Abstract
Chromatin remodelling events play an important role in the secondary metabolism of filamentous fungi. Previously, we showed that a bacterium, Streptomyces rapamycinicus, is able to reprogram the histone-modifying Spt-Ada-Gcn5-acetyltransferase/ADA (SAGA/ADA) complex of the model fungus Aspergillus nidulans. Consequently, the histone H3 amino acids lysine 9 and lysine 14 at distinct secondary metabolism genes were specifically acetylated during the bacterial fungal interaction, which, furthermore, was associated with the activation of the otherwise silent orsellinic acid gene cluster. To investigate the importance of the histone modifications for distinct gene expression profiles in fungal secondary metabolism, we exchanged several amino acids of histone H3 of A. nidulans. These amino acids included lysine residues 9, 14, 18, and 23 as well as serine 10 and threonine 11. Lysine residues were replaced by arginine or glutamine residues, and serine/threonine residues were replaced by alanine. All generated mutant strains were viable, allowing direct analysis of the consequences of missing posttranslational histone modifications. In the mutant strains, major changes in the expression patterns at both the transcriptional and metabolite levels of the penicillin, sterigmatocystin, and orsellinic acid biosynthesis gene clusters were detected. These effects were due mainly to the substitution of the acetylatable lysine 14 of histone H3 and were enhanced in a lysine 14/lysine 9 double mutant of histone H3. Taken together, our findings show a causal linkage between the acetylation of lysine residue 14 of histone H3 and the transcription and product formation of secondary metabolite gene clusters.
INTRODUCTION
Filamentous fungi have an enormous potential to produce multifaceted secondary metabolites. These low-molecular-weight molecules have been suggested to play important roles in the interaction of microbes (1). Aside from their ecological relevance, many fungal secondary metabolites have been shown to be of paramount importance for human health (1). These include the antibiotic penicillin and the cholesterol-lowering compound lovastatin. However, some of these compounds, like ochratoxin and aflatoxin, were classified as mycotoxins because of their detrimental effects on humans (1).
Most biosynthesis enzymes for fungal secondary metabolites are encoded by genes that are located in clusters. In an effort to manipulate the expression of these gene clusters and to understand the mechanisms that underlie their regulation and the spatial and temporal distribution of the generated molecules, complex regulatory processes have been discovered (1, 2). In about one-half of the gene clusters, pathway-specific transcription factor-encoding genes are located within or directly adjacent to the biosynthesis gene clusters. They govern the expression of one or more gene clusters (3–5). In addition, globally acting transcription factors, often observed as being involved in the regulation of central metabolic processes, have been shown to control the expression of gene clusters (1, 2).
Studies in recent years indicated that chromatin-modifying enzymes play a major role in the regulatory circuits of fungal secondary metabolism. Chromatin is defined as the complex of DNA and associated proteins inside the eukaryotic nucleus. Its basic units are nucleosomes, consisting of an octamer of histone proteins wrapped almost twice by the DNA double helix. The chromatin condensation state determines the accessibility of the DNA for all kinds of proteins interacting with the DNA. Histone modifications such as acetylation, methylation, and phosphorylation potentially provoke chromatin rearrangements and thus influence the ability of enzymes to access DNA (6). Furthermore, histone-modifying enzymes can establish a pattern of modifications giving characteristic marks to distinct chromosomal regions. These marks can attract or prevent binding of further enzymes (6, 7).
For filamentous fungi, it has been shown that histone acetyl- and methyltransferases as well as histone-binding proteins are essential for the expression of secondary metabolite biosynthesis gene clusters (7–9). These findings were underlined by the detection of distinct acetylation and methylation patterns of histone amino acids during secondary metabolite production phases. In agreement with a function of these patterns, genetic engineering of fungal strains impaired in proper chromatin modification led to both the activation of otherwise silent secondary metabolite biosynthesis gene clusters and the silencing of active gene clusters (10, 11).
Previously, we showed that the Spt-Ada-Gcn5-acetyltransferase/ADA (SAGA/ADA) complex containing the GcnE histone acetyltransferase and AdaB is required for the activation of several secondary metabolite gene clusters in the filamentous fungus Aspergillus nidulans. In particular, we revealed that the SAGA/ADA complex is essential for the induction of the orsellinic acid gene cluster during coincubation of A. nidulans with the bacterium Streptomyces rapamycinicus (8). The acetylation level of the histone H3 amino acids lysine 9 and lysine 14 in promoters of member genes of several secondary metabolite biosynthesis gene clusters increased during their expression. This increase of acetylation depended on the GcnE histone acetyltransferase. The SAGA/ADA complex is highly conserved throughout the eukaryotes and consists of ∼20 proteins (12). In addition to its acetylation activity, this complex is involved in the recruitment of the general transcription machinery to genes and the deubiquitination of histones. It is also linked to mRNA export and physically interacts with the 19S regulatory particle, as was shown for Saccharomyces cerevisiae (13–15).
In all studies on the impact of histone modification on secondary metabolism, direct evidence for the importance of single amino acids of histone H3 was lacking. Therefore, here we investigated the direct impact of histone H3 amino acids on secondary metabolism of A. nidulans. We generated A. nidulans strains with altered amino acid sequences of histone H3. The mutant strains were viable and allowed to investigate in detail the effect of amino acid exchange on secondary metabolism.
MATERIALS AND METHODS
Bacterial and fungal strains.
Bacterial and fungal strains are listed in Table 1. The histone H3 substitution strains were obtained by transformation of A. nidulans A1153 with a linear DNA fragment. The DNA fragment carried the histone H3 gene (ANIA_00733) harboring the respective substitutions, flanked by two 1.5-kb sequences homologous to the upstream and downstream regions of the histone H3 gene and, as a selectable marker, the argB gene of A. nidulans (Fig. 1B). The reference strain for this study was obtained by transformation of A. nidulans A1153 with the above-mentioned DNA fragment containing the wild-type histone H3 gene without amino acid substitutions. The DNA constructs were amplified by fusion PCR (16, 17). The oligonucleotides used are listed in Table 2.
Table 1.
