Abstract
Intestinal colonization is influenced by the ability of the bacterium to inhabit a niche, which is based on the expression of colonization factors. Escherichia coli carries a broad range of virulence-associated genes (VAGs) which contribute to intestinal (inVAGs) and extraintestinal (exVAGs) infection. Moreover, initial evidence indicates that inVAGs and exVAGs support intestinal colonization. We developed new screening tools to genotypically and phenotypically characterize E. coli isolates originating in humans, domestic pigs, and 17 wild mammal and avian species. We analyzed 317 isolates for the occurrence of 44 VAGs using a novel multiplex PCR microbead assay (MPMA) and for adhesion to four epithelial cell lines using a new adhesion assay. We correlated data for the definition of new adhesion genes. inVAGs were identified only sporadically, particularly in roe deer (Capreolus capreolus) and the European hedgehog ( Erinaceus europaeus). The prevalence of exVAGs depended on isolation from a specific host. Human uropathogenic E. coli isolates carried exVAGs with the highest prevalence, followed by badger (Meles meles) and roe deer isolates. Adhesion was found to be very diverse. Adhesion was specific to cells, host, and tissue, though it was also unspecific. Occurrence of the following VAGs was associated with a higher rate of adhesion to one or more cell lines: afa-dra, daaD, tsh, vat, ibeA, fyuA, mat, sfa-foc, malX, pic, irp2, and papC. In summary, we established new screening methods which enabled us to characterize large numbers of E. coli isolates. We defined reservoirs for potential pathogenic E. coli. We also identified a very broad range of colonization strategies and defined potential new adhesion genes.
INTRODUCTION
Bacterial intestinal colonization of a host is influenced by the ability of the bacterium to inhabit an intestinal niche which is not occupied by competing bacteria. Resident colonization mainly depends on the bacterium's ability to adhere to the mucosa as well as to its competing successfully with the established microbiota (1, 2).
Escherichia coli is a commensal bacterium in mammals and birds, though it can also be the cause of intestinal and extraintestinal disease (3). The bacterium's ability to adapt its genotype and phenotype to a changing environment renders the strict differentiation between pathogenic and nonpathogenic E. coli somewhat complicated (4). The basis for this rapid adaptation is the genome plasticity of E. coli, a feature which gives rise to an increased diversity of E. coli populations (5).
The infection of a host by E. coli is facilitated by virulence factors, which are coded by virulence-associated genes (VAGs) (6, 7). E. coli can express a broad variety of virulence factors involved in the colonization, adhesion, invasion, and survival of host defenses. These factors are adhesins, invasins, toxins, iron acquisition systems (siderophores), and protectins (Table 1). VAGs which are involved in the pathogenesis of infections with extraintestinal pathogenic E. coli (ExPEC) isolates—so-called exVAGs—and VAGs which are frequently detected in intestinal pathogenic E. coli (InPEC) isolates originating in diarrheic hosts—so-called inVAGs—are often located on mobile genetic elements. This enables horizontal transfer of the respective genes between strains (8, 9).
Table 1.
Description of eight mPCRsa
| VAG | MPMA | Gene/operon | Function | Description |
|---|---|---|---|---|
| exVAGs | mPCR 1 | afa-dra | Adhesin | Central region of afimbrial/Dr antigen-specific adhesin operons |
| sfa-foc | Adhesin | Central region of S (sialic acid specific) and F1C fimbrial operons | ||
| pic | Miscellaneous | Serine protease autotransporter | ||
| hra | Adhesin | Heat-resistant agglutinin | ||
| hlyA | Toxin | Hemolysin A | ||
| mPCR 2 | ibeA | Invasin | Invasion of brain endothelium | |
| traT | Protectin | Transfer protein/surface exclusion, serum survival | ||
| sitchr | Siderophore | Salmonella iron transport system gene, chromosomally localized | ||
| ompA | Protectin | Outer membrane protein A | ||
| iroN | Siderophore | Catecholate siderophore (salmochelin) receptor | ||
| sitep | Siderophore | Salmonella iron transport system gene, episomally localized | ||
| mPCR 3 | vat | Toxin | Vacuolating autotransporter toxin | |
| tsh | Adhesin | Temperature-sensitive hemagglutinin | ||
| iucD | Siderophore | Aerobactin synthesis | ||
| cvi-cva | Protectin | Structural genes of colicin V operon (microcin colicin V) | ||
| papC | Adhesin | Pyelonephritis-associated fimbria, chaperone | ||
| iss | Protectin | Increased serum survival | ||
| astA | Toxin | Heat-stable cytotoxin associated with enteroaggregative E. coil | ||
| mPCR 4 | cnf1-cnf2 | Toxin | Cytotoxic necrotizing factors 1 and 2 | |
| iutA | Siderophore | Ferric aerobactin receptor (iron uptake/transport) | ||
| mat | Adhesin | Meningitis-associated and temperature-regulated fimbriae | ||
| fyuA | Siderophore | Ferric yersinia uptake (yersiniabactin receptor) | ||
| sat | Toxin | Secreted autotransporter toxin | ||
| malX | Miscellaneous | Pathogenicity-associated island marker CFT073 | ||
| csgA | Adhesin | Curlin sigma S-dependent growth | ||
| mPCR 5 | fimC | Adhesin | Type 1 fimbriae (d-mannose-specific adhesin), chaperone | |
| irp2 | Siderophore | Iron-repressible protein (yersiniabactin synthesis) | ||
| ireA | Siderophore | Iron-responsive element (catecholate siderophore receptor) | ||
| tia | Invasin | Toxigenic invasion locus in enterotoxigenic E. coil strains | ||
| inVAGs | mPCR 6 | estA | Toxin | Heat-stable toxin I |
| estB | Toxin | Heat-stable toxin II | ||
| fanA | Adhesin | Biogenesis of K99 fimbriae | ||
| eltB-Ip | Toxin | Heat-labile toxin Ib subunit | ||
| fedA | Adhesin | F18 fimbrial adhesin | ||
| fasA | Adhesin | F6 fimbrial adhesin/987P | ||
| faeG | Adhesin | F4 fimbrial adhesin/K88 | ||
| fimF41a | Adhesin | F41 fimbrial adhesin | ||
| stx2e | Toxin | Shiga toxin IIe | ||
| mPCR 7 | stx1 | Toxin | Shiga toxin I | |
| stx2 | Toxin | Shiga toxin II | ||
| estA | Toxin | Heat-stable toxin Ia | ||
| estB | Toxin | Heat-stable toxin II | ||
| eltB | Toxin | Heat-labile toxin b subunit | ||
| aggR | Miscellaneous | Transcriptional regulator protein | ||
| daaD | Adhesin | Afimbrial/Dr adhesin F1845 | ||
| eaeA | Adhesin | Membrane protein Intimin | ||
| ipaH | Invasin | Invasion plasmid antigen H | ||
| ECoR | mPCR 8 | chuA | Siderophore | Heme receptor gene (E. coli heme utilization) |
| yjaA | Unknown | Identified in E. coli K-12 | ||
| TSPE4C2 | Anonymous DNA fragment |
Virulence-associated genes (VAGs) which are involved in the E. coli pathogenesis of extraintestinal infections (exVAGs) and VAGs frequently detected in E. coli isolated from diarrheic hosts (inVAGs) were identified. Associated functions reported in the literature are included. ECoR, E. coli Reference Collection.
Adhesion is the first step in colonization or infection and is facilitated by fimbrial or afimbrial adhesins which bind to host cell receptors. Tissue and host specificity is then determined on the basis of bacterial expression (1, 10, 11). This means that the different adhesion factors of E. coli result in different colonization and infection mechanisms. For example, S fimbriae and F1C fimbriae are filamentous surface structures generally associated with uropathogenic E. coli (UPEC) and are anchored to the bacterial outer membrane with morphological similarity and sequence homologies. They are encoded by the sfa and the foc gene cluster but display distinct receptor specificities (12, 13). It has been demonstrated that F1C fimbriae also support cell adhesion and subsequent colonization and biofilm formation in the intestine (14, 15).
inVAGs as well as exVAGs can also be found in E. coli isolates from clinically healthy hosts (16–19). Several inVAGs are adhesins, and their role in colonization has been comprehensively investigated. Initial data have shown that other inVAGs, which are not classic adhesins, can also support bacterial adhesion. An example of this is the heat-labile E. coli enterotoxin (20). Since the intestine of clinically healthy hosts is capable of harboring ExPEC (21), it remains unclear whether the intestine is merely a reservoir for such ExPEC isolates or whether exVAGs are involved in the colonization of healthy hosts. On the basis of initial studies and observations, it has been assumed that exVAGs, such as those for iron acquisition systems, also support intestinal colonization by E. coli (16, 18, 22).
