Abstract
Studies have shown that certain opportunistic pathogenic species of nontuberculous mycobacteria (NTM) can be present in distributed drinking water. However, detailed information about NTM population composition in drinking water is lacking. Therefore, NTM communities in unchlorinated drinking water from the distribution system of five treatment plants in the Netherlands were characterized using 454 pyrosequencing of the hsp65 gene. Results showed high diversities in unchlorinated drinking water, with up to 28 different NTM operational taxonomic units (OTUs) in a single sample. Each drinking water sample had a unique NTM community, and most (81.1%) OTUs were observed only once. One OTU was observed in 14 of 16 drinking water samples, indicating that this NTM species is well adapted to unchlorinated drinking water conditions. A clear influence of season, source type (groundwater, surface water), easily assimilable organic carbon (AOC) concentration, biofilm formation rate, and active biomass in treated water on the establishment of an NTM community in drinking water was not observed. Apparently, local conditions are more important for the development of a specific NTM community in the drinking water distribution system. A low (4.2%) number of hsp65 gene sequences showed more than 97% similarity to sequences of the opportunistic pathogens M. avium, M. genavense, and M. gordonae. However, most (95.8%) NTM hsp65 gene sequences were related to not-yet-described NTM species that have not been linked to disease, indicating that most NTM species in unchlorinated drinking water from distribution systems in the Netherlands have a low public health significance.
INTRODUCTION
Several species of the genus Mycobacterium are described as nontuberculous mycobacteria (NTM), and these NTM species can be opportunistic pathogens, causing disease in immunocompromised humans (1). The drinking water environment provides niches for certain NTM species, since some species are capable of multiplying in biofilms, protozoa that graze on biofilms, and sediments (2–7). As a result, studies using cultivation methods have identified different NTM species isolated from drinking water (8–11). Moreover, several studies have suggested that NTM isolates from drinking water and from patients have the same genotype (12–17). Thus, drinking water can be a route of transmission of opportunistic pathogenic NTM species to immunocompromised humans.
A large proportion of the NTM species in drinking water cannot be cultivated with the currently used culture methods (18). Consequently, cultivation-based studies provide a limited view of NTM communities in drinking water. A more complete characterization of the NTM populations in drinking water can be achieved by employing molecular methods such as PCR and sequencing, but such studies are still scarce (19, 20). As a result, the NTM diversity in drinking water is still largely unexplored. A possible reason for this is that the generally used 16S rRNA gene sequence analysis is not suitable to investigate NTM populations, because the 16S rRNA gene sequences of different Mycobacterium species can be 100% identical (21). However, NTM communities can be characterized by analyzing the hsp65 gene, since it has been reported that (i) the resolving power for differentiation among NTM species is higher for hsp65 gene sequences than for 16S rRNA gene sequences and (ii) NTM species with (approximately) 100% 16S rRNA gene sequence identity can be clearly differentiated by hsp65 gene sequence analysis (21).
Regrowth of microorganisms in drinking water in the Netherlands is not limited by a disinfectant residual but by reducing biodegradable organic carbon concentrations to the μg C liter−1 level in treated water (22). However, since NTM species can grow under oligotrophic conditions (5, 23), distribution of drinking water without a disinfection residual might increase the risk of NTM growth during distribution of the drinking water. On the other hand, studies have demonstrated that certain NTM species are especially resistant to monochloramine and that shifts from chlorine to monochloramine disinfection in drinking water treatment resulted in enhanced numbers of NTM in the drinking water distribution system (24–26). Research in the 1980s demonstrated the occurrence of M. kansasii in drinking water from the Rotterdam area in the Netherlands (27). Phage typing showed that M. kansasii strains from drinking water were identical to patient strains (27). However, drinking water in the Rotterdam area was still distributed with a disinfectant residual (chlorine) at that time. More recently, van Ingen et al. (10) isolated several NTM species from unchlorinated drinking water sampled in two different regions in the Netherlands. Most of these strains were related to NTM strains that are generally not observed among patient strains in the Netherlands (10).
Although several studies, using cultivation methods, have described NTM communities in drinking water, a thorough and complete description, using molecular methods, of the NTM communities in drinking water is lacking. The aim of our study was to characterize the NTM communities in unchlorinated drinking water, using 454 pyrosequence analyses of the hsp65 gene. In addition, the influence of sources used for drinking water production (groundwater versus surface water), season, easily assimilable organic carbon (AOC), the biofilm formation rate (BFR), and the amount of active biomass in the water on the NTM populations in drinking water was elucidated.
MATERIALS AND METHODS
Sample locations.
The selection of drinking water treatment plants was based on the type of source water used for drinking water production, total organic carbon (TOC) content, easily assimilable organic carbon (AOC) concentration, and biofilm formation rate (BFR) of the treated water (Table 1). The unchlorinated distributed drinking water of four plants that used surface water (plants SW1, SW2, SW3, and SW4) and one plant that used groundwater (plant GW1) was analyzed. The TOC content of the treated water at the surface water plants is normal in the Netherlands, whereas the TOC concentration of the treated water at plant GW1 is relatively high. In addition, AOC levels above 10 ng liter−1 and BFR levels above 10 pg ATP cm−2 day−1 are considered relatively high in the Netherlands.
Table 1.