List of strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| Aspergillus nidulans | ||
| A1153 | yA 1 pabaA1 argB2 pyroA4 nkuA::bar | Nayak et al. (37) |
| A1153H3 | yA1 pabaA1 pyroA4 nkuA::bar h3-argB2 | This study |
| A1153H3K9R | yA1 pabaA1 pyroA4 nkuA::bar h3-K9R-argB2 | This study |
| A1153H3K14R | yA1 pabaA1 pyroA4 nkuA::bar h3-K14R-argB2 | This study |
| A1153H3K14Q | yA1 pabaA1 pyroA4 nkuA::bar h3-K14Q-argB2 | This study |
| A1153H3K9/14R | yA1 pabaA1 pyroA4 nkuA::bar h3-K9RK14R-argB2 | This study |
| A1153H3K18R | yA1 pabaA1 pyroA4 nkuA::bar h3-K18R-argB2 | This study |
| A1153H3K23R | yA1 pabaA1 pyroA4 nkuA::bar h3-K23R-argB2 | This study |
| A1153H3S10A | yA1 pabaA1 pyroA4 nkuA::bar h3-S10A-argB2 | This study |
| A1153H3T11A | yA1 pabaA1 pyroA4 nkuA::bar h3-T11A-argB2 | This study |
| Bacillus calidolactis C953 | Brakhage et al. (21) | |
| Streptomyces rapamycinicus | Kumar and Goodfellow (38) |
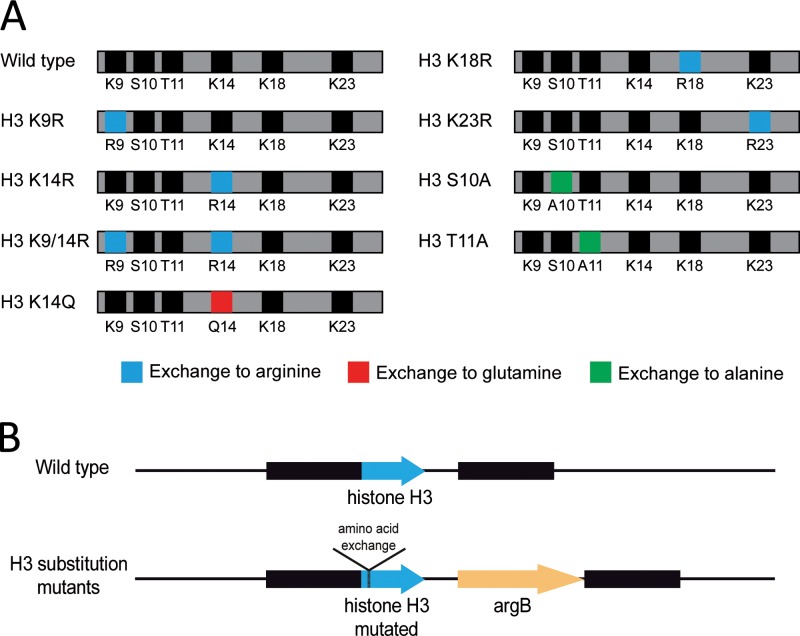
Fig 1.
Histone H3 substitution strains of A. nidulans. (A) Schematic overview of the amino acid substitutions present in histone H3 mutant proteins. Indicated in gray is the histone H3 protein; amino acid residues marked in black were not replaced. The exchange of amino acids to arginine, glutamine, and alanine is marked in blue, red, and green, respectively. (B) Generation of histone H3 substitution strains of A. nidulans. Schematic representation of the genomic H3 locus of wild-type and mutant strains. Arrows mark genes, and the dashed line indicates the region of H3 in which substitutions were introduced. Black boxes mark ∼1,500-bp-long regions serving for homologous recombination.
Table 2.
List of primers used in this study
| Name | Sequence (5′–3′) |
|---|---|
| For generation of constructs for transformation | |
| H3T11→A11rev | CTTGCCACCGGCAGACTTGCCTG |
| H3T11→A11for | CAGGCAAGTCTGCCGGTGGCAAG |
| H3K14→R14rev | ACGGGGAGCACGGCCACCAGT |
| H3K14→R14for | ACTGGTGGCCGTGCTCCCCGT |
| H3K14→Q14rev | ACGGGGAGCCTGGCCACCAGT |
| H3K14→Q14for | ACTGGTGGCCAGGCTCCCCGT |
| H3K18→R18rev | CGCGAGCTGACGACGGGGAGC |
| H3K18→R18for | GCTCCCCGTCGTCAGCTCGCG |
| H3K23→R23rev | ACGGGCAGCACGGGACGCGAG |
| H3K23→R23for | CTCGCGTCCCGTGCTGCCCGT |
| H3K9→R9rev | CACCAGTAGAACGGCCTGTTGG |
| H3K9→R9for | CCAACAGGCCGTTCTACTGGTG |
| H3S10→A10rev | TGCCACCAGTGGCCTTGCCTGT |
| H3S10→A10for | ACAGGCAAGGCCACTGGTGGCA |
| H3lbrevarg | GCAAACAGAACTTTGACTCCCATCAGGCAGAACTTATGCTAAGC |
| H3rbforarg | ATTGACATGTTCTCTCGCTTCCTGTGCTGTTTCGTTCCTGCTA |
| H3lbfor | CGAGCATTACTAGGAGCAAGG |
| H3rbrevQ | CTCACACGGCATCTAAGCTG |
| H3lbforQ | GTATTCATATGACCGCGTCTC |
| H3rbrev | GAGTCAAAGCTGGGTTCCAG |
| qRT-PCR | |
| Qacnfwd | CACCCTTGTTCTTGTTTTGCTC |
| Qacnrev | AAGTTCGCTTTGGCAACGC |
| qaflRfwd | GCGCGAAGAAGACTTCAAC |
| qaflRrev | TGCAATAACTGCCGACGAC |
| qstcOfwd | ATGTGTGAGATTGCCAGCC |
| qstcOrev | TAGCAAGAACCCAGCCAAC |
| qstcAfwd | AACGCGCCCAAAATGAAAG |
| qstcArev | CAAACCACATCCCACCCATC |
| Qaatafwd | TATCCCAACCAGAACCAGCC |
| Qaatarev | ACTGAAACTTGTCGTTCACCC |
| Qipnafwd | TGGCACATCTCACAAACAAC |
| Qipnarev | TTGACGAAGAATGGCAGGG |
| DNA sequencing | |
| pH3seqfor | CTTATGGTGTGCTGGTTTCCG |
| TH3seqrev | CACTGCATATTCGATCATGGAC |
| H3seqfor | CAAAGGAAAGGGCGCTAAGAC |
| H3seqrev | CAGAACTTTGCGTCAAGCACAG |
Media and cultivation of strains.
Standard cultivation of A. nidulans strains was performed in Aspergillus minimal medium (AMM) at 37°C and 200 rpm (18). Required supplements were added as follows: arginine (50 μM), p-aminobenzoic acid (3 μg ml−1), or pyridoxine HCl (5 μg ml−1). AMM was inoculated with 2 × 108 conidia ml−1. For production of orsellinic acid, A. nidulans was precultivated overnight in AMM and transferred into fresh medium inoculated with S. rapamycinicus, as described by Schroeckh et al. (19). Samples for RNA extraction of mycelia for expression analysis of the orsellinic acid gene cluster and sterigmatocystin gene cluster were taken after 3 h and 36 h, respectively. Samples for high-pressure liquid chromatography (HPLC) analysis of orsellinic acid production were taken after 24 h and for sterigmatocystin production after 36 h and 48 h. For the analysis of penicillin production, RNAs of mycelia and preparation of supernatants for compound isolation were obtained from A. nidulans cultures grown in fermentation media as previously described (20). Samples for RNA extraction were taken after 12 h of cultivation and for the penicillin bioassay after 36 h.
