Abstract
Lactococcus lactis subsp. lactis strain A12 was isolated from sourdough. Combined genomic, transcriptomic, and phenotypic analyses were performed to understand its survival capacity in the complex sourdough ecosystem and its role in the microbial community. The genome sequence comparison of strain A12 with strain IL1403 (a derivative of an industrial dairy strain) revealed 78 strain-specific regions representing 23% of the total genome size. Most of the strain-specific genes were involved in carbohydrate metabolism and are potentially required for its persistence in sourdough. Phenotype microarray, growth tests, and analysis of glycoside hydrolase content showed that strain A12 fermented plant-derived carbohydrates, such as arabinose and α-galactosides. Strain A12 exhibited specific growth rates on raffinose that were as high as they were on glucose and was able to release sucrose and galactose outside the cell, providing soluble carbohydrates for sourdough microflora. Transcriptomic analysis identified genes specifically induced during growth on raffinose and arabinose and reveals an alternative pathway for raffinose assimilation to that used by other lactococci.
INTRODUCTION
Lactococcus lactis is a mesophilic Gram-positive species, related to Streptococcacae (1), which is subdivided into two subspecies: lactis and cremoris. Lactococci have been extensively studied for their role in dairy products manufacturing, but the subspecies lactis is able to colonize various environments other than milk, such as plant and animal material (2–5).
L. lactis subsp. lactis is occasionally found among the sourdough microflora used in traditional Italian (6), Portuguese (7), French (8), and Mexican (9) bread-making. Sourdough is a mixture of flour and water that is fermented by a heterogeneous population of lactic acid bacteria (LAB) and yeast and is used for the production of bread and sweet leavened baked goods. The LAB microflora is 100 times more abundant than yeast (10, 11) and is very diverse. LAB detected in sourdough are often obligate heterofermentative lactobacilli (Lactobacillus brevis, Lactobacillus reuteri, or Lactobacillus sanfranciscensis). They can also include less prevalent facultative heterofermentative and obligate homofermentative genera such as Lactobacillus, Pediococcus, Streptococcus, Enterococcus, and Lactococcus spp. or obligate heterofermentative genera such as Leuconostoc and Weissella spp. (10). Analysis of bacterial community dynamics revealed that L. lactis is found at the start of back-slopped fermentation and can predominate in some cases (12). Its functional role remains unknown. Sourdough biodiversity and species stability are influenced by various determinants including the cereal type used (flour composition), fermentation process (temperature, pH, redox potential, dough hydratation, additional ingredients, or back-slopping methods), and metabolic activities resulting from the cooperation and competitiveness of microbiota (13–17). Studies on the most frequent LAB-yeast associations (Lactobacillus sanfranciscensis and Kazachstania exigua [syn. Saccharomyces exigua] or Saccharomyces cerevisiae) or LAB-LAB associations (Lactobacillus sanfranciscensis and Lactobacillus plantarum) reveal trophic and energetic relationships (16, 18, 19); this complex ecosystem has a very diverse set of metabolic activities. Sourdough is expected to enhance properties of end product by developing its sensory qualities (aroma, taste, and texture), increasing the shelf-life, enhancing preservation and food safety by inhibiting pathogens or fungi, improving its nutritive value, and removing undesired compounds (10).
This study aims to understand the genetic features involved for adaptation and persistence of L. lactis in sourdough (an atypical niche for this species), by characterizing the genetic and metabolic properties of a strain isolated from traditional French sourdough (8). Strain A12 belongs to the group of environmental strains; these are more genetically diverse than domesticated/dairy strains (5). We investigate here the specific features of strain A12 that are relevant in a sourdough ecosystem by comparing its genome content and phenotype to the laboratory strain IL1403 (20). Strain IL1403 is a derivative of a dairy strain (21) which has been extensively studied at the physiological level (22, 23). The genes specifically activated by strain A12 on raffinose and arabinose, two sugars relevant for sourdough fermentation, were investigated by transcriptional analysis.
MATERIALS AND METHODS
Bacterial strains.
L. lactis subsp. lactis strain LBAE A12, isolated from a traditional French wheat sourdough (8), belongs to the LBAE-UPS (Auch, France) culture collection and has been previously described in the Genoferment collection (5).
A12 genome sequencing, annotation, and comparison.
A draft genome was obtained by a whole-genome DNA shotgun strategy with 454/Roche GS FLX pyrosequencing (Eurofins MWG Operon, France). We obtained 147 contigs (228,605 reads, 2,727,544 bp, mean coverage sequencing depth of 18 X) using the GS De Novo Assembler software from Newbler package (454 Life Sciences); the 42 contigs of >1 kb were deposited in public databases. Automatic annotation of DNA sequences was performed using the Agmial platform (INRA Jouy-en-Josas, France) (24), and specific genes were manually checked. Genes are described according to 1 of 15 functional categories (defined by Bolotin et al. [20]), designated as follows: amino acid biosynthesis (AMI), biosynthesis of cofactors, prosthetic groups, and carriers (COF), cell envelope (ENV), cellular processes (CEL), central intermediary metabolism (INT), energy metabolism (NRJ), fatty acid and phospholipid metabolism (FAT), purines, pyrimidines, nucleosides, and nucleotides (PUR), regulatory functions (REG), replication (REP), transcription (TRS), translation (TRD), transport and binding proteins (TSP), other categories (OTH), and unknown (UNK). Determination of strain-specific DNA regions was performed using Panseq v2.0 software (25) with default options. All A12-specific regions were individually compared to the IL1403 chromosome by local homology searches (FastScan method) using clone Manager 9.2 (Sci-Ed Software, Cary, NC). Glycoside hydrolases (GH) of sequenced Lactococci genomes listed in the CAZy database (http://www.cazy.org/) (26) allowed us to detect putative active carbohydrate enzymes (CAZymes) in strain A12 using NCBI-BLASTP. We used the InterPro collection of protein signature databases to classify new GH, which were found in strain A12 by automatic annotation. To detect possible sequence errors due to pyrosequencing, the nucleotide sequence of the LL2138 central region was verified by Sanger sequencing.
Global phenotypic analysis.
The Phenotype Microarray (Biolog, Hayward, CA) was used for global phenotypic analysis of strains IL1403 and A12 (27). Two replicates of each plate, PM1 and PM2A (carbon substrates), and one replicate for PM9 (osmotic sensitivity), PM10 (pH), and PM13 (oxidizing agents) were inoculated from isolated colonies on BUG B medium. We added 100 μl of cell suspension at 81% of transmittance to each well according to the manufacturer's instructions. Microplates were incubated at 33°C in Omnilog plate reader, which recorded the Dye G opacity, for 72 h. The data were analyzed using OmniLog Phenotype MicroArray software (release 1.2). The substrate was considered as being specifically metabolized by the strain A12 when the corresponding height value of the color-coded kinetic graphs exceeded that of the strain IL1403, with a threshold of 80.
Effect of oxygen on growth.
The specific growth rates (μmax) under aerobic and anaerobic conditions were measured (four replicates). For anaerobic conditions, strains were grown at 30°C in M17 medium (Merck, Germany) supplemented with glucose (10 g/liter; GM17) in tubes under an N2 atmosphere. For aerobic conditions, cultures were grown with the same medium, under an air gas-phase in 200-ml Erlenmeyer flasks at 30°C and 200 rpm. The optical density at 580 nm (OD580) was recorded approximately every 30 min to estimate the maximal growth rate, which was calculated from the slope of the exponential growth phase.