The influence on adhesion and on tissue and host specificity exercised by a number of the inVAGs and by most of the exVAGs remains unclear. In order to elucidate the nature of this influence, we characterized the VAG profile and the phylogenetic affiliation of 317 nonhemolytic E. coli isolates collected from humans, domestic pigs, 13 wild mammal species, and 4 wild avian species. Hemolytic E. coli isolates were not included, as isolates of this type destroy cell line monolayers, thereby rendering the adhesion of such isolates nonquantifiable. Using the imaging platform VideoScan, as developed in our lab at Brandenburgische Technische Universität Cottbus-Senftenberg, we established innovative fluorescence-based multiplex PCR (mPCR) microbead assays (MPMAs) for the automatic detection of 44 VAGs and to determine phylogenetic origins. The VideoScan platform was also adapted to quantify the adhesion of E. coli to human intestinal Caco-2 and urinary bladder 5637 epithelial cells, as well as to porcine intestinal IPEC-J2 and kidney PK-15 epithelial cells. The adhesion of fluorescence-stained bacteria and host cells was quantified using automatic imaging and data analysis. The final discussion provides information about the distribution of VAGs in E. coli and host-specific and tissue-specific adhesion. This will assist in providing a more detailed understanding of the distribution of and colonization by E. coli, as well as of disease risk and the transmission of pathogens.
MATERIALS AND METHODS
Bacteria.
The study investigated animal and human isolates as well as isolates from diseased and clinically healthy hosts (Table 2). For the isolation, identification, and confirmation of E. coli, rectal swab specimens in the case of mammals and cloacal swab specimens in the case of birds were streaked directly onto CHROMagar orientation plates (Mast Diagnostics, United Kingdom) (23). Pink colonies from CHROMagar were plated onto Gassner agar to provide further confirmation. Pink coliform colonies on CHROMagar appearing as a blue/green/yellow coliform on Gassner agar were assumed to be E. coli, as discussed in various other studies (24–26). One single pink colony was subcultured twice on CHROMagar orientation plates and was then stored in 10% glycerol at −80°C until further processing. Finally, isolates were verified to be E. coli on the basis of the presence of E. coli VAGs. The hemolysis of E. coli was tested by transferring isolates onto 5% sheep blood agar. Hemolytic E. coli isolates were excluded from further analysis, as these isolates destroy cell culture monolayers, thus rendering adhesion assays impossible. This study was approved by the Ministry of Environment, Health and Consumer Protection of the Federal State of Brandenburg, Germany (V3-2347-8-39-1-2011).
Table 2.
E. coli isolates
| Species | Description | No. of isolates | % isolates from the indicated ECoR group |
|||
|---|---|---|---|---|---|---|
| A | B1 | B2 | D | |||
| Human | ||||||
| Homo sapiens | Human, healthy | 19a | 37 | 11 | 42 | 11 |
| Homo sapiens | Human, urinary tract infection | 20b | 5 | 25 | 45 | 25 |
| Domestic mammals | ||||||
| Sus scrofa domestica | Domestic pig, healthy | 22c | 73 | 5 | 5 | 18 |
| Sus scrofa domestica | Domestic pig, urinary tract infection | 15d | 53 | 47 | 0 | 0 |
| Wild mammals | ||||||
| Capreolus capreolus | Roe deer | 23e,f | 4 | 39 | 17 | 39 |
| Erinaceus europaeus | European hedgehog | 22e | 27 | 41 | 5 | 27 |
| Lepus europaeus | European hare | 8e | 13 | 75 | 0 | 13 |
| Lutra lutra | European otter | 7e | 43 | 29 | 14 | 14 |
| Martes sp. | Marten | 19e | 32 | 16 | 26 | 26 |
| Meles meles | European badger | 7e | 0 | 57 | 29 | 14 |
| Mus musculus | House mouse | 9e | 0 | 78 | 0 | 22 |
| Oryctolagus cuniculus | European rabbit | 6e | 33 | 17 | 17 | 33 |
| Procyon lotor | Raccoon | 22e | 23 | 27 | 23 | 27 |
| Rattus norvegicus | Brown rat | 4e | 0 | 50 | 25 | 25 |
| Sciurus vulgaris | Red squirrel | 17e | 6 | 29 | 47 | 18 |
| Sus scrofa | Wild boar | 22e,f | 23 | 50 | 14 | 14 |
| Vulpes vulpes | Red fox | 21e | 14 | 57 | 14 | 14 |
| Wild birds | ||||||
| Accipiter nisus | Eurasian sparrow hawk | 13e | 8 | 31 | 31 | 31 |
| Asio otus | Long-eared owl | 5e | 0 | 20 | 0 | 80 |
| Buteo buteo | Common buzzard | 14e | 7 | 29 | 21 | 43 |
| Turdus merula | Common blackbird | 22e | 9 | 36 | 27 | 27 |
Sampled by Thomas Wex.
Samples from urine of patients with urinary tract infections collected in a hospital in 2009 by Steffen Vogel.
Samples from 18 different pig production units in Eastern Germany in 2009 and 2010.
Sampled by Per Klemm from pigs with pyelonephritis in an animal clinic.
Samples collected in the Lausitz (Lusatia), a region in southeastern Germany, taken from the rectum (mammals) or cloaca (birds) of dead animals which were collected directly after they were discovered as accident victims or which were delivered to Hermann Ansorge and Olaf Zinke between 2007 and 2011.
Rectal samples which were taken during several hunts between 2007 and 2010.
mPCRs.
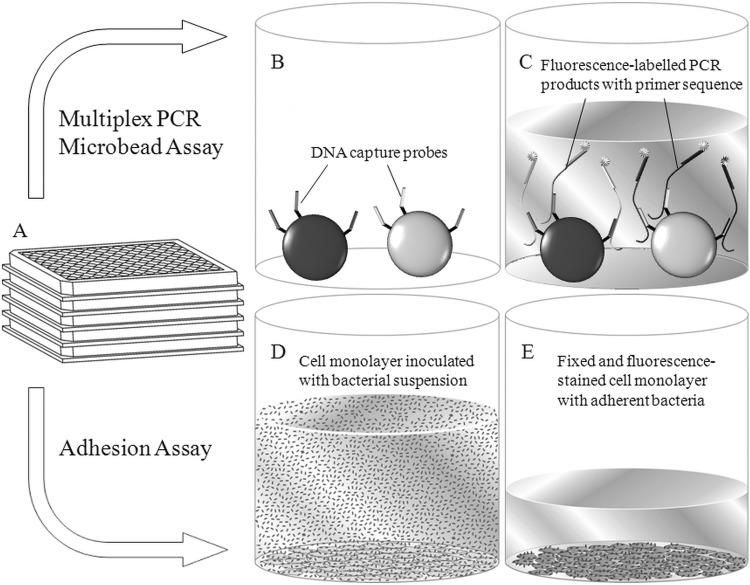
A new MPMA system was developed. This MPMA system was applied to eight mPCRs whose function included the detection of 14 inVAGs, 30 exVAGs, and 2 DNA sequences for the determination of the phylogenetic origin (Table 1). The basic principle of the MPMA is represented schematically in Fig. 1. Microbeads consisting of polymethylmethacrylate (PMMA) were thermotolerant, carboxylated, and coded by size (diameters, between 10 and 20 μm) and by two fluorescence dyes (PolyAn GmbH, Germany). 5′-Amino-modified DNA capture probes with poly(dT)10 spacers were cross-linked to the microbead surface using 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide (EDC) (27). Microbeads with bound capture probes were fixed to the bottom of poly-l-lysine (Sigma-Aldrich, Germany)-coated 96-well NucleoLink plates (Nunc, Germany). The PCR master mix and bacterial lysates were pipetted into these plates, and PCR was carried out (Mastercycler EP Gradient, Eppendorf, Germany). The reverse primers were Cy5 fluorescence labeled, and these strands of the PCR products were conclusively fluorescence labeled. After PCR fluorescence labeling, PCR strands hybridized to the respective DNA capture probes. Hybridization resulted in a fluorescence corona around a microbead. To remove nonincorporated fluorescence-labeled primers and unspecific PCR products, the microbeads were washed three times with 1× phosphate-buffered saline (PBS) with 0.02% Tween 20. Finally, the microbeads were analyzed using the VideoScan technology (see “VideoScan fluorescence imaging technology” below).
Fig 1.
Principle of MPMA and the adhesion assay. (A) Assays were prepared in 96-well plates. (B and C) MPMA. (B) DNA capture probes were coupled to different microbead populations. Microbeads were fixed to the bottom of a plate. (C) E. coli gene fragments were amplified from bacterial lysates in a multiplex format (three to nine PCR products per mPCR) in the presence of microbeads. During amplification, PCR products were labeled by one Cy5-labeled primer. Subsequently, PCR fluorescence-labeled DNA strands hybridized to gene-specific capture probes. Reagents and nonhybridized PCR products were removed by washing the microbeads with 1× PBS. The success of hybridization of the PCR products to DNA capture probes (and, therefore, amplification of a gene) was quantified by measuring the red-fluorescent corona around a microbead. The intensity of this corona (refMFI) correlated to the success of amplification and hybridization. Microbeads were analyzed using the VideoScan system. (D and E) Adhesion assay. (D) A confluent monolayer of a cultured epithelial cell line was inoculated with a bacterial suspension. (E) The supernatant with nonadherent bacteria was removed. After washing with 1× PBS, adherent E. coli cells were fixed with 4% paraformaldehyde and fluorescence stained with propidium iodide. Adherent bacteria were examined by a special data-imaging-processing algorithm of VideoScan.