Water source used for drinking water production, total organic carbon, easily assimable organic carbon, ATP concentration, and biofilm formation rate in the treated drinking water of five treatment plantsa
| Plant | Water source | TOC (mg C liter−1) | AOC (μg C liter−1) | BFR (pg ATP cm−2 day−1) | ATP concn (ng liter−1) |
|---|---|---|---|---|---|
| SW1 | Surface water | 1.6 ± 0.1 | 14.7 ± 4.0 | 26.0 | 4.2 ± 2.4 |
| SW2 | Surface water | 2.3 ± 0.3 | 4.5 ± 0.9 | 0.64 | 1.2 ± 0.6 |
| SW3 | Surface water | 3.1 ± 0.5 | 20.4 ± 7.7 | 4.5 | 4.4 ± 1.3 |
| SW4 | Surface water | 2.5 ± 0.7 | 18.4 ± 6.9 | 15.2 | 4.2 ± 1.0 |
| GW1 | Anoxic groundwater | 7.9 ± 0.6 | 14.8 ± 5.9 | 33.1 | 6.2 ± 1.2 |
TOC, total organic carbon; AOC, easily assimilable organic carbon; BFR, biofilm formation rate; SW, surface water treatment plant; GW, groundwater treatment plant.
Sampling.
Drinking water samples (1,000 ml) were taken from treated water at the plant and at approximately 10 different locations in the distribution system of the five treatment plants in the winter and summer of 2010. Before samples were taken, taps were flushed until the water temperature remained stable for 30 s. The exception to this sampling strategy was drinking water samples from the distribution system of plant SW1 in the summer, which were taken directly from the tap. Water samples were transported and stored at 4°C. Filtration of the water sample for DNA analyses was performed within 24 h after samples were collected.
DNA isolation.
Each water sample (1,000 ml) was filtered using a 25-mm-diameter polycarbonate filter (type GTTP; Millipore, The Netherlands) (0.22 μm pore size). Subsequently, the filter was added to phosphate and MT buffer of a FastDNA Spin kit for soil (Qbiogene) and a DNA fragment of an internal control was included before the buffer with filter was stored at −20°C. The internal control was used to determine the recovery efficiency of DNA isolation and PCR analysis (28, 29). DNA was isolated using a FastDNA Spin kit for soil according to the supplier's protocol and eluted in 200 μl elution buffer.
qPCR analyses.
To determine the hsp65 gene copy numbers of Mycobacterium spp., a quantitative PCR (qPCR) protocol using the TB11 and TB12 primers published by Telenti et al. (30) was used. In short, reaction mixtures of 50 μl for PCR analyses contained 25 μl of 2× iQ SYBR green Supermix (Bio-Rad Laboratories BV, The Netherlands), 10 pmol of forward and reverse primer, 20 μg of bovine serum albumin, and 10 μl of the DNA template. The amplification program consisted of 2 min at 95°C; 43 cycles of 30 s at 95°C, 1 min at 60°C, and 1 min at 72°C; and 7 min at 72°C. Amplification, detection, and data analysis were performed in an iCycler IQ real-time detection system (Bio-Rad Laboratories BV, The Netherlands). The PCR cycle after which the fluorescence signal of the amplified DNA was detected (threshold cycle [CT]) was used to quantify the gene copy concentration. Quantification was based on comparison of the sample CT value with the CT values of a calibration curve based on known copy numbers of the hsp65 gene of M. avium subsp. avium.
Sequence analysis.
A part of the hsp65 gene (441 bp) was amplified from two to three drinking water samples from the distribution system and/or treated water taken in the summer (plants SW3, SW4, and GW1) or in the winter and summer (plants SW1 and SW2) using the TB11 and TB12 primers with identifiable sample bar codes and the PCR conditions described above. 454 pyrosequencing of the amplified hsp65 genes was performed using a 454 Life Sciences GS FLX series genome sequencer (Roche, The Netherlands). Returned hsp65 gene sequences were trimmed, aligned, and assigned to operational taxonomic units (OTUs) using the mothur pipeline software tool (31). In short, sequences were trimmed (only sequences with lengths between 400 and 440 bp and with both primer sequences were selected) and aligned using a reference file with hsp65 gene sequences of 34 different Mycobacterium species, 6 different Nocardia species, 1 Streptomyces species, and 1 Bifidobacterium species (outgroup). Sequences were assigned to the genus Mycobacterium when they showed more than 83.1% similarity to a hsp65 gene sequence from a cultivated Mycobacterium species (21). Subsequently, hsp65 gene sequences from NTM were assigned to operational taxonomic units (OTUs) using a 97% cutoff value (32). The NTM diversity was estimated by calculating the Shannon and Simpson diversity indices (33, 34). Similarity between samples was determined by comparing OTU presence and abundance using the Morisita index for similarity, and cluster analysis based on the Morisita similarity index was done with the unweighted-pair group method using average linkages and the PAST software tool (35, 36). OTU identification was done by comparing the hsp65 gene sequence of each NTM OTU with the hsp65 gene sequences in the GenBank database.
Nucleotide sequence accession numbers.
The GenBank accession numbers of the hsp65 gene sequences obtained in our study are KC832034 to KC832289.
RESULTS
hsp65 gene copies numbers in unchlorinated drinking water.
The hsp65 gene was amplified from all drinking water samples. The geometric gene copy numbers did not differ considerably between the different distribution systems in the winter and summer (1.7 × 104 to 6.3 × 104 gene copies liter−1; Fig. 1). Only the hsp65 gene copy numbers in the drinking water samples from the distribution system of plant SW1 in the summer were significantly higher than the hsp65 gene copy numbers observed in the distribution system of the other plants (analysis of variance [ANOVA] with Bonferroni post hoc test; P < 0.01).
Fig 1.
