Abstract
A collection of 81 isolates of enteropathogenic Escherichia coli (EPEC) was obtained from samples of bulk tank sheep milk (62 isolates), ovine feces (4 isolates), sheep farm environment (water, 4 isolates; air, 1 isolate), and human stool samples (9 isolates). The strains were considered atypical EPEC organisms, carrying the eae gene without harboring the pEAF plasmid. Multilocus sequence typing (MLST) was carried out with seven housekeeping genes and 19 sequence types (ST) were detected, with none of them having been previously reported for atypical EPEC. The most frequent ST included 41 strains isolated from milk and human stool samples. Genetic typing by pulsed-field gel electrophoresis (PFGE) resulted in 57 patterns which grouped in 24 clusters. Comparison of strains isolated from the different samples showed phylogenetic relationships between milk and human isolates and also between milk and water isolates. The results obtained show a possible risk for humans due to the presence of atypical EPEC in ewes' milk and suggest a transmission route for this emerging pathogen through contaminated water.
INTRODUCTION
Enteropathogenic Escherichia coli (EPEC) is an important group of diarrheagenic E. coli, being responsible for outbreaks of infant diarrhea in developing countries, with fatality rates as high as 30% (1), and increasingly being implicated as a causative agent of diarrhea in developed countries, affecting all age groups (2, 3). This epidemiological difference between developing and developed countries is linked to the characteristics of EPEC strains. Typical EPEC organisms, defined as strains of E. coli that produce an attaching and effacing lesion in intestinal cells, carry the EAF virulence plasmid, and do not produce Shiga-like toxins, are predominant in developing countries, whereas atypical EPEC isolates, which are similar to typical EPEC isolates but do not possess the EAF virulence plasmid, are emerging pathogens in developed countries (2, 4, 5).
The reservoir of typical EPEC strains is thought to be human carriers (3), and those strains are rarely isolated from animals (2). In contrast, atypical EPEC strains are readily isolated from humans and animals; there is no confirmation of direct transmission from animals to humans, but the fact that strains belonging to the same serogroups have been found in animal and human diseases suggests that animals can be an important reservoir of atypical EPEC that can be transmitted to humans (5) and also from humans to animals (6). The isolation from food production animals and farm environments has been reported (7–9), and milk and dairy products appear to be regularly contaminated with EPEC strains (10–14), even though other studies failed to isolate EPEC from milk samples (7, 15). Taking into account that farm animals and milk and dairy products carry atypical EPEC strains, it can be presumed that a possible route of transmission is from animals to food and then to humans.
The aim of this study was to analyze the presence of atypical EPEC strains in sheep milk and in the environment of sheep farms and to compare the phylogenetic relationships of isolates obtained from these samples and isolates obtained from humans to search for possible routes of transmission of EPEC to humans.
MATERIALS AND METHODS
Sample collection.
Bulk tank ewes' milk samples were obtained from 388 farms located along 10 different milk collection routes in northwest Spain. Milk samples of 100 ml were aseptically taken in sterile containers.
Additional samples from air, water, feed, and feces were taken from 10 selected farms (one farm at the end of each collection route). Ten-liter air samples were collected using a microbial air sampler (Biotest Hycon, Dreieich, Germany) fitted with a tryptone soya agar (TSA; Oxoid, Basingstoke, United Kingdom) strip. Water troughs and feed (silage) samples were taken in sterile containers of 500 ml and 250 g, respectively. Sampling of feces in farm premises was done with boot swabs by following the protocol outlined by the EU for monitoring the reduction of Salmonella in laying hens. In brief, boot swabs, moistened with a solution of 0.1% peptone and 0.9% NaCl, were used to walk through the premises using a route that produced a sample for all parts of the farm. A minimum of four samples (two pairs of boot swabs) were collected, with the swabs being changed every 100 steps. Boot swabs were removed carefully and stored in sterile zipper storage bags for transport to the laboratory.
Milk and farm environment samples were immediately transported to the laboratory in an insulated cooler. Temperature on arrival to the laboratory was always under 8°C.
Suspected diarrheagenic E. coli isolates from stool samples of patients with gastrointestinal disease were kindly provided by the Microbiology Service of the University Hospital of León.
Sample processing.
