Abstract
Candida albicans is an opportunistic fungal pathogen that can cause disseminated infection in patients with indwelling catheters or other implanted medical devices. A common resident of the human microbiome, C. albicans responds to environmental signals, such as cell contact with catheter materials and exposure to serum or CO2, by triggering the expression of a variety of traits, some of which are known to contribute to its pathogenic lifestyle. Such traits include adhesion, biofilm formation, filamentation, white-to-opaque (W-O) switching, and two recently described phenotypes, finger and tentacle formation. Under distinct sets of environmental conditions and in specific cell types (mating type-like a [MTLa]/alpha cells, MTL homozygotes, or daughter cells), C. albicans utilizes (or reutilizes) a single signal transduction pathway—the Ras pathway—to affect these phenotypes. Ras1, Cyr1, Tpk2, and Pde2, the proteins of the Ras signaling pathway, are the only nontranscriptional regulatory proteins that are known to be essential for regulating all of these processes. How does C. albicans utilize this one pathway to regulate all of these phenotypes? The regulation of distinct and yet related processes by a single, evolutionarily conserved pathway is accomplished through the use of downstream transcription factors that are active under specific environmental conditions and in different cell types. In this minireview, we discuss the role of Ras signaling pathway components and Ras pathway-regulated transcription factors as well as the transcriptional regulatory networks that fine-tune gene expression in diverse biological contexts to generate specific phenotypes that impact the virulence of C. albicans.
INTRODUCTION
Candida albicans is a common commensal microbe of the gastrointestinal and female reproductive tracts, but it can be a devastating pathogen in humans with compromised immune systems. It can colonize and thrive in virtually every tissue of the human body, which means that it must adapt to a variety of host conditions to survive and grow. Candida has a well-characterized set of phenotypic traits, described in detail below, that are important for its pathogenesis, such as cell adhesion, biofilm formation, “white-opaque” (W-O) switching, and a complex system of morphogenesis, some of which promote survival and infection in the host.
Many different microbes can grow in drug-resistant surface-associated communities called biofilms (1–5). C. albicans forms robust biofilms on catheters (6, 7), dental implants (8), and prosthetic devices (7) in vivo. Since biofilms are associated with antifungal drug resistance in the clinical setting (9–11), the ability to form biofilms is considered a pathogenic trait of C. albicans (12). Recent work on Candida biofilms, described below, is shedding light on the regulatory network that controls biofilm formation.
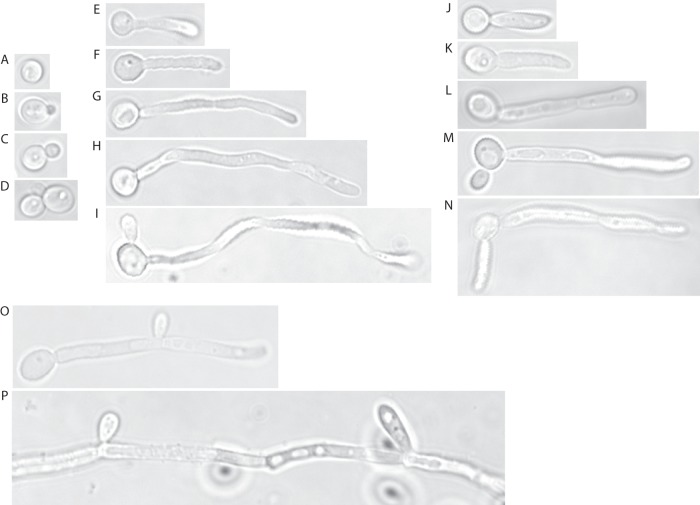
In response to a variety of environmental stimuli, C. albicans undergoes a reversible morphological transition from a round budding yeast form (Fig. 1A to D) to elongated filaments (Fig. 1E to N) (for reviews, see references 13, 14, 15, 16, and 17). If these filaments are then shifted back into yeast-form growth conditions (such as in yeast extract-peptone-dextrose [YPD] at 30°C), new cells in the yeast form will bud from the subapical cell just behind the septum (Fig. 1O) (see reference 18). After prolonged exposure to these stimuli, under conditions such as overnight growth in serum, Candida filaments undergo the reverse morphological transition and revert to producing yeast-form daughter cells from the filaments (Fig. 1P) rather than producing more filamentous cells. One possibility underlying this observation is that farnesol, a quorum-sensing molecule produced by C. albicans that accumulates when cell densities are high and arrests germ tube formation under hypha-inducing conditions (19–21), may accumulate in these overnight filamentous cultures, arresting germ tube formation and causing new cells to be produced in the yeast form. It is also possible that the hypha-inducing stimulus becomes depleted after long-term growth. Another possibility is that C. albicans eventually adapts to the filament-inducing serum and reverts to growth in the yeast form. These possibilities are not mutually exclusive.
Fig 1.
Morphogenesis of C. albicans. (A) A stationary-phase yeast cell from a culture grown overnight at 30°C. (B to D) Yeast cells growing in logarithmic-phase culture in YPD medium at 30°C. (E to N) Induction of hyphal filaments by growth in YPD medium with 10% serum at 37°C (E to I) or pseudohyphal filaments by growth in YPD medium at 37°C (J to N) at 1 h (E and J), 2 h (F and K), 3 h (G and L), 4 h (H and M), and 5 h (I and N) postinoculation of stationary-phase cells into YPD medium at 37°C with (E to I) or without (J to N) 10% serum. Note that pseudohyphae have constrictions at the sites of cell division whereas hyphae lack these constrictions. Serum-induced hyphae were grown for 4 h before transfer to YPD medium at 30°C to promote reversion to yeast-form growth. (O) A yeast cell emerging from the subapical hyphal cell is shown. (P) Serum-induced filaments grown overnight in YPD medium with 10% serum at 37°C revert to producing yeast cells from the hyphal filaments. The SC5314 strain was used in this experiment.
The ability to transition between the yeast and filamentous states is considered a virulence trait of C. albicans (22–24). During biofilm formation, which is intimately connected to changes in cell morphology, and under filament-inducing conditions, C. albicans expresses specific sets of genes that are important for aspects of the related traits of adhesion, morphogenesis, and biofilm formation (25, 26). The W-O switch is also related to changes in cell morphology. This switch occurs in mating type-like (MTL) homozygous strains and is a prerequisite for mating of C. albicans (27). High CO2 levels, such as are found in the human gastrointestinal tract (28), trigger C. albicans cells to develop multicellular structures called “fingers” and “tentacles,” both of which require filament formation as part of these developmental processes, on solid media (29).
C. albicans utilizes the Ras/cyclic AMP (cAMP)/protein kinase A (PKA) signal transduction pathway to regulate each of the above-described traits in response to specific combinations of environmental stimuli and cell types. In order to control these distinct traits, the Ras pathway and its downstream components target dozens or more of downstream transcription factors to modulate the expression of a specific repertoire of genes required for the particular trait or traits needed under each set of conditions. In this minireview, we highlight the most recent developments in the characterization of the transcriptional regulatory programs that act in concert with Ras signaling to control pathogenesis, cell adhesion, biofilm formation, morphogenesis, W-O switching, and the recently described processes of multicellular finger and tentacle formation, distinct but interrelated phenotypic traits that contribute to the pathogenic lifestyle of C. albicans.
Ras/cAMP/PKA SIGNALING IN C. ALBICANS
Ras proteins are a highly conserved family of small eukaryotic GTP-activating proteins (GTPases) that play a central role in sensing and responding to environmental cues in organisms that range from fungi to humans. RAS genes are among the most frequently mutated genes in human cancers (30, 31), and in fungi, Ras signaling has been linked to regulation of cell shape (32), nutrient sensing (33), response to stress (34), and mating (35). Ras is activated by a conformational change that occurs upon binding to GTP and is inactive in its GDP-bound conformation. Ras-GTPase-activating factors (Ras-GAPs) and guanine nucleotide exchange factors (Ras-GEFs) catalyze the exchange of GDP and GTP, creating a cycle between the inactive and the active state (Fig. 2). In addition, Ras GTPases are often localized to membranes by C-terminal farnesylation and palmitoylation and this localization regulates their function (36, 37).
Fig 2.