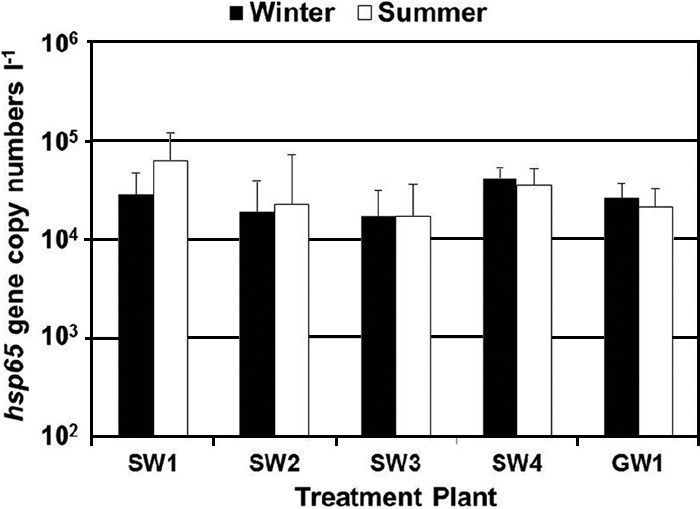
Geometric means (with geometric standard deviations) of the hsp65 gene copy numbers per liter (liter−1) in unchlorinated drinking water samples from the distribution systems of five different treatment plants in the Netherlands.
hsp65 gene sequences.
The 454 pyrosequencing of the hsp65 gene amplified from drinking water resulted in approximately 10,000 hsp65 gene sequences from the 16 samples that were analyzed. Comparing these gene sequences with gene sequences from the GenBank database demonstrated that the similarity between 6,678 (69%) hsp65 gene sequences obtained in our study and the hsp65 gene sequences of defined Mycobacterium species was below the cutoff value (83.1%) for the genus Mycobacterium (21). Thus, bacteria carrying these hsp65 gene sequences belonged to genera other than Mycobacterium. On average, 32% of the hsp65 gene sequences obtained in our study belonged to NTM, but percentages differed between drinking water samples (Fig. 2).
Fig 2.
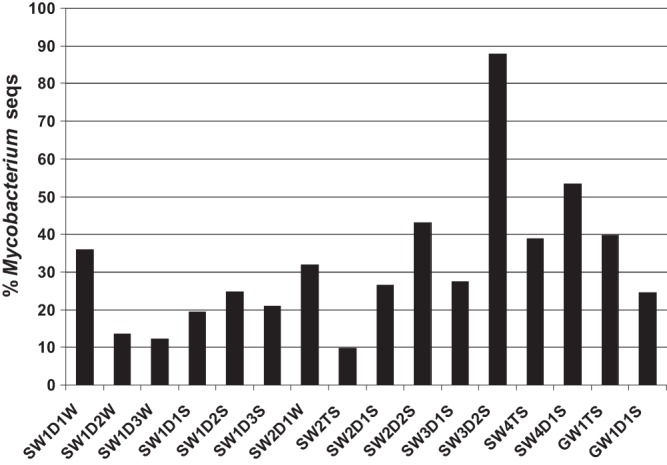
Percentages of hsp65 gene sequences (seqs) that belong to the genus Mycobacterium. SW1, SW2, SW3, SW4, and GW1, treatment plants; D1, D2, and D3, location in the distribution system; T, treated water at plant; S, summer; W, winter.
NTM communities in drinking water.
The 2,943 hsp65 gene sequences from NTM were used for further analyses. The numbers of different NTM OTUs in the drinking water samples ranged between 4 and 28 but were in general higher than 10 (Table 2). Concomitantly, the diversity indices of the NTM community in most drinking water samples were relatively high (Table 2). The numbers of NTM OTUs and NTM diversity indices in drinking water from plants SW1 and SW2 were in general lower in winter (9.2 ± 4.4°C) than in summer (20.3 ± 1.5°C) (Table 2). In addition, the NTM OTU numbers and NTM diversity indices were higher in distributed water than in treated water of plants SW2, SW4, and GW1 (Table 2). A pronounced difference in NTM OTU numbers or NTM diversity indices between water samples from the different plants was not observed.
Table 2.
Numbers of sequences analyzed and operational taxonomic units and Shannon and Simpson diversity indices for the hsp65 gene sequences of NTM in the different drinking water samplesa
| Treatment plant | Location | Season | No. of sequences | No. of OTUs | Diversity index |
|
|---|---|---|---|---|---|---|
| Shannon | Simpson | |||||
| SW1 | DS 1 | W | 227 | 28 | 2.2 | 0.81 |
| SW1 | DS 2 | W | 114 | 16 | 1.6 | 0.64 |
| SW1 | DS 3 | W | 78 | 8 | 1.2 | 0.54 |
| SW1 | DS 1 | S | 96 | 26 | 2.4 | 0.85 |
| SW1 | DS 2 | S | 112 | 21 | 2.2 | 0.82 |
| SW1 | DS 3 | S | 91 | 11 | 1.5 | 0.70 |
| SW2 | DS 1 | W | 114 | 7 | 1.1 | 0.58 |
| SW2 | T | S | 86 | 4 | 0.9 | 0.51 |
| SW2 | DS 1 | S | 167 | 14 | 1.8 | 0.78 |
| SW2 | DS 2 | S | 121 | 18 | 2.0 | 0.78 |
| SW3 | DS 1 | S | 139 | 21 | 2.3 | 0.84 |
| SW3 | DS 2 | S | 665 | 20 | 1.2 | 0.51 |
| SW4 | T | S | 171 | 7 | 1.1 | 0.56 |
| SW4 | DS 1 | S | 161 | 23 | 1.9 | 0.72 |
| GW1 | T | S | 329 | 8 | 0.2 | 0.072 |
| GW1 | DS 1 | S | 272 | 25 | 1.6 | 0.62 |
OTUs, operational taxonomic units; SW, surface water treatment plant; GW, groundwater treatment plant; DS, distribution system; T, treated water; W, winter; S, summer.
Most NTM OTUs were observed in only one drinking water sample (142 of 175 OTUs) or in the distribution system and/or drinking water of one plant (153 of 175 OTUs) (Table 3). Still, one NTM OTU was observed in 14 of the 16 analyzed drinking water samples which came from the distribution system and/or treated water from all five treatment plants (Table 3). In general, this OTU constituted more than 10% of the hsp65 gene sequences that were observed in the drinking water samples and that belonged to the genus Mycobacterium (see Table S1 in the supplemental material). The other NTM OTUs were present in eight or fewer drinking water samples, although four NTM OTUs were observed in the distribution system and/or treated water of four different plants.