Penicillin assay.
Penicillin bioassays were essentially performed as previously described (21). Bacillus calidolactis C953 was used as an indicator organism. The penicillin titer was correlated to the dry weight of the biomass.
qRT-PCR.
To quantify transcript levels by quantitative real-time (qRT)-PCR, total RNA was purified with the RiboPure-Yeast kit (Applied Biosystems) according to the manufacturer's instructions. cDNA synthesis was performed with 15 μg of DNase I-treated RNA for 3 h at 46°C using Superscript III reverse transcriptase (Invitrogen). qRT-PCR was carried out on an Applied Biosystems StepOnePlus Real-Time PCR system in triplicate for each sample. The A. nidulans β-actin gene ANIA_06542 was used as an internal standard for calculation of expression levels. MyTaq HS mix 2× (Bioline) in combination with EvaGreen (Biotium) was used for cDNA amplification with gene-specific primers (Table 2). The cycling parameters included an initial DNA denaturation step at 95°C for 2 min, followed by 45 cycles with DNA denaturation at 95°C for 5 s and primer annealing and extension at 62°C for 15 s. qRT-PCR results were analyzed as described previously (19).
Preparation of chromosomal DNA, Southern blot analysis, and DNA sequencing.
Genomic DNA from A. nidulans mycelia was isolated using the MasterPure Yeast DNA purification kit (Epicentre Biotechnologies) according to a modified isolation protocol (22). Fungal mycelium was disrupted by zirconia beads (Ambion) using FastPrep-24 (MP Biomedicals) for 30 s with a rotation speed of 6,500 rpm. Southern blot analysis was carried out by using a nonradioactively labeled DNA probe as previously described (19). Primers for amplification of probes are listed in Table 2. For DNA sequence analysis, the histone H3 gene of each strain was PCR amplified and the purified DNA products were sequenced by the company MWG-Biotech (Ebersberg, Germany). Two sequencing reactions with primers upstream and downstream of the H3 gene were carried out for each strain. The primers used are listed in Table 2.
Extraction of compounds and HPLC analysis.
Culture broth and fungal mycelia were extracted with ethyl acetate (2 × 100 ml), dried with sodium sulfate, and concentrated under reduced pressure. Prior to extraction, the cultures were homogenized using an Ultra-Turrax batch disperser to blend mycelia. For HPLC analysis, the dry extracts were dissolved in 2 ml of methanol. Analytical HPLC was performed using a Shimadzu LC-10Avp series HPLC system consisting of an autosampler, high-pressure pumps, column oven, and photo diode array (PDA) detector. HPLC conditions were as follows: C18 column, Eurospher 100-5, 250 × 4.6 mm; gradient elution, acetonitrile (MeCN)–0.1% (vol/vol) trifluoroacetic acid (TFA) (H2O), 0.5:99.5 in 30 min, to MeCN–0.1% (vol/vol) TFA, 100:0, and 100% MeCN (vol/vol) for 10 min; flow rate, 1 ml min−1; injection volume, 50 μl. Compounds were identified by comparison with an authentic standard and quantified by integrating the peak area using Shimadzu Class-VP software (version 6.14 SP1).
RESULTS
A. nidulans mutants with substituted lysine and serine/threonine residues of histone H3.
Previously, we have shown that the histone-acetylating complex SAGA/ADA is required for the activation of secondary metabolite biosynthesis gene clusters such as penicillin, sterigmatocystin, terrequinone A, and orsellinic acid in A. nidulans (8). Our data indicated that the complex is targeted to these gene clusters and, furthermore, that the acetylation of the lysine residues 9 and 14 at the N terminus of histone H3 coincides with the activation of the ors gene cluster (8). To prove the importance of the individual amino acids and thus of their modification for secondary metabolism, we generated A. nidulans strains with substitutions at these positions (9 and 14) in the histone H3 protein. Furthermore, the amino acids lysine 18 and lysine 23 as well as serine 10 and threonine 11 of histone H3 were also chosen as additional targets for amino acid substitutions. Lysine 18 and 23 were previously reported to be acetylated by the SAGA/ADA complex in Saccharomyces cerevisiae (12, 23). Serine 10 and threonine 11 were selected because the posttranslational modification of both amino acids was shown to affect the acetylation level of histone H3 in S. cerevisiae (24, 25).
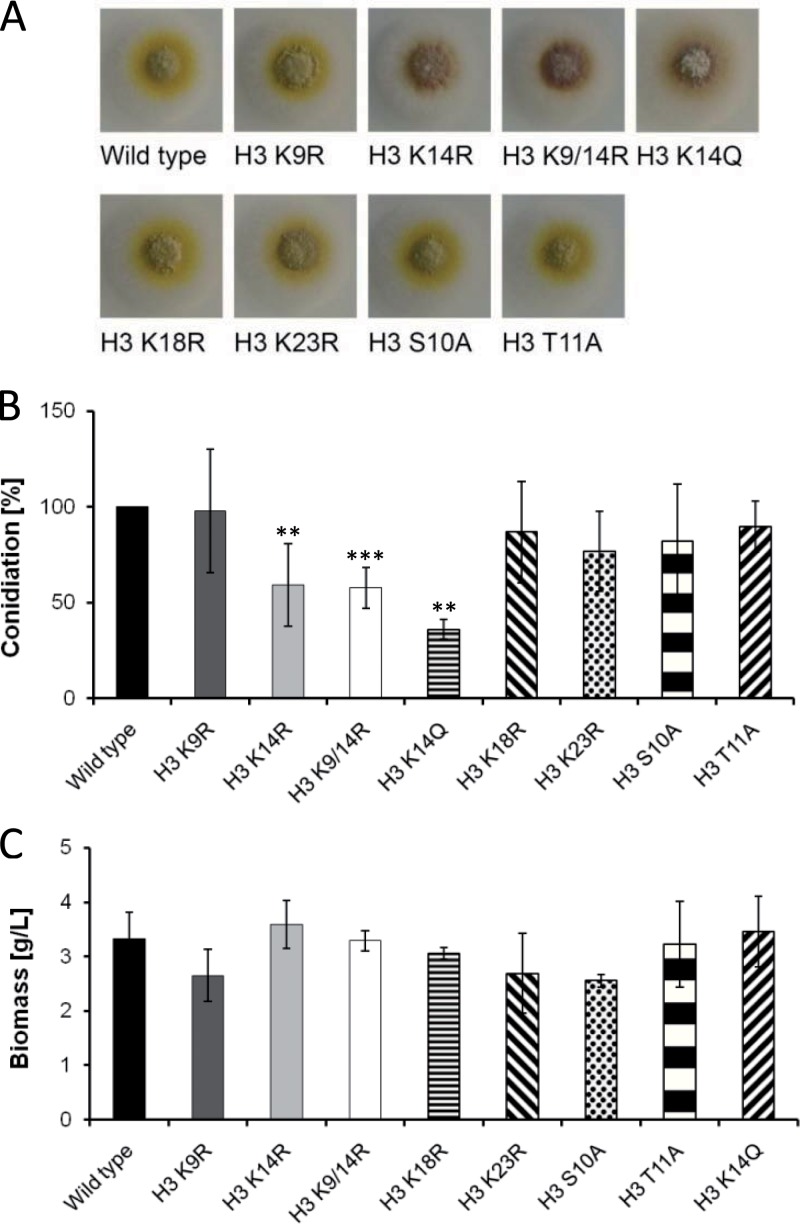