Fifty milliliters of milk was cultured in 450 ml of tryptone soya broth (TSB; Oxoid) plus 0.6% yeast extract (TSBYE; Oxoid) at 42°C for 18 h. An aliquot of the enriched sample was streaked onto CT-SMAC (Oxoid) plates and incubated at 37°C for 24 h. Pink (sorbitol fermenters) and colorless (nonfermenters) colonies were picked and preserved at −40°C in TSB plus 40% glycerol for further characterization.
TSA strips from the air sampling were homogenized with 50 ml of TSBYE in a Masticator blender (IUL SA, Barcelona, Spain). Water samples (250 ml) were filtered through sterile 0.45-μm filters, which were then incubated in 50 ml TSBYE. Twenty five g of feed pellets was blended with 225 ml of TSBYE. Boot swabs were unpacked and placed in 225 ml of TSBYE. All cultures were incubated and plated onto CT-SMAC as described above.
PCR detection of enteropathogenic E. coli.
Presumptive EPEC isolates were cultured in TSB at 37°C for 18 h, and DNA was released by heat lysis. The template DNA was screened by PCR for the presence of the target genes stx1, stx2, eae, and bfpA, as well as plasmid pEAF, using the primers described in Table 1. PCR was carried out in a Mastercycler Personal (Eppendorf-Netheler-Hinz GmbH, Hamburg, Germany) in a final volume of 25 μl and an annealing temperature of 56°C. PCR products (5 μl) were analyzed by 1% agarose gel electrophoresis and viewed after ethidium bromide staining under UV light.
Table 1.
Target genes and primer sequences for the PCR detection of EPEC gene markers
| Target gene or plasmid and primer | Sequence (5′→3′) | Amplicon size (bp) | Reference |
|---|---|---|---|
| Genes | |||
| stx1 | |||
| Stx1F | ATAAATCGCCATTCGTTGACTAC | 180 | 24 |
| Stx1R | AGAACGCCCACTGAGATCATC | ||
| stx2 | |||
| Stx2F | GGCACTGTCTGAAACTGCTCC | 255 | |
| Stx2R | TCGCCAGTTATCTGACATTCTG | ||
| eae | |||
| SK1 | CCCGAATTCGGCACAAGCATAAGC | 881 | 25 |
| SK2 | CCCGGATCCGTCTCGCCAGTATTCG | ||
| bfpA | |||
| EP1 | ATTGGTGCTTGCGCTTGCTGC | 326 | 27 |
| EP2 | GCCGCTTTATCCAACCTGGTA | ||
| Plasmid | |||
| pEAF | |||
| EAF1 | CAGGGTAAAAGAAAGATGATAA | 397 | 26 |
| EAF25 | TATGGGGACCATGTATTATCA |
MLST.
Multilocus sequence typing (MLST) was carried out by amplifying and sequencing seven housekeeping genes (arcA, cyaA, fadD, icdA, lysP, mtlD, and rpoS), as described by Moura et al. (6), by following the protocol detailed at the EcMLST website (http://shigatox.net/new/tools/ecmlst.html). PCR products were purified with a NucleoSpin gel and PCR cleanup kit (Macherey-Nagel, Düren, Germany). Both strands were sequenced in a Megabace 500 sequencer (Amersham Biosciences, Piscataway, NJ).
Raw sequences were visually reviewed and edited using the Chromas Lite 2.1 software (Technelysium, South Brisbane, Queensland, Australia) and aligned with the ClustalW algorithm of the MEGA5 software (16). Each gene locus was assigned an allele number by searching the EcMLST database. New allele numbers were assigned by the curator of the database upon submission and revision of sequence chromatograms. Each isolate was assigned an arbitrary sequence type (ST) number according to the allele profile.
Lineage assignment and phylogenetic analysis.
Grouping of isolates into clonal complexes according to the number of single-locus and double-locus variants was done with the eBursts algorithm implemented in the START2 program (17). Split decomposition analysis was performed with Splitstree software for Windows (18).
A concatenated sequence constructed from the individual gene sequences (in the order of arcA, cyaA, fadD, icdA, lysP, mtlD, rpoS) was prepared for one representative strain of each ST. Concatenated sequences were aligned and the phylogenetic tree was constructed using the neighbor-joining (NJ) method, with the distances estimated by the Kimura 2-parameter model and a bootstrapping of 1,000 replications using MEGA5 software.
PFGE.