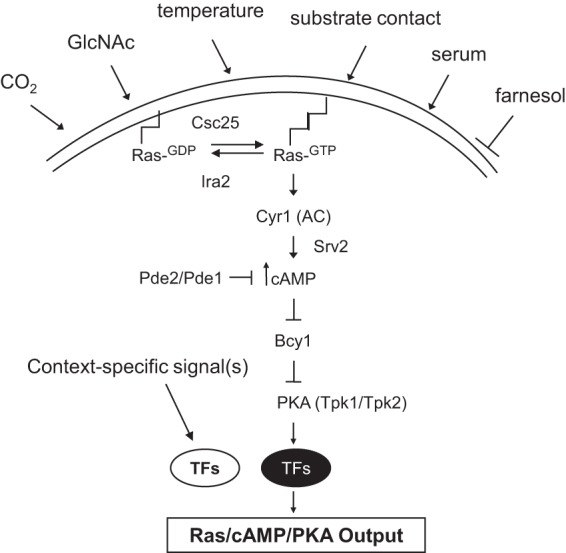
Diagram of the central Ras/cAMP/PKA signaling pathway components and examples of signals to which the Ras signaling pathway responds. TFs, transcription factors.
Activation of C. albicans Ras1 by the guanine exchange factor, Csc25 (commonly known as Cdc25 [orf19.6926]), stimulates the adenylyl cyclase (AC) activity of Cyr1 (also called Cdc35 [orf19.5148]), which works with the AC-associated protein Srv2 (also known as Cap1 [orf19.505]) to increase the intracellular concentration of cAMP, a highly active molecule (38, 39) (Fig. 2). A Ras-GTPase activator, most likely Ira2 (orf19.5219), counteracts Ras signaling by catalyzing the conversion of Ras1 to its inactive GDP-bound state. Activation of Ras1 signaling through Cyr1 occurs through physical interaction of Cyr1 with the Ras1 RA domain (38). The resulting increase in the intracellular concentration of cAMP results in the binding of cAMP to Bcy1 (orf19.2014), the regulatory subunit of protein kinase A (PKA). This in turn, causes the release or activation of the PKA catalytic subunit (encoded by either TPK2 or TPK1 [orf19.2277 or orf19.4892, respectively]) from Bcy1. Tpk1 or Tpk2 then activates transcription factors, such as Efg1, by phosphorylation (40). cAMP levels are also negatively regulated by a high-affinity phosphodiesterase, Pde2 (orf19.2972), and a low-affinity phosphodiesterase, Pde1 (orf19.4235), that increase the rate of cAMP breakdown. Increased PDE2 expression in hyphae counteracts the Srv2-dependent synthesis of cAMP and limits filament formation (41), while loss-of-function mutations in PDE2 increase cAMP levels and lead to constitutive activation of the pathway and increased filament formation (42–44). Pde1 mediates repression of cAMP signaling in response to glucose and intracellular acidification (45). pde1 mutants filament normally; however, in combination with a pde2 mutation, the pde1 mutation exacerbates the virulence defects of pde2, suggesting a minor role for Pde1 in virulence in C. albicans but not in morphogenesis.
In C. albicans, green fluorescent protein (GFP)-tagged Ras1 is localized uniformly to the plasma membrane through farnesyl and palmitoyl groups and this localization is critical for its activity (36). Mutants that lack these modifications, such as a cytosol-localized Ras1 mutant (GFP-Ras1C288S), are unable to undergo hyphal morphogenesis or stimulate farnesylation- and palmitoylation-independent signaling (hyphal inhibition by farnesol) through AC, except in the presence of a Ras1-activating mutation such as RAS1G13V (36). The presence of the activated mutant allele, RAS1G13V, results in constitutive activation of Ras signaling and leads to increased filamentous growth and increased cell adhesion (38). The filamentous-growth defect of ras1 mutants can be suppressed either by the presence of exogenous cAMP or by overexpressing components of a mitogen-activated protein kinase (MAPK) cascade (Hst7 or the transcription factor Cph1) (46), indicating that Ras signaling activates both the AC/PKA pathway and a conserved MAPK pathway (Fig. 3B), each of which controls its own set of transcription factors to promote filamentous growth in C. albicans.
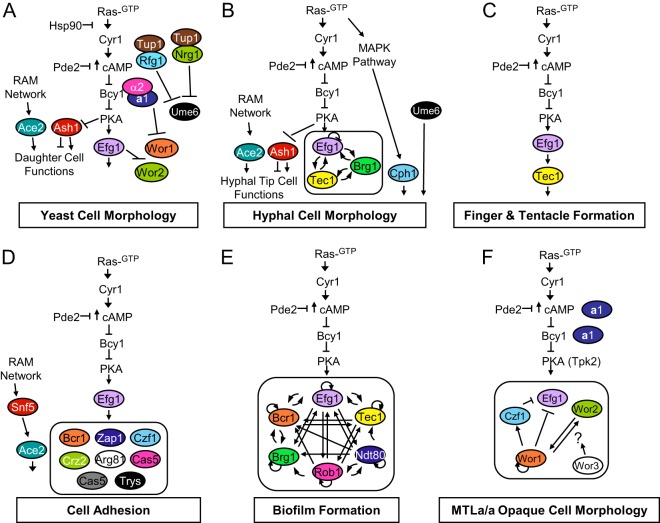
Fig 3.
Diagram of models for regulation of multiple pathogenic traits of C. albicans. (A and B) Yeast (A) and hyphal cell (B) morphology. (C) Finger and tentacle formation. (D) Cell adhesion. (E) Biofilm formation. (F) Regulation of the opaque state. Chromatin modifying factors are not shown.
Regulated proteolysis of Ras1 has also been shown to affect its activity during hypha formation and in response to farnesol (47). The levels of membrane-associated, full-length Ras1 are higher in hyphae than in yeast, and yeast possess a shorter, soluble Ras1 species that results from cleavage. Ras1 mutants that are unable to be cleaved more rapidly convert to hyphae and are delayed in the transition from hypha to yeast. In response to the presence of farnesol, which inhibits Cyr1 and represses filamentation, the levels of cleaved Ras1 increase (47); thus, regulation of Ras1 activity by proteolysis appears to favor the yeast form and the full-length membrane-associated Ras1 is associated with hypha formation. A role for regulated proteolysis of Ras1 during other processes such as biofilm formation, the W-O switch, and finger and tentacle formation is yet to be determined.
Initial studies of Candida Ras function suggested that C. albicans possessed a single nonessential Ras gene, RAS1 (38). Since then, a second Ras gene, RAS2 (orf19.5902), has been identified and is reported to encode an atypical Ras protein that shares poor sequence identity with other Ras GTPases and contains differences in conserved motifs thought to be required for usual Ras-associated activities (48). Strikingly, RAS2 is expressed at a fraction of the level of RAS1 and appears to have the effects on intracellular cAMP levels that contrast with those of RAS1. The deletion of RAS2 alone causes a minor defect in filamentous growth on solid Spider medium and under embedded conditions and yet produces no defect in liquid medium. However, deletion of RAS2 in the ras1 null mutant background actually aggravates the ras1 filamentous-growth defect, suggesting a partial role for Ras2 in promoting filamentous growth and morphogenesis (48). The fact that Ras2 functions to decrease cAMP levels and yet appears to play a minor role in promoting filamentous growth is seemingly at odds with the idea that increased cAMP levels promote filamentous growth of C. albicans. Studies on the interaction of Ras2 with other members of the Ras/cAMP/PKA signaling pathway, such as Pde1, Pde2, and PKA, are currently lacking and could shed light on the details of this apparent conundrum.
Ras/cAMP/PKA SIGNALING CONTROLS KNOWN AND PUTATIVE PATHOGENIC TRAITS OF C. ALBICANS
Ras1, Cyr1, and the transcription factor downstream of Tpk2, Efg1, are each required for wild-type levels of virulence in a mouse model for systemic candidiasis (46, 49, 50), while TPK2, the gene encoding one of two partially redundant catalytic subunits of PKA, is required for the ability to invade and damage oral epithelial cells in vitro (49). The role for signaling through the Ras/cAMP/PKA pathway in virulence of C. albicans has been clearly established and thoroughly reviewed (51, 52). Cutting-edge research on this fungal pathogen has revealed important new insights into the transcriptional regulatory networks that act in concert with Ras-regulated transcription factors to control cell adhesion (53), biofilm formation (54), morphogenesis (55), and W-O switching (56–58), processes in which Ras/cAMP/PKA signaling plays a central but not a sole role (Fig. 2).