A. nidulans strains with altered histone H3 amino acid sequence were obtained by exchange of the wild-type H3 gene against modified gene versions that encode substituted amino acids, as indicated in Fig. 1A; see also Fig. S1 in the supplemental material. Lysine residues were replaced by arginine or glutamine residues, and serine/threonine residues were replaced by alanine. The exchange of lysine to arginine or serine/threonine to alanine mimics a nonacetylatable or nonphosphorylatable residue, respectively. In contrast, the exchange of lysine to glutamine mimics an acetylated lysine residue. In addition to single substitutions, we also generated a double amino acid substitution mutant, which had replaced lysines 9 and 14 by arginine residues (Fig. 1A). To the DNA constructs encoding the H3 amino acid exchanges, the auxotrophic selection marker gene argB was added (Fig. 1B). After transformation of the arginine auxotrophic A. nidulans strain A1153, transformants were isolated, each carrying a gene construct replacing the endogenous wild-type H3 gene. As a control, the wild-type gene was replaced by the wild-type gene fused with the argB cassette. All transformant strains were analyzed by Southern blotting (see Fig. S1 in the supplemental material) and sequencing of the histone H3 locus to prove the integration of a single-copy gene at the H3 genomic locus (data not shown). As shown in Fig. 2A, all mutant strains were viable, which was surprising given the importance of the amino acids as targets for histone modifications. Mutant strains carrying a single substitution of lysine 14 (H3 K14R) and also a double substitution of lysines 9 and 14 (H3 K9/14R) showed a slight reduction in growth and conidium formation (Fig. 2). All other mutant strains encoding the different amino acid exchanges did not reveal visible changes compared to the wild type (Fig. 2). It is worth noticing that the exchange of lysine 14 by the uncharged amino acid glutamine did not overcome the reduced conidiation phenotype although glutamine instead of lysine mimics the acetylation of lysine (26) (see Discussion). The biomass formation for all strains was measured in 20-ml AMM experimental cultures after 24 h of cultivation at 37°C. None of the strains showed a significantly altered growth in comparison to the wild type. The generation of stress caused by rapamycin, Congo red, NaCl, hydroxyurea, camptothecin, methyl methane sulfonate, and hydrogen peroxide and growth on the nitrogen and carbon sources glutamine and ethanol, respectively, did not lead to obvious differences between the wild type and the mutant strains (data not shown). Also, compared to the wild type, there was no difference in the sensitivity of the mutant strains against reactive oxygen/reactive nitrogen intermediate-generating compounds like diamide, diethylenetriamine-nitric oxide (deta-NO), and menadione (data not shown).
Fig 2.
Colony phenotype on agar plates, number of conidia, and biomass formation in experimental cultures of histone H3 mutant strains of A. nidulans. (A) Growth phenotype. The mutant strains were grown on AMM agar plates for 3 days at 37°C. (B) Number of conidia produced by the different strains. Fresh conidia were plated on AMM agar plates, and conidia were harvested after 3 days of incubation at 37°C. Statistical significance of data is given by the P value (**, P < 0.01; ***, P < 0.001). (C) Biomass formation in experimental cultures after 24 h of cultivation at 37°C in 20 ml of AMM.
Lysine residue 14, alone and in combination with lysine residue 9, contributes to secondary metabolite production.
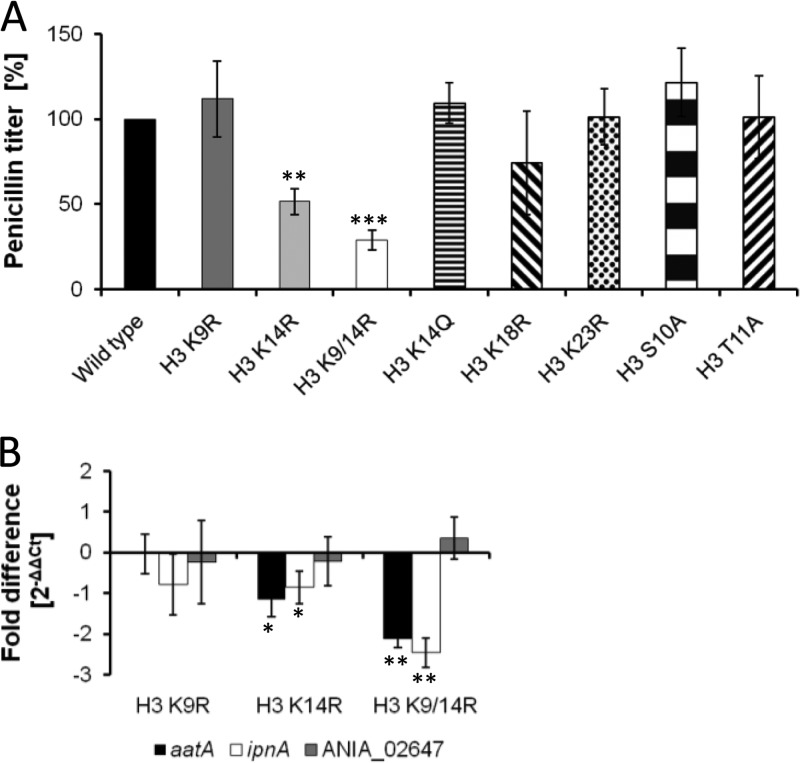
To shed light on the general importance of the selected histone H3 amino acids on secondary metabolism of A. nidulans, we analyzed three secondary metabolism pathways affected by the GcnE histone acetyltransferase and whose biosynthesis genes exhibited acetylation marks at H3 K9 and K14. These include the penicillin, sterigmatocystin, and orsellinic acid biosynthesis pathways. As shown in Fig. 3A, replacement of acetylatable lysine 14 by arginine and double replacement of lysines 9 and 14 by arginine residues led to a significantly decreased penicillin titer. All other single substitutions of histone H3 did not affect the penicillin titer. In contrast to the conidiation phenotype, this included the exchange of H3 K14 by the uncharged glutamine, which overcame the phenotype of the H3 K14R substitution, suggesting that the lack of the charge due to acetylation caused the major effect (26). Interestingly, a smaller amount of penicillin was detected in the H3 K9/14R double mutant strain than in the H3 K14R single substitution mutant. This was surprising, as no altered penicillin titer was measured in the H3 K9R single substitution mutant (Fig. 3A). The reduced penicillin titers in the H3 K14R single mutant and H3 K9/14R double mutant were accompanied by reduced mRNA steady-state levels of the respective biosynthesis genes. This conclusion was based on qRT-PCR results demonstrating that the two penicillin biosynthesis genes ipnA and aatA measured here showed reduced mRNA steady-state levels after 12 h of incubation of A. nidulans in fermentation medium (Fig. 3B).
