Abstract
CTX-M-producing Escherichia coli is the predominant type of extended-spectrum β-lactamase (ESBL)-producing E. coli worldwide. In this study, molecular typing was conducted for 139 CTX-M-producing E. coli isolates, phenotypically positive for ESBLs, isolated from environmental water, swine, healthy humans, and hospitalized patients in Hangzhou, China. The antibiotic resistance profiles of the isolates for the cephalosporins and fluoroquinolones were determined. The isolates showed 100% resistance to cefotaxime and ceftriaxone while maintaining relatively high susceptibility to cefoxitin, cefepime, and ceftazidime. A total of 61.9% (86/139) of the isolates, regardless of origin, showed high resistance to fluoroquinolones. PCRs and DNA sequencing indicated that blaCTX-M-14 was the most prevalent CTX-M-9 group gene and that blaCTX-M-15 and blaCTX-M-55 were the dominant CTX-M-1 group genes. Isolates from all sources with CTX-M types belonging to the CTX-M-1 or CTX-M-9 group were most frequently associated with epidemics. Molecular homology analysis of the isolates, conducted by phylogenetic grouping, pulsed-field gel electrophoresis (PFGE), and multilocus sequence typing (MLST), demonstrated that the dominant clones belonged to B2-ST131, D-ST648, D-ST38, or A-CC10. These four sequence types (STs) were discovered in E. coli isolates both from humans and from environmental water, suggesting frequent and continuous intercompartment transmission between humans and the aquatic environment. Seven novel sequence types were identified in the current study. In conclusion, this study is the first to report the molecular homology analysis of CTX-M-producing E. coli isolates collected from water, swine, and healthy and hospitalized humans, suggesting that pathogens in the environment might originate both from humans and from animals.
INTRODUCTION
Although Escherichia coli normally exists in the intestinal tracts of humans and animals, it is also frequently associated with various intestinal and extraintestinal diseases, such as urinary tract infection (UTI), bacteremia, and meningitis (1). Enterobacteriaceae producing extended-spectrum β-lactamases (ESBLs) have increasingly emerged due to the widespread use of cephalosporins. In China, the prevalence of ESBL-producing E. coli remained above 50.0% during 2009 to 2011, according to the national resistance surveillance program (2–4).
CTX-M-type ESBLs have recently become predominant among ESBL subtypes (5). Since the first report of CTX-M ESBL from a strain of E. coli in 1989 (6), the CTX-M family of ESBLs has spread in many countries, and the global spread of CTX-M-producing Enterobacteriaceae is a major concern (7, 8). Among the different CTX-M subtypes, those of the CTX-M-1 and CTX-M-9 groups were the most common in Asia as well as worldwide (9). In recent years, ESBL-producing E. coli strains have been identified not only in the community (10, 11) but also in animals (12–16), even in food-producing animals (17).
As a matter of fact, in China, 6,000 tons of antibiotics, about half of the world's total consumption, have been used as feed additives yearly (18). Such a huge usage of antibiotics has exerted strong selection pressure for resistant bacteria, especially for zoonotic pathogens, such as E. coli, Staphylococcus spp., and Salmonella spp. We have previously conducted surveys of fluoroquinolone-resistant Citrobacter freundii isolates and their gene subtypes from the aquatic environment in Hangzhou in comparison with those from clinical isolates, and we found that plasmids isolated from environmental and clinical C. freundii isolates appeared to be homogenous (19). Although there are several reports on the hypothesis that animals might become infection sources or even serve as reservoirs for the spread of these bacteria (9), few studies have focused on ESBL subtypes from zoonotic pathogens isolated from the aquatic environment, farm animals, healthy humans, and human patients.
Molecular typing of ESBL-producing E. coli is useful for hospital epidemiologists, microbiologists, and clinicians in guiding surveillance studies, monitoring outbreak situations, and tracking the spread of emerging pathogens. Multilocus sequence typing (MLST) and pulsed-field gel electrophoresis (PFGE) are the methods most widely used to analyze the molecular typing of strains (20). Thus, in the present study, we used MLST and PFGE to analyze the molecular homology of the CTX-M subtypes from E. coli strains collected from the aquatic environment, hospitalized patients, and feces from swine and healthy humans.
MATERIALS AND METHODS
Bacterial isolates.
For microbiological characterization, a total of 139 E. coli isolates phenotypically positive for ESBL production were obtained from Hangzhou, China. Among the 139 isolates, 31 were obtained from swine feces during 2012 to 2013, 26 were recovered from environmental water during the same period, 46 were obtained from physical examination of asymptomatic persons in 2012, and the other 36 clinical isolates were randomly recovered from previously stored, nonduplicated ESBL-producing E. coli isolates collected from hospitalized patients in the Second Affiliated Hospital of Zhejiang University from 2010 to 2012. The clinical isolates were from urine, blood, sputum, and body secretions; none were from fecal samples. All the isolates were identified by the Vitek 2 Compact system (bioMérieux, Hazelwood, MO, USA) and were screened for ESBL production by Etest (AB Biodisk, Solna, Sweden) according to the manufacturer's instructions.
Water samples were collected from 12 distinct water sources in Hangzhou (Huajiachi Lake, East River, West Lake, Xixi Wetland, Jinghang Grand Canal, Qiantang River, Jiefang River, Nine Creeks, Tiesha River, a fountain at the Second Affiliated Hospital of Zhejiang University, the sewer surrounding a pig farm, and a tributary of the Qiantang River) during 2012 to 2013. Two to three representative sites in each locality were selected for water collection. We selected those sampling locations to represent the whole aquatic environment. Bacteria in water samples (1-liter samples) were concentrated by vacuum filtration through a filter membrane (<0.2 μm). The membrane was then washed and was suspended in 10 ml 0.45% saline solution, and 200 μl of the suspension was inoculated onto blood agar, MacConkey agar, and Salmonella-Shigella agar plates. All the colonies on MacConkey medium were isolated and identified.
The swine and healthy-human feces were collected with sterile swabs and were then inoculated onto blood agar, MacConkey agar, and Salmonella-Shigella agar plates within 2 h. Colonies were picked and were identified by the Vitek 2 Compact system. If two or more strains of E. coli were collected from the same swine or human fecal specimen, genotypic characterization was conducted to exclude homologous isolates from the same sample.
PCR and sequence analysis.
To determine the genotypes of ESBLs, we performed PCR using primers specific to the TEM, SHV, CTX-M-1, CTX-M-2, CTX-M-8, and CTX-M-9 groups and AmpC, as reported previously (21, 22). Considering the high prevalence of qnr and aac(6′)-Ib-cr in the aquatic environment of Citrobacter freundii (19), plasmid-mediated quinolone resistance (PMQR) genes, including qnrA (23), qnrB (23), qnrC (24), qnrD (25), qnrS (23), and aac(6′)-Ib (26), were amplified through a TPersonal cycler (Biometra, Germany). PCR products were sequenced using an ABI 3730 sequencer (Applied Biosystems, Foster City, CA), and the sequences were then compared with the reported sequences from GenBank (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Sequence type (ST) determination and phylogenetic grouping.