The capture probes and primers (BioTez, Berlin, Germany) are listed in Table 3. The mPCRs were carried out according to the following protocol: a 20-μl reaction mixture included 2 μl 10× PCR buffer, 2 μl 50 mM MgCl2, 2 U (for exVAGs and phylogenetic determination) or 4 U (for inVAGs) BioTherm DNA polymerase (Rapidozym, Germany), 0.5 μl of each 10 mM deoxynucleoside triphosphate, 0.1 μl (100 pmol) primer pair, and 3 μl E. coli heat lysate supplemented with the appropriate volumes of water. The PCR conditions are included in footnote a of Table 3. Finally, PCR products were hybridized to capture probes for 120 min at 50°C.
Table 3.
Primer and capture probe sequences
| mPCR no.a and gene/operon | Primer or probeb | Primer or probe sequence (5′–3′) | Fragment length (bp) | GenBank accession no. | Reference or source of primer |
|---|---|---|---|---|---|
| mPCR 1 | |||||
| afa-dra | f | TAAGGAAGTGAAGGAGCGTG | 210 | FM955460.1 | 44 |
| r | CCGCCCTGAAGAAGTATCAC | This study | |||
| p | TAAGTGGCGGGCTGACAGAT | This study | |||
| sfa-foc | f | CGGAGAACTGGGTGCATCTTA | 189 | CP000247.1 | 44 |
| r | ACGCATGTGCTTCATCATG | This study | |||
| p | GGCAGATATAATGCCATTTAATTAAGGCTG | This study | |||
| pic | f | ACTGGATCTTAAGGCTCAGG | 168 | AF097644.1 | 44 |
| r | TTCCCCCACAGAGTGTTAC | This study | |||
| p | ATATTCTGAAATCTGCCAGC | This study | |||
| hra | f | GTAACTCACACTGCTGTCACCT | 139 | CP000247.1 | 44 |
| r | CGAATCGTTGTCACGTTCAG | This study | |||
| p | GGTTACGTCATAGCGGACAC | This study | |||
| hlyA | f | GTCCATTGCCGATAAGTTT | 120 | AE014075.1 | 44 |
| r | ATAGCTCCTGTTTCTTTGTG | This study | |||
| p | TGGTGACAGTTTACTTGCTGC | This study | |||
| mPCR 2 | |||||
| sitep | f | CCAGGGGCAGTCACTAAATG | 190 | CP001856.1 | This study |
| r | TACGGCAGTGAGAGCAGAGA | This study | |||
| p | AGCACCGCCGGACATCAGCGC | This study | |||
| ompA | f | GGTGTTGCCAGTAACCGG | 177 | CP002797.2 | 44 |
| r | GACGGTTCCGTAGTTGTTCTG | This study | |||
| p | ATAAGCGTCAGAACCGATGC | This study | |||
| iroN | f | ATCCTCTGGTCGCTAACTG | 167 | CP002212.1 | 44 |
| r | TTTTTAATATCCTCGCTGGTAA | This study | |||
| p | CAGCAGCCGGGCGTGTCGGT | This study | |||
| sitchr | f | GCCGGTTGTACGTTCTTCAT | 140 | CP002212.1 | This study |
| r | ATCAGGCTTGCCACATCC | This study | |||
| p | CGGTTTTGATCCGCGAGCTTG | This study | |||
| traT | f | GTGGTGCGATGAGCACAG | 124 | JN232517.1 | 44 |
| r | TCAGACGTGTTTTTGATCTGC | This study | |||
| p | CGCCAGCGAACGCACGGTAT | This study | |||
| ibeA | f | TGGAACCCGCTCGTAATATAC | 113 | CP002167.1 | 44 |
| r | AGTAGCTGCGCCTTCACG | This study | |||
| p | TGCCGCAGCAATGAGTGCCGC | This study | |||
| mPCR 3 | |||||
| vat | f | GTGTCAGAACGGAATTGTC | 230 | CP000468 | 44 |
| r | GGGTATCTGTATCATGGCAAG | 27 | |||
| p | CCGGGGTTGCTTTATTTGAGAAATTAATATTTCCC | 27 | |||
| tsh | f | TGGTGCAATGCCCATTTATGG | 215 | DQ381420 | 27 |
| r | CTTCCGATGTTCTGAACGT | 44 | |||
| p | TAATGAAGACAATGATGCCC | 27 | |||
| iucD | f | CCTGATCCAGATGATGCTC | 193 | CP002797 | 44 |
| r | CTGGATGAGCAGAAAATGACA | 27 | |||
| p | TGGTCAGTAAAGAATCGGCAGTGATGCC | 27 | |||
| cvi-cva | f | CGCAGCATAGTTCCATGCT | 172 | CP000836 | 44 |
| r | GCAATTTGTTGCAGGAGGA | 27 | |||
| p | TCATATATTGCACCTCCAGCCACACCCC | 27 | |||
| papC | f | GCTCCATGGTCATATAGTTTCG | 155 | CP002797.2 | 27 |
| r | TGATATCACGCAGTCAGTAGC | 44 | |||
| p | ACGTTCTCTCTCCCTCAATACG | 27 | |||
| iss | f | GGACAAGAGAAAACTGTTGATGC | 129 | FJ824853.1 | 27 |
| r | CAGCGGAGTATAGATGCCA | 44 | |||
| p | CCAGTTATGCATCGTGCATATGG | 27 | |||
| astA | f | TGCCATCAACACAGTATATCC | 116 | FN554767.1 | 44 |
| r | TCAGGTCGCGAGTGACGGC | 44 | |||
| p | CCAGTTATGCATCGTGCATATGG | 27 | |||
| mPCR 4 | |||||
| cnf1-cnf2 | f | CTTTACAATATTGACATGCTG | 446 | CP001162.1 | 44 |
| r | TCGTTATAAAATCAAACAGTG | 44 | |||
| p | GATCCCCATACTGGAAGCAATAATCCCC | This study | |||
| iutA | f | GGCTGGACATCATGGGAACTGG | 300 | CU928164.2 | 54 |
| r | CGTCGGGAACGGGTAGAATCG | 54 | |||
| p | CTTTTCTGGGACAGGAGCTG | This study | |||
| mat | f | TATACGCTGGACTGAGTCGTG | 257 | AF325731.1 | 44 |
| r | GCCTGGAGTTTACTGAACCAA | This study | |||
| p | GCAGAGGCCAAGCTGTACTC | This study | |||
| fyuA | f | GCGACGGGAAGCGATGACTTA | 243 | CU928163.2 | 44 |
| r | GTATTTCCCACTCAGGGTCTGG | This study | |||
| p | CCGTAAGCTGTCGATCAGCGATGG | This study | |||
| sat | f | GTTCACAAATGGGGGAAATG | 220 | CU928164.2 | This study |
| r | TTGAACATTCAGAGTACCGGG | 44 | |||
| p | GTAAACTGGAATGCATTGTATTCCAGTG | This study | |||
| malX | f | ACACTTAGCAGAGCAATGGG | 200 | AE014075.1 | This study |
| r | TTGAACGGTATCTGTATGCC | This study | |||
| p | TTAGTCCATTAATACTTTGGTACGAA | This study | |||
| csgA | f | GCTGCGTTACGATGGAAAGT | 118 | CP001637.1 | This study |
| r | GATAACAGCGTATTTACGTGGG | This study | |||
| p | CTCACCCGCGCGTTGTATTTTTCTTTTTTAGTTCATACC | This study | |||
| mPCR 5 | |||||
| fimC | f | GTTTTATCGTGACGCCACCT | 402–408 | CP000468 | This study |
| r | TTCCTGCATCAGAAGGCAAT | This study | |||
| p | ACAGAGTTGAATGCCGGAACTCGG | This study | |||
| irp2 | f | GCCATCAGGAGGAAGAATGA | 353–357 | CP000468 | This study |
| r | GGCGTACTTTCGGTCATGTT | This study | |||
| p | CGGAAGAAGAAACGCCGCCC | This study | |||
| ireA | f | GTTTCAGGTCCCTGTTTCCA | 237 | CP000468 | This study |
| r | ATGCAGCAACGGATTCTCTC | This study | |||
| p | CATGATTATCGAGACGCAGTCCTCCAG | This study | |||
| tia | f | CGAGTAATGCAACGCAGAAA | 113 | CP000468 | This study |
| r | CATGGGTGTCTGTTGGACTG | This study | |||
| p | GTATATGTTGCCTTATGATGTATCCGTGC | This study | |||
| mPCR 6 | |||||
| stx2e | f | GGAGCGTTTTGACCATCTTC | 247 | M21534.1 | This study |
| r | CGCCAGATATGATGAAACCA | This study | |||
| p | CAAATCAGTCGTCACTCACT | This study | |||
| fasA | f | CTGCCAAGTCATTTGCAGTT | 225 | M35257.1 | This study |
| r | AATCCGCCATTTGATACCAC | This study | |||
| p | GCTGTGGAAATTGGTACTGA | This study | |||
| eltB-Ip | f | TTCGGAATATCGCAACACAC | 214 | M17873.1 | This study |
| r | TGGTCTCGGTCAGATATGT | 55 | |||
| p | GCCATTGAAAGGATGAAGGAC | This study | |||
| fimF41a | f | CAAAAGCTGAAATGGCTGGT | 200 | M21788.1 | This study |
| r | GACCCGCAACATCCTTATTT | This study | |||
| p | GGTGTTTCTGCAGGTAGTAC | This study | |||
| faeG | f | ACCAATGGTGGAACCAAACT | 173 | M35954.1 | This study |
| r | TTCACCATCAGGGTTTCTGA | This study | |||
| p | TGAAGGAGCTTCTGTAGTAC | This study | |||
| fedA | f | TGCTTCCTTGTCCACTAAAACA | 157 | M61713.1 | This study |
| r | TGCTATTCGACGCCTTAACC | This study | |||
| p | AAACCGTGAACGGTAAAACAC | This study | |||
| estA | f | CAACTGAATCACTTGACTCTTC | 147 | V00612.1 | This study |
| r | CAGCACAGGCAGGATTACAA | This study | |||
| p | GAACAACACATTTTACTGCTGT | This study | |||
| estB | f | CTATTGCTACAAATGCCTATGC | 126 | M35586.1 | This study |
| r | CTCCAGCAGTACCATCTCTA | 55 | |||
| p | GACAAATAGCCAAGGAAAGTTG | This study | |||