Growth tests on different carbon sources.
Carbon source utilization by strains A12 and IL1403 was tested by inoculating strains in 10 ml of the chemically defined medium (CDM) (28) supplemented with one carbon source (at 2 g/liter), followed by incubation for 24 h. At least two biological replicates were performed for each condition. We tested 28 carbon sources belonging to the following groups: monosaccharides such as aldohexoses (glucose and galactose), cetohexose (fructose), aldopentoses (l-arabinose and d-xylose) and aldohexoses (rhamnose), disaccharides (saccharose, d-melibiose, gentiobiose, d-cellobiose, d-maltose, tréhalose, palatinose, and lactose), trisaccharides (d-raffinose, maltotriose, and melizitose), tetrasaccharides (stachyose), polysaccharides (dextran, pectin, xylan, hydroxyl-cellulose, inuline, and amidon), acid sugars (d-glucuronate, d-galacturonate, and d-gluconate), and aryl-monosaccharides (amygdalin). We measured growth by recording the OD580 with a Spectronic spectrophotometer (Milter Roy).
Hydrolase activity.
Enzymatic activities of A12 and IL1403 strains were determined using insoluble chromogenic polysaccharides: AZCL-β-glucan, AZCL-xylan, AZCL-amylose, AZCL-arabinoxylan, AZCL-galactan, AZCL-rhamnogalacturonan, or cellulose-AZUR (Megazyme, Bray, Ireland). Each strain was incubated in M17 medium supplemented with glucose (0.5%) and an insoluble AZCL-polysaccharide (0.1%). After incubation at 30°C for 48 h, polysaccharide digestion by endohydrolases was detected by complete pellet solubilization. To test arabinofuranosidase activity, bacterial strains were plated on GM17 agar containing the substrate 5-bromo-4-chloro-3-indolyl-α-l-arabinofuranoside (X-ara; 100 μM; Carbosynth, United Kingdom). Enzymatic activity was indicated by blue colonies.
Fermentation culture and metabolic analyses.
Strain A12 was grown under a nitrogen atmosphere in a 500-ml fermentor (Sartorius, Germany) for 24 h, at 30°C and agitated at 150 rpm. The automatic addition of KOH (10 N) maintained a pH of 6.6. CDM (28) was supplemented with either 10 g of glucose, 10 g of arabinose, 10 g of fructose, 10 g of galactose, or 9 g of raffinose/liter (equivalent to 330 mmol C/liter for each). Two independent replicates were carried out for each growth condition.
Spectrophotometric measurements at 580 nm (Hitachi U1100; 1 U of absorbance is equivalent to 0.3 g of biomass liter−1 for L. lactis) were taken to estimate bacterial growth. Raffinose, glucose, galactose, fructose, arabinose, lactate, acetate, formate, and ethanol concentrations were measured by high-pressure liquid chromatography (Hewlett-Packard 1050; Waters 717 autosampler; Bio-Rad Microguard; H+ column, Bio-Rad HPX87H; Perkin-Elmer LC90Bio UV detector; Hewlett-Packard 1047A refractometer). High-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD) was used to study the breakdown of raffinose. A 4- by 250-mm Dionex Carbo-pack PA100 was used for separation. A gradient of sodium acetate (in 150 mM NaOH) was applied at a flow rate of 1 ml/min: from 0 to 300 mM in 30 min, from 300 to 450 mM in 1 s, from 450 to 0 mM in 5 min, and then at 0 mM for 10 min.
The maintenance energy coefficient (m) corresponds to the specific rate of sugar consumption when the growth rate is zero. This was calculated during the deceleration phase from the specific substrate consumption rate q (mmol C/gX/h, where gX is grams of biomass) as a function of the growth rate (μ), a linear relationship, as described previously (29).
Transcriptomic analysis.
A nylon array, containing 1,948 PCR fragments (Eurogentec) that correspond to 84% of the IL1403 annotated open reading frame (ORF) (accession number NC_002662), was supplemented with 433 PCR amplicons (ClinEurodiag, Belgium) corresponding to A12-specific genes. Partial prophage, transposases, and coding DNA sequences (CDS) smaller than 200 bp were omitted from the macroarray. Membrane spotting and analytical support were provided by the GeT platform (Genopole, Toulouse, France).
The equivalent of 3 mg of dried cells was harvested during the early exponential growth phase (OD580 = 1) of the fermentation process for transcriptional analysis. Cell lysis, RNA extraction, and hybridization with radiolabeled cDNA were carried out as previously described (23). Membranes were exposed to a phosphorimager screen for 5 days and scanned with a phosphofluorimager (Storm 860; Molecular Dynamics).
Hybridization signals were quantified from four replicates (two biological and two technical replicates), assigned to gene names, and statistically treated with Bioplot software (GeT platform). The local background was removed, and the signals were normalized to the mean membrane intensity. The expression ratios for each sugar pairing were calculated; they were estimated as statistically different from 1 when the false discovery rate was under 10% (as previously described [22]) (see Table S1 in the supplemental material).
Nucleotide sequence accession numbers.
The GenBank accession numbers assigned to the L. lactis strain A12 nucleotide sequence are CBLU010000001 to CBLU010000042.
RESULTS
The dispensable genome of strain A12 reveals a particular adaptation for sugar metabolism.
A draft sequence of the L. lactis subsp. lactis A12 genome was established by shotgun pyrosequencing (Roche/454). Of the 147 contigs generated, the 42 that were larger than 1 kbp (227,901 reads, 2,700,005 bp) were retained for further analysis. A12 strain-specific DNA regions, thought to contain genes required for adaptation to sourdough ecosystem, were determined by comparison to the chromosome of strain IL1403. These specific regions represent nearly 630 kb (23% of total A12 genetic content) and are distributed in 78 DNA fragments ranging from 513 bp to 57 kbp. Comparison with the chromosome and plasmids of strain KF147, a nondairy strain isolated from mung bean sprouts (30), revealed nearly 560 kbp of A12-specific regions distributed in 100 fragments ranging from 507 bp to 57 kbp in size. This suggests high diversity of genomic content among L. lactis subsp. lactis, even between strains from plant origin. The functional categories of the specific genes were manually classified according to the method of Bolotin et al. (20): 40% of A12-specific CDS encoded unknown functions and 27% encoded protein related to prophages. The remaining CDS corresponded mostly to adaptive functions such as transporters (36 CDS: 22 involved in sugars transport), sugar energetic metabolism (35 CDS), cell envelope constitution (33 CDS), and regulation (17 CDS). A large proportion of genes identified are involved in sugar metabolism and sugar transport; analysis of specific glycoside hydrolases (GH) was therefore performed. GH, listed in the CAZy (Carbohydrate-Active Enzyme) database (26), catalyze the hydrolysis of the glycoside bond between two or more carbohydrates or between a carbohydrate and a noncarbohydrate moiety. All 32 GH coded in the IL1403 genome were also found in the A12 genome. However, the A12 genome encoded eight additional GH: LL1981 (family GH 1), SucP (GH 13), XylS (GH 31), Aga (GH 36), LL2113 (GH 65), LL1819 (GH 65), LL498 (GH 113), and LL2114 (GH 101). In contrast to other GH, the genetic sequence for LL2114 was not found in the genomes of the two nondairy strains KF147 (30) and IO-1 (31; data not shown). The GH 101 family contains enzymes with an endo-α-N-acetylgalactosaminidase activity and therefore represents a new activity in L. lactis subsp. lactis.
