Abstract
Clostridium thermocellum encodes a cellulosomal, modular, and thermostable serine protease inhibitor (serpin), PinA. PinA stability but not inhibitory activity is affected by the Fn(III) and Doc(I) domains, and PinA is a broad inhibitor of subtilisin-like proteases and may play a key role in protecting the cellulosome from protease attack.
TEXT
Much of the organic carbon and nitrogen in soil is sequestered in cellulosic and protein biomass, respectively, and microorganisms have consequently evolved specialized strategies to facilitate the release and assimilation of these biomaterials. Clostridium thermocellum is an anaerobic, thermophilic soil bacterium that produces a large extracellular multisubunit protein complex termed the cellulosome that functions to deliver organic carbon to the cell. The extracellular location of the cellulosome leaves it vulnerable to protease attack, and bacterial and fungal cellulosomes have been shown to carry specific protease inhibitors, including serpins and cyspins, that can confer protection against protease attack (1–4). Serpins function as metastable suicide substrates for their cognate protease(s) with cleavage of the reactive center loop resulting in the formation of an inactive covalently linked serpin-protease complex. Clostridium thermocellum ATCC 27405 produces a cellulosomally targeted modular thermostable serpin termed PinA (2) (GenBank accession number YP_001036624); however, the contribution of the modular domains to serpin functionality remains unknown, as does the potential broader ecological role of PinA.
To address these issues, we expressed a nearly full-length recombinant protein lacking the N-terminal signal sequence (rPinA75) and a truncated protein lacking the N-terminal signal sequence, Fn(III)-like, and Doc(I) domains (rPinA614) (Fig. 1). The stoichiometry of inhibition (SI) of rPinA75 (1.3 ± 0.1) against subtilisin type VIII from Bacillus licheniformis was similar to that previously reported (2), indicating that the N-terminal signal sequence does not affect inhibitory activity. Similarly, the SI for rPinA614 (1.4 ± 0.3; Student's independent t test, P > 0.05) was not significantly different from that for rPinA75, indicating that the Fn(III)-like and Doc(I) domains do not contribute to inhibitory activity.
Fig 1.
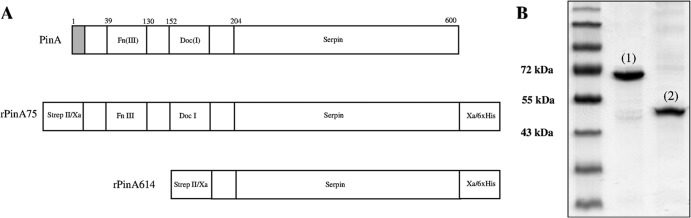
(A) Schematic representation of the modular structure of PinA with the rPinA75 and the rPinA614 constructs as indicated. Briefly, pinA was PCR amplified from pET21-SK2 (2) and cloned into pET-28c(+) (Novagen), placing the gene under the control of the vector-borne T7 promoter and facilitating the fusion of a C-terminal 6×His tag. The final rPinA75- and rPinA614-expressing plasmid constructs were confirmed by Sanger sequencing. The PCR primers used facilitated the addition of an N-terminally located Strep II tag and a factor Xa cleavage site to the serpin and a C-terminally located factor Xa cleavage site. The signal sequence (gray), Strep II tag/factor Xa site (Strep II/Xa), factor Xa site/6×His tag (Xa/6XHis), fibronectin III (Fn III), dockerin (Doc I), and serpin regions are indicated. (B) SDS-PAGE analysis of rPinA75 (1) and rPinA614 (2).