MLST was performed as reported previously; seven housekeeping genes, including adk (encoding adenylate kinase), fumC (fumarate hydratase), gyrB (DNA gyrase), icd (isocitrate dehydrogenase), mdh (malate dehydrogenase), purA (adenylosuccinate synthetase), and recA (ATP/GTP binding motif), were used (27). Clonal complexes (CC) were compared with those of the E. coli MLST website (http://mlst.ucc.ie/mlst/dbs/Ecoli). Phylogenetic groups were determined using a multiplex PCR assay of chuA, yjaA, and tspE4.C2 as described previously (28).
PFGE profiles.
The chromosomal genome was prepared in agarose blocks and was digested with the restriction enzyme XbaI. DNA fragments were separated by use of a Rotaphor System 6.0 instrument (Whatman Biometra, Goettingen, Germany). Salmonella enterica serovar Braenderup H9812 was used as a size marker. The pulse times were increased from 1 to 30 s over 24 h at a voltage of 6 V and an angle of 120°. A dendrogram was generated from the homology matrix with a coefficient of 2.0% by the unweighted pair-group method using arithmetic averages (UPGMA) to describe the relationships among PFGE profiles. Isolates were considered to belong to the same PFGE group if their Dice similarity index was ≥80% (29).
Antibiotic susceptibility testing.
In vitro testing of susceptibility to ampicillin, ceftazidime, cefotaxime, ceftriaxone, aztreonam, cefoxitin, cefepime, levofloxacin, moxifloxacin, piperacillin-tazobactam, cefoperazone-sulbactam, imipenem, meropenem, and ertapenem was performed by the Mueller-Hinton agar dilution method, while the MICs of amikacin and ciprofloxacin were determined by the Etest (AB Biodisk, Solna, Sweden) according to the manufacturer's instructions. Susceptibility results were interpreted based on the recommended breakpoints of the CLSI (30). E. coli ATCC 25922 was used for quality control.
Statistical analysis.
Fisher's exact t test was used to analyze the significant differences in resistance rates by using the SPSS for Windows software package (version 17.0; SPSS Inc., Chicago, IL, USA).
RESULTS
Bacteria isolated from healthy humans, animals, and the aquatic environment.
A total of 208 E. coli isolates were collected from water samples. No more than two E. coli isolates were collected from the same fecal sample. Ninety-seven E. coli isolates were obtained from 105 swine (E. coli was not isolated from 18 swine, and 20 E. coli isolates were isolated from 10 samples of swine feces), and 263 E. coli isolates were obtained from physical examination of 200 asymptomatic persons (for 63 samples, 2 E. coli strains each were isolated).
Among the isolates from water samples, swine feces, and feces from healthy humans, 26, 31, and 46 isolates, respectively, were confirmed as ESBL-producing E. coli. The prevalences of ESBLs were 12.5% (26/208) in the aquatic environment, 32.0% (31/97) in the swine feces, and 17.5% (46/263) in the feces from healthy humans. Twenty-six aquatic-environment ESBL-producing E. coli isolates were recovered from Huajiachi Lake (4 isolates), Xixi Wetland (3 isolates), Jinghang Grand Canal (10 isolates), and East River (9 isolates).
PCR classification of CTX-M-type β-lactamases and PMQR genes.
Among the 139 ESBL-positive E. coli isolates (26 from water, 31 from swine feces, 46 from healthy humans, and 36 from hospitalized patients), 33 carried CTX-M-1 group ESBL genes, while 99 carried CTX-M-9 group ESBL genes and 7 carried both CTX-M-1 and CTX-M-9 group ESBL genes. blaCTX-M-14 was the most prevalent CTX-M-9 group gene, and blaCTX-M-55 and blaCTX-M-15 were the most prevalent CTX-M-1 group genes. In addition, blaCTX-M-123 was detected for the first time in clinical E. coli isolates, while blaCTX-M-121 and blaCTX-M-104 were detected for the first time in pig feces. The distributions of CTX-M gene types were different in different sources. blaCTX-M-24 was found in all isolates except for those from the aquatic environment, while blaCTX-M-3 and blaCTX-M-15 were discovered in all isolates except for those collected from swine feces. Additionally, one isolate for which the cefoxitin MIC value was 256 μg/ml harbored CMY-2. None of the other types of AmpC were detected. Furthermore, among the PMQR genes, four E. coli isolates from swine feces and feces from healthy humans harbored the qnrS1 gene, and one isolate from environmental water possessed the qnrS2 gene. The aac(6′)-Ib-cr gene was detected in nine isolates collected from all sources except healthy-human feces.
Sequence type distribution and phylogenetic grouping.
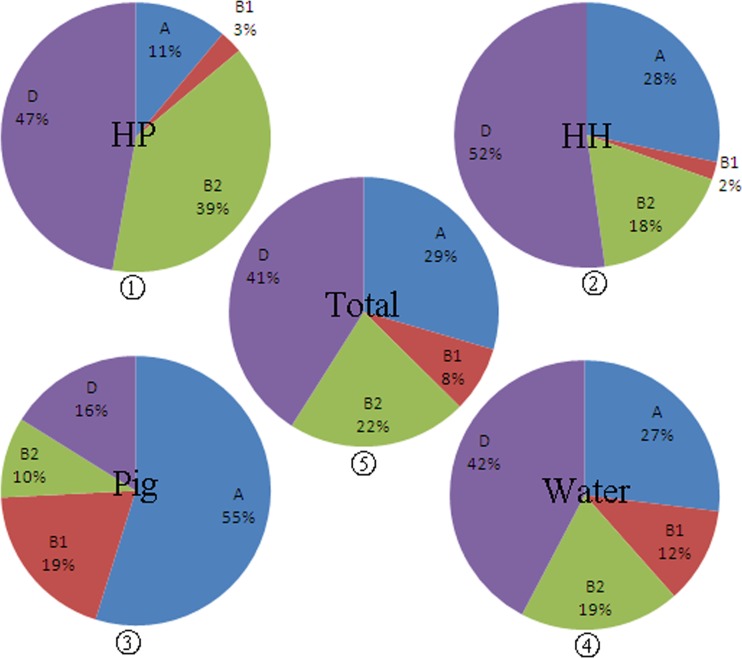
Fifty-one different sequence types, including 17 clonal complexes and 28 singletons, were identified among the 139 E. coli isolates. Among these, ST131 was the most prevalent sequence type in the current study, accounting for 14.4% of isolates (20 isolates), followed by ST648 (13.7%; 19 isolates), CC10 (13.7%; 19 isolates), and ST38 (11.5%; 16 isolates). CC10 was found in water, humans, and animals, while ST131, ST648, and ST38 were found only in water and humans. E. coli strains isolated from swine feces exhibited a much higher diversity of sequence types than isolates from other sources. ST3484, ST3485, ST3725, and ST3744 were novel STs first identified in the current study and were isolated from swine feces. In addition, one ST3724 isolate from a healthy human and two other isolates, with ST3745 and ST3746, from environmental water also represented novel STs found in the present study. Besides CC10, ST405 and ST46 were found in swine feces, healthy humans, and hospitalized patients (see Table S1 in the supplemental material). The CTX-M-9 producers were distributed among 15 ST complexes (48 isolates) and 25 singletons (58 isolates), while the CTX-M-1-producers were distributed among 11 ST complexes (22 isolates) and 11 singletons (18 isolates). Among the 139 ESBL-producing E. coli isolates, phylogenetic groups A, B1, B2, and D were all detected in both the CTX-M-9-producing and the CTX-M-1-producing E. coli group (see Table S2 in the supplemental material); subgroup D was predominant (57 isolates; 41.0%), followed by subgroups A (41 isolates; 29.5%), B2 (30 isolates; 21.6%), and B1 (11 isolates; 7.9%). E. coli isolates from different sources exhibited totally different distributions among phylogenetic groups. Isolates from hospitalized patients belonged mainly to subgroups B2 and D, whereas isolates from swine belonged mainly to subgroups A and B1 (Fig. 1). On the other hand, the phylogenetic subtype distribution of the ESBLs from environmental water was intermediate, with proportions in each group close to the averages for total isolates (Fig. 1).