Genomic DNA preparation for pulsed-field gel electrophoresis (PFGE) analysis was carried out according to the procedure of PulseNet (19). DNA was digested with 30 U XbaI (Fermentas, Thermo Fisher Scientific, Waltham, MA), and the fragments were resolved in a 1% Seakem Gold agarose gel (Lonza, Rockland, ME) in a contour-clamped homogeneous electric field (CHEF) DRIII apparatus (Bio-Rad, Hercules, CA) during 19 h with the following conditions: 14°C, ramp of 2.2 to 54.2 s, 120° angle, and voltage of 6 V/cm. Comparison of PFGE profiles was done with the GelCompar 6.1 software (Applied Maths, St. Martens Latem, Belgium). Similarities were obtained using the Dice coefficient at 0.5% optimization and 1.5% tolerance, and a dendrogram was constructed with the unweighted-pair group method using average linkages (UPGMA) clustering method.
RESULTS
EPEC isolation and identification.
A total of 62 suspected isolates were obtained from 55 of the 388 bulk tank milk samples (14.17%). All of the isolates were considered atypical EPEC, carrying the eae gene without amplifying any of the additional target genes (stx1, stx2, and bfpA) or pEAF. Ten more isolates were recovered from the environmental samples taken from 5 out of 10 farms (50%): five isolates were obtained from water trough samples taken from four different farms, four isolates were from feces from four different farms, and one isolate was obtained from an air sample; moreover, 9 atypical EPEC isolates were confirmed from human stools.
Multilocus sequence typing and phylogenetic analysis.
Sequences of seven housekeeping genes were obtained from all 81 isolates, and 19 ST were detected (Table 2); none of them were previously reported for atypical EPEC in studies carried out with the same genes (6, 20). The most frequent ST was ST 14, including 41 isolates, all of them obtained from milk and clinical samples. The allele frequency of the housekeeping genes ranged from 6 to 12 alleles per locus, with the most variable being loci icdA and mltD with 12 alleles each. Ten new alleles were detected in five different genes (Table 2).
Table 2.
Allele profile and relative frequency of ST detected after MLST analysis of 81 EPEC isolates
| ST | Allele no. of: |
Relative frequency | Origin of isolates | ||||||
|---|---|---|---|---|---|---|---|---|---|
| arcA | cyaA | fadD | icdA | lysP | mtlD | rpoS | |||
| 1 | 2 | 36a | 115 | 8 | 17 | 45a | 71a | 1 | Clinical |
| 2 | 3 | 2 | 13 | 4 | 1 | 13 | 1 | 1 | Milk |
| 3 | 3 | 3 | 2 | 2 | 1 | 3 | 1 | 1 | Clinical |
| 4 | 3 | 3 | 2 | 2 | 1 | 4 | 1 | 6 | Milk and water |
| 5 | 3 | 3 | 13 | 4 | 1 | 13 | 1 | 1 | Milk |
| 6 | 3 | 3 | 13 | 39 | 1 | 13 | 17 | 1 | Feces |
| 7 | 3 | 3 | 13 | 203a | 1 | 13 | 69a | 2 | Milk and water |
| 8 | 3 | 3 | 20 | 23 | 1 | 31 | 1 | 8 | Milk |
| 9 | 3 | 3 | 20 | 23 | 1 | 31 | 70a | 1 | Water |
| 10 | 3 | 3 | 49 | 4 | 1 | 13 | 1 | 1 | Milk |
| 11 | 3 | 11 | 13 | 39 | 1 | 43a | 1 | 3 | Milk and water |
| 12 | 6 | 1 | 4 | 3 | 2 | 11 | 8 | 1 | Milk |
| 13 | 7 | 26 | 13 | 54 | 35 | 34 | 46 | 1 | Milk |
| 14 | 8 | 2 | 1 | 15 | 1 | 2 | 1 | 41 | Milk and clinical |
| 15 | 8 | 3 | 2 | 4 | 1 | 13 | 1 | 4 | Milk, air, and feces |
| 16 | 9 | 2 | 1 | 1 | 1 | 41 | 1 | 3 | Milk and clinical |
| 17 | 9 | 3 | 2 | 4 | 1 | 13 | 1 | 3 | Feces and clinical |
| 18 | 24 | 32 | 6 | 75 | 4 | 42 | 26 | 1 | Milk |
| 19 | 27a | 35a | 110 | 136 | 78 | 44a | 17 | 1 | Water |
New alleles.