Fig 3.
Penicillin titers and mRNA steady-state levels of penicillin biosynthesis genes of histone H3 mutant strains. Wild-type and histone mutant strains were grown in fermentation media and harvested. (A) Penicillin titers measured after 36 h are given as relative values, with the amount measured for the wild type set at 100%. Statistical significance of data is given by the P value (**, P < 0.01; ***, P < 0.001). (B) Fold difference of the mRNA steady-state level of penicillin biosynthesis genes ipnA and aatA and the nonrelated gene ANIA_2647 measured by qRT-PCR after 12 h of incubation of strains. The β-actin gene of A. nidulans was used as an internal standard. Statistical significance of data is given by the P value (*, P < 0.05; **, P < 0.01).
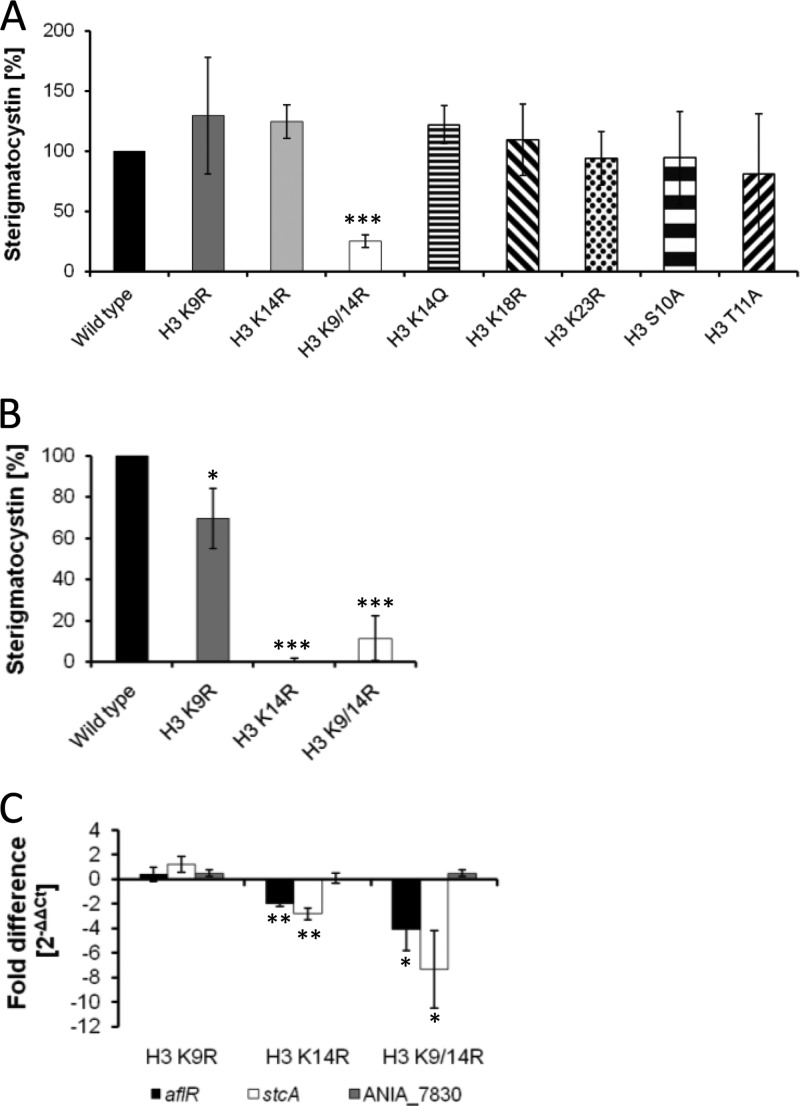
Similar observations were made for the production of sterigmatocystin (Fig. 4A). After 48 h of incubation, only the double substitution of acetylatable lysines 9 and 14 in the H3 K9/14R double mutant by arginine residues led to a significantly decreased sterigmatocystin production. However, when the sterigmatocystin titer was measured after 36 h, the H3 K9R mutant showed a slight reduction of sterigmatocystin, whereas both the H3 K14R mutant and the H3 K9/14R double mutant displayed a strong reduction of the sterigmatocystin titer, indicating that the histone marks are more relevant at the onset of the production of the metabolite (Fig. 4B). Consistent with the sterigmatocystin titers after 36 h, the steady-state mRNA levels of the sterigmatocystin cluster genes aflR and stcA were reduced in both the H3 K14R single substitution mutant and the H3 K9/14R double mutant. The single substitution of H3 K9 did not lead to a reduction of gene expression (Fig. 4C).
Fig 4.
Sterigmatocystin titer and mRNA steady-state levels of sterigmatocystin cluster genes of histone H3 mutant strains grown in AMM. Sterigmatocystin titers are given as relative values, with the amount measured for the wild type set at 100%. Statistical significance of data is given by the P value (**, P < 0.01; *, P < 0.05; ***, P < 0.001). (A) Sterigmatocystin titers after 48 h. (B) Sterigmatocystin titers after 36 h. (C) qRT-PCR analysis of sterigmatocystin cluster genes aflR and stcA, the gene ANIA_7830, located next to the sterigmatocystin cluster, and the β-actin-encoding acn gene used as an internal standard. Gene expression was measured after 36 h of cultivation of strains.
We also tested the H3 K9 and K14 substitution strains for expression of members of the gene cluster and metabolite production under noninducing conditions. There was no difference in the expression of penicillin and sterigmatocystin biosynthesis genes in the mutant strains compared to the wild type (see Fig. S2 in the supplemental material; also data not shown).
Substitution of lysine 14 by arginine abolished the inducibility of the silent A. nidulans orsellinic acid gene cluster by Streptomyces rapamycinicus.