The large set of strain-specific genes found in L. lactis subsp. lactis suggests substantial potential for adaptation in a complex ecosystem. The genome of strain A12 contained genetic sequences relating to more extensive variation in carbohydrate metabolism than that of strain IL1403. The functionality of these genes was examined by phenotypic analysis.
Strain A12 is able to metabolize plant-derived carbohydrates.
The global phenotype of the strains A12 and IL1403 was investigated using Phenotype Microarray (Biolog). A phenotypic profile comparison of these two strains enabled us to determine specific capacities of strain A12 possibly related to its sourdough survival. The Biolog method measures the reduced coenzyme NADH production capacity during sugar fermentation, but this is not necessarily correlated with biomass production (27). Carbon source utilization was the most apparent phenotypic difference between the two strains (see Table S1 in the supplemental material); we detected only minor differences in responses to osmotic sensitivity, pH variation, and oxidizing agent sensitivity. Growth tests were carried out in liquid CDM supplemented with various sugars. We tested 28 carbon sources displaying diverse levels of complexity (monosaccharides, disaccharides, trisaccharides, tetrasaccharides, and polysaccharides) (Table 1). The effect of oxygen on IL1403 and A12 growth was investigated to optimize culture conditions. Strain IL1403 exhibited similar maximum specific growth rates (μmax) under anaerobic and aerobic conditions; strain A12 had a 40% higher μmax under anaerobic conditions (data not shown). All physiological studies were therefore performed in anaerobic conditions, i.e., conditions that also prevail during sourdough fermentation.
Table 1.
Growth rates (μmax) of strains IL1403 and A12 on different carbon sources under anaerobic conditions
| Substrate | Family | Mean growth rate (μmax [h−1]) ± SDa |
|
|---|---|---|---|
| IL1403 | A12 | ||
| Glucose | Aldohexose | 0.63 ± 0.07 | 0.34 ± 0.04 |
| Galactose | Aldohexose | – | 0.13 ± 0.03 |
| Fructose | Cetohexose | 0.35 ± 0.04 | 0.12 ± 0.04 |
| l-Arabinose | Aldohexose | – | 0.18 ± 0.09 |
| d-Xylose | Aldohexose | – | – |
| Rhamnose | Aldohexose | – | – |
| Sucrose | Disaccharide | – | 0.06 ± 0.04 |
| d-Melibiose | Disaccharide | – | 0.36 ± 0.06 |
| Gentiobiose | Disaccharide | 0.03 ± 0.01 | 0.30 ± 0.03 |
| d-Cellobiose | Disaccharide | 0.54 ± 0.03 | 0.40 ± 0.07 |
| d-Maltose | Disaccharide | 0.35 ± 0.05 | 0.09 ± 0.06 |
| Trehalose | Disaccharide | 0.66 ± 0.02 | 0.17 ± 0.00 |
| Palatinose | Disaccharide | – | – |
| Lactose | Disaccharide | – | – |
| d-Raffinose | Trisaccharide | – | 0.37 ± 0.06 |
| Maltotriose | Trisaccharide | 0.36 ± 0.01 | 0.26 ± 0.06 |
| Melizitose | Trisaccharide | – | – |
| Stachyose | Tetrasaccharide | – | 0.30 ± 0.08 |
| Dextran | Polysaccharide | – | – |
| Pectin | Polyoside | – | – |
| Xylan | Polyoside | – | – |
| Hydroxycellulose | Polysaccharide | – | – |
| Inulin | Polysaccharide | – | – |
| Starch | Polysaccharide | – | – |
| d-Glucuronate | Sugar acid | – | – |
| d-Galacturonate | Sugar acid | – | – |
| d-Gluconate | Sugar acid | 0.06 ± 0.03 | 0.05 ± 0.03 |
| Amygdalin | Aryl-disaccharide | – | – |
Two to four replicates were performed for each growth test. –, no growth.
Neither strain A12 nor IL1403 were able to grow on d-xylose, rhamnose, palatinose, lactose, melizitose, and complex polysaccharide carbon sources (Table 1). A lack of growth on complex sugars by strain A12 correlated with a lack of enzymes responsible for the cleavage of the commercial chromogenic polysaccharides: β-glucans, xylans, amylose, arabinoxylan, and cellulose (Table 2). Strain A12 exhibited a growth rate lower than that of strain IL1403 for other sugars (glucose, fructose, d-cellobiose, d-maltose, trehalose, and maltotriose). The cellobiose metabolism was efficient since the strain A12 displayed the highest growth rate on this plant derivative substrate (Table 1). Only strain A12 was able to grow on galactose, l-arabinose, sucrose, d-melibiose, gentiobiose, d-raffinose, and stachyose (strain IL1403 could grow at a very low μmax on gentiobiose). These carbon sources are plant-derived carbohydrates: gentiobiose is the glucose moiety of the amygdalin found in some vegetables (32), l-arabinose and d-galactose are hemicellulose constituents of plant cell walls (33), and d-raffinose, stachyose, and the related disaccharide melibiose are important soluble α-galactosides in plant tissues (34, 35). Strain A12 cleaved chromogenic galactan and rhamnogalacturonan, showing the presence of an active galactanase that can hydrolyze α-galactoside bonds (Table 2).
Table 2.
Enzymatic activities of strains IL1403 and A12
| Substrate | Detected enzyme | Enzymatic activitya |
|
|---|---|---|---|
| IL1403 | A12 | ||
| AZCL-β-glucan | β-Glucanase | – | – |
| AZCL-xylan | Endo-xylanase | – | – |
| AZCL-amylose | α-Amylase | – | – |
| AZCL-arabinoxylan | Endo-xylanase | – | – |
| Cellulose-AZUR | Cellulase | – | – |
| AZCL-galactan | Endo-1,4-β-galactanase | – | + |
| AZCL-rhamnogalacturonan | Rhamnogalacturonan hydrolase | – | + |
| X-ara | Arabinofuranosidase | – | – |
+, hydrolysis of a chromogenic polysaccharide by a related enzyme; –, no activity.
Selective advantage of strain A12 for growth on arabinose and raffinose.