Table 3.
Occurrence of operational taxonomic units in one or more drinking water samples or in one or more treatment plants
| No. of OTUsa | No. of drinking water samples | No. of OTUs | No. of treatment plants |
|---|---|---|---|
| 142 | 1 | 153 | 1 |
| 16 | 2 | 11 | 2 |
| 9 | 3 | 5 | 3 |
| 4 | 6 | 4 | 4 |
| 1 | 5 | 1 | 5 |
| 1 | 7 | ||
| 1 | 8 | ||
| 1 | 14 |
OTUs, operational taxonomic units.
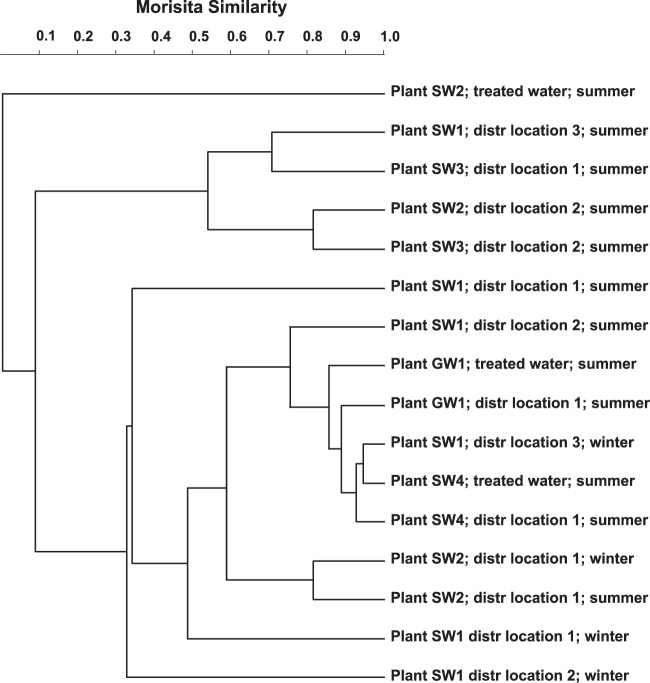
The Morisita similarity index demonstrated that each drinking water sample had a unique NTM community (Fig. 3). Consequently, the similarity of the NTM OTU communities of the sampled locations in each distribution system of plants SW1, SW2, and SW3 was low (Fig. 3), indicating that the NTM communities differed considerably between different locations in the distribution system of these plants. None of the NTM OTUs in the treated water of plant SW2 were observed in the distributed drinking water (see Table S1 in the supplemental material), indicating that the NTM community in treated water differs completely from the NTM community in distributed water. In contrast, the NTM communities in the treated water and in the distribution system of plants SW4 and GW1 were relatively similar (86% to 94% similarity; Fig. 3), although the NTM diversity was greater in the distributed water of these two plants than in the treated water (Table 2). Thus, the dominant NTM OTUs in the treated water of plants SW4 and GW1 were also observed in the distributed water, but the distributed water contained additional NTM OTUs (Table S1). The NTM communities at a specific location in the distribution system of plant SW1 differed considerably between summer and winter (31% to 67% similarity; Fig. 3). In contrast, the NTM communities at location 1 in the distribution system of plant SW2 were not that different between summer and winter (81% similarity). The NTM communities in treated water and distribution system of a treatment plant were in general different from the NTM communities at other plants (Fig. 3). However, similarities between NTM communities at a certain location in the distribution system of plants SW1 and SW3, SW2 and SW3, SW1 and GW1, and SW4 and GW1 were observed.
Fig 3.
UPGMA (unweighted-pair group method using average linkages) cluster analysis based on the Morisita index of similarity between the operational taxonomic units of the hsp65 gene sequences of nontuberculous mycobacteria in the different drinking water samples.
NTM identification in drinking water.
All NTM hsp65 gene sequences observed in water samples from the distribution system and/or treated water of plants SW1, SW4, and GW1 had less than 97% similarity to hsp65 gene sequences of defined NTM species (see Table S1 in the supplemental material). This indicates that the dominant NTM species in the distribution system of these plants are not-yet-described NTM species. hsp65 gene sequences with more than 97% similarity to those of defined NTM species were observed in a few drinking water samples of the other plants. A total 4 of 86 hsp65 gene sequences from the treated water of plant SW2 were related to M. salmoniphilum (98.9% sequence similarity). hsp65 gene sequences in drinking water (2 of 114 sequences in winter and 16 of 167 sequences in summer) at location 1 in the distribution system of plant SW2 were related to those of M. gordonae (98.5% to 99.8% sequence similarity). hsp65 gene sequences (6 of 121 sequences) related to M. gordonae (99.8% sequence similarity) were also observed in the drinking water sample from the other location in the distribution system of plant SW2. In addition, 8 hsp65 gene sequences were related to M. llatzerense (97.5% sequence similarity) and 14 sequences to M. avium (97.5% sequence similarity) in this sample. hsp65 gene sequences (61 of 665 sequences) were also related to M. gordonae (99.8% sequence similarity) in drinking water sampled from one location in the distribution system of plant SW3. Thirteen of the 129 hsp65 gene sequences were related to M. genavense (97.3% sequence similarity) in drinking water from the other location in the distribution system of plant SW3.
DISCUSSION
Lack of hsp65 PCR primer specificity.
The objective of our study was to investigate the NTM community in unchlorinated drinking water in the Netherlands by sequencing the hsp65 gene after PCR amplification using primers TB11 and TB12 (30). The results showed that the majority of the hsp65 gene sequences obtained from drinking water samples were not related to bacteria from the genus Mycobacterium. Thus, primer pair TB11 and TB12 cannot be used for the quantitative detection of Mycobacterium in drinking water. It is, therefore, better to use other developed qPCR assays for the quantitative detection of NTM in the drinking water environment (18, 37). One of these qPCR assays has also been used for the quantitative detection of NTM in unchlorinated drinking water in the Netherlands (38).