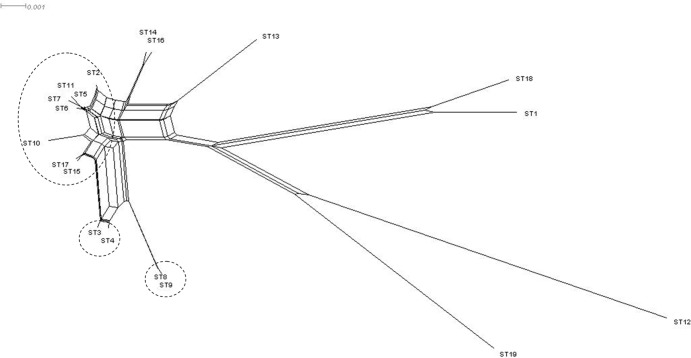
Three clonal complexes were revealed by eBurst analysis, grouping 7, 2, and 2 ST, respectively. Clonal group 1 was centered on ST 5, with ST 2 and ST 10 as a single-locus variant (SLV) and ST 6, ST 7, ST 15, and ST 17 as a double-locus variant (DLV). Clonal group 2 contained ST 3 and ST4, whereas clonal group 3 included ST 8 and ST 9. The remaining 8 ST were considered singletons, including the most frequent ST, ST 14. A SplitsTree graph (Fig. 1) shows the relative distances between STs.
Fig 1.
Phylogenetic splits network obtained after allele profile analysis with eBURST algorithm. Clonal complexes are indicated by broken lines.
Phylogenetic analysis of the supergene is shown in Fig. 2, showing a clustering of STs similar to that obtained with SplitsTree, with high bootstrapping values.
Fig 2.
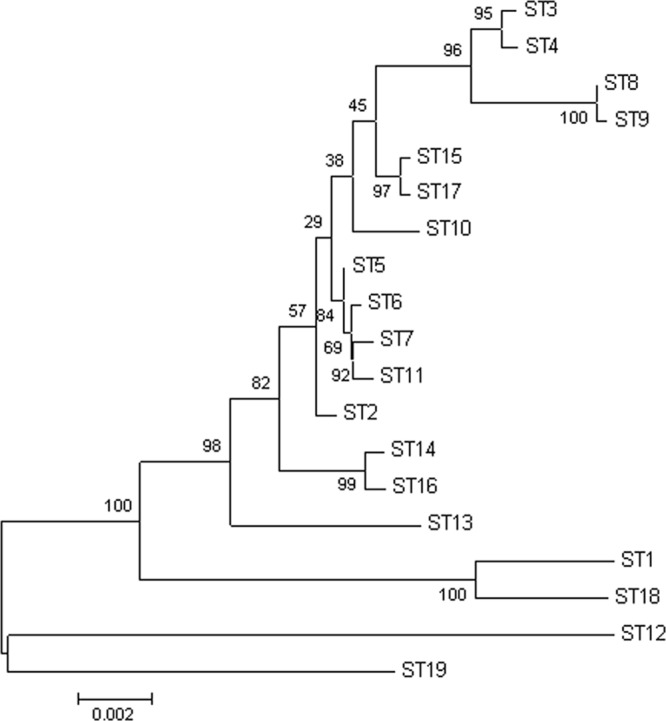
Neighbor-joining tree obtained from the phylogenetic analysis of the combined nucleotide sequences of the seven housekeeping genes in representatives from each ST. Bootstrapping values are shown in branch nodes.
Some STs grouped together isolates from different origins, as in ST 4, which grouped five isolates obtained from milk produced on five farms with a water strain isolated from a different farm. ST 7 included water and milk samples isolated from different locations, ST 11 was composed of one water and two milk isolates, ST 15 included two isolates from milk, one from air, and one from animal feces samples taken in different farms, ST 16 was composed of two clinical and one milk isolate, and ST 17 included two isolates from animal feces and one clinical isolate. An interesting finding was the absence of environmental isolates from ST 14 (Table 3).
Table 3.