CELL ADHESION
Cell adhesion is a pathogenic trait of C. albicans that is important for normal filamentous growth, biofilm formation, and the ability to adhere to host tissues. Ras signaling is linked to cell adhesion through its role in regulating both filamentous growth and biofilm formation, likely through the activation of Efg1 and Tec1 (and Bcr1 during biofilm formation), which control the expression of genes involved in cell adhesion (59–61). During both biofilm formation and filamentous growth, the genes from the ALS (agglutinin-like sequence) adhesin gene family are highly induced (62). This family, and Als3 in particular, is involved in cell adhesion and mediates attachment to endothelial cells (63), epithelial cells, and extracellular matrix proteins (64). In a mouse model of oropharyngeal candidiasis and during biofilm growth, Bcr1 (biofilm and cell wall regulator) regulates the expression of ALS3 (60, 65), which is considered a core hypha-specific gene (26, 66, 67). In spite of the importance of ALS3 expression during biofilm formation and filamentous growth, Als3 is dispensable for virulence during disseminated infection in mice (68), suggesting that additional cell adhesion factors must influence pathogenesis during these two processes. HWP1 may be one such factor. While ALS3 expression is Bcr1 dependent in vivo, HWP1 expression is not. HWP1 is expressed exclusively in filaments and is regulated by Ras/cAMP/PKA signaling via Efg1 in concert with other transcriptional regulators (69) (described below).
Finkel et al. recently identified at least 30 transcription factors that control the adherence of yeast cells to a silicone substrate (53). Their findings revealed the existence of a large regulatory network that links 12 of these transcription factors. Thus, while signaling through Ras1 to regulate cell adhesion in the context of contact with a solid substrate or with host tissues is important, other Ras-independent transcription factors come into play to provide additional regulation of expression of genes involved in cell adhesion during contact with an abiotic substrate such as silicone discs. Notably, this study identified Ace2 and the RAM network as being a major network that regulates cell adhesion in the context of a silicone substrate (Fig. 3D). Ace2 is a daughter cell- and hyphal tip cell-localized transcription factor that is required for adherence, cell separation, virulence in mice, and negative regulation of morphogenesis. Thus, aspects of cell adhesion are likely to require daughter cell-specific functions in C. albicans.
BIOFILM FORMATION
Biofilms are surface-associated microbial colonies that are embedded within a matrix of extracellular polymers (70, 71). In humans, C. albicans infections are often associated with biofilms growing on medical devices such as indwelling catheters and mechanical devices or on acrylic dental implants (72). This strategy for colonization is thought to enable microbes to maintain a protected antimicrobial, drug-resistant environment for growth and dissemination (5, 9, 10, 70). Biofilm formation begins with the attachment (adhesion) of yeast cells to a substrate. The yeast cells then form filaments that extend perpendicularly to the substrate. Extracellular matrix material is then secreted and provides protection from chemical and immunological insults. Finally, planktonic daughter cells released from the biofilm filaments (a form of filament-yeast transition) within the host promote further colonization of tissues and disseminated disease (for reviews, see references 73 and 74). The pathogenic biofilms formed by wild-type C. albicans are refractory to antifungal drugs (70, 72), which complicates the treatment of infections involving biofilms (for a recent review, see reference 73). Biofilms are also poorly penetrated by human polymorphonuclear leukocytes (PMNs) (75), another property that makes biofilm growth an effective microbial strategy for enhanced proliferation within a host.
The formation of pathogenic Candida biofilms, with the expected pathogenic traits (increased resistance to drugs, PMN invasion, and a predominantly filamentous architecture), is dependent on Ras1, Cyr1, and Tpk2, the transcription factors Efg1 and Tec1, and a major transcriptional regulator of biofilm development, Bcr1 (51, 60, 75). In contrast, homozygous MTLa/a or MTLα/α strains form biofilms that lack most pathogenic traits and are controlled by a Ras-independent mechanism (75). Loss-of-function mutations in genes encoding any of the above components of the Ras1 signaling pathway or its transcriptional effectors (Tec1, Efg1, and Bcr1) severely compromise biofilm adhesion, accumulation of biomass, β-glucan release from the matrix, and biofilm architecture and reduce the expression of HWP1 and ALS3 (described above), which is induced during pathogenic biofilm development (25). RNA-Seq studies identified 1,006 genes that were induced during biofilm formation (54) and at least 11 Bcr1-regulated, cell wall-associated genes (as determined using a 2-fold cutoff; see Supplementary Table S3 in reference 54), 19 cell wall-associated genes regulated by Efg1, and 10 Tec1-regulated cell wall-associated genes, several of which show that their promoters are bound by these transcription factors, but not exclusively; these genes are in fact bound and regulated by multiple factors. Notably, ALS1 is bound by all three Ras-regulated transcription factors (Bcr1, Efg1, and Tec1) to induce its expression during biofilm formation, suggesting a critical role for Als1 during some aspect of biofilm formation such as cell-cell interactions, cell adhesion, or architectural integrity within the biofilm matrix. These data also show that ACE2 gene expression was upregulated in those biofilm experiments (see Table S3 in reference 54), implying a role for daughter cell functions in biofilm development such as the separation of daughter cells from the mature biofilms during the process of dissemination.
By epistasis analysis, transcription factors Efg1, Tec1, and Bcr1 (in that order) were determined to function downstream of the Ras1/cAMP pathway of C. albicans (75). For example, loss-of-function mutations in EFG1 lead to decreased biofilm formation, whereas its overexpression rescues the biofilm defects of the ras1, cyr1, and tpk2 mutants, indicating that Efg1 functions downstream of these genes to control biofilm formation in MTLa/alpha strains (75). An analogous method was used to place Tec1 and Bcr1 downstream of Ras1, Cyr1, Tpk2, and Efg1. Thus, the Ras/cAMP/PKA signaling pathway is critical for the formation of biofilms as well as for the pathogenic traits of wild-type biofilms and the three Ras-regulated transcription factors Efg1, Tec1, and Bcr1 are absolutely required for wild-type biofim formation. The studies by Nobile et al. (54) also showed that at least three additional transcription factors (Ndt80, Brg1, and Rob1) control biofilm formation in a complex, interwoven transcriptional regulatory circuit where all 6 of these transcription factors directly regulate each other (Fig. 3E) as well as both distinct and overlapping sets of target genes to regulate pathogenic biofilm formation. The fact that Efg1, Tec1, and Ndt80 also function during morphogenesis (76) links biofilm formation to morphogenesis. However, BCR1 and ROB1 mutants filament normally (54) and thus appear to have functions distinct from the morphogenetic aspects of biofilm formation. Brg1 recruits the Hda1 histone deacetylase to hypha-specific promoters during hyphal cell elongation under conditions of reduced Tor1 signaling (77) and therefore may play a role during hyphal elongation in biofilm formation. Overall, these results suggest that the Ras pathway has been wired into a more recently evolved transcriptional regulatory circuit in the context of biofilm formation, in which signaling inputs distinct from the Ras pathway may differentially influence these transcription factors to control one or more of the multiple stages of development required for mature biofilm development.
MORPHOGENESIS
Hyphal morphogenesis has long been associated with virulence of C. albicans (for reviews, see references 50 and 78), and the RAS/cAMP/PKA pathway is central to this process (46). Much attention has been focused on the switch in growth mode from budding as a yeast cell (Fig. 1A to D) to growth as elongated filaments (Fig. 1E to N). However, both the forward (yeast-to-filament) and reverse (filament-to-yeast) transitions are important for normal biofilm formation. The forward transition is important for normal biofilm architecture, and the reverse transition is presumably important for the production of yeast-form daughter cells from the biofilm filaments and their subsequent release and dissemination within a host.