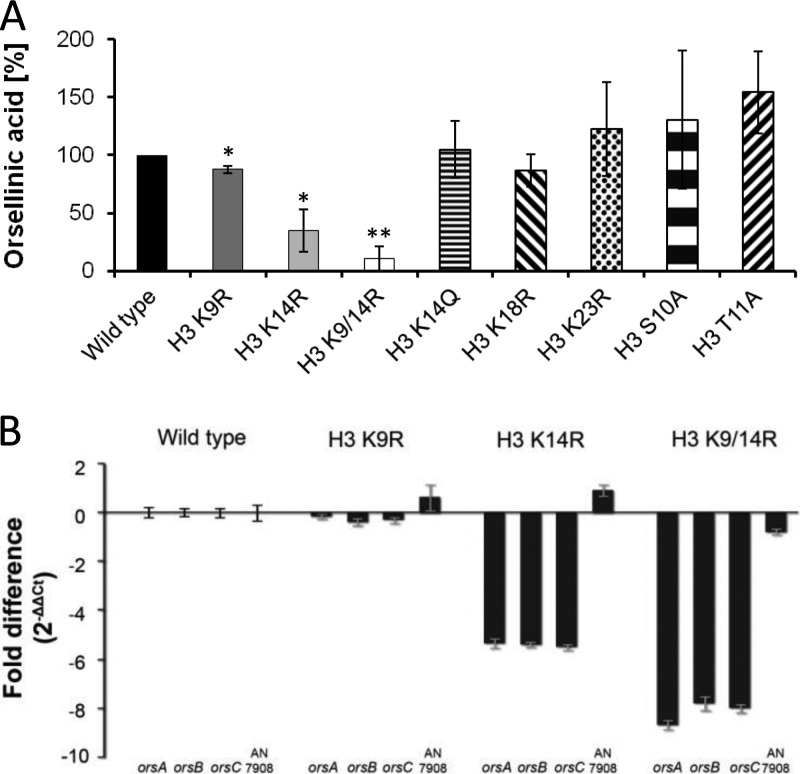
Previously, we showed that the SAGA/ADA complex is required for the activation of the orsellinic acid gene cluster during the interaction of A. nidulans with the streptomycete S. rapamycinicus (8, 19). The SAGA/ADA complex led to increased acetylation of lysine 9 and lysine 14 of histone H3. Further experiments suggested that lysine 9 acetylation was specifically associated with active secondary metabolism gene promoters, whereas lysine 14 acetylation increased not only within gene cluster boundaries but also at promoters of genes outside the gene clusters (8). Because the causal link of acetylated lysine residues and the expression of secondary metabolism genes and production of metabolites was lacking, we analyzed the effect of S. rapamycinicus on the various histone mutant strains of A. nidulans and measured the activation of the ors gene cluster as a readout. In accordance with the results obtained for penicillin and sterigmatocystin, we observed a drastic reduction of production of orsellinic acid in the H3 K14R single substitution mutant and the H3 K9/14R double mutant (Fig. 5A). All other histone mutant strains exhibited orsellinic acid levels similar to those of the wild type. In agreement with the reduced orsellinic acid, a strong decrease of the mRNA steady-state level of the ors genes was detected in the mutant strains H3 K14R and H3 K9/14R in comparison to the wild type (Fig. 5B). The H3 K9/14R double substitution again resulted in a more pronounced reduction of mRNA steady-state levels and orsellinic acid production (Fig. 5B). As also noted for both the penicillin and sterigmatocystin biosynthesis genes, single substitution of lysine 9 did not lead to an alteration of the transcription rate of the analyzed genes of the ors cluster.
Fig 5.
(A) Orsellinic acid production during interaction of S. rapamycinicus with the histone H3 substitution strains of A. nidulans after 24 h of coincubation. (B) qRT-PCR analysis of ors genes orsA, orsB, and orsC and of the nonrelated gene AN7908. Fold difference is given according to the 2−ΔΔCT method. Expression levels were calculated with the A. nidulans β-actin gene as an internal standard. Mycelia for RNA isolation were taken after 3 h of coincubation of A. nidulans strains with S. rapamycinicus.
DISCUSSION
Previously, we showed that in A. nidulans the activity of genes of several secondary metabolism biosynthesis gene clusters was associated with increased acetylation of lysines 9 and 14 of histone H3 during expression of these genes (8). These secondary metabolites included penicillin, sterigmatocystin, and orsellinic acid. Furthermore, the SAGA/ADA complex containing the GcnE histone acetyltransferase was shown to be required for this acetylation (8). Although compelling, these findings allowed only an association between histone modification, deletion of histone modifier-encoding genes, and regulation of secondary metabolism. It was conceivable that secondary effects of the generated gcnE and adaB (also encoding a subunit of SAGA/ADA) mutant strains or other missing activities of deleted genes led to alterations of secondary metabolism. For example, the importance of the SAGA/ADA complex for gene cluster expression could be due to activities other than its function for histone acetylation (12). Because the causal link of lysine acetylation of histone H3 and secondary metabolism was lacking, we addressed this problem by analyzing the effect of the exchange of the respective amino acids of histone H3 against other amino acids. Substitution of histone amino acids is an established tool to test the individual impact of histone residues on distinct cellular processes. In S. cerevisiae, mutant libraries with substitutions in each histone residue have been generated (27, 28). Histones in higher eukaryotic model organisms, such as Neurospora crassa, Tetrahymena thermophila, and Drosophila melanogaster, were the targets for individual amino acid exchanges (29–31). A common initial concern in these studies was the viability of the mutated organisms, because the high evolutionary conservation of histone proteins and their function as an acceptor of various modifications suggest their essential importance for cells. However, results obtained for S. cerevisiae showed that the vast majority of histone amino acids can be changed (27, 28). That this does not hold true for all eukaryotes was elegantly shown in N. crassa, where among other substitutions lysines 9 and 14 of histone H3 were shown to be essential (29). All A. nidulans histone mutant strains generated here were viable and thus allowed us to investigate the importance of distinct amino acids and their potential to become posttranslationally modified for fungal secondary metabolism. For all three secondary metabolism pathways, we could unequivocally show that lysine 14 and its acetylation are of major importance for the production of penicillin, sterigmatocystin, and upon cocultivation with S. rapamycinicus, orsellinic acid. The qRT-PCR analyses fully supported these findings, as reduced transcription of the respective gene clusters was detected in the H3 K14R mutant strain. Therefore, this lysine residue and its posttranslational modification play a major role for the expression of these secondary metabolite biosynthesis gene clusters. Interestingly, the time-dependent measurements made for sterigmatocystin production indicate that the H3 K14 mark is of particular importance for the initial activation of the gene cluster. The drastic reduction of metabolite titer due to the exchange of lysine against arginine was overcome at later stages of cultivation. Similar observations were made for the penicillin and orsellinic acid gene clusters (data not shown).
In general, histone H3 acetylation appears to function in two, non-mutually exclusive manners: (i) to provide a molecular tag for the recruitment of chromatin-modifying complexes and (ii) to directly change the chromatin structure by weakening histone-DNA contacts (26, 32, 33). The finding that the exchange of K14 by the uncharged glutamine residue did not affect the expression of the gene clusters suggests that the neutralization of the charge of lysine 14 by acetylation is of major importance rather than the recruitment of other proteins upon acetylation of K14. However, the unaffected metabolite and transcript levels in the lysine-to-glutamine exchange strain also showed that a more neutral charged histone tail alone is not sufficient to activate secondary metabolite gene clusters under noninducing conditions. Most likely, a lack of transcriptional activators cannot be compensated.