Serpins fold to a metastable state, and we next examined the effect of the N-terminal truncation on the conformational stability and functionality of rPinA. Protein samples were harvested in a longitudinal manner, and native PAGE analysis revealed that two rPinA614 species, rPinA614 and rPinA614-L, as identified by liquid chromatography electrospray ionization tandem mass spectrometry (LC-EIS MS/MS), were evident after 5 h with a gradual accumulation up to 24 h where each species accounted for ∼50% of the total recombinant serpin produced (Fig. 2A). We observed a single species for rPinA75 over 24 h as determined by native PAGE; we thus hypothesized that the PinA614-L species represented a cleaved or latent state, and we sought to discriminate between these two possibilities as previously outlined by Dafforn et al. (5). First, samples containing both rPinA614 and rPinA614-L were analyzed by SDS-PAGE and revealed the presence of a single serpin band at the time points examined, suggesting that the two species had similar mobilities and that rPinA614-L was therefore uncleaved (Fig. 2B). Serpins in the latent conformation are more thermostable than those in the metastable conformation, and we next investigated the thermal stability of rPinA614 harvested at 9 h to allow discrimination between the two forms. Native PAGE indicated that rPinA614 underwent thermal denaturation at 70°C over a period of 180 min, resulting in the accumulation of rPinA614-L (Fig. 2C). Subsequent SDS-PAGE analysis indicated that these differences in mobility were not due to protein cleavage and/or degradation. Finally, the SI of rPinA614 harvested at 24 h was significantly different (2.5 ± 0.3; P < 0.01), further suggesting that rPinA614-L was in a noninhibitory latent conformation. Taken together, these results suggest that the truncation of the Fn(III)-like and Doc(I) domains is not necessary for rPinA614 to fold to the metastable state but it may leave the serpin domain prone to assuming a latent conformation. Thus, the N-terminal region of PinA may function in a manner analogous to that of the N-terminal domain of the thermostable serpin tengpin from Thermoanaerobacter tengcongensis, where an N-terminal domain that is not required for folding to the metastable state functions to stabilize the serpin and maintain it in that conformation (6).
Fig 2.

(A) Native PAGE analysis of recombinant rPinA614 harvested at 4, 5, 6, 7, 8, 9, and 24 h. The metastable (rPinA614) and latent (rPinA614-L) forms are indicated. The identity of rPinA614 and rPinA614-L from 5 to 24 h was confirmed by LC-EIS MS/MS. (B) SDS-PAGE analysis of recombinant rPinA614 harvested at 5, 6, 7, 8, 9, and 24 h. (C) Native PAGE analysis of the thermostability profile of rPinA614 heated at 70°C and harvested as indicated. The identity of rPinA614 and rPinA614-L at all time points was confirmed by LC-EIS MS/MS with the exception of rPinA614-L at 60 min. (D) Thermostability analysis of rPinA75 and rPinA614. The thermal stability of rPinA was determined essentially as described by Kang et al. (2) except that the reactions were performed at 60°C. The thermostability analyses were independently performed at least twice per sample.
We next sought to determine whether the modular nature of PinA contributed to its thermostability. As expected, the thermostability profile of rPinA75 at 60°C was similar to that previously reported by Kang et al. (2), with a calculated half-life for the serpin of approximately 180 min (Fig. 2D). Surprisingly, deletion of the Fn(III)-like and Doc(I) domains enhanced the thermostability profile of rPinA at 60°C with no significant loss in activity observed over 180 min. These results are consistent with previous reports that revealed that protein thermostability can be influenced by interdomain interactions and that truncated protein domains can exhibit enhanced thermostability (7, 8). Native PAGE did not reveal any change to the migration profile of either rPinA75 or rPinA614 over the 180 min (data not shown), suggesting that the overall protein conformations were unaffected following incubation at 60°C. Thermostable multidomain proteins can undergo cooperative unfolding during thermal denaturation (9–11), and the Fn(III)-like and Doc(I) domains may affect the thermostability of the reactive center loop in rPinA75, although the overall protein structure remains unaffected. Thus, while the N-terminal domains of PinA may stabilize the protein in the metastable state, they may also increase its thermolability.