We assessed the biomass and fermentation end products of strain A12, when grown on various plant sugars, to investigate the potential selective advantage of this strain during plant sugar fermentation. Cultures were performed in a bioreactor where pH, agitation, and anaerobic atmosphere were controlled. Five sugars were selected: a plant pentose (arabinose), a complex sugar belonging to the α-galactoside family (raffinose), and three related monosaccharides (galactose, glucose, and fructose). The physiological characteristics of strain A12 differed according to the carbon source used (Table 3). The metabolism of glucose and fructose was homolactic; lactate represented the major fermentation end product (>90%). Deviation from homolactic to mixed acid fermentation was observed on galactose, raffinose, and, in particular, on arabinose (where 33.3% ± 0.5% of carbon flux was routed toward formate, acetate, and ethanol production). This was twice as great as when raffinose (16.08% ± 0.03%) or galactose (18.53% ± 2.24%) were used as the carbon source. Such a metabolic shift has been previously associated with low growth rates, either when sugar is metabolized slowly (e.g., galactose) or in sugar-limited chemostat cultures, and would be mainly controlled by the NADH/NAD+ ratio (36, 37). Our results were consistent with this notion for galactose and arabinose carbon sources, where strain A12 exhibited lower μmax values (0.18 ± 0.00 h−1 and 0.28 ± 0.01 h−1, respectively, versus 0.53 ± 0.01 h−1 for glucose) (Table 3). However, strain A12 had a high μmax on raffinose (0.46 ± 0.01 h−1), as well as favoring a mixed acid pathway, suggesting an original metabolic pathway control in this strain. The molar ratios between lactate and acetate, a relevant parameter for aromatic quality of bread, were 1.4 on arabinose, 5 on galactose, and 6.5 on raffinose. These values were similar to the optimal values found in rye, spelt, and wheat sourdough (ratios between 1.5 and 4) (38, 39).
Table 3.
Growth characteristics of strain A12 on different sugars
| Substrate | Structure | Mean μmax (h−1) ± SD | Maximum rate sugar of consumption (mmol C/gX/h) | Mean concn (mM) ± SD of fermentation end products at 24 h |
Mean % carbon routed through the mixed acid pathway at 24 h (%) ± SDa | Estimated maintenance energy coefficientb (mmol C/gX/h) | |||
|---|---|---|---|---|---|---|---|---|---|
| Lactate | Formate | Acetate | Ethanol | ||||||
| Glucose | C6H12O6 | 0.53 ± 0.01 | 128.4 ± 32.1 | 79.9 ± 1.9 | 7.8 ± 1.3 | 4.3 ± 0.9 | 8.3 ± 0.7 | 7.3 ± 0.1 | 24.9 |
| Fructose | C6H12O6 | 0.17 ± 0.00 | 44.6 ± 0.2 | 86.6 ± 0.8 | 6.3 ± 0.6 | 3.9 ± 0.1 | 7.1 ± 0.2 | 6.0 ± 0.0 | 37.0 |
| Galactose | C6H12O6 | 0.18 ± 0.00 | 34.7 ± 2.7 | 59.2 ± 5.7 | 15.4 ± 0.8 | 12.4 ± 0.6 | 14.5 ± 2.1 | 18.5 ± 2.2 | 32.0 |
| Arabinose | C5H10O5 | 0.28 ± 0.01 | 73.6 ± 0.3 | 52.0 ± 0.6 | 10.6 ± 0.9 | 36.7 ± 0.6 | 15.3 ± 1.2 | 33.31 ± 0.5 | 19.4 |
| Raffinose | C18H32O16 | 0.46 ± 0.01 | 163.6 ± 27.9 | 52.2 ± 2.7 | 10.4 ± 4.0 | 7.6 ± 2.0 | 12.4 ± 3.0 | 16.1 ± 0.0 | 5.7 |
Calculated according to the following formula: 3([acetate] + [ethanol])/{6[lactate] + 3([acetate] + [ethanol])}.
Determined as described in Materials and Methods.
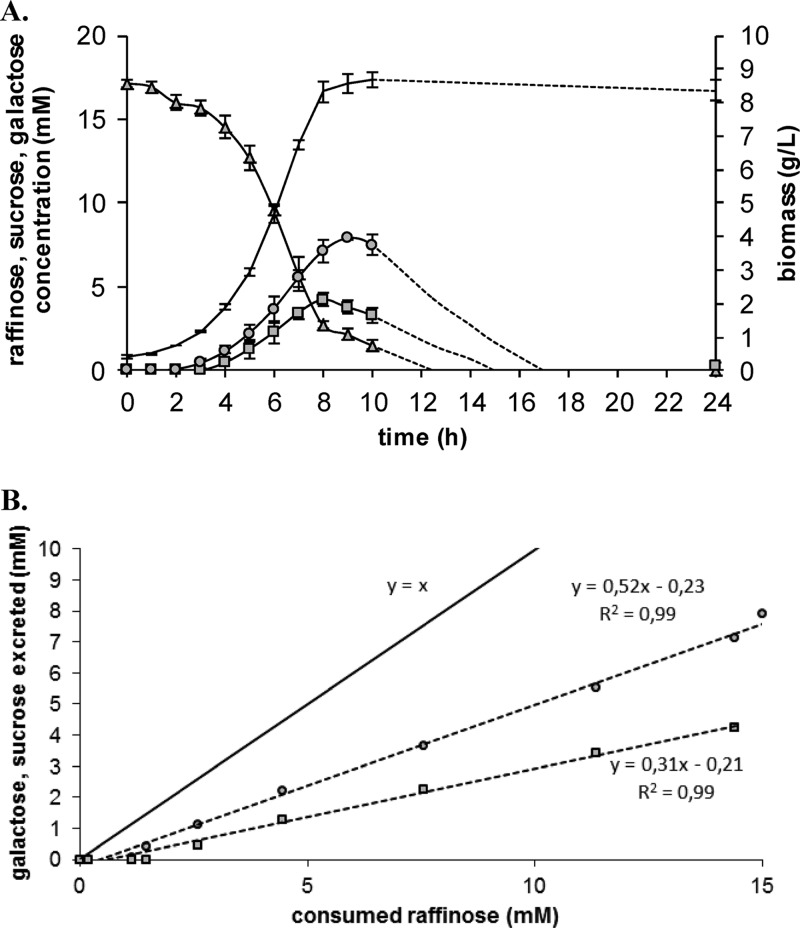
Physiological maintenance, corresponding to the minimum substrate necessary to maintain cell survival, was estimated for strain A12 on each substrate (these were estimated using the instantaneous growth rates and measures of sugar consumption during the decelerating phase [see Materials and Methods]). On glucose, fructose, and galactose, maintenance coefficients were ∼30 mmol C/gX/h, corresponding to the value calculated for IL1403 cultivated on glucose (22). On plant-derived carbohydrates, the maintenance coefficient significantly decreased to 19 mmol C/gX/h (R2 = 0.92) on arabinose and 6 mmol C/gX/h (R2 = 0.97) on raffinose. This suggests that strain A12 is metabolically efficient on arabinose and raffinose: it needs little energy to ensure its maintenance and may use the majority of carbon provided to produce biomass. The cleavage of raffinose (α-d-galactosyl-α-d-glucosyl-β-d-fructose) into individual units can occur in two ways: α-1,6 linkage hydrolysis (generating sucrose and galactose) or β-1,2 linkage hydrolysis (generating melibiose and fructose). We used high-pressure liquid chromatography, with culture supernatants, to monitor the concentrations of raffinose, fructose, galactose, glucose, melibiose, and sucrose in culture supernatants. Raffinose was consumed from the beginning of fermentation and had a maximal specific rate of consumption of 163.6 ± 27.9 mmol C/gX/h (as high as the glucose assimilation rate; Table 3). Sucrose and galactose were accumulated transiently during growth, but there was no trace of fructose, glucose, or melibiose in the supernatant (Fig. 1A). Galactose excretion has been observed during lactose catabolism of Lactococcus lactis subsp. cremoris (40) and other LAB (41, 42). Here, galactose and sucrose were excreted after 2 and 3 h of fermentation, respectively. Their concentrations reached a maximum of 7 mM (galactose) and 4 mM (sucrose) after 8 to 9 h of growth and then fell until reaching total depletion at 24 h. The amount of released sucrose and galactose was proportional to that of metabolized raffinose, but lower than the theoretical maximum, which would correspond to the absence of bacterial consumption (Fig. 1B). The slopes of linear correlations showed that, in a medium containing raffinose, ca. 50% of galactose and 70% of sucrose were directly used by cell. This suggests that sucrose was more efficiently used through raffinose metabolism (μmax = 0.46 h−1) than alone (μmax = 0.06 h−1, Table 2). Intracellular sucrose is well metabolized, but not when it is provided as the sole carbon source; sucrose transport was probably the limiting step for sucrose metabolism by strain A12. Indeed, the described gene for sucrose transport (scrA) is lacking in the A12 genome.