Multiple environmental stimuli, including serum, in combination with 37°C temperature (79–81), neutral pH (82), the presence of N-acetylglucosamine (GlcNAc) (83–85), and high CO2 (39, 86), induce C. albicans to grow as filaments, whereas low temperatures (30°C or below), low pH, and glucose-rich media favor growth in the yeast form (18). (For a recent review of hyphal morphogenesis in C. albicans, see reference 13.) Ras1 and components of the Ras signaling pathway (Csc25, Cyr1, Srv2, and Tpk1/Tpk2) are required for the filamentous-growth response to a variety of these filament-inducing stimuli (Fig. 3). Filamentous growth via the Ras pathway is also stimulated by chemical or genetic depletion of Hsp90, a molecular chaperone that represses the Ras pathway at temperatures of 30°C or below (87). Thus, the serum-induced yeast-to-filament transition (typically in 5% to 10% serum) is stimulated in combination with elevated temperature (usually 37°C), which relieves Hsp90-mediated repression of the Ras pathway. In contrast, ras1, csc25, and cyr1 null mutants are completely blocked in the morphogenetic response to Hsp90 inhibition (87), as are cells depleted in both TPK1 and TPK2, the two partially redundant PKA isoforms, which demonstrates that Hsp90 negatively regulates morphogenesis by controlling Ras signaling.
Another environmental factor that affects filamentous growth is cell density. C. albicans secretes farnesol, an extracellular quorum-sensing molecule that inhibits the induction of hyphal growth by inhibiting Cyr1 (19, 20). Preformed filamentous cells transition to growth as yeast cells in response to farnesol, and strains with increased cAMP signaling, such as the pde2 mutant, exhibit greater resistance to farnesol (19). These data show that the Ras/cAMP/PKA pathway is involved in the maintenance of filamentous growth and that farnesol induces the filament-to-yeast transition by inhibiting this pathway.
Kadosh and Johnson showed that the hyphal induction program is under negative regulation by the transcription factors Tup1, Nrg1, and Rfg1 and that these factors control the expression of UME6, which encodes another negative transcriptional regulator of morphogenesis that, in turn, regulates the expression of genes associated with filamentation (55, 67, 88, 89). This negative regulatory program has a dramatic impact on morphogenesis, and the temporal regulation of NRG1 expression is dependent upon the presence of an intact Ras/cAMP/PKA pathway (90). The major Ras pathway-regulated transcription factor that controls morphogenesis is Efg1 (Fig. 3A and B). Genome-wide chromatin immunoprecipitation (ChIP) experiments performed with Efg1 show that the promoter sites bound by Efg1 in yeast-form cells are dramatically different from the sites bound upon filament induction (91). Interestingly, Efg1 negatively regulates its own expression during morphogenesis but is positively autoregulated in another biological context, namely, during biofilm formation (54) (described below). This provides support for the idea that in different environmental contexts, condition-dependent regulation of even a single transcription factor can generate different phenotypic outcomes.
Transcriptional profiling experiments performed on ras1, cyr1, and efg1 mutants in yeast and hyphae revealed that ASH1 (orf19.5343), which encodes a GATA-like transcription factor localized in daughter cells and hyphal tips, is negatively regulated by signaling though AC, independently of Efg1 (92). In other words, transcriptional regulation through Ras signaling can occur independently of Efg1. Presumably, the regulation of ASH1 occurs at the level of regulation by PKA or other Ras-dependent, Efg1-independent factors. ASH1 is required for virulence and dissemination in mice and for filamentation on solid media (93) and likely directs the expression of a yeast daughter cell- and hyphal tip cell-regulatory program to activate filamentous growth and promote pathogenesis in mice. Ash1 can function as both a transcriptional repressor and an activator of gene expression (93); thus, Ras1 may utilize Ash1 independently of Efg1 to impact gene expression (either positively or negatively) during filamentation in specialized cell types such as daughter cells and hyphal tip cells. Note that Ace2 and Ash1 are both daughter and hyphal tip cell-localized factors that function during morphogenesis (Fig. 3A and B) and promote virulence of C. albicans (93, 94). The identification of target genes regulated by these two transcription factors during morphogenesis or other related processes would shed light on the distinct and/or overlapping functions of daughter cells and hyphal tip cells during cell adhesion and morphogenesis.
Another component important to the regulation of gene expression during hyphal development is the regulation of chromatin accessibility, which can dramatically impact gene expression and is critical for regulating hyphal morphogenesis and hypha-specific gene expression (90, 95). Regulation of gene expression by the Hda1 and Rpd3 histone deacetylases was initially observed to play a role in the W-O switch (96). More recently, recruitment of Hda1 to hypha-specific gene promoters was shown to require Brg1 (77), one of the 6 major regulators of biofilm formation. The promoters of hypha-specific target genes are inaccessible to Brg1 in yeast cells due to the positioning of nucleosomes over putative Brg1 binding sites (72). In contrast, Nrg1, which is transiently removed from cells during the first 2 h of hypha induction with serum, has binding sites in nucleosome-free regions during growth in the yeast form, thus allowing Nrg1 to bind its target genes and maintain the yeast state by repressing hypha development (77). The temporal pattern of Nrg1 removal during the early stage of hyphal induction depends on Ras signaling, which indicates that both transcription factors and chromatin-modifying enzymes interact with the Ras pathway to orchestrate a complex temporal pattern of gene expression during hyphal morphogenesis.
The Set3C histone deacetylase complex, which can deacetylate histone H4 and is composed of the Set3 and Hos2 subunits, is present at highly transcribed genes and is also important for inhibiting the yeast-to-filament transition in a cAMP/PKA pathway-dependent manner (97). Recent work by Hnisz et al., using a combination of RNA-seq and ChIP-seq, showed that Set3C modulates the expression of key regulators of hyphal morphogenesis, including genes for the transcription factors NRG1, EFG1, BRG1, and TEC1 (95). Those authors showed that, as is the case in biofilm formation, the yeast- and hypha-specific Set3C target transcription factor genes form a core, interwoven transcriptional circuit that regulates hyphal morphogenesis. Remarkably, this work revealed that 5 of the 6 major biofilm regulators (all but BCR1) participate in this circuit, indicating extensive overlap between the regulation of hyphal morphogenesis and biofilm formation.
WHITE-TO-OPAQUE SWITCHING
The white-to-opaque (W-O) switch involves dramatic and reversible changes in colony morphology (98). White-form cells (considered the default growth state) convert to opaque-form cells that display alterations in cellular morphology (99), environmental responses (100), gene expression (101), host immune cell interactions (102, 103), and the ability to mate (27). White cells are also more virulent than opaque cells in a mouse model of oropharyngeal candidiasis (104). W-O switching of natural isolates is controlled by the configuration of the mating type locus; homozygous MTLa/a or MTLα/α strains can undergo W-O switching, whereas wild-type heterozygous MTLa/α strains cannot (105). The Ras/cAMP/PKA pathway is known to be involved in the control of this switch (106), though few papers currently address its role in this regulation.
Both CO2 and GlcNAc are potent inducers of the switch from white to opaque cells (39, 106, 107), while the opaque-to-white transition is blocked under these conditions. Because of the well-established link between the response to CO2 and the Ras signaling pathway, it was first hypothesized that mutants in the Ras pathway might be defective for the GlcNAc-induced W-O switch (106). To test whether GlcNAc induction of W-O switching was mediated by the Ras pathway, Huang and colleagues directly tested whether a ras1 mutant could undergo GlcNAc-induced high-frequency W-O switching. In this mutant, switching was dramatically reduced (11.2% in the mutant compared to 90.5% in the wild-type strain). Alternatively, overexpression of the constitutively activated RAS1 allele, RAS1G13V, causes the near-complete induction of opaque-sectored colonies (107), suggesting that it plays a major (but not exclusive) role in regulating the W-O switch.