Fig 1.
Proportions of bacteria from each source belonging to phylogenetic groups A, B1, B2, and D. HP, hospitalized patients; HH, healthy humans. Pie charts 1, 2, 3, and 4 represent the distributions of isolates from hospitalized patients, healthy humans, pigs, and water, respectively. Pie chart 5 displays the proportion of all 139 CTX-M-producing E. coli strains in each phylogenetic group.
PFGE.
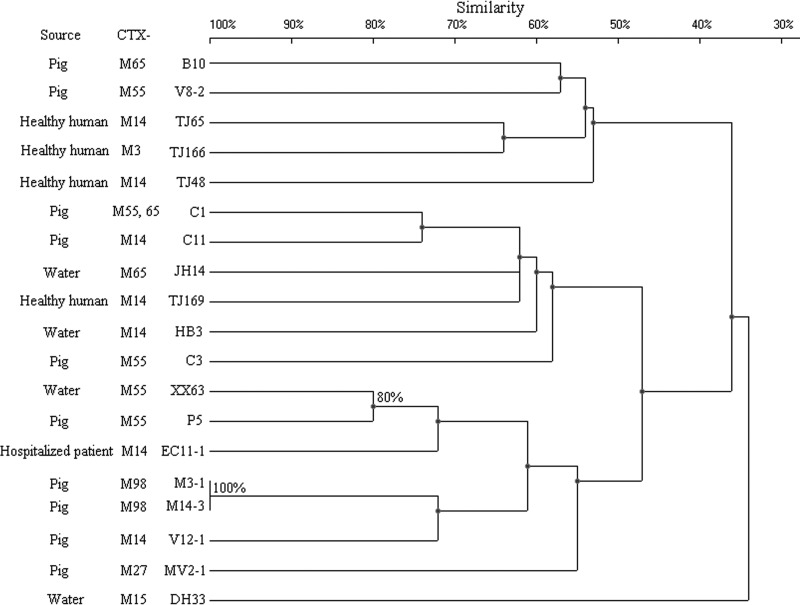
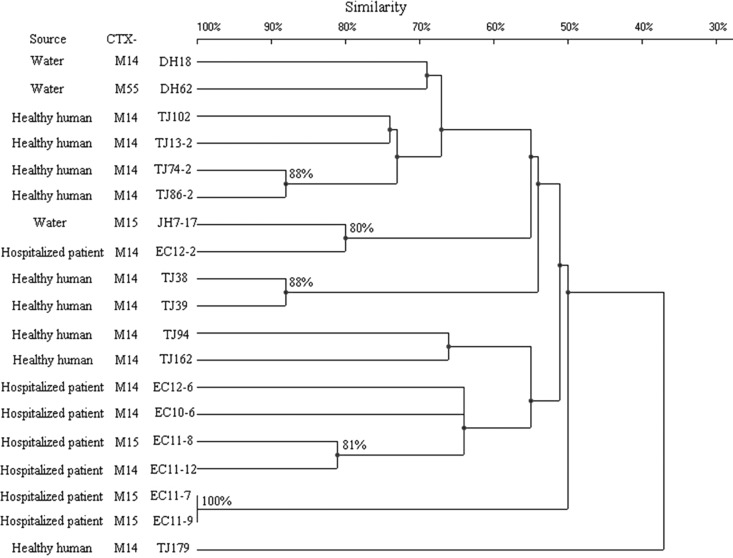
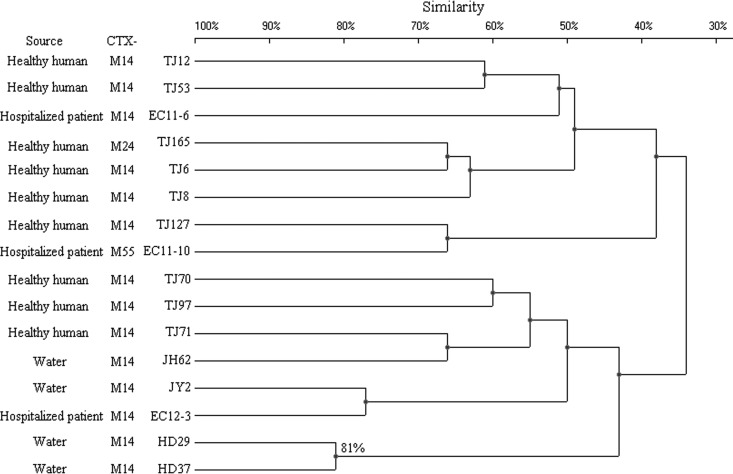
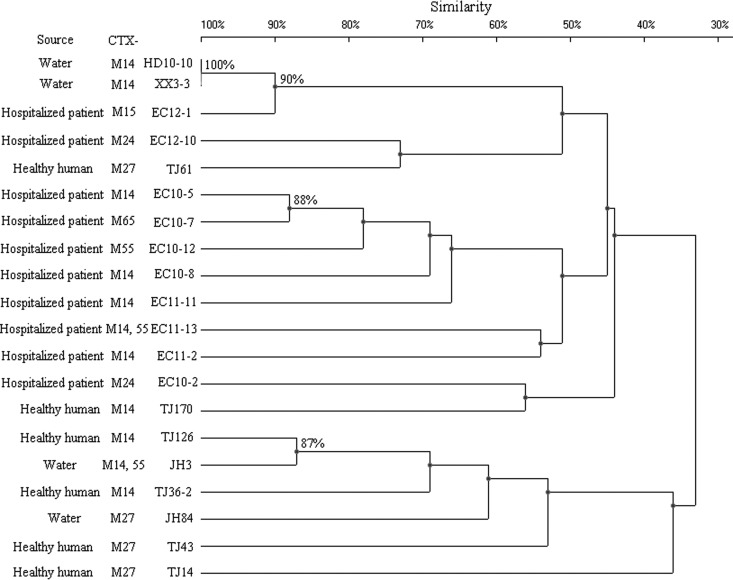
Some strains that belonged to the same ST exhibited quite different PFGE patterns. Compared with the ST131, CC10, and ST38 groups, the ST648 group constituted one large cluster (defined at the 50% similarity level), aside from strain TJ179 (Fig. 2 to 5). The other three groups, in turn, were distributed among several clusters with a similarity level of <50%. When 80% similarity was used as the cutoff point, HD10-10, XX3-3, and EC12-1 (90% similarity), EC10-5 and EC10-7 (88% similarity), and TJ126 and JH3 (87% similarity) within the ST131 group were considered to be related. Ten isolates within the ST648 group, 4 within the CC10 group, and 2 within the ST38 group were regarded as related (Fig. 2 to 5). The remaining isolates were defined as unrelated.