NTM communities in drinking water.
We obtained 2,943 hsp65 gene sequences of NTM in our study, which is a quantity adequate to describe and compare the NTM communities from the different drinking water samples. Overall, the results demonstrate that each drinking water sample had a unique NTM composition, as was also observed with cultivation methods for drinking water samples in the United States (39). Most NTM OTUs were present in only one drinking water sample, indicating that local conditions are important drivers for the establishment of NTM species in the drinking water distribution systems. Still, one NTM OTU was observed in the distributed and/or treated water of all five plants. Consequently, this NTM species seems well adapted to the unchlorinated drinking water environment in the Netherlands. Unfortunately, this NTM species is a not-yet-described NTM species and the hsp65 gene sequence of this species has not been deposited in the GenBank database before.
The number of NTM OTUs and the Shannon and Simpson diversity indices were noticeably higher in drinking water samples from the distribution system than in treated water. These results demonstrate that NTM species that are not present or are present at low numbers in the treated water are able to establish themselves and multiply in the drinking water distribution system. However, the dominant NTM OTUs in treated water of plant SW4 and GW1 were also observed in the distributed drinking water. Only the NTM OTUs in treated water of plant SW2 were not observed in the distributed drinking water of that plant. Plant SW2 is the only one of these three plants that uses slow sand filtration as the last step in the treatment train. Perhaps conditions in the slow sand filters select for NTM species that are not well adapted to growth in the distribution system.
Angenent et al. (19) observed a substantial NTM diversity in the air and water from a hospital therapy pool, although OTU numbers or diversity indices were not given. Still, the NTM diversity observed in that study is difficult to compare with the NTM diversity observed in our study, because they analyzed 16S rRNA gene sequences instead of hsp65 gene sequences (21). A much lower number of NTM species (normally up to 8 in a drinking water sample) was observed in drinking water samples when cultivation-based methods were used (9–11, 18, 39–45). These cultivation-based methods normally analyze about 100 to 1,000 ml of drinking water. The amount of drinking water volume analyzed in our PCR corresponds to 50 ml of drinking water, since only a small (5%) portion of the isolated DNA was used in the PCR. Consequently, the NTM diversity and number of OTUs might have been even higher if 100 to 1,000 ml of drinking water had been used for analysis. Our results indicate that numerous NTM species in drinking water have not yet been cultivated, an interpretation confirmed by our observation that most hsp65 gene sequences of NTM were related to not-yet-described species. Consequently, cultivation-based methods provide a limited view of NTM diversity in drinking water. The observation that most NTM in unchlorinated drinking water have not yet been cultivated, and may not be cultivable, is consistent with observations made for other microorganisms in unchlorinated drinking water such as Legionella (29), protozoa (46), and fungi (37).
The source used for drinking water production (groundwater versus surface water) had no or a minor influence on the establishment of a specific NTM community, since more than 90% similarity between the NTM communities in drinking water from plant GW1 and plant SW1 or SW4 was observed. This similarity value was higher than the similarity between some NTM communities in drinking water produced at different surface water plants (e.g., SW3 and SW4). In the United States and France, a clear difference between the NTM communities in drinking water from groundwater and surface water was observed when cultivation-based methods were used (41, 43). In addition, NTM could not be cultivated from drinking water in 64% to 69% of the investigated plants in the United States and in 28% of the investigated plants in France. The discrepancy between their and our results reconfirms the need for DNA-based methods to identify factors that affect NTM communities in drinking water.
A distinct seasonal influence on the NTM community was observed for the samples of plant SW1, but a less noticeable seasonal effect was observed for samples from plant SW2. However, this apparent disagreement can also be explained by the different sampling methods used for the winter and summer samples of plant SW1. Winter samples were obtained after flushing the tap until the water temperature was stable for 30 s (assuming that the drinking water sample came from the distribution system), whereas summer samples were obtained directly from the tap (assuming that the drinking water sample came from the premise plumbing system). Consequently, the different NTM populations in winter and summer samples in the distributed drinking water of plant SW1 might indicate that NTM communities differ between the premise plumbing and distribution systems. A seasonal effect on the NTM numbers in distributed drinking water has been observed before (38, 39), as has an effect of the premise plumbing system (25, 38). Therefore, additional experiments are required to determine whether the NTM community in distributed drinking water is affected by the season and/or the premise plumbing system.
The drinking water from plant SW2 has a lower AOC concentration and a lower BFR value than the drinking water from the other four plants (Table 1). As a result, the active biomass concentration (i.e., ATP concentration) in the drinking water from plant SW2 is low compared to that in the drinking water from the other plants (Table 1). The NTM community compositions of drinking water from SW2 did not cluster separately from the NTM community compositions of drinking water from the other plants. Moreover, the drinking water NTM community at one location in the distribution system of plant SW2 was relatively similar to the drinking water NTM community at a location in the distribution system of plant SW3 (82% similarity). These results show that a low BFR, AOC, and ATP concentration in drinking water has limited impact on the NTM community composition in the drinking water distribution system. A previous study had shown that the AOC and/or BFR of the drinking water did not affect the 16S rRNA gene copy numbers of NTM in unchlorinated drinking water in the Netherlands (38). It can be concluded from our studies that both the NTM numbers and NTM community composition cannot be controlled by further reduction of the microbial activity, AOC concentration, and/or BFR in unchlorinated drinking water in the Netherlands. Obviously, certain NTM species are well adapted to the intense oligotrophic conditions in the drinking water environment.
NTM identification in drinking water.