Fig 1.
Raffinose metabolism by L. lactis subsp. lactis strain A12. (A) Kinetics of carbohydrates quantified in supernatant culture; (B) sucrose and galactose excreted, as a function of the raffinose metabolized. The line y = x represents the theoretical maximum of excreted sugar without any consumption of sugar. Data for raffinose (△), galactose (○), and sucrose (□) are shown. No melibiose, fructose, or glucose was detected.
Few genes displayed a specific expression variation related to arabinose and raffinose metabolism in strain A12.
The global transcriptional response of strain A12 was investigated during the early exponential growth phase, in the conditions described above, to identify genes involved in arabinose and raffinose metabolism and to analyze their expression levels.
In order to highlight the first steps of the sugar pathway or an original pathway in strain A12, the transcriptomic responses on arabinose and raffinose were compared to those on the three simple sugars, galactose, glucose and fructose (Fig. 2). This allowed us to exclude genes whose expression was modified by fermentation type (homolactic or mixed) or by growth rate variation (see Table S2 in the supplemental data). Analysis of the transcriptomic responses of strain IL1403, in continuous cultures at different growth rates, revealed that nearly 700 genes displayed variation in their expression (22). Our comparison method detected only a small number of differentially expressed, sugar-specific genes: three genes were identified during growth on raffinose (LL2138, aga, and sucP), and nine were detected during growth on arabinose (araR, araT, araB, araA, araD, ptk, xylX, xylT, and arcB). The method also detected genes specific to glucose, fructose, and galactose metabolism (1, 15, and 24 genes, respectively [see Table S3 in the supplemental material]).
Fig 2.

Venn diagrams representing the number of strain A12 genes with variable expression in different sugar substrates: combined comparison of raffinose (raff) (A) or arabinose (ara) (B) with the three simple sugars (glucose [glu], fructose [fru], and galactose [gal]), highlighting raffinose- and arabinose-specific genes. The variation of gene expression was considered significant when the false discovery rate was <10%.
Raffinose metabolism was investigated using pairwise comparisons of the transcriptomic response on this substrate and that of other simple carbon substrates (Fig. 2). In addition to the raffinose-specific activation of LL2138, sucP, and aga genes, strain A12 induced a substantial number of other genes. The genes galK, galM, galT, lacZ, yugA, and yugB, specific to galactose metabolism, were upregulated on raffinose compared to glucose or fructose. The gal genes encode enzymes of the Leloir pathway, which facilitate the conversion of galactose into glucose-1-phosphate further routed toward glycolytic pathway (43). No genes involved in the tagatose-6-phosphate pathway, the alternative pathway for galactose conversion, were induced; the plasmid-borne lactose operon (involved in the first steps of the tagatose-6-phosphate pathway) is not found in strain A12 (5). Expression of fruC was greater on raffinose than on glucose or galactose. This gene, originally identified as the tagatose-6-phosphate kinase encoding lacC, has been re-annotated as the 1-phosphofructokinase fruC and is involved in fructose metabolism (44). Increased expression of genes specifically related to galactose and fructose pathways is indicative of the metabolism of both raffinose subunits: this result is supported by the physiological data.
An original gene cluster encoding raffinose metabolism in strain A12.
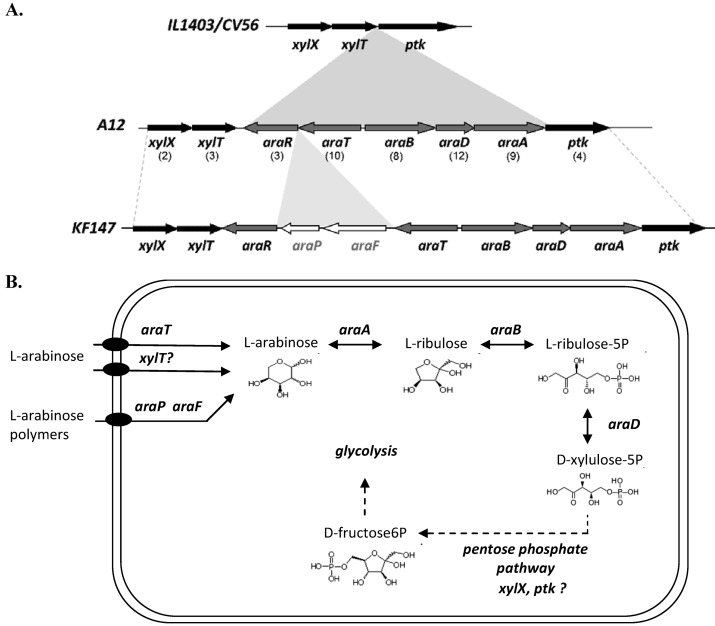
We investigated the genetic organization of induced genes when strain A12 was cultivated on arabinose and raffinose. The genes araR, araT, araB, araA, and araD, induced when strain A12 was cultivated on arabinose, corresponded to a genomic island previously detected by comparative genome hybridization in another L. lactis subsp. lactis strain isolated from plant material (45). The ara gene cluster had a different organization compared to that found in strain KF147 but was determined to be functional by transcriptomic analysis. It is likely that ptk, xylX, and xylT (found in the core genome of L. lactis and on either side of the arabinose cluster when it was present), were involved in arabinose metabolism (Fig. 3).
Fig 3.
Arabinose metabolism by L. lactis subsp. lactis. (A) Localization and genetic organization of the arabinose operon in sequenced genomes; (B) diagram of the related metabolic pathway. The numbers in brackets correspond to the average ratio values obtained by transcriptomic analysis for arabinose versus that of simple sugars. xylX, acetyltransférase; xylT, d-xylose proton-symporter; araA, l-arabinose isomerase; araD, l-ribulose-5-phosphate 4-epimerase; araB, l-ribulokinase; araT, arabinose-proton symporter; araF, α-N-arabinofuranosidase; araP, disaccharide permease; araR, GntR family arabinose operon repressor; ptk, phosphoketolase.