Fig 2.
PFGE of XbaI-digested DNA from 19 A-CC10 E. coli isolates. An UPGMA dendrogram based on Dice similarity coefficients was generated using UVI-Band software (Bio-Rad). Eighty percent similarity was used as the cutoff point.
Fig 5.
PFGE of XbaI-digested DNA from 19 D-ST648 E. coli isolates. An UPGMA dendrogram based on Dice similarity coefficients was generated using UVI-Band software (Bio-Rad). Eighty percent similarity was used as the cutoff point.
Fig 3.
PFGE of XbaI-digested DNA from 16 D-ST38 E. coli isolates. An UPGMA dendrogram based on Dice similarity coefficients was generated using UVI-Band software (Bio-Rad). Eighty percent similarity was used as the cutoff point.
Fig 4.
PFGE of XbaI-digested DNA from 20 B2-ST131 E. coli isolates. An UPGMA dendrogram based on Dice similarity coefficients was generated using UVI-Band software (Bio-Rad). Eighty percent similarity was used as the cutoff point.
Antimicrobial susceptibility profiles.
All the ESBL-producing E. coli isolates showed resistance to ampicillin, cefotaxime, and ceftriaxone. In contrast, ESBL-producing E. coli isolates showed high susceptibility to carbapenems (100% susceptible), followed by piperacillin-tazobactam (1.6% resistant) and amikacin (4.3% resistant). Among the cephalosporin antibiotics, there was high susceptibility to cefoxitin (6.5% resistant), followed by cefepime (19.4% resistant) and ceftazidime (24.4% resistant). On the other hand, the isolates showed quite high resistance (61.9%) to the fluoroquinolones. In addition, antimicrobial resistance differed between the CTX-M-1- and CTX-M-9-producing groups with regard to ceftazidime (P < 0.001), aztreonam (P < 0.001), cefoxitin (P = 0.003), and cefepime (P < 0.001); the rates of resistance of the CTX-M-1-producing group to these antibiotics were much higher than those of the CTX-M-9-producing group (Table 1). For most antibiotics (except piperacillin-tazobactam and amikacin), the resistance rates of isolates from different sources differed significantly (Table 2). The resistance rates of isolates from hospitalized patients were higher than those of the other three groups, and the isolates from healthy humans appeared to be much more sensitive than the other groups. Isolates of different sequence types differed significantly in susceptibility to fluoroquinolones. The rates of resistance of the ST38 group to fluoroquinolones were lower than those of the other ST groups (Table 1). There was no significant difference among isolates of different phylogenetic groups (Table 2). For human hosts, significant differences between isolates from healthy humans and hospitalized patients (P < 0.001) were identified for all the antibiotics other than three to which the isolates were relatively sensitive (cefoxitin, piperacillin-tazobactam, and amikacin) (see Table S3 in the supplemental material).
Table 1.
Resistance rates of ESBL-producing E. coli isolates by sequence type and ESBL type
| Antimicrobial agent | Rate of resistance (%) by sequence type (no. of isolates) |
P value for sequence typesa | Rate of resistance (%) by ESBL type (no. of isolates) |
P value by ESBL typeb | |||||
|---|---|---|---|---|---|---|---|---|---|
| ST131 (20) | ST648 (19) | ST10 complex (19) | ST38 (16) | Other STs (65) | CTX-M1 group (33) | CTX-M9 group (99) | |||
| Ceftazidime | 30.0 | 26.3 | 15.8 | 6.3 | 29.2 | 0.298 | 69.7 | 6.1 | 0.000 |
| Cefotaxime | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | —b | 100.0 | 100.0 | — |
| Ceftriaxone | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | — | 100.0 | 100.0 | — |
| Aztreonam | 65.0 | 31.6 | 63.2 | 18.8 | 63.1 | 0.003 | 87.9 | 36.4 | 0.000 |
| Cefoxitin | 0.0 | 5.3 | 5.3 | 12.5 | 7.7 | 0.665 | 18.2 | 2.0 | 0.003 |
| Cefepime | 25.0 | 26.3 | 10.5 | 12.5 | 20.0 | 0.680 | 51.5 | 8.1 | 0.000 |
| Levofloxacin | 75.0 | 100.0 | 68.4 | 18.8 | 55.4 | 0.000 | 75.8 | 51.5 | 0.016 |
| Moxifloxacin | 75.0 | 100.0 | 68.4 | 18.8 | 58.5 | 0.000 | 75.8 | 54.5 | 0.040 |
| Piperacillin-tazobactam | 0.0 | 0.0 | 5.3 | 6.3 | 0.0 | 0.131 | 0.0 | 2.0 | 1.000 |
| Cefoperazone-sulbactam | 30.0 | 26.3 | 5.3 | 18.8 | 10.8 | 0.099 | 3.0 | 18.2 | 0.042 |
| Ciprofloxacin | 75.0 | 100.0 | 68.4 | 18.8 | 55.4 | 0.000 | 72.7 | 57.6 | 0.150 |
| Amikacin | 0.0 | 0.0 | 10.5 | 0.0 | 6.2 | 0.400 | 3.0 | 4.0 | 1.000 |
P values of <0.01, indicating significant differences, are shown in boldface. —, no P value calculated, since the data did not meet the criteria of the Fisher exact t test.
For 7 subgroups of the CTX-M-1 and CTX-M-9 groups, P values were not calculated due to the small number of strains.
Table 2.