| fanA | f | TTGTTCAGGGAGAAACTTGGTT | 110 | X05797.1 | This study |
| r | AGGGCGTAAATACCCCTAGAA | This study | |||
| p | GTGGAGTTATCAAGTATACGTAG | This study | |||
| mPCR 7 | |||||
| ipaH | f | GTTCCTTGACCGCCTTTCCGATACCGTC | 619 | M32063 | 56 |
| r | GCCGGTCAGCCACCCTCTGAGAGTAC | 56 | |||
| p | CCATGGCATGCTGTACTGAA | This study | |||
| daaD | f | TGAACGGGAGTATAAGGAAGATG | 371 | AY525531.1 | 56 |
| r | GTCCGCCATCACATCAAAA | 56 | |||
| p | GGAAGGGGGTGTCGAGGCCCGGT | This study | |||
| eltB | f | TCTCTATGTGCATACGGAGC | 322 | EU113252.1 | 56 |
| r | CCATACTGATTGCCGCAAT | 56 | |||
| p | GGAATAATAAAACCCCCAATTC | This study | |||
| eaeA | f | CCTGGTTACAACATTATGGAACG | 287 | AJ308550.1 | This study |
| r | TGAAATAGTCTCGCCAGTATTCG | This study | |||
| p | GGGCTATAACGTCTTCATTGATC | This study | |||
| stx2 | f | GGCACTGTCTGAAACTGCTCC | 255 | FN252459 | 56 |
| r | TCGCCAGTTATCTGACATTCTG | 56 | |||
| p | CGCCGTGAATGAAGAGAGTCAA | This study | |||
| aggR | f | CGTAAGCCGGGTATGAAAGA | 188 | Z32523.1 | This study |
| r | GCCAGTTCAGAAGCAGGAAC | This study | |||
| p | CTGGAAGGAACTCTCAATGCTG | This study | |||
| estA | f | TTTCCCCTCTTTTAGTCAGTCAA | 159 | AY342057.1 | 56 |
| r | GCAGGATTACAACACAATTCACAGCAG | 56 | |||
| p | TCAGAAAATATGAACAACACATTT | This study | |||
| stx1 | f | CTGGATTTAATGTCGCATAGTG | 150 | HM367099.1 | 56 |
| r | AGAACGCCCACTGAGATCATC | 56 | |||
| p | CAGAGGGGATTTCGTACAACAC | This study | |||
| estB | f | CTATTGCTACAAATGCCTATGC | 126 | M35586.1 | This study |
| r | CTCCAGCAGTACCATCTCTA | 55 | |||
| p | GACAAATAGCCAAGGAAAGTTG | This study | |||
| mPCR 8 | |||||
| chuA | f | TGCCGCCAGTACCAAAGACA | 121 | CP002797.2 | 57 |
| r | TTCAGCATTACTGTATGGCAGTG | This study | |||
| p | GGAGATCACTCCACCCAGCG | This study | |||
| yjaA | f | CTGCCTTCAGTAACCAGCG | 106 | CP002797.2 | This study |
| r | AAAGAATGCCAGGTTGAACG | This study | |||
| p | GCAAGATCTTGTTCTGCAACTC | This study | |||
| TSPE4C2 | f | GAGTAATGTCGGGGCATTCA | 90 | CP002212 | 57 |
| r | GAGACAGAAACGCGGGTAGA | This study | |||
| p | CAAGTTCGATAGTCTGAATA | This study |
PCR conditions were as follows: for mPCR 1, mPCR 3, and mPCR 4, 3 min at 94°C; 40 cycles of 30 s at 94°C, 30 s at 52°C, and 1 min at 72°C; and 5 min at 72°C; for mPCR 2, 3 min at 94°C; 35 cycles of 30 s at 94°C, 30 s at 53°C, and 1 min at 72°C; and 5 min at 72°C; for mPCR 5, 3 min at 94°C; 35 cycles of 30 s at 94°C, 30 s at 57°C, and 1 min at 72°C; and 5 min at 72°C; for mPCR 6 and mPCR 7, 3 min at 94°C; 35 cycles of 60 s at 94°C, 60 s at 55°C, and 90 s at 72°C; and 5 min at 72°C; for mPCR 8 (ECoR), 3 min at 94°C; 30 cycles of 30 s at 94°C, 30 s at 56.5°C, and 1 min at 72°C; and 5 min at 72°C.
f, forward; r, reverse (5′-Cy5); p, probe (5′-aminohexyl plus 10 T residues).
E. coli heat lysates were prepared by inoculating a single E. coli colony in 1 ml of LB medium and incubation at 37°C and 180 rpm overnight. Bacteria were centrifuged, and the supernatants were discarded. Bacterial pellets were resuspended in 300 ml double-distilled H2O (ddH2O), boiled for 10 min, and incubated on ice. After centrifugation (14,000 rpm), supernatants were stored at −20°C until further use. Lysates contained approximately 230 ng DNA/μl.
Adhesion assays. (i) Cell culture conditions.
All the cells were grown in 96-well plates (Nunc, Germany) in an atmosphere of 5% CO2 at 37°C and were used when they reached confluence, which usually occurred after 2 to 4 days. Caco-2 cells were grown in Dulbecco modified Eagle medium (DMEM)–Ham's F-12 medium (1:1), and 5637 cells were grown in RPMI 1640 supplemented with 1 mM Na pyruvate. Both media were supplemented with 10% fetal calf serum (FCS) and 2 mM l-glutamine. IPEC-J2 (28) and PK-15 cells were grown in DMEM–Ham's F-12 medium (1:1) supplemented with 5% FCS and 2 mM l-glutamine. All cell culture media were from Biochrom (Germany).
(ii) E. coli adhesion assays.
All E. coli adhesion assays were carried out as previously described (2). The basic principle is represented schematically in Fig. 1. E. coli cells were grown overnight in LB medium to an optical density at 600 nm (OD600) of 0.8 to 1.2. Confluent cell monolayers were inoculated with E. coli with an infection dose of 62,500 bacteria per mm2 of a cell monolayer using a conversion of 3 × 108 bacteria/ml/OD600. This unit was necessary due to the different sizes of cells of different cell lines. However, the infection dose corresponded to a multiplicity of infection (MOI) of 100:1 E. coli cells to host cells according to the results for IPEC-J2 cells. Epithelial (Caco-2, IPEC-J2, 5637, PK-15) cells were incubated with the respective E. coli isolate for 4 h at 37°C. Cells were washed three times with 1× PBS to remove nonadherent E. coli. Cells and adherent bacteria were fixed with 4% paraformaldehyde in ddH2O and kept at 4°C until further use. After fixation, the wells were washed three times with 1× PBS and all cells and bacteria were stained with propidium iodide (10 μg/ml in ddH2O) for 15 min. All determinations were carried out three times, and tests were repeated at least three times.
The sampling of E. coli from different host species was the first stage in the process and was defined as species specificity. The cell line origin (human or porcine cells) was defined as host specificity. The tissue type of a cell line (intestinal or urinary tract epithelial cells) was defined as tissue specificity. The case of a bacterium adhering to all cell lines was defined as unspecific adhesion.
Bacterial adhesion was classified into nonadherent (less than 1,000 bacteria/mm2 of a cell monolayer) and adherent (more than 1,000 bacteria/mm2). Adherent bacteria were then subclassified into low-level-adherent (1,000 to 4,999 bacteria/mm2), medium-level-adherent (5,000 to 9,999 bacteria/mm2), and strongly adherent (>10,000 bacteria/mm2) bacteria. Less than 1,000 bacteria/mm2 is equivalent to less than one bacterium per epithelial cell and also the background noise of negative controls (cell monolayers without bacteria). The background was defined on the basis of the analysis of 300 wells of each cell line not containing bacteria.
VideoScan fluorescence imaging technology.
The VideoScan technology implemented in the Aklides system (GA Generic Assays GmbH, Dahlewitz, Germany) is a versatile fluorescence microscope imaging technology for the analysis of fluorescent objects (27, 29–31). The VideoScan instrument was automatically guided to each well of a 96-well plate, where it focused on microbeads or cell monolayer surfaces and took images, which were saved as .bmp files. These images were analyzed by a microbead-analyzing or a cell/bacterium-analyzing VideoScan software module.