When strain A12 was cultivated on raffinose, LL2138, aga, and sucP were specifically induced. These genes are clustered upstream of ORF LL2157 that encodes a putative transcriptional regulator of the AraC family (Fig. 4). The aga and LL2138 genes were expressed on raffinose at rates 10 times greater than on glucose or fructose and five times greater than on galactose (see Table S3 in the supplemental material). sucP was expressed approximately four times more on raffinose than on the other sugars. The activation of aga and sucP is correlated with the cleavage of raffinose in our physiological study. The aga gene encodes an α-galactosidase (EC 3.2.1.22) of 725 amino acids that belongs to the family GH 36 and is responsible for the hydrolysis of the α-1,6-galactosyl bond (characteristic of the α-galactoside family). No signal peptide was observed in the aga gene sequence of strain A12, suggesting that Aga is probably an intracellular protein. The sucP gene encodes a 489-amino-acid GH with an α-amylase domain that belongs to the family GH13. This sucrose phosphorylase (EC 2.4.1.7) is known to cleave sucrose into d-fructose and α-d-glucose-1-phosphate. The SucP protein is relatively well conserved in LAB and has a protein similarity to Leuconostoc mesenteroides, Streptococcus mutans, and other L. lactis strains that is >80%.
Fig 4.
Comparison of the organization of genes involved in raffinose metabolism in Lactococcus raffinolactis ATCC 43920, Lactococcus lactis subsp. lactis KF147, and Lactococcus lactis subsp. lactis A12. Arrows correspond to the different genes: sugar catabolism genes are indicated by hatched arrows, sugar transporter genes are represented in gray, regulatory genes are represented in black, and strain-specific or unknown genes are represented in white. Numbers indicate the percent identities of nucleotide sequences of common CDS identified.
Lactococcus genes relating to raffinose metabolism have only been previously described for the strains KF147 and Lactococcus raffinolactis ATCC 43920 (data not shown). A comparison of the raffinose gene clusters found in the three strains (A12, KF147, and Lactococcus raffinolactis ATCC 43920) suggests that the ability of strain A12 to use raffinose is associated with genes of xenologous origin (Fig. 4). The location of the raffinose gene cluster differs for strain A12: it is located on a plasmid in strain A12 but on the chromosome in strains KF147 (30, 45, 46) and ATCC 43920 (47), close to the galKT genes cluster involved in the galactose metabolism. This was confirmed by hybridization of the PFGE fingerprint using LL2138 as a probe (data not shown). There are also differences in the nucleotide sequences of aga genes: those in KF147 and ATCC 43920 strains are 93% identical, but the A12 aga gene sequence shares only 52% identity with those in the other strains (Fig. 4). The characteristic motifs of family GH36 are present in all strains, but some amino acids were found to be specific to strain A12 (see Fig. S1 in the supplemental material). The sucP genes of strains A12 and KF147 share only 65% identity, a value substantially less than that observed among L. lactis subsp. lactis strains (98% [data not shown]) or between cremoris and lactis subspecies (88% [data not shown]). Strain A12 also differed in that it contains neither the putative raffinose ABC transporter (encoded by agaA, agaB, and agaC in KF147 strain) nor the putative PTS system (encoded by orf2, orf3, and orf4 in the ATCC 43920 strain). It was found to contain a particular transporter encoded by the LL2138 gene. This gene encodes a 856-amino-acid protein, corresponding to the translational fusion of permease and kinase domains. The first 450 N-terminal amino acids display 61% identity (78% similarity) with a glycoside-pentoside-hexuronide (GPH):cation symporter of Leuconostoc mesenteroides, and the 406 C-terminal amino acids display 54% identity (70% similarity) to a galactokinase of Lactococcus garviae. Such fusion has not been described before and could indicate a previously undescribed raffinose transporter.
DISCUSSION
Sourdough constitutes an ecological niche that is highly suitable for lactic acid bacteria (LAB), particularly Lactobacillus. In some cases, other LAB such as Pediococcus, Leuconostoc, or Lactococcus are found in traditional sourdough (18), but their functional role remains elusive. Here, L. lactis subsp. lactis strain A12 (isolated from French traditional sourdough) was genetically, physiologically, and metabolically characterized. We investigated its role and adaptation in sourdough, the unusual ecosystem in which it is found.
Genome comparison with the sequenced L. lactis subsp. lactis dairy strain IL1403 revealed that 23% of the A12 genome corresponds to strain-specific genes. Their annotation revealed that more than half of these genes coded proteins for unknown function or phages proteins and that others encoded proteins mainly involved in sugar metabolic pathways. Survival in sourdough is therefore likely to be achieved by its adaptations for metabolizing carbohydrates. The specific ability of strain A12 to metabolize plant sugars suggests that it is an autochthonous strain, brought into sourdough fermentation on the raw materials.
Our work suggests that trophic relationships in the sourdough ecosystem are managed by microbial interactions; microbial relationships related to maltose have been extensively described in sourdough (16, 18, 19). Here, we show new possible trophic interaction types. Strain A12 showed a slow fermentation rate of the sugars fructose, sucrose, and maltose, which are, along with glucose, the main soluble carbohydrate sources of sourdough, but a much higher fermentation rate of raffinose and arabinose. These two carbon sources are, at best, weakly metabolized by most lactobacilli and yeasts (6), revealing a competitive advantage for strain A12 in utilizing these sourdough carbon sources. Strain A12 was able to grow on some plant sugars (such as raffinose, melibiose, and gentiobiose) with growth rates similar to those observed on glucose. Its metabolism on raffinose and arabinose was particularly efficient, requiring between two and four times less energy for its own physiological maintenance than on fructose, glucose, and galactose. During raffinose fermentation by strain A12, by-products of sucrose and galactose were released into the medium and were thus available for other microorganisms to use. As well as trophic cooperation with other organisms, properties of strain A12 can influence the final product. Strain A12 produces non-negligible amounts of diacetyl with a very low carbon requirement (48) which may enhance the flavor of bread. Like other L. lactis strains isolated from sourdough, strain A12 also produces bacteriocins that differ from nisin and lactococcin (49; data not shown). A putative bacteriocin gene, similar to LLKF2208 in strain KF147, was detected in the A12 genome (89% sequence identity [data not shown]). Due to its capacity to ferment raffinose and other α-galactosides such as stachyose and melibiose, strain A12 may enhance the digestibility of plant-based foods, such as legumes, which are particularly rich in α-galactosides. These nondigestible oligosaccharides contribute to flatulence and other gastrointestinal disorders when they are fermented by intestinal gas-producing microorganisms in mammals lacking α-galactosidase (50). Engineered strains of L. lactis that express a heterologous α-galactosidase were tested for their ability to improve the digestibility of soymilk in rodents (51). Therefore, the strain A12 could act as a “digestibility enhancer” and may deliver a useful probiotic effect.
The unusual plasmid location of the raffinose gene cluster in strain A12 could facilitate the transfer of the α-galactosidase gene (or the complete cluster) to other strains. This plasmid would be especially useful if it was conjugative, as is the case for the sole raffinose megaplasmid described in Enterococcus faecium (52). To investigate this further, isolation of the plasmid and characterization of its size and genetic content is required. Combined genetic, physiological, and transcriptomic analysis allowed the A12 raffinose pathway to be partly elucidated; this metabolic pathway has not previously been described in L. lactis or other LAB. We have identified an original transport system, encoded by LL2138, which contains both permease and kinase domains. This represents a new type of fusion for permease transport, although various combinations of two or three domains fusion were previously reported in the phosphotransferase system (PTS). These include fusion between PTS component proteins or between PTS and non-PTS proteins (for example, EIIAGlc and Na+/melibiose symporter protein in Lactobacillus subsp.) (53). Such a fusion could be related to a decrease in energetic requirements for cells; in the case of LL2138, it may contribute to the low maintenance cost observed during growth on raffinose. Transcriptomic analysis revealed the simultaneous metabolism of glucose, fructose, and galactose (by the Leloir pathway); this was confirmed by the transient accumulation of sucrose and galactose during raffinose metabolism. We showed activity of the raffinose pathway and hydrolysis of α-1,6 linkage via the sucrose pathway. Further investigation is required to completely elucidate raffinose catabolism and demonstrate whether strain A12 has β-1,2 hydrolysis activity.