Resistance rates of ESBL-producing E. coli isolates by source and phylogenetic group
| Antimicrobial agent | Rate of resistance (%) by source (no. of isolates) |
P value for sourcesa | Rate of resistance (%) by phylogenetic group (no. of isolates) |
P value for phylogenetic groups | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Water (26) | Pig feces (31) | Hospitalized patients (36) | Healthy humans (46) | A (41) | B1 (11) | B2 (30) | D (57) | |||
| Ceftazidime | 34.6 | 16.1 | 47.2 | 6.5 | 0.000 | 17.1 | 36.4 | 30.0 | 24.6 | 0.437 |
| Cefotaxime | 100.0 | 100.0 | 100.0 | 100.0 | — | 100.0 | 100.0 | 100.0 | 100.0 | — |
| Ceftriaxone | 100.0 | 100.0 | 100.0 | 100.0 | — | 100.0 | 100.0 | 100.0 | 100.0 | — |
| Aztreonam | 69.2 | 61.3 | 78.4 | 19.6 | 0.000 | 56.1 | 90.9 | 60.0 | 42.1 | 0.018 |
| Cefoxitin | 19.2 | 0.0 | 11.1 | 0.0 | 0.001 | 2.4 | 0.0 | 0.0 | 14.0 | 0.036 |
| Cefepime | 19.2 | 9.7 | 47.2 | 4.3 | 0.000 | 12.2 | 9.1 | 23.3 | 24.6 | 0.367 |
| Levofloxacin | 76.9 | 48.4 | 88.9 | 37.0 | 0.000 | 53.7 | 36.4 | 73.3 | 64.9 | 0.115 |
| Moxifloxacin | 76.9 | 48.4 | 88.9 | 41.3 | 0.000 | 61.0 | 36.4 | 73.3 | 64.9 | 0.193 |
| Piperacillin-tazobactam | 0.0 | 0.0 | 6.3 | 0.0 | 0.148 | 2.4 | 0.0 | 0.0 | 1.8 | 1.000 |
| Cefoperazone-sulbactam | 15.4 | 3.2 | 41.7 | 4.3 | 0.000 | 4.9 | 9.1 | 26.7 | 19.3 | 0.051 |
| Ciprofloxacin | 76.9 | 48.4 | 88.9 | 41.3 | 0.000 | 58.5 | 27.3 | 73.3 | 64.9 | 0.059 |
| Amikacin | 0.0 | 12.9 | 5.6 | 0.0 | 0.023 | 7.3 | 0.0 | 0.0 | 5.3 | 0.524 |
P values of <0.01, indicating significant differences, are shown in boldface. —, no P value calculated, since the data did not meet the criteria of the Fisher exact t test.
DISCUSSION
ESBL-producing E. coli strains seem to be the emergent cause of urinary tract ailments and other serious infections of humans in different countries. Among E. coli clinical isolates, CTX-M types belonging to the CTX-M-1 and CTX-M-9 groups were the most prevalent. It is surprising that these two groups were also prevalent in E. coli strains isolated from the aquatic environment and swine feces. CTX-M-15 and CTX-M-14 were used to represent the epidemic CTX-M-1 and CTX-M-9 groups in humans in Asia, respectively (9). Nevertheless, among the 17 CTX-M-1-producing human isolates in the present study, 8 isolates harbored the blaCTX-M-55 gene, while only 6 isolates possessed the blaCTX-M-15 gene. blaCTX-M-55 was first discovered in community-onset ESBL-producing E. coli, hospital-acquired ESBL-producing E. coli, or hospital-acquired ESBL-producing Klebsiella pneumoniae infections in Thailand in 2007; this gene carried an Ala-77-Val substitution relative to blaCTX-M-15 and had reduced susceptibility to ceftazidime (31). Ewers et al. (32) had reported that blaCTX-M-15 was the epidemic gene type of the CTX-M-1 group in pigs. However, in our study, the 11 E. coli strains collected from swine feces that produced CTX-M-1 group enzymes all had blaCTX-M-55, which was reported to be the dominant gene type in companion animals in Asia (9). In consideration of the study of Leverstein-van Hall et al. reporting that Dutch patients, retail chicken meat, and poultry share the same ESBL genes, plasmids, and strains (33), the rising proportions of blaCTX-M-55 both in humans and in pigs in the present study suggested two possibilities. One is the possible transmission of the pathogen carrying the gene, or the gene itself, between humans and animals or the transmission of ESBL genes through the food chain. The other possibility is that the blaCTX-M-15 gene may mutate to blaCTX-M-55 by an Ala-77-Val substitution during the course of transmission. Meanwhile, a clinical isolate possessing blaCTX-M-123 was obtained in this study; this gene was first discovered in a pig feces isolate from Guangzhou, China, in 2012 and had never been reported in a human specimen. This finding also revealed the possibility of cross-transmission between humans and pigs.
In the current study, the prevalence of ESBL-producing E. coli isolates in swine feces was 32.0%, much higher than in previous studies (10.7% in 2007; 21.5% in 2008 to 2009) (16, 34). However, in E. coli strains isolated from the feces of healthy humans, the prevalence of ESBLs was 17.5%, much lower than that in swine feces. The dramatic increase in the prevalence of ESBLs in swine feces may be due to the increasing use of antibiotics in recent years. In China, pig feed is usually mixed with a small amount of amoxicillin and quinolones for growth promotion and prophylaxis. Long-term exposure to small doses of antibiotics might have caused the very high proportion of ESBL-producing Enterobacteriaceae isolated from the feces of sows and piglets. Thus, the control of antimicrobial resistance may require monitoring of antimicrobial use and surveillance of drug-resistant pathogens in the veterinary environment.
The proportions of isolates from hospitalized patients with phylogenetic subgroups A, B1, B2, and D in the current study were similar to those reported previously (35), while the proportions for the other three sources were slightly different, especially those for pigs. Subgroups B2 and D were the main phylogenetic groups for hospitalized patients, whereas subgroups A and B1 were the prominent groups for pig feces. The phylogenetic subtype distribution of the ESBLs in isolates from environmental water is intermediate between those for hospitalized patients and pigs, close to the average distribution of total isolates, suggesting that the pathogen in environmental water might originate from both humans and animals. The phylogenetic grouping results are in accordance with the study of Johnson and Stell (36), which demonstrated that isolates in phylogenetic groups A and B1 were generally commensal strains, while subgroups D and B2 were associated with extraintestinal infections. Although there is insufficient data to determine whether the CTX-M-type-producing E. coli strains found in healthy humans or hospitalized patients have acquired these genes from food animals via the food chain, the high prevalence of CTX-M-type ESBLs among commensal E. coli isolates strongly suggests a significant role for commensal E. coli strains as ESBL gene reservoirs, which poses an additional risk to humans. Therefore, monitoring of the spread of CTX-M-producing E. coli strains in foods is urgently needed for the protection of public health.