Genotypic characteristics and origin (farm registration number and type of sample) of 81 EPEC isolates studied
| Strain | PFGE type | ST | Farm no. | Type of sample |
|---|---|---|---|---|
| M271VO | 1 | 4 | 158 | Milk |
| M277aVO | 1 | 4 | 160 | Milk |
| M277bVO | 1 | 4 | 160 | Milk |
| M289VO | 1 | 4 | 167 | Milk |
| F15VO | 1 | 4 | 187 | Water |
| M387VO | 1 | 4 | 234 | Milk |
| C156VO | 2 | 3 | Clinical | |
| C289VO | 3 | 16 | Clinical | |
| M39VO | 4 | 8 | 31 | Milk |
| M77VO | 4 | 8 | 87 | Milk |
| M312VO | 4 | 8 | 179 | Milk |
| M367VO | 4 | 8 | 210 | Milk |
| M370VO | 4 | 8 | 213 | Milk |
| M434VO | 4 | 8 | 275 | Milk |
| M443VO | 4 | 8 | 287 | Milk |
| M523VO | 4 | 8 | 361 | Milk |
| F12VO | 4 | 9 | 42 | Water |
| F23VO | 5 | 6 | 66 | Feces |
| M407VO | 5 | 15 | 243 | Milk |
| F20VO | 6 | 15 | 64 | Air |
| M313VO | 6 | 15 | 179 | Milk |
| F16VO | 6 | 15 | 187 | Feces |
| M278VO | 6 | 16 | 160 | Milk |
| C294VO | 6 | 16 | Clinical | |
| F21VO | 8 | 17 | 64 | Feces |
| F14VO | 9 | 17 | 157 | Feces |
| C322VO | 9 | 17 | Clinical | |
| M361VO | 10 | 14 | 207 | Milk |
| M385VO | 10 | 14 | 233 | Milk |
| M337VO | 11 | 14 | 189 | Milk |
| M229VO | 12 | 14 | 77 | Milk |
| M232VO | 12 | 14 | 91 | Milk |
| M251VO | 12 | 14 | 106 | Milk |
| M124VO | 12 | 14 | 223 | Milk |
| M138VO | 12 | 14 | 261 | Milk |
| M142VO | 12 | 14 | 314 | Milk |
| M487VO | 12 | 14 | 336 | Milk |
| M176VO | 12 | 14 | 375 | Milk |
| M330VO | 13 | 14 | 185 | Milk |
| M341VO | 13 | 14 | 191 | Milk |
| M42VO | 14 | 14 | 32 | Milk |
| M51VO | 14 | 14 | 58 | Milk |
| M54VO | 14 | 14 | 59 | Milk |
| M59VO | 14 | 14 | 61 | Milk |
| M78VO | 14 | 14 | 87 | Milk |
| M242VO | 14 | 14 | 98 | Milk |
| M247VO | 14 | 14 | 103 | Milk |
| M82VO | 14 | 14 | 114 | Milk |
| M90VO | 14 | 14 | 120 | Milk |
| M92VO | 14 | 14 | 125 | Milk |
| M93VO | 14 | 14 | 125 | Milk |
| M100VO | 14 | 14 | 127 | Milk |
| M99VO | 14 | 14 | 127 | Milk |
| M105VO | 14 | 14 | 145 | Milk |
| M106VO | 14 | 14 | 148 | Milk |
| M108VO | 14 | 14 | 151 | Milk |
| M110VO | 14 | 14 | 156 | Milk |
| M111VO | 14 | 14 | 157 | Milk |
| M126VO | 14 | 14 | 223 | Milk |
| M130VO | 14 | 14 | 224 | Milk |
| M186VO | 14 | 14 | 381 | Milk |
| M187VO | 14 | 14 | 382 | Milk |
| M344VO | 15 | 12 | 193 | Milk |
| C147VO | 15 | 14 | Clinical | |
| C171VO | 16 | 14 | Clinical | |
| C198VO | 16 | 14 | Clinical | |
| C212VO | 16 | 14 | Clinical | |
| F22VO | 17 | 11 | 66 | Water |
| M448aVO | 17 | 11 | 296 | Milk |
| M532VO | 17 | 11 | 388 | Milk |
| F17VO | 18 | 7 | 42 | Water |
| M102VO | 18 | 7 | 133 | Milk |
| M508VO | 19 | 14 | 354 | Milk |
| M151VO | 19 | 14 | 364 | Milk |
| M254aVO | 20 | 2 | 107 | Milk |
| M29aVO | 20 | 5 | 24 | Milk |
| M442aVO | 20 | 10 | 286 | Milk |
| M36VO | 21 | 18 | 29 | Milk |
| M438aVO | 22 | 13 | 281 | Milk |
| F13VO | 23 | 19 | 157 | Water |
| C22VO | 24 | 1 | Clinical |
Pulsed-field gel electrophoresis.