These findings constitute a preliminary study of specific carbohydrate metabolism probably involved in the survival of the strain A12 in sourdough and provide a basis for further exploration of this metabolism in such ecosystems. More generally, an integrated approach of functional genomics can help to explain the adaptation of a strain to a complex ecosystem and be used to assess its ecological impact.
Supplementary Material
Footnotes
Published ahead of print 19 July 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01560-13.
REFERENCES
- 1.Teuber M, Geis A. 2006. The genus Lactococcus, p 205–228 In Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E. (ed), The prokaryotes. Springer, New York, NY [Google Scholar]
- 2.Gutiérrez-Méndez N, Rodríguez-Figueroa JC, González-Córdova AF, Nevárez-Moorillón GV, Rivera-Chavira B, Vallejo-Cordoba B. 2010. Phenotypic and genotypic characteristics of Lactococcus lactis strains isolated from different ecosystems. Can. J. Microbiol. 56:432–439 [DOI] [PubMed] [Google Scholar]
- 3.Klijn N, Weerkamp AH, de Vos WM. 1995. Detection and characterization of lactose-utilizing Lactococcus spp. in natural ecosystems. Appl. Environ. Microbiol. 61:788–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nomura M, Kobayashi M, Narita T, Kimoto-Nira H, Okamoto T. 2006. Phenotypic and molecular characterization of Lactococcus lactis from milk and plants. J. Appl. Microbiol. 101:396–405 [DOI] [PubMed] [Google Scholar]
- 5.Passerini D, Beltramo C, Coddeville M, Quentin Y, Ritzenthaler P, Daveran-Mingot M-L, Le Bourgeois P. 2010. Genes but not genomes reveal bacterial domestication of Lactococcus lactis. PLoS One 5:e15306. 10.1371/journal.pone.0015306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corsetti A, Lavermicocca P, Morea M, Baruzzi F, Tosti N, Gobbetti M. 2001. Phenotypic and molecular identification and clustering of lactic acid bacteria and yeasts from wheat (species Triticum durum and Triticum aestivum) sourdoughs of Southern Italy. Int. J. Food Microbiol. 64:95–104 [DOI] [PubMed] [Google Scholar]
- 7.Rocha JM, Malcata FX. 1999. On the microbiological profile of traditional Portuguese sourdough. J. Food Prot. 62:1416–1429 [DOI] [PubMed] [Google Scholar]
- 8.Robert H, Gabriel V, Fontagné-Faucher C. 2009. Biodiversity of lactic acid bacteria in French wheat sourdough as determined by molecular characterization using species-specific PCR. Int. J. Food Microbiol. 135:53–59 [DOI] [PubMed] [Google Scholar]
- 9.Escalante A, Wacher C, Farrés A. 2001. Lactic acid bacterial diversity in the traditional Mexican fermented dough pozol as determined by 16S rDNA sequence analysis. Int. J. Food Microbiol. 64:21–31 [DOI] [PubMed] [Google Scholar]
- 10.De Vuyst L, Vrancken G, Ravyts F, Rimaux T, Weckx S. 2009. Biodiversity, ecological determinants, and metabolic exploitation of sourdough microbiota. Food Microbiol. 26:666–675 [DOI] [PubMed] [Google Scholar]
- 11.Lattanzi A, Minervini F, Di Cagno R, Diviccaro A, Antonielli L, Cardinali G, Cappelle S, De Angelis M, Gobbetti M. 2013. The lactic acid bacteria and yeast microbiota of eighteen sourdoughs used for the manufacture of traditional Italian sweet leavened baked goods. Int. J. Food Microbiol. 163:71–79 [DOI] [PubMed] [Google Scholar]
- 12.Weckx S, Van der Meulen R, Allemeersch J, Huys G, Vandamme P, Van Hummelen P, De Vuyst L. 2010. Community dynamics of bacteria in sourdough fermentations as revealed by their metatranscriptome. Appl. Environ. Microbiol. 76:5402–5408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brandt MJ, Hammes WP, Gänzle MG. 2004. Effects of process parameters on growth and metabolism of Lactobacillus sanfranciscensis and Candida humilis during rye sourdough fermentation. Eur. Food Res. Technol. 218:333–338 [Google Scholar]
- 14.Gänzle MG, Ehmann M, Hammes WP. 1998. Modeling of growth of Lactobacillus sanfranciscensis and Candida milleri in response to process parameters of sourdough fermentation. Appl. Environ. Microbiol. 64:2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hammes WP, Brandt MJ, Francis KL, Rosenheim J, Seitter MFH, Vogelmann SA. 2005. Microbial ecology of cereal fermentations. Trends Food Sci. Technol. 16:4–11 [Google Scholar]
- 16.Vogelmann SA, Hertel C. 2011. Impact of ecological factors on the stability of microbial associations in sourdough fermentation. Food Microbiol. 28:583–589 [DOI] [PubMed] [Google Scholar]
- 17.Vrancken G, Rimaux T, Weckx S, Leroy F, De Vuyst L. 2011. Influence of temperature and backslopping time on the microbiota of a type I propagated laboratory wheat sourdough fermentation. Appl. Environ. Microbiol. 77:2716–2726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Vuyst L, Neysens P. 2005. The sourdough microflora: biodiversity and metabolic interactions. Trends Food Sci. Technol. 16:43–56 [Google Scholar]
- 19.Gobbetti M, De Angelis M, Corsetti A, Di Cagno R. 2005. Biochemistry and physiology of sourdough lactic acid bacteria. Trends Food Sci. Technol. 16:57–69 [Google Scholar]
- 20.Bolotin A, Wincker P, Mauger S, Jaillon O, Malarme K, Weissenbach J, Ehrlich SD, Sorokin A. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis subsp. lactis IL1403. Genome Res. 11:731–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chopin A, Chopin MC, Moillo-Batt A, Langella P. 1984. Two plasmid-determined restriction and modification systems in Streptococcus lactis. Plasmid 11:260–263 [DOI] [PubMed] [Google Scholar]
- 22.Dressaire C, Redon E, Milhem H, Besse P, Loubiere P, Cocaign-Bousquet M. 2008. Growth rate regulated genes and their wide involvement in the Lactococcus lactis stress responses. BMC Genomics 9:343. 10.1186/1471-2164-9-343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Redon E, Loubière P, Cocaign-Bousquet M. 2005. Role of mRNA stability during genome-wide adaptation of Lactococcus lactis to carbon starvation. J. Biol. Chem. 280:36380–36385 [DOI] [PubMed] [Google Scholar]
- 24.Bryson K. 2006. AGMIAL: implementing an annotation strategy for prokaryote genomes as a distributed system. Nucleic Acids Res. 34:3533–3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laing C, Buchanan C, Taboada EN, Zhang Y, Kropinski A, Villegas A, Thomas JE, Gannon VP. 2010. Pan-genome sequence analysis using Panseq: an online tool for the rapid analysis of core and accessory genomic regions. BMC Bioinformatics 11:461. 10.1186/1471-2105-11-461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. 2009. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37:D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bochner BR. 2009. Global phenotypic characterization of bacteria. FEMS Microbiol. Rev. 33:191–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poolman B, Konings WN. 1988. Relation of growth of Streptococcus lactis and Streptococcus cremoris to amino acid transport. J. Bacteriol. 170:700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tempest DW, Neijssel OM. 1984. The status of YATP and maintenance energy as biologically interpretable phenomena. Annu. Rev. Microbiol. 38:459–486 [DOI] [PubMed] [Google Scholar]