Multiple papers have reported that ST131 was the ESBL-producing E. coli sequence type most frequently associated with epidemics (32, 35, 37–39). In 2008, an MLST investigation revealed a pandemic clone, B2-O25b:H4-ST131 CTX-M-15, with high extraintestinal virulence, causing urinary tract infections, bacteremia, urinary sepsis, and neonatal sepsis (38, 39). Soon after the first discovery of human clinical isolates of E. coli producing ST131 ESBLs, they disseminated to various animal species, including poultry, cattle, pigs, wildlife, and companion animals (9). ST131 is also a major ST in the present study, but only one E. coli isolate, EC12-1, with B2-ST131 CTX-M-15 was collected, from the bile specimen of a 68-year-old woman with choledocholithiasis. The patient eventually recovered through rational treatment. Meanwhile, D-ST648, D-ST38, and A-CC10 are becoming the other most prevalent sequence types. E. coli D-ST648 strains have been reported to produce CTX-M-type ESBLs or NDM carbapenemase and have been isolated from humans or poultry in recent years (40–42). E. coli D-ST38 and A-CC10 strains have also been reported to produce CTX-M-type ESBLs in clinical patients (43, 44). Thus, in addition to the reported pandemic B2-O25b:H4-ST131 CTX-M-15 clone, attention should be paid to the rising of E. coli D-ST648, D-ST38, and A-ST10 strains.
PFGE indicated that there is diversity among the human, water, and swine strains (data not shown). Only limited similarity was found. Nevertheless, the main STs—ST131, ST648, ST38, and CC10–were identified in the E. coli strains isolated from the aquatic environment. Besides, some E. coli strains isolated from the aquatic environment were closely related to isolates collected from hospitalized patients. Additionally, E. coli isolates HD10-10 and XX3-3, both belonging to D-ST131, were obtained from two distinct water sources, whereas their resistance profiles, ESBL gene types, sequence types, and PFGE patterns were all the same. These two water sources were 12.8 km away from each other. One was located on the campus of Zhejiang University, and the other was located in a pond in a scenic spot. All these results suggest possible frequent and continuous interspecies transmission, and water may be an important vehicle for the spread of ESBL genes. However, more discriminative typing tools are necessary to obtain data to support this hypothesis.
Nonetheless, the sequence types of the ESBLs from swine feces were much more diversified than those of ESBLs in isolates from environmental water. Four of seven novel STs reported in the current study were from swine feces. Among the four main STs, E. coli isolates from swine feces belong only to CC10. However, 10 of 19 CC10 strains were from swine. Although only one isolate from a hospitalized patient was found to belong to CC10, the emergence of CTX-M-type-producing E. coli strains with A-CC10 should be tracked and monitored carefully to guard against a A-CC10 emergency in patients and even the general community.
The fact that E. coli strains with the same phylogenetic group and sequence type were distributed among various PFGE patterns indicated that the molecular evolution of epidemic CTX-M-producing E. coli occurred not as a recent emergence but by long-term interspecies cross-transmission. Furthermore, the CTX-M-type seems to have no relationship with the ST, since isolates with the four main STs had both CTX-M-1 and CTX-M-9 enzymes and were in different clusters. This result is somewhat different from those of a previous study (45).
With regard to antibiotic resistance, the rates of resistance of CTX-M-1 group isolates to ceftazidime, aztreonam, and cefepime were significantly higher than those of CTX-M-9 group isolates, probably due to the blaCTX-M-55 gene, the main gene that caused ceftazidime resistance (31). Not only clinical E. coli isolates but also E. coli strains collected from environmental water, pig feces, and healthy humans had high rates of resistance to fluoroquinolone. The high rate of resistance of E. coli strains from healthy humans to fluoroquinolone may be due to previous abuse of antibiotics. As with the high resistance rate of the isolates from pig feces, long-term small-dose feeding of antibiotics to animals may be the primary cause. This observation further supports the idea that the use of antimicrobial agents as growth promoters in animal food contributes to the initial selection of resistant organisms. The resistance rates of the hospitalized-patient group were significantly higher than those of the other three groups due to antibiotic use in clinical therapy, further proving this selection pressure hypothesis. Only 11 isolates harboring the aac(6′)-Ib-cr or qnrS gene were detected. In contrast to the high prevalences of the qnr and aac(6′)-Ib-cr genes in both waterborne environmental bacteria and clinical isolates of C. freundii reported previously (19), the proportions of these genes in E. coli isolates in the current study were much lower. This demonstrates that it may be much easier for C. freundii than for E. coli to carry qnr genes in the aquatic environment, and the most probable mechanism for the high rates of resistance to fluoroquinolone may be chromosome-mediated resistance, such as point mutations in the gyrase and topoisomerase IV genes (25). Otherwise, the ST648 group was 100% resistant to fluoroquinolone, yet the resistance rate of the ST38 group was only 18.8%. Although the ST38 and ST648 groups originated mainly from humans, hospitalized patients accounted for 36.8% (7/19) of isolates in the ST648 group compared with 18.8% (3/16) in the ST38 group. ST648 isolates possessing β-lactamases have been identified worldwide (40, 42, 46), suggesting that ST648 ESBLs may contribute to the spread of the β-lactamase gene and that these isolates may be more likely than others to cause extraintestinal infections, ultimately resulting in multidrug resistance.
To the best of our knowledge, this is the first report on molecular homology analysis of CTX-M-producing E. coli isolates collected from water, swine, and healthy and hospitalized humans. The study implied that these pathogens in the environment might originate from both humans and animals and can serve as a reservoir and medium for the spread of resistant pathogens.
Supplementary Material
Footnotes
Published ahead of print 26 July 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01740-13.