Gene clusters are rarely found in eukaryotes (1, 2). A prominent exception is secondary metabolism gene clusters. Most, if not all, fungal secondary metabolism genes are found in clusters. For example, in A. nidulans more than 45 gene clusters were found based on an in silico analysis of the genome (1). Several arguments have been put forward to explain gene clusters in eukaryotes. Recently, an increasing number of reports on the specific regulation of gene clusters by histone modification (8–10, 34, 35) suggests that chromatin-based regulation through histone acetylation or methylation could be a good reason for clustering of secondary metabolism genes (1). Histone acetylation or methylation has the advantage that it can be restricted to defined regions of the chromosome spanning only a few genes.
Interestingly, smaller amounts of secondary metabolites were detected in the H3 K9/14R double mutant strain than in the H3 K14R single substitution mutant. This was surprising, as the penicillin and orsellinic acid titers did not change in the H3 K9R single substitution mutant and the sterigmatocystin production rate decreased only very slightly. Previously, two modifications were assigned to H3 K9. Methylation of K9 was associated with repression of several secondary metabolism gene clusters (9) and acetylation of K9 with activation of gene clusters (8). It is thus conceivable that the lack of both posttranslational marks at K9 leads to an unchanged phenotype in the single substitution strain. Another explanation could be that the loss of K9 acetylation can be mostly overcome by the K14 acetylation, and thereby the effect of exchange of K9 can be seen only in an H3 K14R mutant background. However, the observations based on the K9/K14 double exchange clearly suggest a role of H3 K9 for gene expression. Furthermore, it can be expected that interdependence of K14 and K9 exists. Consistently, as previously shown by Nützmann et al. (8), lysine 9 acetylation was specifically induced in promoters of genes located in the analyzed clusters. In contrast, lysine 14 acetylation seems to be increased in a more genome-wide fashion (8). We can speculate that the more unspecific K14 acetylation mediated by GcnE is a prerequisite for a specific K9 acetylation and full activation of the gene clusters. In S. cerevisiae, reports suggest such a direct link between H3 K14 acetylation and H3 K4 methylation. Missing K14 acetylation led to increased activity of H3 K4 demethylases and subsequently to decreased methylation levels (36). It was not in the scope of our analysis to measure the effect of the amino acid substitutions on other histone amino acid modifications. However, it would be interesting to analyze the histone H3 modification pattern, e.g., the H3 K9 and H3 K4 methylation levels, in the K14 substitution strain generated here.
The exchange of H3 K14 by arginine led to a reduced conidiation phenotype of the mutant strain. This finding agrees well with the observation that the gcnE mutant strain of A. nidulans also exhibited reduced conidiation (8). However, it is worth noticing that the exchange of K14 by the uncharged amino acid glutamine did not overcome the reduced conidiation phenotype, although it was suggested that glutamine instead of lysine mimics the acetylation of lysine (26). It is thus conceivable that for conidiation not only the neutralization of the charge of K14 by acetate groups is required but also the recruitment of additional chromatin-modifying complexes to histone H3.
In conclusion, our study closes a missing link in the field of chromatin genetics and its importance for fungal secondary metabolism. Individual reports on deletion of chromatin modifier genes, measurement of histone modification states, and substitution of histone amino acids, as investigated here, can provide only a limited general view on the ongoing regulatory processes and cannot exclude indirect effects caused by the studied histone acetyltransferase. However, in this study we underline the previous finding that the SAGA/ADA complex acetylates histone H3 lysine residues and thereby regulates secondary metabolism in A. nidulans with the observation that individual histone amino acids are indeed essential for the activation of secondary metabolite biosynthesis gene clusters.
Supplementary Material
ACKNOWLEDGMENTS
We thank Christina Täumer and Carmen Karkowski for technical assistance and V. Schroeckh for helpful discussions.
Financial support by the Excellence Graduate School Jena School for Microbial Communication (JSMC), the International Leibniz Research School for Microbial and Biomolecular Interactions as part of the JSMC, the Leibniz Association (Pact for Research and Innovation), and the HKI is gratefully acknowledged.
Footnotes
Published ahead of print 26 July 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01578-13.
REFERENCES
- 1.Brakhage AA. 2013. Regulation of fungal secondary metabolism. Nat. Rev. Microbiol. 11:21–32 [DOI] [PubMed] [Google Scholar]
- 2.Yin W, Keller NP. 2011. Transcriptional regulatory elements in fungal secondary metabolism. J. Microbiol. 49:329–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergmann S, Funk AN, Scherlach K, Schroeckh V, Shelest E, Horn U, Hertweck C, Brakhage AA. 2010. Activation of a silent fungal polyketide biosynthesis pathway through regulatory cross talk with a cryptic nonribosomal peptide synthetase gene cluster. Appl. Environ. Microbiol. 76:8143–8149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergmann S, Schuemann J, Scherlach K, Lange C, Brakhage AA, Hertweck C. 2007. Genomics-driven discovery of PKS-NRPS hybrid metabolites from Aspergillus nidulans. Nat. Chem. Biol. 3:213–217 [DOI] [PubMed] [Google Scholar]
- 5.Payne GA, Nystrom GJ, Bhatnagar D, Cleveland TE, Woloshuk CP. 1993. Cloning of the afl-2 gene involved in aflatoxin biosynthesis from Aspergillus flavus. Appl. Environ. Microbiol. 59:156–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bannister AJ, Kouzarides T. 2011. Regulation of chromatin by histone modifications. Cell Res. 21:381–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gacek A, Strauss J. 2012. The chromatin code of fungal secondary metabolite gene clusters. Appl. Microbiol. Biotechnol. 95:1389–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nützmann HW, Reyes-Dominguez Y, Scherlach K, Schroeckh V, Horn F, Gacek A, Schumann J, Hertweck C, Strauss J, Brakhage AA. 2011. Bacteria-induced natural product formation in the fungus Aspergillus nidulans requires Saga/Ada-mediated histone acetylation. Proc. Natl. Acad. Sci. U. S. A. 108:14282–14287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reyes-Dominguez Y, Bok JW, Berger H, Shwab EK, Basheer A, Gallmetzer A, Scazzocchio C, Keller N, Strauss J. 2010. Heterochromatic marks are associated with the repression of secondary metabolism clusters in Aspergillus nidulans. Mol. Microbiol. 76:1376–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bok JW, Chiang YM, Szewczyk E, Reyes-Dominguez Y, Davidson AD, Sanchez JF, Lo HC, Watanabe K, Strauss J, Oakley BR, Wang CC, Keller NP. 2009. Chromatin-level regulation of biosynthetic gene clusters. Nat. Chem. Biol. 5:462–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scherlach K, Nützmann HW, Schroeckh V, Dahse HM, Brakhage AA, Hertweck C. 2011. Cytotoxic pheofungins from an engineered fungus impaired in posttranslational protein modification. Angew. Chem. Int. Ed. Engl. 50:9843–9847 [DOI] [PubMed] [Google Scholar]