- 30.Siezen RJ, Bayjanov J, Renckens B, Wels M, van Hijum SAFT, Molenaar D, van Hylckama Vlieg JET. 2010. Complete genome sequence of Lactococcus lactis subsp. lactis KF147, a plant-associated lactic acid bacterium. J. Bacteriol. 192:2649–2650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kato H, Shiwa Y, Oshima K, Machii M, Araya-Kojima T, Zendo T, Shimizu-Kadota M, Hattori M, Sonomoto K, Yoshikawa H. 2012. Complete genome sequence of Lactococcus lactis IO-1, a lactic acid bacterium that utilizes xylose and produces high Levels of l-lactic acid. J. Bacteriol. 194:2102–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dumville J, Fry S. 2003. Gentiobiose: a novel oligosaccharin in ripening tomato fruit. Planta 216:484–495 [DOI] [PubMed] [Google Scholar]
- 33.Zykwinska A, Thibault J-F, Ralet M-C. 2007. Organization of pectic arabinan and galactan side chains in association with cellulose microfibrils in primary cell walls and related models envisaged. J. Exp. Bot. 58:1795–1802 [DOI] [PubMed] [Google Scholar]
- 34.Trugo LC, Farah A, Cabral L. 1995. Oligosaccharide distribution in Brazilian soya bean cultivars. Food Chem. 52:385–387 [Google Scholar]
- 35.Gänzle MG, Follador R. 2012. Metabolism of oligosaccharides and starch in lactobacilli: a review. Front. Microbiol. 3:340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garrigues C, Loubiere P, Lindley ND, Cocaign-Bousquet M. 1997. Control of the shift from homolactic acid to mixed-acid fermentation in Lactococcus lactis: predominant role of the NADH/NAD+ ratio. J. Bacteriol. 179:5282–5287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomas TD, Turner KW, Crow VL. 1980. Galactose fermentation by Streptococcus lactis and Streptococcus cremoris: pathways, products, and regulation. J. Bacteriol. 144:672–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coda R, Nionelli L, Rizzello CG, De Angelis M, Tossut P, Gobbetti M. 2010. Spelt and emmer flours: characterization of the lactic acid bacteria microbiota and selection of mixed starters for bread making. J. Appl. Microbiol. 108:925–935 [DOI] [PubMed] [Google Scholar]
- 39.Corsetti A, Settanni L. 2007. Lactobacilli in sourdough fermentation. Food Res. Int. 40:539–558 [Google Scholar]
- 40.Benthin S, Nielsen J, Villadsen J. 1994. Galactose expulsion during lactose metabolism in Lactococcus lactis subsp. cremoris FD1 due to dephosphorylation of intracellular galactose 6-phosphate. Appl. Environ. Microbiol. 60:1254–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hickey MW, Hillier AJ, Jago GR. 1986. Transport and metabolism of lactose, glucose, and galactose in homofermentative lactobacilli. Appl. Environ. Microbiol. 51:825–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hutkins RW, Ponne C. 1991. Lactose uptake driven by galactose efflux in Streptococcus thermophilus: evidence for a galactose-lactose antiporter. Appl. Environ. Microbiol. 57:941–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grossiord BP, Luesink EJ, Vaughan EE, Arnaud A, de Vos WM. 2003. Characterization, expression, and mutation of the Lactococcus lactis galPMKTE genes, involved in galactose utilization via the Leloir pathway. J. Bacteriol. 185:870–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barrière C, Veiga-da-Cunha M, Pons N, Guédon E, van Hijum SAFT, Kok J, Kuipers OP, Ehrlich DS, Renault P. 2005. Fructose utilization in Lactococcus lactis as a model for low-GC gram-positive bacteria: its regulator, signal, and DNA-binding site. J. Bacteriol. 187:3752–3761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siezen RJ, Bayjanov JR, Felis GE, van der Sijde MR, Starrenburg M, Molenaar D, Wels M, van Hijum SAFT, van Hylckama Vlieg JET. 2011. Genome-scale diversity and niche adaptation analysis of Lactococcus lactis by comparative genome hybridization using multi-strain arrays. Microb. Biotechnol. 4:383–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Machielsen R, Siezen RJ, van Hijum SAFT, van Hylckama Vlieg JET. 2011. Molecular description and industrial potential of Tn6098 conjugative transfer conferring alpha-galactoside metabolism in Lactococcus lactis. Appl. Environ. Microbiol. 77:555–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boucher I, Vadeboncoeur C, Moineau S. 2003. Characterization of genes involved in the metabolism of α-galactosides by Lactococcus raffinolactis. Appl. Environ. Microbiol. 69:4049–4056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Passerini D, Laroute V, Coddeville M, Le Bourgeois P, Loubière P, Ritzenthaler P, Cocaign-Bousquet M, Daveran-Mingot M-L. 2013. New insights into Lactococcus lactis diacetyl- and acetoin-producing strains isolated from diverse origins. Int. J. Food Microbiol. 160:329–336 [DOI] [PubMed] [Google Scholar]
- 49.Corsetti A, Settanni L, van Sinderen D. 2004. Characterization of bacteriocin-like inhibitory substances (BLIS) from sourdough lactic acid bacteria and evaluation of their in vitro and in situ activity. J. Appl. Microbiol. 96:521–534 [DOI] [PubMed] [Google Scholar]
- 50.Quigley ME, Hudson GJ, Englyst HN. 1999. Determination of resistant short-chain carbohydrates (non-digestible oligosaccharides) using gas-liquid chromatography. Food Chem. 65:381–390 [Google Scholar]
- 51.LeBlanc JG, Silvestroni A, Connes C, Juillard V, de Giori GS, Piard J-C, Sesma F. 2004. Reduction of non-digestible oligosaccharides in soymilk: application of engineered lactic acid bacteria that produce alpha-galactosidase. Genet. Mol. Res. 3:432–440 [PubMed] [Google Scholar]
- 52.Zhang X, Vrijenhoek JEP, Bonten MJM, Willems RJL, van Schaik W. 2011. A genetic element present on megaplasmids allows Enterococcus faecium to use raffinose as carbon source. Environ. Microbiol. 13:518–528 [DOI] [PubMed] [Google Scholar]
- 53.Barabote RD, Saier MH., Jr 2005. Comparative genomic analyses of the bacterial phosphotransferase system. Microbiol. Mol. Biol. Rev. 69:608–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.