Since Ras1 positively regulates Cyr1 and the resulting increase in cAMP levels is kept in check by Pde2, it was reasoned that deletion of CYR1 should reduce GlcNAc-induced high-frequency switching whereas the deletion of PDE2 should enhance it (106). As expected, strains deleted for CYR1 did show reduced GlcNAc-induced switching, whereas pde2 mutants displayed a dramatic increase in switching. The two isoforms of the catalytic subunit of PKA, Tpk1 and Tpk2, have been shown to play different roles in filamentation, depending upon environmental conditions (108, 109). Attempts to delete both TPK1 and TPK2 have been unsuccessful, suggesting such a mutant is inviable in C. albicans (108). By overexpressing Tpk1 and Tpk2, it was demonstrated that Tpk1 has no effect on W-O switching, while Tpk2 does appear to have a major impact on GlcNAc-induced signal transduction (106). Therefore, the major components of the Ras/cAMP/PKA pathway contribute to GlcNAc-induced high-frequency W-O switching of C. albicans; this signal is then propagated downstream by the master switch regulator, Wor1. When WOR1 is overexpressed in ras1, pde2, cyr1, tpk1, or tpk2 mutant backgrounds, the opaque state results, showing that Wor1 is the transcriptional effector downstream of the Ras signaling components that regulates the W-O switch (106). Other studies have identified additional transcription factors that control the W-O switch in concert with Wor1, namely, Czf1, Efg1, Wor2 (58, 110), the recently described Wor3 (56), and the mating-type proteins a1 and α2 (110) that create the genetic environment required for the W-O switch (Fig. 3E). While a direct association with Ras signaling and Czf1, Wor2, or Wor3 has not been described, these transcription factors act in concert in a transcriptional regulatory-feedback loop with Ras-regulated Efg1 and Wor1. Similarly to the complex regulatory circuit that controls biofilm formation, Czf1, Wor1, Wor2, and Wor3 participate in an interlocking regulatory-feedback loop with Efg1 to control the W-O switch (56, 110) and thus provide additional support for the idea of fine-tuning of Ras signaling by a complex, context-specific, transcriptional regulatory network that controls phenotypic switching in C. albicans. This transcriptional circuit further is regulated in a complex manner by the regulation of chromatin structure, as the phenotype of the hda1 mutant suggests that the expression of key switch regulators is impacted by Hda1-mediated histone positioning (96).
FINGERS AND TENTACLES
Under conditions of high levels of CO2 (5 to 20%) in vitro, MTL-heterozygous strains (MTLa/α) plated on well-dried modified Lee's agar form multicellular structures called fingers and tentacles (29). Fingers are elongated structures that extend aerially from colony surfaces and are comprised of a central core hypha surrounded by a sheath of yeast cells. They are attached to their substrates through a basal monolayer of yeast cells. Electron micrographs reveal a yeast-lined crater in the agar that forms from the fragile interface at the bulb-like base of an incipient finger and the agar (29). Tentacles are multicellular structures that also form under 20% CO2 on the surface of agar containing modified Lee's medium. Tentacles bear a morphology distinct from that of fingers and yet develop under the same environmental conditions. Scanning electron microscopy images of tentacles show budding yeast cells surrounding a central, core hyphae that presumably extends along the length of the tentacle (29). Because of the presence of a single core hypha surrounded by yeast cells and the link between Ras signaling, the CO2 response, and filamentation, it was predicted that components of the Ras pathway would be critical for the formation of the central hypha, upon which development of both the finger and tentacle structures likely depends (29). Indeed, experiments have shown that both finger formation and tentacle formation are dependent upon the Cyr1 and Tpk2 Ras/cAMP/PKA pathway components and the Tec1 transcription factor (Fig. 3C). Whether or not additional regulatory circuitry has developed to control finger and tentacle development is currently not known, but given the complexity of regulation of other traits of C. albicans, it is possible that a complex regulatory circuit also governs the development of these multicellular structures. It has been suggested that the finger may function as a multicellular dispersal mechanism produced in host niches that contain high levels of CO2 such as that of the gastrointestinal tract or within host tissues (29), and as such, the function of biofilms as a dispersal mechanism bears some similarity to the potential function of the finger. It is of significant interest to know whether formation of fingers and/or tentacles plays an important role in vivo.
CONCLUSIONS
Ras/cAMP/PKA signaling plays a central role in the pathogenesis of C. albicans and orchestrates the expression of distinct but interrelated traits. Ras1, Cyr1, Pde2, and Efg1 are the only C. albicans proteins that play a role in cell adhesion, biofilm formation, filamentation, W-O switching, and finger and tentacle formation and are also required for pathogenesis. Cell adhesion contributes to multiple pathogenic traits of C. albicans, and both forward morphogenesis and reverse morphogenesis are required to complete the biofilm life cycle. Because both switching and filamentation are triggered by high levels of either CO2 or GlcNAc and both processes involve the remodeling of cell shape, W-O switching shares characteristics with morphogenesis. It is tempting to speculate that finger and tentacle formation may also be linked to W-O switching and filamentation within the gut, as these processes are triggered by growth under the same conditions and are dependent upon the same signaling pathways. It would be interesting to know whether fingers can form in vivo and whether they play a role during W-O switching, and thus mating, of C. albicans.
One set of environmental signals can activate Ras signaling and elicit a phenotypic response in one genetic context and yet in other genetic backgrounds can produce a different response. Both wild-type (MTLa/α) biofilm formation and W-O switching (MTLa/a or MTLα/α strains) depend upon Ras signaling; however, at least one of the downstream transcriptional effectors that regulate these processes differs for each process. Bcr1, Tec1, and Efg1 regulate cell adhesion proteins and (along with Brg1, Ndt80, and Rob1) biofilm formation, while the positive regulation of morphogenesis requires Tec1, Efg1, and Brg1 as well as additional daughter cell-specific factors Ace2 and Ash1. W-O switching is also regulated through a complex, interwoven network that interacts with other W-O-specific transcription factors and histone deacetylases that are active in particular environmental and cell type-specific processes. The fact that temporal regulation of chromatin accessibility by histone-modifying enzymes dramatically impacts both morphogenesis and W-O switching suggests that chromatin-level regulation of gene expression is an important area for future investigation of complex developmental processes such biofilm formation and finger and tentacle formation. Given the central role that Ras signaling plays in regulating major pathogenic traits of C. albicans, targeting this pathway in vivo might improve clinical outcomes of patients with Candida infections.
ACKNOWLEDGMENTS
We thank David R. Soll and Marek S. Skrzypek and two anonymous reviewers for very helpful comments on the manuscript. We especially thank Suzanne Noble and her laboratory for the use of cell culture equipment and Anita Sil and her laboratory for use of the Zeiss Axioplan inverted microscope for imaging C. albicans.
This review was supported by the National Institute of Dental and Craniofacial Research at the U.S. National Institutes of Health (grant no. R01 DE015873 to G.S.).