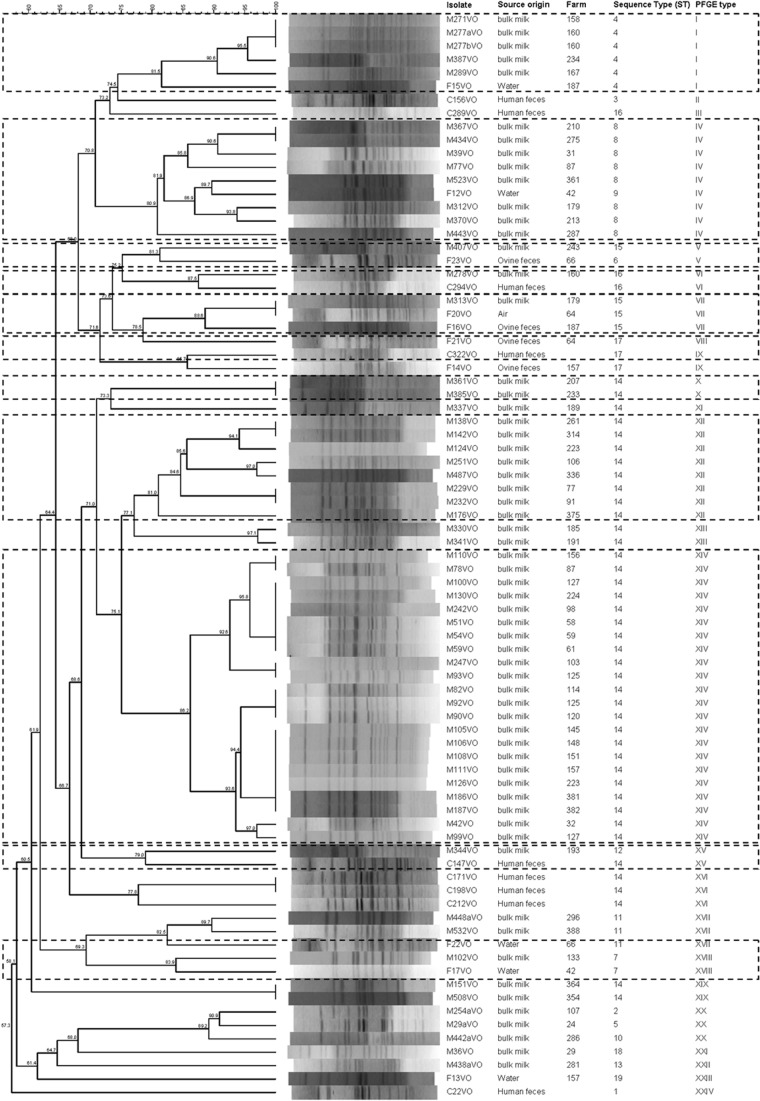
PFGE typing of 81 isolates resulted in 57 patterns, which grouped in 24 clusters with a minimum cutoff value of 74.5% (Fig. 3). Identical profiles were detected in isolates from milk (34 strains in pulsotypes I, IV, X, XII, XIV, and XIX) and clinical samples (2 strains in pulsotype XVI) and between one isolate from milk and one isolate from air (pulsotype VII). All of the isolates with indistinguishable PFGE profiles belong to the same ST. Nine clusters grouped isolates from different origins: milk and water (I, IV, XVII, and XVIII), milk and ovine feces (V), milk, air and ovine feces (VII), clinical samples and ovine feces (IX), and milk and clinical samples (VI and XV).
Fig 3.
PFGE profiles of 81 atypical EPEC isolates obtained from ewes' milk, farm environment, and clinical samples. The corresponding sequence types (ST) are also listed. Main groups are marked by broken lines.
Table 3 summarizes the features of the 81 strains investigated in this study.
DISCUSSION
Humans have long been considered the main reservoir of EPEC (3), but this assumption is being revised in light of the characteristics of atypical EPEC strains, which are regularly found in animals (2, 5). The increasing isolation of atypical EPEC strains from foods of animal origin (10, 12, 14, 21) suggests that different animal species act as reservoirs for these microorganisms and represent a source of infections for humans, as already proposed by other authors (6, 8).