The quantities of microbead/probe-hybridized PCR products were calculated by measuring the intensity of the Cy5 fluorescence corona around a microbead. The ultimate parameter, the referenced mean fluorescence intensity (refMFI), was referenced to the intensity of the dyes encoding the microbeads and correlated with the amount of hybridized PCR products and, therefore, with the quantity of a PCR product (27, 32).
Each image from an adhesion assay was divided into 108 square tiles (12 by 9 tiles), with each tile having a side length of 37 μm. If brightness (the mean gray value of a tile) was above a threshold value, this tile was considered to be overexposed. Such areas displayed high unspecific background staining. Blurred areas resulted from inhomogeneous three-dimensional cell growth and the limited depth of field of the objective (about 5 μm). If the numerical value of focus of a tile was below a defined threshold, the tiles were defined as blurred. Defects in monolayers were also defined in terms of the focus threshold. The focus threshold was determined empirically on one occasion for one cell line and remained constant throughout all measurements. Tiles with overexposed and blurred areas, as well as areas not containing cells and their neighboring tiles, were excluded from further processing. Moreover, the exclusion of neighboring tiles permitted good analysis of partially valid images.
The calculation of adherent bacteria on the cell monolayer was examined using a special image-data-processing algorithm of VideoScan. The definition of a single bacterium depended on its shape and size. A nonlinear image filter specifically reinforced all structures of this dimension and dampened any other image structures. The resulting intermediate images underwent conventional object recognition with dynamically adjusted binarization. All objects displaying an intensity of shape, size, or color unusual for E. coli were excluded, and any remaining interferences were eliminated from all further calculations. The number of objects remaining therefore represented the number of E. coli cells. A minimum cell monolayer area of 0.3 mm2 per well was screened for bacterial adhesion. The bacteria were quantified, and adhesion data were exported as the number of bacteria/mm2. Approximately 150,000 images were analyzed for the purposes of this study.
Statistical analysis.
The P values with regard to the significance of phylogenetic groups were calculated using Pearson's chi-square test (implemented in the statistical software SPSS Statistics, version 17.0; SPSS, Chicago, IL). On the basis of the Wilcoxon signed-rank test, the adhesion preference of isolates from certain species, as well as the ratio of the occurrence of the VAGs to adhesion to epithelial cell lines, was analyzed using the RKWard program, version 0.57, using either dedicated R packages and custom-made scripts or RKWard plug-ins (33). Probability values of less than 0.05 were considered statistically significant.
RESULTS
We have established multiplex PCR microbead assays (MPMAs) which enable the clear and rapid identification of PCR products. In the course of the confirmatory tests, we defined MPMA cutoff values for each gene and classified MPMA results into three categories: positive, negative, and questionable. If the MPMA results for single genes of an isolate proved to be inconclusive during two mPCRs, the result for the gene in question was independently validated by two single PCRs and agarose gel electrophoresis (AGE). Comparing the MPMA with AGE, which was done for about 3,000 PCRs, the MPMA results could be confirmed, with two exceptions. The malX gene was detectable in five isolates and the csgA gene was detectable in one isolate via AGE but not via MPMA. For clarification, the respective PCR products were sequenced. We found mismatches in the reverse DNA strands of the bacterial lysates either in the area of capture probe sequences or in the primer region responsible for the missing of detection via MPMA. In contrast to conventional PCR, which is defined by primers only, the specificity of the MPMA is increased by using the primer and the capture probe to exclude unspecific PCR products. The MPMA is more sensitive than AGE analysis for the detection of PCR products. While both detection systems report identical results with template DNA concentrations higher than 0.83 ng, VideoScan was still able to exactly detect PCR products below this concentration.
exVAGs.
Each exVAG was found in at least one isolate, with ompA (99.7%), fimC (95.3%), and csgA (90.9%) having the greatest prevalence and afa-dra (0.6%, in human isolates only), cnf1, cnf2 (0.6%), and sat (2.8%) having the lowest prevalence. Detailed information is given in Table 4 and 5 and in Table S1 in the supplemental material. Human UPEC isolates had the highest number of exVAGs per isolate (mean ± standard deviation, 13.3 ± 3.8 exVAGs), followed by badger (Meles meles) isolates (10.4 ± 5.1), roe deer (Capreolus capreolus) isolates (9.8 ± 2.5), human intestinal isolates (9.7 ± 5.9), red squirrel (Sciurus vulgaris) isolates (9.4 ± 4.3), and sparrow hawk (Accipiter nisus) isolates (9.4 ± 4.8). In contrast to human UPEC isolates, porcine UPEC isolates exhibited a relatively low number of exVAGs per isolate (8.7 ± 3.6). The isolate with the lowest number of exVAGs was from a hedgehog (Erinaceus europaeusnt>), with only one exVAG (ompA). The isolates containing the highest number of exVAGs came from a single human UPEC isolate and a single sparrow hawk isolate, each having 20 exVAGs.
Table 4.
Prevalence and diversity of VAGs in E. coli isolates from 19 different species
| Species | exVAGs |
inVAGs |
||||
|---|---|---|---|---|---|---|
| No. of isolates | All VAGs: mean % | Mean % | Diversity (no. of exVAGs) | Mean % | Diversity (no. of inVAGs) | |
| Human, healthy | 19 | 9.8 | 9.7 | 26 | 0.1 | 2 |
| Human, UPEC | 20 | 13.4 | 13.3 | 30 | 0.1 | 2 |
| Domestic pig, healthy | 22 | 8.3 | 8.3 | 25 | 0 | 1 |
| Domestic pig, UPEC | 15 | 8.7 | 8.7 | 18 | 0 | 0 |
| Roe deer | 23 | 10.3 | 9.8 | 18 | 0.5 | 3 |
| European hedgehog | 22 | 5.3 | 5.3 | 15 | 0 | 1 |
| European hare | 8 | 5.4 | 5 | 10 | 0.4 | 1 |
| European otter | 7 | 7 | 7 | 19 | 0 | 0 |
| Marten | 19 | 6.5 | 6.4 | 19 | 0.2 | 1 |
| European badger | 7 | 10.6 | 10.4 | 21 | 0.1 | 1 |
| House mouse | 9 | 7.1 | 7.1 | 12 | 0 | 0 |
| European rabbit | 6 | 5.2 | 5 | 9 | 0.2 | 1 |
| Raccoon | 22 | 7 | 7 | 23 | 0.1 | 1 |
| Brown rat | 4 | 9.3 | 9.3 | 16 | 0 | 0 |
| Red squirrel | 17 | 9.5 | 9.4 | 20 | 0.1 | 1 |
| Wild boar | 22 | 5.4 | 5.4 | 16 | 0 | 0 |
| Red fox | 21 | 6.1 | 6 | 18 | 0.1 | 2 |
| Eurasian sparrow hawk | 13 | 9.4 | 9.4 | 25 | 0 | 0 |
| Long-eared owl | 5 | 7 | 7 | 15 | 0 | 0 |
| Common buzzard | 14 | 8.4 | 8.4 | 19 | 0 | 0 |
| Common blackbird | 22 | 8.3 | 8.3 | 22 | 0 | 0 |
Table 5.