REFERENCES
- 1.Donnenberg M. 2002. Escherichia coli: virulence mechanisms of a versatile pathogen. Elsevier Science, San Diego, CA [Google Scholar]
- 2.Wang F, Zhu DM, Hu FP, Ruan FY, Ni YX, Sun JY, Xu YC, Zhang XJ, Hu YJ, Ai XM, Yu YS, Yang Q, Sun ZY, Li L, Jia B, Huang WX, Zhuo C, Su DH, Wei LH, Wu L, Zhang ZX, Ji P, Wang CQ, Xue JC, Zhang H, Li WH, Xu YH, Shen JL, Shan B, Du Y. 2010. CHINET 2009 surveillance of bacterial resistance in China. Chin. J. Infect. Chemother. 10:325–334 (In Chinese.) [Google Scholar]
- 3.Zhu DM, Wang F, Hu FP, Jiang XF, Ni YX, Sun JY, Xu YC, Zhang XJ, Hu YJ, Ai XM, Yu YS, Yang Q, Sun ZY, Chen ZJ, Jia B, Huang WX, Zhuo C, Su DH, Wei LH, Wu L, Zhang ZX, Ji P, Wang CQ, Wang AM, Zhang H, Kong J, Xu YH, Shen JL, Shan B, Du Y. 2011. CHINET 2010 surveillance of bacterial resistance in China. Chin. J. Infect. Chemother. 11:321–329 (In Chinese.) [Google Scholar]
- 4.Hu FP, Zhu DM, Wang F, Jiang XF, Yang Q, Xu YC, Zhang XJ, Sun ZY, Chen ZJ, Wang CQ, Wang AM, Ni YX, Sun JY, Yu YS, Lin J, Shan B, Du Y, Xu YH, Shen JL, Zhang H, Kong J, Zhuo C, Su DH, Zhang ZX, Ji P, Hu YJ, Ai XM, Huang WX, Jia B, Wei LH, Wu L. 2012. CHINET 2011 surveillance of bacterial resistance in China. Chin. J. Infect. Chemother. 12:321–329 (In Chinese.) [Google Scholar]
- 5.Canton R, Coque TM. 2006. The CTX-M beta-lactamase pandemic. Curr. Opin. Microbiol. 9:466–475 [DOI] [PubMed] [Google Scholar]
- 6.Bauernfeind A, Grimm H. 1990. A new plasmidic cefotaximase in a clinical isolate of Escherichia coli. Infection 18:294–298 [DOI] [PubMed] [Google Scholar]
- 7.Bonnet R. 2004. Growing group of extended-spectrum beta-lactamases: the CTX-M enzymes. Antimicrob. Agents Chemother. 48:1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tzouvelekis LS, Tzelepi E, Tassios PT, Legakis NJ. 2000. CTX-M-type beta-lactamases: an emerging group of extended-spectrum enzymes. Int. J. Antimicrob. Agents 14:137–142 [DOI] [PubMed] [Google Scholar]
- 9.Ewers C, Bethe A, Semmler T, Guenther S, Wieler LH. 2012. Extended-spectrum β-lactamase-producing and AmpC-producing Escherichia coli from livestock and companion animals, and their putative impact on public health: a global perspective. Clin. Microbiol. Infect. 18:646–655 [DOI] [PubMed] [Google Scholar]
- 10.Coque TM, Baquero F, Canton R. 20 November 2008. Increasing prevalence of ESBL-producing Enterobacteriaceae in Europe. Euro Surveill. 13(47):pii:19044 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19044 [PubMed] [Google Scholar]
- 11.Livermore DM, Canton R, Gniadkowski M, Nordmann P, Rossolini GM, Arlet G, Ayala J, Coque TM, Kern-Zdanowicz I, Luzzaro F, Poirel L, Woodford N. 2007. CTX-M: changing the face of ESBLs in Europe. J. Antimicrob. Chemother. 59:165–174 [DOI] [PubMed] [Google Scholar]
- 12.Bortolaia V, Guardabassi L, Trevisani M, Bisgaard M, Venturi L, Bojesen AM. 2010. High diversity of extended-spectrum beta-lactamases in Escherichia coli isolates from Italian broiler flocks. Antimicrob. Agents Chemother. 54:1623–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cloeckaert A, Praud K, Lefevre M, Doublet B, Pardos M, Granier SA, Brisabois A, Weill FX. 2010. IncI1 plasmid carrying extended-spectrum-beta-lactamase gene blaCTX-M-1 in Salmonella enterica isolates from poultry and humans in France, 2003 to 2008. Antimicrob. Agents Chemother. 54:4484–4486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Girlich D, Poirel L, Carattoli A, Kempf I, Lartigue MF, Bertini A, Nordmann P. 2007. Extended-spectrum beta-lactamase CTX-M-1 in Escherichia coli isolates from healthy poultry in France. Appl. Environ. Microbiol. 73:4681–4685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meunier D, Jouy E, Lazizzera C, Kobisch M, Madec JY. 2006. CTX-M-1- and CTX-M-15-type beta-lactamases in clinical Escherichia coli isolates recovered from food-producing animals in France. Int. J. Antimicrob. Agents 28:402–407 [DOI] [PubMed] [Google Scholar]
- 16.Tian GB, Wang HN, Zou LK, Tang JN, Zhao YW, Ye MY, Tang JY, Zhang Y, Zhang AY, Yang X, Xu CW, Fu YJ. 2009. Detection of CTX-M-15, CTX-M-22, and SHV-2 extended-spectrum beta-lactamases (ESBLs) in Escherichia coli fecal-sample isolates from pig farms in China. Foodborne Pathog. Dis. 6:297–304 [DOI] [PubMed] [Google Scholar]
- 17.Blanc V, Mesa R, Saco M, Lavilla S, Prats G, Miró E, Navarro F, Cortés P, Llagostera M. 2006. ESBL- and plasmidic class C beta-lactamase-producing E. coli strains isolated from poultry, pig and rabbit farms. Vet. Microbiol. 118:299–304 [DOI] [PubMed] [Google Scholar]
- 18.Niu XX, Xing HX, Yao SM. 2010. Effects of antibiotic on the human body. World Health Digest 7:15–16 (In Chinese.) [Google Scholar]
- 19.Zhang R, Ichijo T, Huang YL, Cai JC, Zhou HW, Yamaguchi N, Nasu M, Chen GX. 2012. High prevalence of qnr and aac(6′)-Ib-cr genes in both water-borne environmental bacteria and clinical isolates of Citrobacter freundii in China. Microbes Environ. 27:158–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nemoy LL, Kotetishvili M, Tigno J, Keefer-Norris A, Harris AD, Perencevich EN, Johnson JA, Torpey D, Sulakvelidze A, Morris JG, Stine OC. 2005. Multilocus sequence typing versus pulsed-field gel electrophoresis for characterization of extended-spectrum beta-lactamase-producing Escherichia coli isolates. J. Clin. Microbiol. 43:1776–1781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perez-Perez FJ, Hanson ND. 2002. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J. Clin. Microbiol. 40:2153–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu Y, Ji S, Chen Y, Zhou W, Wei Z, Li L, Ma Y. 2007. Resistance of strains producing extended-spectrum beta-lactamases and genotype distribution in China. J. Infect. 54:53–57 [DOI] [PubMed] [Google Scholar]
- 23.Robicsek A, Strahilevitz J, Jacoby GA, Macielag M, Abbanat D, Park CH, Bush K, Hooper DC. 2006. Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat. Med. 12:83–88 [DOI] [PubMed] [Google Scholar]