- 12.Baker SP, Grant PA. 2007. The SAGA continues: expanding the cellular role of a transcriptional co-activator complex. Oncogene 26:5329–5340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee D, Ezhkova E, Li B, Pattenden SG, Tansey WP, Workman JL. 2005. The proteasome regulatory particle alters the SAGA coactivator to enhance its interactions with transcriptional activators. Cell 123:423–436 [DOI] [PubMed] [Google Scholar]
- 14.Rodriguez-Navarro S. 2009. Insights into SAGA function during gene expression. EMBO Rep. 10:843–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodriguez-Navarro S, Fischer T, Luo MJ, Antunez O, Brettschneider S, Lechner J, Perez-Ortin JE, Reed R, Hurt E. 2004. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell 116:75–86 [DOI] [PubMed] [Google Scholar]
- 16.Szewczyk E, Nayak T, Oakley CE, Edgerton H, Xiong Y, Taheri-Talesh N, Osmani SA, Oakley BR. 2006. Fusion PCR and gene targeting in Aspergillus nidulans. Nat. Protoc. 1:3111–3120 [DOI] [PubMed] [Google Scholar]
- 17.Yu JH, Hamari Z, Han KH, Seo JA, Reyes-Dominguez Y, Scazzocchio C. 2004. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 41:973–981 [DOI] [PubMed] [Google Scholar]
- 18.Brakhage AA, Van den Brulle J. 1995. Use of reporter genes to identify recessive trans-acting mutations specifically involved in the regulation of Aspergillus nidulans penicillin biosynthesis genes. J. Bacteriol. 177:2781–2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schroeckh V, Scherlach K, Nützmann HW, Shelest E, Schmidt-Heck W, Schuemann J, Martin K, Hertweck C, Brakhage AA. 2009. Intimate bacterial-fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proc. Natl. Acad. Sci. U. S. A. 106:14558–14563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bergh KT, Litzka O, Brakhage AA. 1996. Identification of a major cis-acting DNA element controlling the bidirectionally transcribed penicillin biosynthesis genes acvA (pcbAB) and ipnA (pcbC) of Aspergillus nidulans. J. Bacteriol. 178:3908–3916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brakhage AA, Browne P, Turner G. 1992. Regulation of Aspergillus nidulans penicillin biosynthesis and penicillin biosynthesis genes acvA and ipnA by glucose. J. Bacteriol. 174:3789–3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wickes B. 2004. DNA isolation from a filamentous fungus using the MasterPure Yeast DNA purification kit. Epicentre Forum 11:6 http://www.epibio.com/docs/default-source/forum-archive/forum-11-6 [Google Scholar]
- 23.Grant PA, Eberharter A, John S, Cook RG, Turner BM, Workman JL. 1999. Expanded lysine acetylation specificity of Gcn5 in native complexes. J. Biol. Chem. 274:5895–5900 [DOI] [PubMed] [Google Scholar]
- 24.Lo WS, Trievel RC, Rojas JR, Duggan L, Hsu JY, Allis CD, Marmorstein R, Berger SL. 2000. Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol. Cell 5:917–926 [DOI] [PubMed] [Google Scholar]
- 25.Shimada M, Niida H, Zineldeen DH, Tagami H, Tanaka M, Saito H, Nakanishi M. 2008. Chk1 is a histone H3 threonine 11 kinase that regulates DNA damage-induced transcriptional repression. Cell 132:221–232 [DOI] [PubMed] [Google Scholar]
- 26.Choi JK, Grimes DE, Rowe KM, Howe LJ. 2008. Acetylation of Rsc4p by Gcn5p is essential in the absence of histone H3 acetylation. Mol. Cell. Biol. 28:6967–6972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dai J, Hyland EM, Yuan DS, Huang H, Bader JS, Boeke JD. 2008. Probing nucleosome function: a highly versatile library of synthetic histone H3 and H4 mutants. Cell 134:1066–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakanishi S, Sanderson BW, Delventhal KM, Bradford WD, Staehling-Hampton K, Shilatifard A. 2008. A comprehensive library of histone mutants identifies nucleosomal residues required for H3K4 methylation. Nat. Struct. Mol. Biol. 15:881–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adhvaryu KK, Berge E, Tamaru H, Freitag M, Selker EU. 2011. Substitutions in the amino-terminal tail of Neurospora histone H3 have varied effects on DNA methylation. PLoS Genet. 7:e1002423. 10.1371/journal.pgen.1002423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pengelly AR, Copur O, Jackle H, Herzig A, Muller J. 2013. A histone mutant reproduces the phenotype caused by loss of histone-modifying factor Polycomb. Science 339:698–699 [DOI] [PubMed] [Google Scholar]
- 31.Wei Y, Yu L, Bowen J, Gorovsky MA, Allis CD. 1999. Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell 97:99–109 [DOI] [PubMed] [Google Scholar]
- 32.Ferreira H, Flaus A, Owen-Hughes T. 2007. Histone modifications influence the action of Snf2 family remodelling enzymes by different mechanisms. J. Mol. Biol. 374:563–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toth K, Brun N, Langowski J. 2006. Chromatin compaction at the mononucleosome level. Biochemistry 45:1591–1598 [DOI] [PubMed] [Google Scholar]
- 34.Shwab EK, Bok JW, Tribus M, Galehr J, Graessle S, Keller NP. 2007. Histone deacetylase activity regulates chemical diversity in Aspergillus. Eukaryot. Cell 6:1656–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soukup AA, Chiang YM, Bok JW, Reyes-Dominguez Y, Oakley BR, Wang CC, Strauss J, Keller NP. 2012. Overexpression of the Aspergillus nidulans histone 4 acetyltransferase EsaA increases activation of secondary metabolite production. Mol. Microbiol. 86:314–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maltby VE, Martin BJ, Brind'Amour J, Chruscicki AT, McBurney KL, Schulze JM, Johnson IJ, Hills M, Hentrich T, Kobor MS, Lorincz MC, Howe LJ. 2012. Histone H3K4 demethylation is negatively regulated by histone H3 acetylation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 109:18505–18510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nayak T, Szewczyk E, Oakley CE, Osmani A, Ukil L, Murray SL, Hynes MJ, Osmani SA, Oakley BR. 2006. A versatile and efficient gene-targeting system for Aspergillus nidulans. Genetics 172:1557–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar Y, Goodfellow M. 2008. Five new members of the Streptomyces violaceusniger 16S rRNA gene clade: Streptomyces castelarensis sp. nov., comb. nov., Streptomyces himastatinicus sp. nov., Streptomyces mordarskii sp. nov., Streptomyces rapamycinicus sp. nov. and Streptomyces ruanii sp. nov. Int. J. Syst. Evol. Microbiol. 58:1369–1378 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.