Prevalence of exVAGs and inVAGs in E. coli isolates from 19 different speciesa
| VAG | Gene/operon | % of the following species with isolates carrying the indicated gene(s): |
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Human, healthy | Human, UPEC | Domestic pig, healthy | Domestic pig, UPEC | Roe deer | European hedgehog | European hare | European otter | Marten | European badger | House mouse | European rabbit | Raccoon | Brown rat | Red squirrel | Wild boar | Red fox | Eurasian sparrow hawk | Long-eared owl | Common buzzard | Common blackbird | ||
| exVAGs | ||||||||||||||||||||||
| Adhesins | afa-dra | 5 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| csgA | 100 | 100 | 86 | 93 | 91 | 86 | 88 | 86 | 74 | 100 | 100 | 83 | 77 | 100 | 100 | 100 | 90 | 92 | 80 | 93 | 91 | |
| fimC | 89 | 90 | 91 | 93 | 100 | 91 | 100 | 100 | 95 | 100 | 100 | 83 | 95 | 100 | 94 | 100 | 100 | 100 | 100 | 100 | 91 | |
| hra | 5 | 30 | 14 | 0 | 57 | 45 | 13 | 29 | 37 | 43 | 11 | 0 | 9 | 25 | 35 | 36 | 52 | 8 | 20 | 79 | 41 | |
| mat | 68 | 100 | 82 | 93 | 96 | 86 | 75 | 71 | 79 | 100 | 100 | 67 | 77 | 100 | 94 | 9 | 95 | 92 | 60 | 93 | 82 | |
| papC | 5 | 35 | 5 | 0 | 0 | 0 | 0 | 14 | 0 | 29 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| sfa-foc | 16 | 20 | 14 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 25 | 6 | 0 | 5 | 23 | 0 | 14 | 0 | |
| tsh | 5 | 15 | 27 | 33 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 33 | 14 | 0 | 0 | 0 | 0 | 15 | 0 | 21 | 23 | |
| Invasins | ibeA | 32 | 15 | 5 | 0 | 0 | 0 | 0 | 14 | 11 | 86 | 22 | 0 | 23 | 50 | 47 | 14 | 5 | 38 | 20 | 36 | 27 |
| tia | 5 | 10 | 0 | 0 | 78 | 0 | 0 | 0 | 11 | 14 | 0 | 0 | 0 | 0 | 24 | 5 | 10 | 15 | 0 | 29 | 18 | |
| Toxins | astA | 0 | 20 | 18 | 7 | 65 | 14 | 13 | 29 | 26 | 29 | 56 | 17 | 27 | 0 | 29 | 36 | 29 | 23 | 60 | 14 | 36 |
| cnf1-cnf2 | 0 | 5 | 0 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| hlyA | 0 | 5 | 0 | 0 | 9 | 5 | 0 | 0 | 0 | 0 | 11 | 17 | 5 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | |
| sat | 16 | 30 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| vat | 26 | 30 | 9 | 0 | 22 | 0 | 0 | 14 | 16 | 0 | 33 | 0 | 23 | 50 | 47 | 0 | 10 | 23 | 20 | 14 | 32 | |
| Protectins | cvi-cva | 21 | 30 | 36 | 33 | 9 | 0 | 0 | 14 | 0 | 43 | 0 | 0 | 9 | 0 | 0 | 0 | 0 | 8 | 0 | 0 | 0 |
| iss | 26 | 45 | 23 | 33 | 4 | 0 | 0 | 14 | 0 | 14 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 8 | 0 | 0 | 0 | |
| ompA | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 95 | 100 | 100 | 100 | 100 | |
| traT | 42 | 70 | 55 | 73 | 65 | 18 | 63 | 57 | 21 | 57 | 78 | 50 | 27 | 75 | 47 | 41 | 24 | 62 | 60 | 21 | 32 | |
| Siderophores | chuA | 53 | 70 | 23 | 0 | 57 | 32 | 25 | 29 | 53 | 43 | 22 | 50 | 50 | 50 | 65 | 27 | 29 | 62 | 80 | 71 | 55 |
| fyuA | 47 | 75 | 23 | 33 | 26 | 9 | 0 | 14 | 21 | 14 | 0 | 0 | 27 | 50 | 53 | 18 | 14 | 46 | 20 | 21 | 55 | |
| ireA | 5 | 15 | 9 | 0 | 87 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 5 | 0 | 6 | 5 | 0 | 8 | 0 | 0 | 9 | |
| iroN | 37 | 55 | 32 | 33 | 0 | 5 | 13 | 0 | 16 | 57 | 0 | 0 | 9 | 25 | 6 | 23 | 5 | 38 | 20 | 21 | 23 | |
| irp2 | 58 | 75 | 32 | 33 | 30 | 5 | 0 | 14 | 21 | 29 | 0 | 0 | 27 | 50 | 47 | 18 | 14 | 46 | 20 | 21 | 36 | |
| iucD | 37 | 65 | 32 | 40 | 0 | 0 | 0 | 14 | 0 | 43 | 0 | 0 | 9 | 0 | 0 | 0 | 0 | 15 | 0 | 0 | 5 | |
| iutA | 32 | 60 | 23 | 40 | 0 | 0 | 0 | 14 | 0 | 43 | 0 | 0 | 9 | 0 | 0 | 0 | 0 | 15 | 0 | 0 | 9 | |
| sitchr | 63 | 75 | 14 | 27 | 13 | 23 | 13 | 29 | 26 | 29 | 0 | 0 | 32 | 50 | 47 | 5 | 10 | 31 | 20 | 36 | 32 | |
| sitep | 37 | 45 | 64 | 93 | 0 | 0 | 0 | 29 | 5 | 43 | 0 | 0 | 9 | 0 | 0 | 0 | 0 | 23 | 0 | 0 | 5 | |
| Miscellaneous | malX | 42 | 30 | 5 | 7 | 39 | 5 | 0 | 0 | 16 | 29 | 0 | 0 | 27 | 25 | 41 | 5 | 10 | 31 | 20 | 21 | 27 |
| pic | 0 | 10 | 9 | 0 | 30 | 0 | 0 | 14 | 5 | 0 | 78 | 0 | 0 | 50 | 41 | 0 | 10 | 15 | 0 | 36 | 5 | |
| inVAGs | ||||||||||||||||||||||
| Adhesins | daaD | 5 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| eaeA | 0 | 0 | 0 | 0 | 0 | 5 | 38 | 0 | 16 | 14 | 0 | 17 | 9 | 0 | 12 | 0 | 5 | 0 | 0 | 0 | 0 | |
| Toxins | estA | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 |
| estB | 0 | 5 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| stx1 | 5 | 0 | 0 | 0 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| stx2e | 0 | 0 | 0 | 0 | 39 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
VAGs not identified (fanA, fedA, fasA, faeG, fimF41a, ipaH, eltB-Ip, aggR) were excluded.
All 30 exVAGs were detectable in human UPEC isolates. On the other hand, many exVAGs in other E. coli groups, such as those from hedgehog or wild boar (Sus scrofa), could not be identified. A total of 15 exVAGs could not be identified in the case of hedgehogs, while 14 exVAGs were not detectable in wild boar. The prevalence of single exVAGs differed markedly between E. coli groups. If one exVAG had a very high prevalence in one group, this gene was not necessarily found in another group. For example, the gene ireA was found in roe deer isolates with a prevalence of 87% but was found with a 0% prevalence in porcine UPEC, red fox (Vulpes vulpes), hedgehog, and common buzzard (Buteo buteo) E. coli isolates. Detailed information is given in Table 4.
Although the prevalences of the exVAGs sitchr (75%), iroN (55%), iucD (65%), papC (35%), iss (45%), iutA (60%), mat (100%), fyuA (75%), sat (30%), and irp2 (75%) were the highest in human UPEC isolates, other exVAGs were more prevalent in other E. coli groups: pic (41%), ibeA (47%), and vat (47%) in the red squirrel; astA (65%), ireA (87%), and tia (78%) in the roe deer; malX (42%) in the human intestine; cvi-cva (36%) in the domestic pig; traT (73%), sitep (93%), and tsh (33%) in porcine UPEC isolates; and hra (79%) and chuA (71%) in the common buzzard. For more details see Table 5.
Human UPEC isolates carried a greater prevalence of papC (P < 0.05) and mat (P < 0.01) than human commensal isolates. Porcine UPEC isolates carried a greater prevalence of sitep (P < 0.05) than porcine commensal isolates. Wild bird isolates (n = 54) carried a greater prevalence of the following exVAGs than wild mammal isolates (n = 187): sfa-foc, ibeA, irp2, tsh, and fyuA(P < 0.05) and iroN and chuA (P < 0.01).
inVAGs.
inVAGs were identified only sporadically: estB (prevalence, 0.6%), estA (0.6%), stx1 (0.9%), stx2e (2.8%), daaD (0.6%), and eaeA (4.4%). All inVAGs were found in intestinal E. coli isolates, with the exception of one human UPEC isolate carrying the estB gene and another human UPEC carrying the daaD gene. All nine stx2e-positive isolates were from roe deer. Both afa-dra- and daaD-positive isolates were from humans.
ECoR groups.
E. coli isolates were classified into phylogenetic groups according to the ECoR (E. coli Reference Collection) system. Most isolates belonged to ECoR group B1 (34.4%), followed by ECoR groups D (23.3%), A (21.8%), and B2 (20.6%). On average, ECoR group B2 isolates carried the most exVAGs per isolate (mean, 13 exVAGs; median 13, exVAGs), followed by isolates of groups D (mean, 8.1 exVAGs; median, 7 exVAGs), B1 (mean, 6.5 exVAGs; median, 6 exVAGs), and A (mean, 5.4 exVAGs; median, 5 exVAGs).
In comparing human commensal isolates (n = 19) with human UPEC isolates (n = 20), it was found that the isolates belonged to ECoR groups A (36.8% as against 5%), B1 (10.5% as against 25%), B2 (42.1% as against 45%), and D (10.5% as against 25%). In the comparison of porcine commensal isolates (n = 22) with porcine UPEC isolates (n = 15), it was found that isolates belonged to ECoR groups A (72.7% as against 53.3%), B1 (4.6% as against 46.7%), B2 (4.6% as against 0%), and D (18.2% as against 0%). Therefore, human intestinal and UPEC isolates belonged predominantly to ECoR group B2 (P < 0.05) and porcine intestinal isolates belonged predominantly to group A (P < 0.05), while porcine UPEC isolates exclusively belonged to groups A and B1. The comparison of wild mammal isolates (n = 187) with wild bird isolates (n = 54) showed that isolates belonged to ECoR groups A (17.6% as against 7.4%), B1 (41.2% as against 31.5%), B2 (18.2% as against 24.1%), and D (23% as against 37%). Wild mammal isolates therefore belonged predominantly to ECoR group B1 (P < 0.05) and wild bird isolates belonged predominantly to ECoR group D (P < 0.05). The relevant information is given in Table 2.
exVAGs such as pic, ibeA, iroN, vat, mat, fyuA, malX, and irp2 were significantly more prevalent in ECoR group B2 isolates than in ECoR group A, B1, and D isolates (P was at least <0.05). traT, sitchr, csgA, fimC, ireA, tia, and chuA were significantly more prevalent in the ECoR group B2 isolates than in group A, B1, or D isolates (P was at least <0.05).
Adhesion.