- 24.Wang M, Guo Q, Xu X, Wang X, Ye X, Wu S, Hooper DC. 2009. New plasmid-mediated quinolone resistance gene, qnrC, found in a clinical isolate of Proteus mirabilis. Antimicrob. Agents Chemother. 53:1892–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu YY, Cai JC, Zhang R, Zhou HW, Sun Q, Chen GX. 2012. Emergence of Proteus mirabilis harboring blaKPC-2 and qnrD in a Chinese hospital. Antimicrob. Agents Chemother. 56:2278–2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang Y, Zhou Z, Qian Y, Wei Z, Yu Y, Hu S, Li L. 2008. Plasmid-mediated quinolone resistance determinants qnr and aac(6′)-Ib-cr in extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in China. J. Antimicrob. Chemother. 61:1003–1006 [DOI] [PubMed] [Google Scholar]
- 27.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MC, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60:1136–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clermont O, Bonacorsi S, Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555–4558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maule J. 1998. Pulsed-field gel electrophoresis. Mol. Biotechnol. 9:107–126 [DOI] [PubMed] [Google Scholar]
- 30.Clinical and Laboratory Standards Institute 2012. Performance standards for antimicrobial susceptibility testing; 22nd informational supplement. Document M100-S22. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 31.Kiratisin P, Apisarnthanarak A, Saifon P, Laesripa C, Kitphati R, Mundy LM. 2007. The emergence of a novel ceftazidime-resistant CTX-M extended-spectrum beta-lactamase, CTX-M-55, in both community-onset and hospital-acquired infections in Thailand. Diagn. Microbiol. Infect. Dis. 58:349–355 [DOI] [PubMed] [Google Scholar]
- 32.Ewers C, Grobbel M, Stamm I, Kopp PA, Diehl I, Semmler T, Fruth A, Beutlich J, Guerra B, Wieler LH, Guenther S. 2010. Emergence of human pandemic O25:H4-ST131 CTX-M-15 extended-spectrum-beta-lactamase-producing Escherichia coli among companion animals. J. Antimicrob. Chemother. 65:651–660 [DOI] [PubMed] [Google Scholar]
- 33.Leverstein-van Hall MA, Dierikx CM, Cohen Stuart J, Voets GM, van den Munckhof MP, van Essen-Zandbergen A, Platteel T, Fluit AC, van de Sande-Bruinsma N, Scharinga J, Bonten MJ, Mevius DJ, National ESBL surveillance group 2011. Dutch patients, retail chicken meat and poultry share the same ESBL genes, plasmids and strains. Clin. Microbiol. Infect. 17:873–880 [DOI] [PubMed] [Google Scholar]
- 34.Tamang MD, Nam HM, Kim SR, Chae MH, Jang GC, Jung SC, Lim SK. 2013. Prevalence and molecular characterization of CTX-M β-lactamase-producing Escherichia coli isolated from healthy swine and cattle. Foodborne Pathog. Dis. 10:13–20 [DOI] [PubMed] [Google Scholar]
- 35.Lee MY, Choi HJ, Choi JY, Song M, Song Y, Kim SW, Chang HH, Jung SI, Kim YS, Ki HK, Son JS, Kwon KT, Heo ST, Yeom JS, Shin SY, Chung DR, Peck KR, Song JH, Ko KS. 2010. Dissemination of ST131 and ST393 community-onset, ciprofloxacin-resistant Escherichia coli clones causing urinary tract infections in Korea. J. Infect. 60:146–153 [DOI] [PubMed] [Google Scholar]
- 36.Johnson JR, Stell AL. 2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 181:261–272 [DOI] [PubMed] [Google Scholar]
- 37.Oteo J, Diestra K, Juan C, Bautista V, Novais A, Perez-Vazquez M, Moya B, Miro E, Coque TM, Oliver A, Canton R, Navarro F, Campos J. 2009. Extended-spectrum beta-lactamase-producing Escherichia coli in Spain belong to a large variety of multilocus sequence typing types, including ST10 complex/A, ST23 complex/A and ST131/B2. Int. J. Antimicrob. Agents 34:173–176 [DOI] [PubMed] [Google Scholar]
- 38.Nicolas-Chanoine MH, Blanco J, Leflon-Guibout V, Demarty R, Alonso MP, Canica MM, Park YJ, Lavigne JP, Pitout J, Johnson JR. 2008. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 61:273–281 [DOI] [PubMed] [Google Scholar]
- 39.Rogers BA, Sidjabat HE, Paterson DL. 2011. Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J. Antimicrob. Chemother. 66:1–14 [DOI] [PubMed] [Google Scholar]
- 40.Sidjabat HE, Paterson DL, Adams-Haduch JM, Ewan L, Pasculle AW, Muto CA, Tian GB, Doi Y. 2009. Molecular epidemiology of CTX-M-producing Escherichia coli isolates at a tertiary medical center in western Pennsylvania. Antimicrob. Agents Chemother. 53:4733–4739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mushtaq S, Irfan S, Sarma JB, Doumith M, Pike R, Pitout J, Livermore DM, Woodford N. 2011. Phylogenetic diversity of Escherichia coli strains producing NDM-type carbapenemases. J. Antimicrob. Chemother. 66:2002–2005 [DOI] [PubMed] [Google Scholar]
- 42.Cortes P, Blanc V, Mora A, Dahbi G, Blanco JE, Blanco M, Lopez Z, Andreu A, Navarro F, Alonso MP, Bou G, Blanco J, Llagostera M. 2010. Isolation and characterization of potentially pathogenic antimicrobial-resistant Escherichia coli strains from chicken and pig farms in Spain. Appl. Environ. Microbiol. 76:2799–2805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weissman SJ, Adler A, Qin X, Zerr DM. 2013. Emergence of extended-spectrum β-lactam resistance among Escherichia coli at a US academic children's hospital is clonal at the sequence type level for CTX-M-15, but not for CMY-2. Int. J. Antimicrob. Agents 41:414–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Izdebski R, Baraniak A, Fiett J, Adler A, Kazma M, Salomon J, Lawrence C, Rossini A, Salvia A, Vidal Samso J, Fierro J, Paul M, Lerman Y, Malhotra-Kumar S, Lammens C, Goossens H, Hryniewicz W, Brun-Buisson C, Carmeli Y, Gniadkowski M, MOSAR WP2 and WP5 Study Groups. 2013. Clonal structure, extended-spectrum β-lactamases, and acquired AmpC-type cephalosporinases of Escherichia coli populations colonizing patients in rehabilitation centers in four countries. Antimicrob. Agents Chemother. 57:309–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blanco M, Alonso MP, Nicolas-Chanoine MH, Dahbi G, Mora A, Blanco JE, Lopez C, Cortes P, Llagostera M, Leflon-Guibout V, Puentes B, Mamani R, Herrera A, Corira MA, Garcia-Garrote F, Pita JM, Blanco J. 2009. Molecular epidemiology of Escherichia coli producing extended-spectrum β-lactamases in Lugo (Spain): dissemination of clone O25b:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 63:1135–1141 [DOI] [PubMed] [Google Scholar]
- 46.Guenther S, Aschenbrenner K, Stamm I, Bethe A, Semmler T, Stubbe A, Stubbe M, Batsajkhan N, Glupczynski Y, Wieler LH, Ewers C. 2012. Comparable high rates of extended-spectrum-beta-lactamase-producing Escherichia coli in birds of prey from Germany and Mongolia. PLoS One 7:e53039. 10.1371/journal.pone.0053039 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.