Footnotes
Published ahead of print 2 August 2013
REFERENCES
- 1.Seneviratne CJ, Wang Y, Jin L, Wong SS, Herath TD, Samaranayake LP. 2012. Unraveling the resistance of microbial biofilms: has proteomics been helpful? Proteomics 12:651–665 [DOI] [PubMed] [Google Scholar]
- 2.Vlamakis H, Chai Y, Beauregard P, Losick R, Kolter R. 2013. Sticking together: building a biofilm the Bacillus subtilis way. Nat. Rev. Microbiol. 11:157–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Verstraelen H, Swidsinski A. 2013. The biofilm in bacterial vaginosis: implications for epidemiology, diagnosis and treatment. Curr. Opin. Infect. Dis. 26:86–89 [DOI] [PubMed] [Google Scholar]
- 4.Islam MS, Richards JP, Ojha AK. 2012. Targeting drug tolerance in mycobacteria: a perspective from mycobacterial biofilms. Expert Rev. Anti Infect. Ther. 10:1055–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Römling U, Balsalobre C. 2012. Biofilm infections, their resilience to therapy and innovative treatment strategies. J. Intern. Med. 272:541–561 [DOI] [PubMed] [Google Scholar]
- 6.Walz JM, Memtsoudis SG, Heard SO. 2010. Prevention of central venous catheter bloodstream infections. J. Intensive Care Med. 25:131–138 [DOI] [PubMed] [Google Scholar]
- 7.Cauda R. 2009. Candidaemia in patients with an inserted medical device. Drugs 69(Suppl 1):33–38 [DOI] [PubMed] [Google Scholar]
- 8.Busscher HJ, Rinastiti M, Siswomihardjo W, van der Mei HC. 2010. Biofilm formation on dental restorative and implant materials. J. Dent. Res. 89:657–665 [DOI] [PubMed] [Google Scholar]
- 9.Mah TF. 2012. Biofilm-specific antibiotic resistance. Future Microbiol. 7:1061–1072 [DOI] [PubMed] [Google Scholar]
- 10.Ramage G, Rajendran R, Sherry L, Williams C. 2012. Fungal biofilm resistance. Int. J. Microbiol. 2012:528521. 10.1155/2012/528521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sardi JC, Scorzoni L, Bernardi T, Fusco-Almeida AM, Mendes Giannini MJ. 2013. Candida species: current epidemiology, pathogenicity, biofilm formation, natural antifungal products and new therapeutic options. J. Med. Microbiol. 62(Pt 1):10–24 [DOI] [PubMed] [Google Scholar]
- 12.Douglas LJ. 2002. Medical importance of biofilms in Candida infections. Rev. Iberoam. Micol. 19:139–143 [PubMed] [Google Scholar]
- 13.Sudbery PE. 2011. Growth of Candida albicans hyphae. Nat. Rev. Microbiol. 9:737–748 [DOI] [PubMed] [Google Scholar]
- 14.Biswas S, Van Dijck P, Datta A. 2007. Environmental sensing and signal transduction pathways regulating morphopathogenic determinants of Candida albicans. Microbiol. Mol. Biol. Rev. 71:348–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whiteway M, Bachewich C. 2007. Morphogenesis in Candida albicans. Annu. Rev. Microbiol. 61:529–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berman J. 2006. Morphogenesis and cell cycle progression in Candida albicans. Curr. Opin. Microbiol. 9:595–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sudbery P, Gow N, Berman J. 2004. The distinct morphogenic states of Candida albicans. Trends Microbiol. 12:317–324 [DOI] [PubMed] [Google Scholar]
- 18.Odds FC. 1985. Morphogenesis in Candida albicans. Crit. Rev. Microbiol. 12:45–93 [DOI] [PubMed] [Google Scholar]
- 19.Lindsay AK, Deveau A, Piispanen AE, Hogan DA. 2012. Farnesol and cyclic AMP signaling effects on the hypha-to-yeast transition in Candida albicans. Eukaryot. Cell 11:1219–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hornby JM, Jensen EC, Lisec AD, Tasto JJ, Jahnke B, Shoemaker R, Dussault P, Nickerson KW. 2001. Quorum sensing in the dimorphic fungus Candida albicans is mediated by farnesol. Appl. Environ. Microbiol. 67:2982–2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han TL, Cannon RD, Villas-Bôas SG. 2012. The metabolic response of Candida albicans to farnesol under hyphae-inducing conditions. FEMS Yeast Res. 12:879–889 [DOI] [PubMed] [Google Scholar]
- 22.Lo HJ, Köhler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink GR. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90:939–949 [DOI] [PubMed] [Google Scholar]
- 23.Saville SP, Lazzell AL, Monteagudo C, Lopez-Ribot JL. 2003. Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during infection. Eukaryot. Cell 2:1053–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carlisle PL, Banerjee M, Lazzell A, Monteagudo C, López-Ribot JL, Kadosh D. 2009. Expression levels of a filament-specific transcriptional regulator are sufficient to determine Candida albicans morphology and virulence. Proc. Natl. Acad. Sci. U. S. A. 106:599–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nobile CJ, Andes DR, Nett JE, Smith FJ, Yue F, Phan QT, Edwards JE, Filler SG, Mitchell AP. 2006. Critical role of Bcr1-dependent adhesins in C. albicans biofilm formation in vitro and in vivo. PLoS Pathog. 2:e63. 10.1371/journal.ppat.0020063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoyer LL, Payne TL, Bell M, Myers AM, Scherer S. 1998. Candida albicans ALS3 and insights into the nature of the ALS gene family. Curr. Genet. 33:451–459 [DOI] [PubMed] [Google Scholar]
- 27.Miller MG, Johnson AD. 2002. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell 110:293–302 [DOI] [PubMed] [Google Scholar]
- 28.Levitt MD, Bond JH., Jr 1970. Volume, composition, and source of intestinal gas. Gastroenterology 59:921–929 [PubMed] [Google Scholar]
- 29.Daniels KJ, Pujol C, Srikantha T, Soll DR. 2012. The “finger,” a unique multicellular morphology of Candida albicans induced by CO2 and dependent upon the Ras1-cyclic AMP pathway. Eukaryot. Cell 11:1257–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baines AT, Xu D, Der CJ. 2011. Inhibition of Ras for cancer treatment: the search continues. Future Med. Chem. 3:1787–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poulogiannis G, Luo F, Arends MJ. 2012. RAS signalling in the colorectum in health and disease. Cell Commun. Adhes. 19:1–9 [DOI] [PubMed] [Google Scholar]
- 32.Cullen PJ, Sprague GF., Jr 2012. The regulation of filamentous growth in yeast. Genetics 190:23–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Broach JR. 2012. Nutritional control of growth and development in yeast. Genetics 192:73–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de la Torre-Ruiz Mozo-Villarías A, Pujol N, Petkova MI. 2010. How budding yeast sense and transduce the oxidative stress signal and the impact in cell growth and morphogenesis. Curr. Protein Pept. Sci. 11:669–679 [DOI] [PubMed] [Google Scholar]
- 35.Alspaugh JA, Cavallo LM, Perfect JR, Heitman J. 2000. RAS1 regulates filamentation, mating and growth at high temperature of Cryptococcus neoformans. Mol. Microbiol. 36:352–365 [DOI] [PubMed] [Google Scholar]
- 36.Piispanen AE, Bonnefoi O, Carden S, Deveau A, Bassilana M, Hogan DA. 2011. Roles of Ras1 membrane localization during Candida albicans hyphal growth and farnesol response. Eukaryot. Cell 10:1473–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prior Hancock IA, JF 2012. Ras trafficking, localization and compartmentalized signalling. Semin. Cell Dev. Biol. 23:145–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fang HM, Wang Y. 2006. RA domain-mediated interaction of Cdc35 with Ras1 is essential for increasing cellular cAMP level for Candida albicans hyphal development. Mol. Microbiol. 61:484–496 [DOI] [PubMed] [Google Scholar]
- 39.Klengel T, Liang WJ, Chaloupka J, Ruoff C, Schröppel K, Naglik JR, Eckert SE, Mogensen EG, Haynes K, Tuite MF, Levin LR, Buck J, Mühlschlegel FA. 2005. Fungal adenylyl cyclase integrates CO2 sensing with cAMP signaling and virulence. Curr. Biol. 15:2021–2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bockmühl DP, Ernst JF. 2001. A potential phosphorylation site for an A-type kinase in the Efg1 regulator protein contributes to hyphal morphogenesis of Candida albicans. Genetics 157:1523–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bahn Y, Staab J, Sundstrom P. 2003. Increased high-affinity phosphodiesterase PDE2 gene expression in germ tubes counteracts CAP1-dependent synthesis of cyclic AMP, limits hypha production and promotes virulence of Candida albicans. Mol. Microbiol. 50:391–409 [DOI] [PubMed] [Google Scholar]