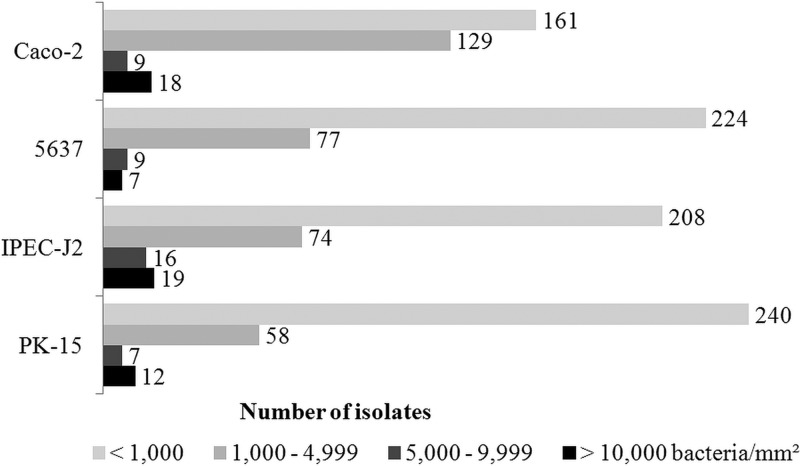
One hundred eight isolates (33.8%) were nonadherent to all cell lines. In comparison, only two isolates (from a squirrel and a roe deer) were strongly adherent to all four cell lines. The highest adhesion numbers were 56,040 bacteria/mm2 for Caco-2 cells, 37,966 bacteria/mm2 for 5637 cells, 64,766 bacteria/mm2 for IPEC-J2 cells, and 58,031 bacteria/mm2 for PK-15 cells. Isolates were strongly adherent to IPEC-J2 (19 isolates, 6.0%), Caco-2 (18 isolates, 5.7%), PK-15 (12 isolates, 3.8%), and 5637 (7 isolates, 2.2%) cells (Fig. 2).
Fig 2.
Rates of adhesion of 317 E. coli isolates to Caco-2, 5637, IPEC-J2, and PK-15 cells. Epithelial cells were incubated with E. coli for 4 h. Adhesion was quantified after removing nonadherent bacteria. Numerical values at the end of each bar represent the number of isolates. Most isolates did not adhere to any cell line. <1,000 bacteria/mm2, nonadherent; 1,000 to 4,999 bacteria/mm2, low level of adherence; 5,000 to 9,999 bacteria/mm2, medium level of adherence; >10,000, strongly adherent.
There were also isolates which were strongly adherent to one cell line but were nonadherent to others. For example, one rabbit isolate adhered to Caco-2 cells (21,689 bacteria/mm2), IPEC-J2 cells (23,036 bacteria/mm2), and PK-15 cells (10,116 bacteria/mm2) but not to 5637 cells. One human UPEC isolate adhered to IPEC-J2 cells (21,060 bacteria/mm2) but not to Caco-2, 5637, and PK-15 cells.
There were no significant differences between the adhesion of human UPEC isolates and human intestinal isolates to Caco-2 and 5637 cells and no significant differences between the adhesion of porcine UPEC isolates and porcine intestinal isolates to IPEC-J2 and PK-15 cells. Wild bird isolates adhered much better than wild mammal isolates to all four mammalian cell lines, especially IPEC-J2 and Caco-2 cells (P < 0.05). This means that there were more nonadherent wild mammal isolates than nonadherent wild bird isolates.
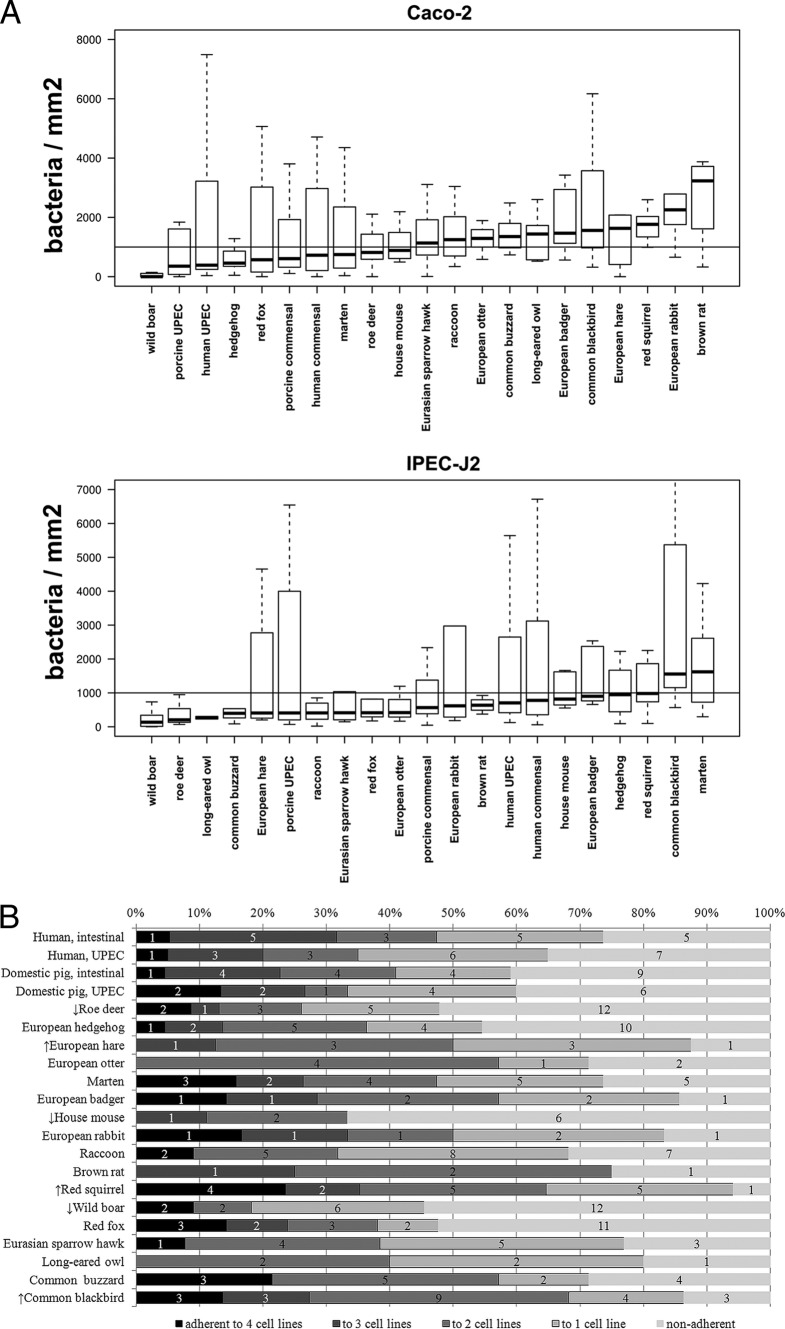
In general, the ECoR group B2 isolates adhered much better than the ECoR group A, B1, and D isolates (P < 0.05). Taking into account the exclusion of nonadherent isolates, wild animal ECoR group A isolates adhered better to Caco-2 cells than the adherent ECoR group D isolates (P < 0.05) and adhered much better to 5637 cells than the adherent ECoR group B2 isolates (P < 0.05). The adherent human UPEC isolates adhered better to 5637 and PK-15 cells than the adherent human commensal isolates (P < 0.05). There was adhesion to all four cell lines specific to the animal groups (Fig. 3).
Fig 3.
(A) Species-specific adhesion visualized by box plots. The median adhesion (bacteria/mm2, y axis) of the isolates from each animal group to Caco-2 and IPEC-J2 cells is shown here as horizontal bold bars in the boxes. Many isolates were nonadherent. This explains why the median is often below the background noise of 1,000 bacteria/mm2 (horizontal line). (B) Species-specific adhesion of E. coli isolates to Caco-2, 5637, IPEC-J2, and PK-15 cells. Bordered gray bars, percentage of bacteria which adhered to four, three, two, or one cell line; nonbordered light gray bars, percentage of nonadherent bacteria. The numbers of isolates are given in the bars. The arrows indicate that the isolates of these species have either the highest (↑) or lowest (↓) average number of adhesion rates.