Although 95.8% of the hsp65 gene sequences were related to not-yet-described NTM species, 4.2% of the hsp65 gene sequences demonstrated more than 97% similarity to hsp65 gene sequences from defined NTM species. Most of these hsp65 gene sequences were related to M. gordonae, which has also been regularly detected in cultivation-based analysis of NTM communities in drinking water in the Netherlands (27, 47) and other countries (9, 11, 18, 39–41, 43–45). In a more recent study, the rapidly growing mycobacteria M. llatzerense, M. chelonae, M. vaccae, M. salmoniphilum, M. peregrinum, and M. septicum were cultivated from drinking water sampled at two locations in the Netherlands (10). Only two of these species (M. llatzerense and M. salmoniphilum) were observed in our study as well. Drinking water samples from plants other than those used in our study were analyzed in that recent study (10). This might explain the differences between their study and ours in the NTM species observed, since our results indicate that each drinking water sample has a unique NTM population. M. genavense had been observed in hospital drinking water in the Netherlands in 1999 using molecular methods (48). That hospital is located in the same geographic area as the single location where hsp65 gene sequences related to M. genavense were found in our study and might indicate a specific geographic distribution or adaptation to a specific water type of this NTM species. Other publications that describe the detection of M. genavense in drinking water could not be found, probably because M. genavense is difficult to cultivate (48).
The observation that 14 hsp65 gene sequences showed more than 97% similarity to those of the hsp65 gene of M. avium was unexpected, because M. avium could not be detected in the same water type with a previously described qPCR protocol (37). The hsp65 gene sequences related to M. avium were observed at only one location in the distribution system of SW2 in the summer, where it constituted 5.0% of the total hsp65 gene sequences. Given the relatively high hsp65 gene copy number (9.2 × 104 liter−1) and the number of hsp65 gene sequences related to M. avium in this drinking water sample, M. avium numbers should have been 10 times above the detection limit of the specific qPCR assay. Therefore, the bacterium carrying this hsp65 gene may represent a separate NTM species closely related to M. avium or the selective PCR method used for M. avium does not detect all M. avium strains.
Our study showed that the majority of NTM species in drinking water are related to not-yet-described species. It is unlikely that these NTM species are of public health significance, because these unidentified NTM species have not been linked to disease. Still, elucidation of the virulence properties of these species is necessary to definitely exclude their role in public health. A small number of hsp65 gene sequences related to M. gordonae, M. avium, M. salmoniphilum, M. genavense, and M. llatzerense were observed in drinking water in our study. Some of these species have been reported to be involved in disease of immunocompromised persons (1, 48). Reporting disease caused by NTM to the health authorities in the Netherlands is not obligatory; therefore, detailed information about NTM species involved in disease in the Netherlands is not available. A recent examination of several clinical NTM cases in the Netherlands indicated that, of the five species observed in drinking water, only M. gordonae and M. avium have been confirmed in clinical cases (49, 50). However, the clinical relevance of M. gordonae in the Netherlands is very low, whereas the clinical relevance of M. avium is moderate (49, 50). Consequently, the results from our study do not indicate that NTM in unchlorinated drinking water from distribution systems in the Netherlands are of important public health significance.
The NTM with the highest clinical relevance in the Netherlands seem to be M. xenopi, M. kansasii, and M. malmoense (49, 50), species that were not observed in our NTM community analysis. Since we analyzed NTM communities in 50-ml samples of drinking water, it remains possible that these three NTM species were present at numbers that were below the detection limit of our sequencing approach. In addition, M. kansasii has been observed in drinking water from the premise plumbing system in the Netherlands but not in drinking water from the treatment plant or distribution system (27). This indicates that premise plumbing systems rather than distribution systems enhance growth of M. kansasii. Therefore, specific qPCR methods are currently being developed for the quantitative detection of M. kansasii, M. xenopi, and M. malmoense in drinking water. Such qPCR assays can subsequently be used to determine whether these three NTM species are present in unchlorinated drinking water in the distribution and/or premise plumbing systems in the Netherlands.
Supplementary Material
ACKNOWLEDGMENT
This work was financed by the joint research program (BTO) of the Dutch drinking water companies.
Footnotes
Published ahead of print 2 August 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01591-13.
REFERENCES
- 1.Falkinham JO., III 1996. Epidemiology of infection by nontuberculous mycobacteria. Clin. Microbiol. Rev. 9:177–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dailloux M, Albert M, Laurain C, Andolfatto S, Lozniewski A, Hartemann P, Mathieu L. 2003. Mycobacterium xenopi and drinking water biofilms. Appl. Environ. Microbiol. 69:6946–6948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goy G, Thomas V, Rimann K, Jaton K, Prod'hom G, Greub G. 2007. The Neff strain of Acanthamoeba castellanii, a tool for testing the virulence of Mycobacterium kansasii. Res. Microbiol. 158:393–397 [DOI] [PubMed] [Google Scholar]