- 42.Bahn YS, Molenda M, Staab JF, Lyman CA, Gordon LJ, Sundstrom P. 2007. Genome-wide transcriptional profiling of the cyclic AMP-dependent signaling pathway during morphogenic transitions of Candida albicans. Eukaryot. Cell 6:2376–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bahn YS, Sundstrom P. 2001. CAP1, an adenylate cyclase-associated protein gene, regulates bud-hypha transitions, filamentous growth, and cyclic AMP levels and is required for virulence of Candida albicans. J. Bacteriol. 183:3211–3223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jung WH, Warn P, Ragni E, Popolo L, Nunn CD, Turner MP, Stateva L. 2005. Deletion of PDE2, the gene encoding the high-affinity cAMP phosphodiesterase, results in changes of the cell wall and membrane in Candida albicans. Yeast 22:285–294 [DOI] [PubMed] [Google Scholar]
- 45.Wilson D, Fiori A, Brucker KD, Van Dijck P, Stateva L. 2010. Candida albicans Pde1p and Gpa2p comprise a regulatory module mediating agonist-induced cAMP signalling and environmental adaptation. Fungal Genet. Biol. 47:742–752 [DOI] [PubMed] [Google Scholar]
- 46.Leberer E, Harcus D, Dignard D, Johnson L, Ushinsky S, Thomas DY, Schröppel K. 2001. Ras links cellular morphogenesis to virulence by regulation of the MAP kinase and cAMP signalling pathways in the pathogenic fungus Candida albicans. Mol. Microbiol. 42:673–687 [DOI] [PubMed] [Google Scholar]
- 47.Piispanen AE, Grahl N, Hollomon JM, Hogan DA. 2013. Regulated proteolysis of Candida albicans Ras1 is involved in morphogenesis and quorum sensing regulation. Mol. Microbiol. 89:166–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu Y, Fang HM, Wang YM, Zeng GS, Zheng XD, Wang Y. 2009. Ras1 and Ras2 play antagonistic roles in regulating cellular cAMP level, stationary-phase entry and stress response in Candida albicans. Mol. Microbiol. 74:862–875 [DOI] [PubMed] [Google Scholar]
- 49.Park H, Myers CL, Sheppard DC, Phan QT, Sanchez AA, E Edwards J, Filler SG. 2005. Role of the fungal Ras-protein kinase A pathway in governing epithelial cell interactions during oropharyngeal candidiasis. Cell. Microbiol. 7:499–510 [DOI] [PubMed] [Google Scholar]
- 50.Rocha CR, Schröppel K, Harcus D, Marcil A, Dignard D, Taylor BN, Thomas DY, Whiteway M, Leberer E. 2001. Signaling through adenylyl cyclase is essential for hyphal growth and virulence in the pathogenic fungus Candida albicans. Mol. Biol. Cell 12:3631–3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hogan DA, Mühlschlegel FA. 2011. Candida albicans developmental regulation: adenylyl cyclase as a coincidence detector of parallel signals. Curr. Opin. Microbiol. 14:682–686 [DOI] [PubMed] [Google Scholar]
- 52.Hogan DA, Sundstrom P. 2009. The Ras/cAMP/PKA signaling pathway and virulence in Candida albicans. Future Microbiol. 4:1263–1270 [DOI] [PubMed] [Google Scholar]
- 53.Finkel JS, Xu W, Huang D, Hill EM, Desai JV, Woolford CA, Nett JE, Taff H, Norice CT, Andes DR, Lanni F, Mitchell AP. 2012. Portrait of Candida albicans adherence regulators. PLoS Pathog. 8:e1002525. 10.1371/journal.ppat.1002525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nobile CJ, Fox EP, Nett JE, Sorrells TR, Mitrovich QM, Hernday AD, Tuch BB, Andes DR, Johnson AD. 2012. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell 148:126–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Carlisle PL, Kadosh D. 2013. A genome-wide transcriptional analysis of morphology determination in Candida albicans. Mol. Biol. Cell 24:246–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lohse MB, Hernday AD, Fordyce PM, Noiman L, Sorrells TR, Hanson-Smith V, Nobile CJ, Derisi JL, Johnson AD. 2013. Identification and characterization of a previously undescribed family of sequence-specific DNA-binding domains. Proc. Natl. Acad. Sci. U. S. A. 110:7660–7665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tuch BB, Mitrovich QM, Homann OR, Hernday AD, Monighetti CK, De La Vega FM, Johnson AD. 2010. The transcriptomes of two heritable cell types illuminate the circuit governing their differentiation. PLoS Genet. 6:e1001070. 10.1371/journal.pgen.1001070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lohse MB, Johnson AD. 2010. Temporal anatomy of an epigenetic switch in cell programming: the white-opaque transition of C. albicans. Mol. Microbiol. 78:331–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Doedt T, Krishnamurthy S, Bockmühl DP, Tebarth B, Stempel C, Russell CL, Brown AJ, Ernst JF. 2004. APSES proteins regulate morphogenesis and metabolism in Candida albicans. Mol. Biol. Cell 15:3167–3180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nobile CJ, Mitchell AP. 2005. Regulation of cell-surface genes and biofilm formation by the C. albicans transcription factor Bcr1p. Curr. Biol. 15:1150–1155 [DOI] [PubMed] [Google Scholar]
- 61.Li F, Palecek SP. 2005. Identification of Candida albicans genes that induce Saccharomyces cerevisiae cell adhesion and morphogenesis. Biotechnol. Prog. 21:1601–1609 [DOI] [PubMed] [Google Scholar]
- 62.Heilmann CJ, Sorgo AG, Siliakus AR, Dekker HL, Brul S, de Koster CG, de Koning LJ, Klis FM. 2011. Hyphal induction in the human fungal pathogen Candida albicans reveals a characteristic wall protein profile. Microbiology 157(Pt 8):2297–2307 [DOI] [PubMed] [Google Scholar]
- 63.Sheppard DC, Yeaman MR, Welch WH, Phan QT, Fu Y, Ibrahim AS, Filler SG, Zhang M, Waring AJ, Edwards JE., Jr 2004. Functional and structural diversity in the Als protein family of Candida albicans. J. Biol. Chem. 279:30480–30489 [DOI] [PubMed] [Google Scholar]
- 64.Nobbs AH, Vickerman MM, Jenkinson HF. 2010. Heterologous expression of Candida albicans cell wall-associated adhesins in Saccharomyces cerevisiae reveals differential specificities in adherence and biofilm formation and in binding oral Streptococcus gordonii. Eukaryot. Cell 9:1622–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fanning S, Xu W, Solis N, Woolford CA, Filler SG, Mitchell AP. 2012. Divergent targets of Candida albicans biofilm regulator Bcr1 in vitro and in vivo. Eukaryot. Cell 11:896–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Martin R, Albrecht-Eckardt D, Brunke S, Hube B, Hünniger K, Kurzai O. 2013. A core filamentation response network in Candida albicans is restricted to eight genes. PLoS One 8:e58613. 10.1371/journal.pone.0058613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kadosh D, Johnson AD. 2005. Induction of the Candida albicans filamentous growth program by relief of transcriptional repression: a genome-wide analysis. Mol. Biol. Cell 16:2903–2912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cleary IA, Reinhard SM, Miller CL, Murdoch C, Thornhill MH, Lazzell AL, Monteagudo C, Thomas DP, Saville SP. 2011. Candida albicans adhesin Als3p is dispensable for virulence in the mouse model of disseminated candidiasis. Microbiology 157(Pt 6):1806–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sharkey LL, McNemar MD, Saporito-Irwin SM, Sypherd PS, Fonzi WA. 1999. HWP1 functions in the morphological development of Candida albicans downstream of EFG1, TUP1, and RBF1. J. Bacteriol. 181:5273–5279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chandra J, Kuhn DM, Mukherjee PK, Hoyer LL, McCormick T, Ghannoum MA. 2001. Biofilm formation by the fungal pathogen Candida albicans: development, architecture, and drug resistance. J. Bacteriol. 183:5385–5394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mukherjee PK, Zhou G, Munyon R, Ghannoum MA. 2005. Candida biofilm: a well-designed protected environment. Med. Mycol. 43:191–208 [DOI] [PubMed] [Google Scholar]
- 72.Chandra J, Mukherjee PK, Leidich SD, Faddoul FF, Hoyer LL, Douglas LJ, Ghannoum MA. 2001. Antifungal resistance of candidal biofilms formed on denture acrylic in vitro. J. Dent. Res. 80:903–908 [DOI] [PubMed] [Google Scholar]
- 73.Fox EP, Nobile CJ. 2012. A sticky situation: untangling the transcriptional network controlling biofilm development in Candida albicans. Transcription 3:315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Soll DR. 2008. Candida biofilms: is adhesion sexy? Curr. Biol. 18:R717–R720 [DOI] [PubMed] [Google Scholar]