Subtilisin-like proteases are among the most diverse and prevalent proteases found in soil environments, where they have been proposed to play a critical role in the cycling of organic nitrogen (12–14), and we finally investigated the ability of rPinA to function as a general inhibitor of MEROPS family S8, subfamily A proteases. rPinA75 effectively inhibited Savinase (1.2 ± 0.2) and Esperase (1.1 ± 0.1) produced by Bacillus clausii and Bacillus halodurans, respectively (15) (liquid enzyme formulations available from Sigma-Aldrich Corp., St. Louis, MO), and subtilisin type XXIV (1.3 ± 0.3) produced by B. licheniformis (Sigma-Aldrich Corp., St. Louis, MO), and the SI was not significantly different from that for subtilisin type VIII (P > 0.05). In addition, rPinA75 was capable of inhibiting proteinase K from Tritirachium album Limber (1.3 ± 0.2; P > 0.05), although it is more distantly related to subtilisin (16, 17). The ability of PinA to function as a broad inhibitor of MEROPS family S8, subfamily A proteases is of particular interest, as these proteases show significant sequence variability (76%, 70%, and 40% similarity of Savinase, Esperase, and proteinase K, respectively, with subtilisin type VIII; no sequence available for subtilisin type XXIV); however, the protease active site and substrate recognition site are highly conserved, which likely explains the ability of rPinA to effectively inhibit these proteases. Furthermore, specific serpin domains termed exosites also contribute to serpin specificity (reviewed in reference 18) and, as subtilisin-like proteases are characterized by a common tertiary structure (see, e.g., references 19 and 20), it is possible that specific PinA exosites also contribute to the inhibition process.
Our results suggest that PinA functions to protect the cellulosome from attack by MEROPS family S8, subfamily A proteases. C. thermocellum ATCC 27405 encodes a second putative cellulosomal serpin, PinB (2) (GenBank accession number YP_001036625); though its cognate protease has not yet been identified, PinB may also function to protect the cellulosome from attack, and this remains to be examined. Separately, while the role of the cellulosome vis-à-vis carbon has been well described, little has been postulated regarding the potential role of the cellulosome in nitrogen acquisition. The genome sequences of several C. thermocellum strains have revealed the presence of putative cellulosomally targeted subtilisin-like proteases, indicating that the cellulosome may also function to provide organic nitrogen to the cell. C. thermocellum is capable of synthesizing all 20 amino acids (21, 22); however, it also encodes numerous putative peptidases and an oligopeptide transport system, suggesting that it can acquire exogenous amino acids and/or peptides. In this context, the tethering of a protease to the cellulosome may serve to prevent diffusion away from the cell and ensure that any organic nitrogen produced will be located in a position proximal to the host cell and will not be readily available to competing microorganisms. As production of the cellulosome is metabolically demanding, future studies should examine the role of cellulosomal proteases and protease inhibitors in relation to nitrogen bioavailability and their contribution to the ecological fitness of the host.
ACKNOWLEDGMENTS
We gratefully acknowledge the support provided by a CSIRO Science Leader award to M.M.
We thank colleagues for providing critical reading and suggestions to improve the manuscript.
Footnotes
Published ahead of print 19 July 2013
REFERENCES
- 1.Meguro H, Morisaka H, Kuroda K, Miyake H, Tamaru Y, Ueda M. 2011. Putative role of cellulosomal protease inhibitors in Clostridium cellulovorans based on gene expression and measurement of activities. J. Bacteriol. 193:5527–5530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kang S, Barak Y, Lamed R, Bayer EA, Morrison M. 2006. The functional repertoire of prokaryote cellulosomes includes the serpin superfamily of serine proteinase inhibitors. Mol. Microbiol. 60:1344–1354 [DOI] [PubMed] [Google Scholar]
- 3.Fendri I, Tardif C, Fierobe H-P, Lignon S, Valette O, Pagès S, Perret S. 2009. The cellulosomes from Clostridium cellulolyticum. FEBS J. 276:3076–3086 [DOI] [PubMed] [Google Scholar]