- 4.Kazda J. 1973. The importance of water for the spread of potentially pathogenic Mycobacteria. I. Possibilities for the multiplication of Mycobacteria. Zentralbl. Bakteriol. Orig. B 158:161–169 (In German.) [PubMed] [Google Scholar]
- 5.Norton CD, LeChevallier MW, Falkinham JO., III 2004. Survival of Mycobacterium avium in a model distribution system. Water Res. 38:1457–1466 [DOI] [PubMed] [Google Scholar]
- 6.Schulze-Röbbecke R, Fischeder R. 1989. Mycobacteria in biofilms. Zentralbl. Hyg. Umweltmed. 188:385–390 [PubMed] [Google Scholar]
- 7.Strahl ED, Gillaspy GE, Falkinham JO., III 2001. Fluorescent acid-fast microscopy for measuring phagocytosis of Mycobacterium avium, Mycobacterium intracellulare, and Mycobacterium scrofulaceum by Tetrahymena pyriformis and their intracellular growth. Appl. Environ. Microbiol. 67:4432–4439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neumann M, Schulze-Robbecke R, Hagenau C, Behringer K. 1997. Comparison of methods for isolation of mycobacteria from water. Appl. Environ. Microbiol. 63:547–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peters M, Muller C, Rusch-Gerdes S, Seidel C, Gobel U, Pohle HD, Ruf B. 1995. Isolation of atypical mycobacteria from tap water in hospitals and homes: is this a possible source of disseminated MAC infection in AIDS patients? J. Infect. 31:39–44 [DOI] [PubMed] [Google Scholar]
- 10.van Ingen J, Blaak H, de Beer J, de Roda Husman AM, van Soolingen D. 2010. Rapidly growing nontuberculous mycobacteria cultured from home tap and shower water. Appl. Environ. Microbiol. 76:6017–6019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.September SM, Brozel VS, Venter SN. 2004. Diversity of nontuberculoid Mycobacterium species in biofilms of urban and semiurban drinking water distribution systems. Appl. Environ. Microbiol. 70:7571–7573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burns DN, Wallace RJ, Jr, Schultz ME, Zhang YS, Zubairi SQ, Pang YJ, Gibert CL, Brown BA, Noel ES, Gordin FM. 1991. Nosocomial outbreak of respiratory tract colonization with Mycobacterium fortuitum: demonstration of the usefulness of pulsed-field gel electrophoresis in an epidemiologic investigation. Am. Rev. Respir. Dis. 144:1153–1159 [DOI] [PubMed] [Google Scholar]
- 13.Conger NG, O'Connell RJ, Laurel VL, Olivier KN, Graviss EA, Williams-Bouyer N, Zhang Y, Brown-Elliott BA, Wallace RJ., Jr 2004. Mycobacterium simae outbreak associated with a hospital water supply. Infect. Control Hosp. Epidemiol. 25:1050–1055 [DOI] [PubMed] [Google Scholar]
- 14.Hilborn ED, Yakrus MA, Covert TC, Harris SI, Donnelly SF, Schmitt MT, Toney S, Bailey SA, Stelma GN., Jr 2008. Molecular comparison of Mycobacterium avium isolates from clinical and environmental sources. Appl. Environ. Microbiol. 74:4966–4968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kline S, Cameron S, Streifel A, Yakrus MA, Kairis F, Peacock K, Besser J, Cooksey RC. 2004. An outbreak of bacteremias associated with Mycobacterium mucogenicum in a hospital water supply. Infect. Control Hosp. Epidemiol. 25:1042–1049 [DOI] [PubMed] [Google Scholar]
- 16.Marshall HM, Carter R, Torbey MJ, Minion S, Tolson C, Sidjabat HE, Huygens F, Hargreaves M, Thomson RM. 2011. Mycobacterium lentiflavum in drinking water supplies, Australia. Emerg. Infect. Dis. 17:395–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.von Reyn CF, Maslow JN, Barber TW, Falkinham JO, III, Arbeit RD. 1994. Persistent colonisation of potable water as a source of Mycobacterium avium infection in AIDS. Lancet 343:1137–1141 [DOI] [PubMed] [Google Scholar]
- 18.Hussein Z, Landt O, Wirths B, Wellinghausen N. 2009. Detection of non-tuberculous mycobacteria in hospital water by culture and molecular methods. Int. J. Med. Microbiol. 299:281–290 [DOI] [PubMed] [Google Scholar]
- 19.Angenent LT, Kelley ST, St Amand A, Pace NR, Hernandez MT. 2005. Molecular identification of potential pathogens in water and air of a hospital therapy pool. Proc. Natl. Acad. Sci. U. S. A. 102:4860–4865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perkins SD, Mayfield J, Fraser V, Angenent LT. 2009. Potentially pathogenic bacteria in shower water and air of a stem cell transplant unit. Appl. Environ. Microbiol. 75:5363–5572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim H, Kim SH, Shim TS, Kim MN, Bai GH, Park YG, Lee SH, Chae GT, Cha CY, Kook YH, Kim BJ. 2005. Differentiation of Mycobacterium species by analysis of the heat-shock protein 65 gene (hsp65). Int. J. Syst. Evol. Microbiol. 55:1649–1656 [DOI] [PubMed] [Google Scholar]
- 22.van der Kooij D, Veenendaal HR. 2013. Regrowth problems and biological stability assessment in the Netherlands. In van der Kooij D, van der Wielen PWJJ. (ed), Microbial growth in drinking water supplies: problems, causes, controls and research needs, in press IWA Publishing, London, United Kingdom [Google Scholar]
- 23.Torvinen E, Lehtola MJ, Martikainen PJ, Miettinen IT. 2007. Survival of Mycobacterium avium in drinking water biofilms as affected by water flow velocity, availability of phosphorus, and temperature. Appl. Environ. Microbiol. 73:6201–6207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moore MR, Pryor M, Fields B, Lucas C, Phelan M, Besser RE. 2006. Introduction of monochloramine into a municipal water system: impact on colonization of buildings by Legionella spp. Appl. Environ. Microbiol. 72:378–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang H, Edwards M, Falkinham JO, III, Pruden A. 2012. Molecular survey of the occurrence of Legionella spp., Mycobacterium spp., Pseudomonas aeruginosa, and amoeba hosts in two chloraminated drinking water distribution systems. Appl. Environ. Microbiol. 78:6285–6294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang H, Masters S, Hong Y, Stallings J, Falkinham JO, III, Edwards MA, Pruden A. 2012. Effect of disinfectant, water age, and pipe material on occurrence and persistence of Legionella, mycobacteria, Pseudomonas aeruginosa, and two amoebas. Environ. Sci. Technol. 46:11566–11574 [DOI] [PubMed] [Google Scholar]