In this study, bulk tank sheep milk tested positive for 14.17% of the samples, and atypical EPEC strains were also isolated from the environment of the farm and from feces of the animals. A high degree of phylogenetic heterogeneity was observed among the strains, which form 19 sequence types and 24 pulsotypes. The genetic diversity in atypical EPEC appears to be a common trait, as already reported (22). In spite of the heterogeneity, comparison of the strains isolated from milk and farm environments to isolates obtained from human patients showed a clear relationship between milk and human isolates; thus, the main ST (ST 14) was composed of strains of those origins. This relationship was demonstrated by PFGE typing as well (clusters VI and XV) (Fig. 3). Another interesting finding of this study is the similarities observed between water and milk isolates, which grouped together in two ST and four PFGE clusters (Fig. 3).
The results obtained in this study do not clarify the routes of transmission of EPEC but indicate that strains that appear in milk are similar to strains causing disease in humans. Why the milk became contaminated remains unresolved, but similarities between milk and water isolates suggest a way of transmission through water used in the production of milk, either for drinking or for cleaning equipment. Robins-Browne et al. (23) showed that atypical EPEC strains are an important cause of human gastroenteritis acquired through water consumption, and García-Díez et al. (10) reported that molecular characteristics found in atypical EPEC isolates from samples of surface water were similar to those detected in human samples. These pieces of evidence, together with the findings of this study, suggest a possible route of human infection.
In conclusion, this study shows that ewes' milk carries strains of atypical EPEC related to those found in human samples and suggests possible transmission routes for this emerging pathogen.
ACKNOWLEDGMENTS
This work was financed by project AGL2011-26118/ALI of the Spanish R&D program (Ministry of Economy and Competitiveness) and Junta de Castilla y León (Ayuda Grupo de Excelencia GR155). Verónica Otero was supported by a grant from the FPU program of the Spanish government.
We are grateful for the valuable assistance of Silvia Herrera-León and Sergio Sánchez from the Bacteriology Department of the National Microbiology Centre of the Instituto de Salud Carlos III and María Antonia Remacha from the Microbiology Service of the University Hospital of León.
Footnotes
Published ahead of print 19 July 2013
REFERENCES
- 1.Chen DH, Frankel G. 2005. Enteropathogenic Escherichia coli: unravelling pathogenesis. FEMS Microbiol. Rev. 29:83–98 [DOI] [PubMed] [Google Scholar]
- 2.Trabulsi LR, Keller R, Gomes T. 2002. Typical and atypical enteropathogenic Escherichia coli. Emerg. Infect. Dis. 8:508–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nataro JP, Kaper JB. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaper JB, Nataro JP, Mobley HLT. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123–140 [DOI] [PubMed] [Google Scholar]
- 5.Hernandes RT, Elias WP, Vieira M, Gomes T. 2009. An overview of atypical enteropathogenic Escherichia coli. FEMS Microbiol. Lett. 297:137–149 [DOI] [PubMed] [Google Scholar]
- 6.Moura RA, Sircili MP, Leomil L, Matté MHT, Elias LRWP, Irino K, Pestana de Castro AF. 2009. Clonal relationship among atypical enteropathogenic Escherichia coli strains isolated from different animal species and humans. Appl. Environ. Microbiol. 75:7399–7408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cortés C, De la Fuente R, Blanco J, Blanco M, Blanco JE, Dhabi G, Mora A, Justel P, Contreras A, Sanchez A. 2005. Serotypes, virulence genes and intimin types of verotoxin-producing Escherichia coli and enteropathogenic E. coli isolated from healthy dairy goats in Spain. Vet. Microbiol. 110:67–76 [DOI] [PubMed] [Google Scholar]
- 8.Blanco M, Schumacher S, Tasara T, Zweifel C, Blanco J, Dahbi G, Blanco J, Stephan R. 2005. Serotypes, intimin variants and other virulence factors of eae positive Escherichia coli strains isolated from healthy cattle in Switzerland. Identification of a new intimin variant gene (eae-η2). BMC Microbiol. 5:23. 10.1186/1471-2180-5-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farooq S, Hussain I, Mir MA, Bhat MA, Wani SA. 2009. Isolation of atypical enteropathogenic Escherichia coli and Shiga toxin 1 and 2f-producing Escherichia coli from avian species in India. Lett. Appl. Microbiol. 48:692–697 [DOI] [PubMed] [Google Scholar]