- 4.Steenbakkers PJM, Irving JA, Harhangi HR, Swinkels WJC, Akhmanova A, Dijkerman R, Jetten MSM, van der Drift C, Whisstock JC, Op den Camp HJM. 2008. A serpin in the cellulosome of the anaerobic fungus Piromyces sp. strain E2. Mycol. Res. 112:999–1006 [DOI] [PubMed] [Google Scholar]
- 5.Dafforn TR, Pike RN, Bottomley SP. 2004. Physical characterization of serpin conformations. Methods 32:150–158 [DOI] [PubMed] [Google Scholar]
- 6.Zhang Q, Buckle AM, Law RHP, Pearce MC, Cabrita LD, Lloyd GJ, Irving JA, Smith AI, Ruzyla K, Rossjohn J, Bottomley SP, Whisstock JC. 2007. The N terminus of the serpin, tengpin, functions to trap the metastable native state. EMBO Rep. 8:658–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kataeva IA, Seidel RD, Shah IIIA, West LT, Li X-L, Ljungdahl LG. 2002. The fibronectin type 3-like repeat from the Clostridium thermocellum cellobiohydrolase CbhA promotes hydrolysis of cellulose by modifying its surface. Appl. Environ. Microbiol. 68:4292–4300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kataeva IA, Blum DL, Li X-L, Ljungdahl LG. 2001. Do domain interactions of glycosyl hydrolases from Clostridium thermocellum contribute to protein thermostability? Protein Eng. 14:167–172 [DOI] [PubMed] [Google Scholar]
- 9.Kataeva IA, Brewer JM, Uversky VN, Ljungdahl LG. 2005. Domain coupling in a multimodular cellobiohydrolase CbhA from Clostridium thermocellum. FEBS Lett. 579:4367–4373 [DOI] [PubMed] [Google Scholar]
- 10.Kataeva IA, Uversky VN, Brewer JM, Schubot F, Rose JP, Wang BC, Ljungdahl LG. 2004. Interactions between immunoglobulin-like and catalytic modules in Clostridium thermocellum cellulosomal cellobiohydrolase CbhA. Protein Eng. Des. Sel. 17:759–769 [DOI] [PubMed] [Google Scholar]
- 11.Wassenberg D, Schurig H, Liebl W, Jaenicke R. 1997. Xylanase XynA from the hyperthermophilic bacterium Thermotoga maritima: structure and stability of the recombinant enzyme and its isolated cellulose-binding domain. Protein Sci. 6:1718–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mrkonjic Fuka M, Engel M, Gattinger A, Bausenwein U, Sommer M, Munch JC, Schloter M. 2008. Factors influencing variability of proteolytic genes and activities in arable soils. Soil Biol. Biochem. 40:1646–1653 [Google Scholar]
- 13.Gabor E, Niehaus F, Aehle W, Eck J. 2012. Zooming in on metagenomics: molecular microdiversity of subtilisin Carlsberg in soil. J. Mol. Biol. 418:16–20 [DOI] [PubMed] [Google Scholar]
- 14.Kaiser C, Koranda M, Kitzler B, Fuchslueger L, Schnecker J, Schweiger P, Rasche F, Zechmeister-Boltenstern S, Sessitsch A, Richter A. 2010. Belowground carbon allocation by trees drives seasonal patterns of extracellular enzyme activities by altering microbial community composition in a beech forest soil. New Phytol. 187:843–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maurer KH. 2004. Detergent proteases. Curr. Opin. Biotechnol. 15:330–334 [DOI] [PubMed] [Google Scholar]
- 16.Jany K-D, Lederer G, Mayer B. 1986. Amino acid sequence of proteinase K from the mold Tritirachium album Limber: Proteinase K—a subtilisin-related enzyme with disulfide bonds. FEBS Lett. 199:139–144 [Google Scholar]
- 17.Gunkel FA, Gassen HG. 1989. Proteinase K from Tritirachium album Limber. Characterization of the chromosomal gene and expression of the cDNA in Escherichia coli. Eur. J. Biochem. 179:185–194 [DOI] [PubMed] [Google Scholar]
- 18.Gettins PG, Olson ST. 2009. Exosite determinants of serpin specificity. J. Biol. Chem. 284:20441–20445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Betzel C, Klupsch S, Branner S, Wilson KS. 1996. Crystal structures of the alkaline proteases savinase and esperase from Bacillus lentus. Adv. Exp. Med. Biol. 379:49–61 [DOI] [PubMed] [Google Scholar]
- 20.Pähler A, Banerjee A, Dattagupta JK, Fujiwara T, Lindner K, Pal GP, Suck D, Weber G, Saenger W. 1984. Three-dimensional structure of fungal proteinase K reveals similarity to bacterial subtilisin. EMBO J. 3:1311–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roberts SB, Gowen CM, Brooks JP, Fong SS. 2010. Genome-scale metabolic analysis of Clostridium thermocellum for bioethanol production. BMC Syst. Biol. 4:31. 10.1186/1752-0509-4-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson EA, Madia A, Demain AL. 1981. Chemically defined minimal medium for growth of the anaerobic cellulolytic thermophile Clostridium thermocellum. Appl. Environ. Microbiol. 41:1060–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]