- 27.Engel HW, Berwald LG, Havelaar AH. 1980. The occurrence of Mycobacterium kansasii in tapwater. Tubercle 61:21–26 [DOI] [PubMed] [Google Scholar]
- 28.van der Wielen PWJJ, Medema G. 2010. Unsuitability of quantitative Bacteroidales 16S rRNA gene assays for discerning fecal contamination of drinking water. Appl. Environ. Microbiol. 76:4876–4881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wullings BA, Bakker G, van der Kooij D. 2011. Concentration and diversity of uncultured Legionella spp. in two unchlorinated drinking water supplies with different concentrations of natural organic matter. Appl. Environ. Microbiol. 77:634–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Telenti A, Marchesi F, Balz M, Bally F, Bottger EC, Bodmer T. 1993. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J. Clin. Microbiol. 31:175–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McNabb A, Eisler D, Adie K, Amos M, Rodrigues M, Stephens G, Black WA, Isaac-Renton J. 2004. Assessment of partial sequencing of the 65-kilodalton heat shock protein gene (hsp65) for routine identification of Mycobacterium species isolated from clinical sources. J. Clin. Microbiol. 42:3000–3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hill MO. 1973. Diversity and evenness: a unifying notation and its consequences. Ecology 54:427–432 [Google Scholar]
- 34.Haegeman B, Hamelin J, Moriarty J, Neal P, Dushoff J, Weitz JS. 2013. Robust estimation of microbial diversity in theory and in practice. ISME J. 7:1092–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van der Wielen PWJJ, Bolhuis H, Borin S, Daffonchio D, Corselli C, Giuliano L, D'Auria G, de Lange GJ, Huebner A, Varnavas SP, Thomson J, Tamburini C, Marty D, McGenity TJ, Timmis KN. 2005. The enigma of prokaryotic life in deep hypersaline anoxic basins. Science 307:121–123 [DOI] [PubMed] [Google Scholar]
- 36.Wolda H. 1981. Similarity indices, sample size and diversity. Oecologia (Berl) 50:296–302 [DOI] [PubMed] [Google Scholar]
- 37.van der Wielen PWJJ, Italiaander R, Wullings BA, Heijnen L, van der Kooij D. 2013. Opportunistic pathogens in drinking water in the Netherlands. In van der Kooij D, van der Wielen PWJJ. (ed), Microbial growth in drinking water supplies: problems, causes, controls and research needs, in press IWA Publishing, London, United Kingdom [Google Scholar]
- 38.van der Wielen PWJJ, van der Kooij D. 2013. Nontuberculous mycobacteria, fungi, and opportunistic pathogens in unchlorinated drinking water in the Netherlands. Appl. Environ. Microbiol. 79:825–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Falkinham JO, III, Norton CD, LeChevallier MW. 2001. Factors influencing numbers of Mycobacterium avium, Mycobacterium intracellulare, and other mycobacteria in drinking water distribution systems. Appl. Environ. Microbiol. 67:1225–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Castillo-Rodal AI, Mazari-Hiriart M, Lloret-Sánchez LT, Sachman-Ruiz B, Vinuesa P, López-Vidal Y. 2012. Potentially pathogenic nontuberculous mycobacteria found in aquatic systems. Analysis from a reclaimed water and water distribution system in Mexico City. Eur. J. Clin. Microbiol. Infect. Dis. 31:683–694 [DOI] [PubMed] [Google Scholar]
- 41.Covert TC, Rodgers MR, Reyes AL, Stelma GN., Jr 1999. Occurrence of nontuberculous mycobacteria in environmental samples. Appl. Environ. Microbiol. 65:2492–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hilborn ED, Covert TC, Yakrus MA, Harris SI, Donnelly SF, Rice EW, Toney S, Bailey SA, Stelma GN., Jr 2006. Persistence of nontuberculous mycobacteria in a drinking water system after addition of filtration treatment. Appl. Environ. Microbiol. 72:5864–5869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Le Dantec C, Duguet JP, Montiel A, Dumoutier N, Dubrou S, Vincent V. 2002. Occurrence of mycobacteria in water treatment lines and in water distribution systems. Appl. Environ. Microbiol. 68:5318–5325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schulze-Röbbecke R, Janning B, Fischeder R. 1992. Occurrence of mycobacteria in biofilm samples. Tuber. Lung Dis. 73:141–144 [DOI] [PubMed] [Google Scholar]
- 45.Torvinen E, Suomalainen S, Lehtola MJ, Miettinen IT, Zacheus O, Paulin L, Katila ML, Martikainen PJ. 2004. Mycobacteria in water and loose deposits of drinking water distribution systems in Finland. Appl. Environ. Microbiol. 70:1973–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Valster RM, Wullings BA, Bakker G, Smidt H, van der Kooij D. 2009. Free-living protozoa in two unchlorinated drinking water supplies, identified by phylogenic analysis of 18S rRNA gene sequences. Appl. Environ. Microbiol. 75:4736–4746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Groothuis DG, Berwald LG. 1982. Voorkomen van M. kansasii in leidingwater te Amsterdam en te 's-Gravenhage. Rapport nr 128102001. RIVM, Bilthoven, the Netherlands [Google Scholar]
- 48.Hillebrand-Haverkort ME, Kolk AH, Kox LF, Ten Velden JJ, Ten Veen JH. 1999. Generalized Mycobacterium genavense infection in HIV-infected patients: detection of the mycobacterium in hospital tap water. Scand. J. Infect. Dis. 31:63–68 [DOI] [PubMed] [Google Scholar]
- 49.van Ingen J. 2009. Nontuberculous mycobacteria. From gene sequences to clinical relevance. Ph.D. thesis. Radboud University Nijmegen, Nijmegen, the Netherlands [Google Scholar]
- 50.van Ingen J, Bendien SA, de Lange WC, Hoefsloot W, Dekhuijzen PN, Boeree MJ, van Soolingen D. 2009. Clinical relevance of non-tuberculous mycobacteria isolated in the Nijmegen-Arnhem region, The Netherlands. Thorax 64:502–506 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.