- 10.García Díez M, Meindl K, Frässdorf J, Wolf S, Schalch B, Busch U. 2009. Prevalence of enteropathogenic Escherichia coli in food and water in Bavaria in 2007. Arch. Lebensmittelhyg. 60:77–81 [Google Scholar]
- 11.Altalhi AD, Hassan SA. 2009. Bacterial quality of raw milk investigated by Escherichia coli and isolates analysis for specific virulence-gene markers. Food Control 20:913–917 [Google Scholar]
- 12.Holko I, Bisova T, Holkova Z, Kmet V. 2006. Virulence markers of Escherichia coli strains isolated from traditional cheeses made from unpasteurised sheep milk in Slovakia. Food Control 17:393–396 [Google Scholar]
- 13.Carneiro LAM, Lins MC, Garcia FRA, Silva APS, Mauller PM, Alves GB, Rosa ACP, Andrade JRC, Freitas-Almeida AC, Queiroz MLP. 2006. Phenotypic and genotypic characterisation of Escherichia coli strains serogrouped as enteropathogenic E. coli (EPEC) isolated from pasteurised milk. Int. J. Food Microbiol. 108:15–21 [DOI] [PubMed] [Google Scholar]
- 14.da Silva ZN, Cunha AS, Lins MC, Carneiro LAM, Almeida ACF, Queiroz MLP. 2001. Isolation and serological identification of enteropathogenic Escherichia coli in pasteurized milk in Brazil. Rev. Saúde Pública 35:375–379 [DOI] [PubMed] [Google Scholar]
- 15.Son I, Van Kessel JA, Karns JS. 2009. Genotypic diversity of Escherichia coli in a dairy farm. Foodborne Pathog. Dis. 6:837–847 [DOI] [PubMed] [Google Scholar]
- 16.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jolley KA, Feil EJ, Chan MS, Maiden MC.2001. Sequence type analysis and recombinational tests (START). Bioinformatics 17:1230–1231 [DOI] [PubMed] [Google Scholar]
- 18.Huson DH, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23:254–267 [DOI] [PubMed] [Google Scholar]
- 19.Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, Barrett TJ. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 3:59–67 [DOI] [PubMed] [Google Scholar]
- 20.Leomil L, Pestana de Castro AF, Krause G, Schmidt H, Beutin L. 2005. Characterization of two major groups of diarrheagenic Escherichia coli O26 strains which are globally spread in human patients and domestic animals of different species. FEMS Microbiol. Lett. 249:335–342 [DOI] [PubMed] [Google Scholar]
- 21.Bandyopadhyay S, Lodh C, Rahaman H, Bhattacharya D, Bera AK, Ahmed FA, Mahanti A, Samanta I, Mondal DK, Bandyopadhyay S, Sarkar S, Dutta TK, Maity S, Paul V, Ghosh MK, Sarkar M, Baruah KK. 2012. Characterization of Shiga toxin producing (STEC) and enteropathogenic Escherichia coli (EPEC) in raw yak (Poephagus grunniens) milk and milk products. Res. Vet. Sci. 93:604–610 [DOI] [PubMed] [Google Scholar]
- 22.Afset JE, Anderssen E, Bruant G, Harel J, Wieler L, Bergh K. 2008. Phylogenetic backgrounds and virulence profiles of atypical enteropathogenic Escherichia coli strains from a case-control study using multilocus sequence typing and DNA microarray analysis. J. Clin. Microbiol. 46:2280–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robins-Browne RM, Bordun AM, Tauschek M, Bennett-Wood VR, Russell J, Oppedisano F, Lister NA, Bettelheim KA, Fairley CK, Sinclair MI, Hellard ME. 2004. Escherichia coli and community-acquired gastroenteritis, Melbourne, Australia. Emerg. Infect. Dis. 10:1797–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paton AW, Paton JC. 1998. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157. J. Clin. Microbiol. 36:598–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oswald E, Schmidt H, Morabito S, Karch H, Marches O, Caprioli A. 2000. Typing of intimin genes in human and animal enterohemorrhagic and enteropathogenic Escherichia coli: characterization of a new intimin variant. Infect. Immun. 68:64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Franke J, Franke S, Schmidt H, Schwarzkopf A, Wieler LH, Baljer G, Beutin L, Karch H. 1994. Nucleotide sequence analysis of enteropathogenic Escherichia coli (EPEC) adherence factor probe and development of PCR for rapid detection of EPEC harboring virulence plasmids. J. Clin. Microbiol. 32:2460–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gunzburg ST, Tornieporth NG, Riley LW. 1995. Identification of enteropathogenic Escherichia coli by PCR-based detection of the bundle-forming pilus gene. J. Clin. Microbiol. 33:1375–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]