. Hung. 58:359–370 [DOI] [PubMed] [Google Scholar]