- 75.Yi S, Sahni N, Daniels KJ, Lu KL, Srikantha T, Huang G, Garnaas AM, Soll DR. 2011. Alternative mating type configurations (a/α versus a/a or α/α) of Candida albicans result in alternative biofilms regulated by different pathways. PLoS Biol. 9:e1001117. 10.1371/journal.pbio.1001117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sellam A, Askew C, Epp E, Tebbji F, Mullick A, Whiteway M, Nantel A. 2010. Role of transcription factor CaNdt80p in cell separation, hyphal growth, and virulence in Candida albicans. Eukaryot. Cell 9:634–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lu Y, Su C, Liu H. 2012. A GATA transcription factor recruits Hda1 in response to reduced Tor1 signaling to establish a hyphal chromatin state in Candida albicans. PLoS Pathog. 8:e1002663. 10.1371/journal.ppat.1002663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brown AJ, Gow NA. 1999. Regulatory networks controlling Candida albicans morphogenesis. Trends Microbiol. 7:333–338 [DOI] [PubMed] [Google Scholar]
- 79.Odds FC. 1988. Candida and candidosis, 2nd ed. Bailliere Tindall, London, United Kingdom [Google Scholar]
- 80.Taschdjian CL, Burchall JJ, Kozinn PJ. 1960. Rapid identification of Candida albicans by filamentation on serum and serum substitutes. AMA J. Dis. Child. 99:212–215 [DOI] [PubMed] [Google Scholar]
- 81.Berman J, Sudbery PE. 2002. Candida albicans: a molecular revolution built on lessons from budding yeast. Nat. Rev. Genet. 3:918–930 [DOI] [PubMed] [Google Scholar]
- 82.Buffo J, Herman MA, Soll DR. 1984. A characterization of pH-regulated dimorphism in Candida albicans. Mycopathologia 85:21–30 [DOI] [PubMed] [Google Scholar]
- 83.Hrmová M, Drobnica L. 1982. Induction of mycelial type of development in Candida albicans by the antibiotic monorden and N-acetyl-D-glucosamine. Mycopathologia 79:55–64 [DOI] [PubMed] [Google Scholar]
- 84.Mattia E, Carruba G, Angiolella L, Cassone A. 1982. Induction of germ tube formation by N-acetyl-D-glucosamine in Candida albicans: uptake of inducer and germinative response. J. Bacteriol. 152:555–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Simonetti N, Strippoli V, Cassone A. 1974. Yeast-mycelial conversion induced by N-acetyl-D-glucosamine in Candida albicans. Nature 250:344–346 [DOI] [PubMed] [Google Scholar]
- 86.Mock RC, Pollack JH, Hashimoto T. 1990. Carbon dioxide induces endotrophic germ tube formation in Candida albicans. Can. J. Microbiol. 36:249–253 [DOI] [PubMed] [Google Scholar]
- 87.Shapiro RS, Uppuluri P, Zaas AK, Collins C, Senn H, Perfect JR, Heitman J, Cowen LE. 2009. Hsp90 orchestrates temperature-dependent Candida albicans morphogenesis via Ras1-PKA signaling. Curr. Biol. 19:621–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Banerjee M, Thompson DS, Lazzell A, Carlisle PL, Pierce C, Monteagudo C, López-Ribot JL, Kadosh D. 2008. UME6, a novel filament-specific regulator of Candida albicans hyphal extension and virulence. Mol. Biol. Cell 19:1354–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zeidler U, Lettner T, Lassnig C, Müller M, Lajko R, Hintner H, Breitenbach M, Bito A. 2009. UME6 is a crucial downstream target of other transcriptional regulators of true hyphal development in Candida albicans. FEMS Yeast Res. 9:126–142 [DOI] [PubMed] [Google Scholar]
- 90.Lu Y, Su C, Wang A, Liu H. 2011. Hyphal development in Candida albicans requires two temporally linked changes in promoter chromatin for initiation and maintenance. PLoS Biol. 9:e1001105. 10.1371/journal.pbio.1001105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lassak T, Schneider E, Bussmann M, Kurtz D, Manak JR, Srikantha T, Soll DR, Ernst JF. 2011. Target specificity of the Candida albicans Efg1 regulator. Mol. Microbiol. 82:602–618 [DOI] [PubMed] [Google Scholar]
- 92.Harcus D, Nantel A, Marcil A, Rigby T, Whiteway M. 2004. Transcription profiling of cyclic AMP signaling in Candida albicans. Mol. Biol. Cell 15:4490–4499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Inglis DO, Johnson AD. 2002. Ash1 protein, an asymmetrically localized transcriptional regulator, controls filamentous growth and virulence of Candida albicans. Mol. Cell. Biol. 22:8669–8680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kelly MT, MacCallum DM, Clancy SD, Odds FC, Brown AJ, Butler G. 2004. The Candida albicans CaACE2 gene affects morphogenesis, adherence and virulence. Mol. Microbiol. 53:969–983 [DOI] [PubMed] [Google Scholar]
- 95.Hnisz D, Bardet AF, Nobile CJ, Petryshyn A, Glaser W, Schöck U, Stark A, Kuchler K. 2012. A histone deacetylase adjusts transcription kinetics at coding sequences during Candida albicans morphogenesis. PLoS Genet. 8:e1003118. 10.1371/journal.pgen.1003118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Srikantha T, Tsai L, Daniels K, Klar AJ, Soll DR. 2001. The histone deacetylase genes HDA1 and RPD3 play distinct roles in regulation of high-frequency phenotypic switching in Candida albicans. J. Bacteriol. 183:4614–4625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hnisz D, Majer O, Frohner IE, Komnenovic V, Kuchler K. 2010. The Set3/Hos2 histone deacetylase complex attenuates cAMP/PKA signaling to regulate morphogenesis and virulence of Candida albicans. PLoS Pathog. 6:e1000889. 10.1371/journal.ppat.1000889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Slutsky B, Buffo J, Soll DR. 1985. High-frequency switching of colony morphology in Candida albicans. Science 230:666–669 [DOI] [PubMed] [Google Scholar]
- 99.Anderson J, Soll DR. 1987. Unique phenotype of opaque cells in the white-opaque transition of Candida albicans. J. Bacteriol. 169:5579–5588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kolotila MP, Diamond RD. 1990. Effects of neutrophils and in vitro oxidants on survival and phenotypic switching of Candida albicans WO-1. Infect. Immun. 58:1174–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lan CY, Newport G, Murillo LA, Jones T, Scherer S, Davis RW, Agabian N. 2002. Metabolic specialization associated with phenotypic switching in Candida albicans. Proc. Natl. Acad. Sci. U. S. A. 99:14907–14912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Geiger J, Wessels D, Lockhart SR, Soll DR. 2004. Release of a potent polymorphonuclear leukocyte chemoattractant is regulated by white-opaque switching in Candida albicans. Infect. Immun. 72:667–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lohse MB, Johnson AD. 2008. Differential phagocytosis of white versus opaque Candida albicans by Drosophila and mouse phagocytes. PLoS One 3:e1473. 10.1371/journal.pone.0001473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Liu ZH, Li M, Lu XL, She XD, Hu SQ, Chen W, Liu WD. 2010. Higher concentration of CO2 and 37°C stabilize the less virulent opaque cell of Candida albicans. Chin. Med. J. (Engl) 123:2446–2450 [PubMed] [Google Scholar]
- 105.Lockhart SR, Pujol C, Daniels KJ, Miller MG, Johnson AD, Pfaller MA, Soll DR. 2002. In Candida albicans, white-opaque switchers are homozygous for mating type. Genetics 162:737–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Huang G, Yi S, Sahni N, Daniels KJ, Srikantha T, Soll DR. 2010. N-Acetylglucosamine induces white to opaque switching, a mating prerequisite in Candida albicans. PLoS Pathog. 6:e1000806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Huang G, Srikantha T, Sahni N, Yi S, Soll DR. 2009. CO(2) regulates white-to-opaque switching in Candida albicans. Curr. Biol. 19:330–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bockmühl DP, Krishnamurthy S, Gerads M, Sonneborn A, Ernst JF. 2001. Distinct and redundant roles of the two protein kinase A isoforms Tpk1p and Tpk2p in morphogenesis and growth of Candida albicans. Mol. Microbiol. 42:1243–1257 [DOI] [PubMed] [Google Scholar]
- 109.Cloutier M, Castilla R, Bolduc N, Zelada A, Martineau P, Bouillon M, Magee BB, Passeron S, Giasson L, Cantore ML. 2003. The two isoforms of the cAMP-dependent protein kinase catalytic subunit are involved in the control of dimorphism in the human fungal pathogen Candida albicans. Fungal Genet. Biol. 38:133–141 [DOI] [PubMed] [Google Scholar]
- 110.Zordan RE, Miller MG, Galgoczy DJ, Tuch BB, Johnson AD. 2007. Interlocking transcriptional feedback loops control white-opaque switching in Candida albicans. PLoS Biol. 5:e256. 10.1371/journal.pbio.0050256 [DOI] [PMC free article] [PubMed] [Google Scholar]