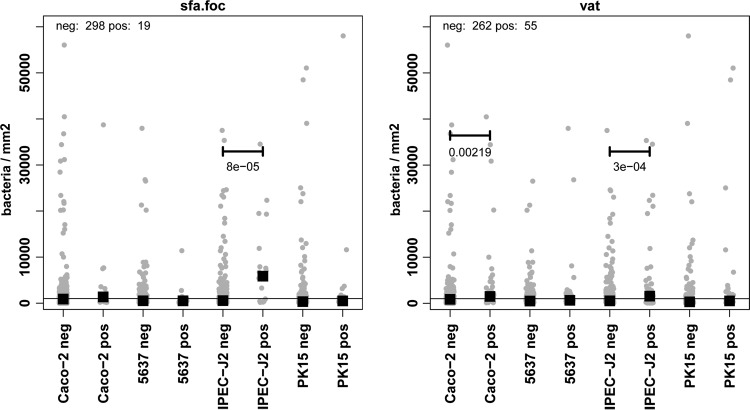
Our goal was to find links between the occurrence of a single VAG and adhesion to a single cell line. We therefore grouped all isolates into positive or negative for this gene and compared the adhesion of these two groups to the cell lines. There were several significant links which met both criteria: first, where the number of adherent bacteria was greater than the background noise of 1 bacterium per epithelial cell, and second, where, on average, adhesion of VAG-positive E. coli isolates differed significantly (P < 0.05) from adhesion of VAG-negative E. coli isolates. All significant associations demonstrated that VAG-positive E. coli isolates are better able to adhere than VAG-negative E. coli isolates, with the following associations being detected: Caco-2 cells with afa-dra and daaD (number of VAG-positive isolates, n = 2; increase from 0 to 24,600 bacteria/mm2), pic (n = 39; increase from 0 to 330 bacteria/mm2), ibeA (n = 57; increase from 0 to 355 bacteria/mm2), vat (n = 55; increase from 0 to 491 bacteria/mm2), tsh (n = 31; increase from 0 to 918 bacteria/mm2), mat (n = 257; increase from 0 to 81 bacteria/mm2), fyuA (n = 94; increase from 0 to 283 bacteria/mm2), malX (n = 62; increase from 0 to 355 bacteria/mm2), and irp2 (n = 94; increase from 0 to 283 bacteria/mm2); 5637 cells with afa-dra and daaD (n = 2; increase from 0 to 2,874 bacteria/mm2), estB (n = 2; increase from 0 to 4,277 bacteria/mm2), and stx1 (n = 3; increase from 0 to 418 bacteria/mm2); and IPEC-J2 cells with sfa-foc (n = 19; increase from 0 to 4,857 bacteria/mm2), vat (n = 55; increase from 0 to 556 bacteria/mm2), and papC (n = 12, increase from 0 to 649 bacteria/mm2). These findings are depicted schematically for sfa-foc and for vat in Fig. 4.
Fig 4.
Correlation between adhesion rates and VAG occurrence. Epithelial cells were incubated with E. coli for 4 h. Adhesion was quantified after removing nonadherent bacteria. All E. coli isolates were grouped into isolates which were negative (neg) or positive (pos) for a VAG, for example, Caco-2 neg with rates of adhesion of sfa-foc-negative bacteria to Caco-2 cells. Black squares, median for each group; horizontal line, background noise of 1,000 bacteria/mm2.
In order to validate these results, which are based on low isolate numbers, we tested some other isolates from our E. coli laboratory collection (Brandenburgische Technische Universität Cottbus-Senftenberg and Institut für Mikrobiologie und Tierseuchen, Freie Universität Berlin) for adhesion. For Caco-2 cells and afa-dra and daaD, we discovered an additional 16 isolates which were positive for these genes (8 human UPEC isolates and 8 porcine intestinal commensal isolates). The mean adhesion of all 18 afa-dra- and daaD-positive isolates on Caco-2 cells increased from 0 to 2,845 bacteria/mm2. In the case of 5637 cells and estB, we found that an additional 6 porcine intestinal commensal isolates tested positive for this gene. The median adhesion of all 8 estB-positive isolates on 5637 cells was below the background noise of 1,000 bacteria/mm2 (horizontal line). With 5637 cells and stx1, we found an additional 10 bovine intestinal commensal isolates which tested positive for this gene. The adhesion of all 13 stx1-positive isolates on 5637 cells was below the background noise of 1,000 bacteria/mm2.
DISCUSSION
Using newly developed screening methods, we characterized the genotypes and phenotypes of a large number of E. coli isolates to study any possible associations between the occurrence of a VAG and adhesion to multiple epithelial cell lines. Genotypes were characterized using our multiplex PCR microbead assay (MPMA). We were able to replace time-consuming agarose gel electrophoresis with an automated high-throughput approach. To the best of our knowledge, there is currently no comparable microbead-based platform for low-density DNA hybridization assays available for routine analysis. Commonly, microbead-based assays are used in suspension with direct hybridization of DNA to capture probes and measurement by flow cytometer-based technologies (34, 35). Suspension-array technologies (SAT) have higher technical demands due to the need for a fluidic system and readout. In contrast, our MPMAs used randomly ordered microbeads which were fixed on the well bottom. Microbeads and multiwell plates for the MPMA can be prepared in bulk with a defined layout ready for high-throughput analysis. In comparison with SATs, we used MPMA primers with covalently linked dyes during the PCRs. The samples were ready for analysis after one washing step. There is therefore no need for postlabeling, as is found in other systems (cf. reference 36). In addition, and again, in contrast to SATs, neither ligand artifacts nor microbead aggregates disturb the assay, as such artifacts or aggregates are well recognized and excluded from analysis by the system. Adhesion phenotypes were characterized by an assay without time-consuming lysis of adherent bacteria and subsequent plating of dilutions, incubation overnight, and counting of the colonies by hand. The results of these combined assays for high-throughput screening enabled an overview of genotypic characteristics in association with the phenotypic characteristics of many E. coli isolates.
In general, there was a low prevalence of inVAGs. This can be regarded as normal, since the E. coli isolates did not come from diarrheic hosts. However, a few exceptions, such as E. coli isolates from roe deer (stx positive) or the European hare (Lepus europaeus) (eae positive), indicate that roe deer are more likely to be a reservoir host for Shiga toxin-producing E. coli (STEC) than other hosts and that the same applies to the European hare with regard to enteropathogenic E. coli (EPEC). The occurrence of STEC in roe deer has already been mentioned (37). The common occurrence of EPEC in the European hare and in other wild animal hosts remains to be verified in more comprehensive studies.
The high prevalence of several exVAGs, such as ompA, fimC, and csgA, reflects their common occurrence in all E. coli strains. The high prevalence of other exVAGs in a specific animal group, such as ireA and tia in roe deer, indicates a certain role of such genes in colonization. The validation of this hypothesis using an appropriate animal infection model (according to species, nutrition, and environment) is nearly impossible. This and all similar hypotheses can be substantiated only by further epidemiological studies.
Differences in the prevalence of VAGs in animal groups were also noticeable when comparing wild bird and wild mammal isolates. Many exVAGs and, in particular, iron acquisition genes (sfa-foc, ibeA, tsh, irp2, iroN, chuA, fyuA) were more prevalent in wild bird isolates. This difference would appear to be quite relevant due to the high number of isolates included (54 wild bird and 187 wild mammal isolates) and indicates a role in colonization and a difference between the two intestinal environments (birds versus mammals).
In general, human UPEC isolates contained the largest amount of exVAGs, a fact which is already known and has been explained on the basis of their extraintestinal colonization/infection (38). Wild animal intestinal E. coli groups (badgers, squirrels) also carried high numbers of exVAGs, and one of the two isolates containing the largest amount of exVAGs was a sparrow hawk isolate with 20 exVAGs (similar to one human UPEC isolate). This shows that ExPEC and wild animal E. coli isolates, especially from wild birds, the European badger, the red squirrel, and the roe deer, can be very similar, and zoonotic risks are conceivable (39, 40).
All 30 exVAGs examined in human UPEC isolates and 26 exVAGs in human intestinal isolates were detectable. Such diversity has not been seen in any other E. coli group, and several isolates, such as the hedgehog or wild boar E. coli isolates, contained only half of the tested exVAGs. This supports the hypothesis that the human intestine is the most probable reservoir for human UPEC (22, 39, 41). The high number and the great diversity of exVAGs in human intestinal isolates compared to the number and diversity in wild animal isolates might also reflect the fact that the human intestinal environment is more adaptive than the wild animal intestinal environment, something that is particularly due to the very diverse nature of human nutrition. Any E. coli isolate in the human intestinal tract has to adapt rapidly to changing conditions and, thus, to a more complex ecological niche, and this need for rapid adaptation is supported by a broad diversity of exVAGs (40).
We can confirm here the findings of several other studies regarding the classification of isolates into ECoR groups. Comparing E. coli isolates from humans with those from animals, it turned out that human isolates belonged predominantly to ECoR group B2, in contrast to animal isolates, which belonged to ECoR group B1 (40). However, the prevalence of ECoR group B2 isolates from human urinary tract infections was lower in our study than in other studies (42, 43). A general absence of ECoR group B2 isolates from porcine urinary tract infections, as seen in our actual study, has to be verified in future studies. Wild birds in temperate climates mostly harbor ECoR group D isolates (40). The ECoR group B2 isolates carried exVAGs, such as pic, ibeA, iroN, vat, mat, fyuA, malX, irp2, traT, sitchr, csgA, fimC, ireA, tia, and chuA, at a greater prevalence than the isolates of all other ECoR groups, a phenomenon which has already been mentioned in other studies (44–46).
Using newly developed adhesion assays, we quantified the adhesion of all 317 isolates to four different epithelial cell lines. Adhesion rates were generally found to be very diverse. Isolates displayed cell-specific, host-specific, tissue-specific, and, indeed, unspecific adhesion. However, one-third of the isolates were nonadherent, and only two isolates were strongly adherent to all cell lines. This indicates a broad variety of different E. coli colonization strategies, including expression of different adhesins.
In general, isolates more easily adhered to human cell lines than to porcine cell lines. This is a phenomenon which we cannot explain at the moment. The better adhesion of wild bird isolates than mammal isolates to mammalian intestinal cells is also something not yet explicable. Greater general adhesion to intestinal cell lines than to urinary tract cell lines can be explained on the basis of testing of intestinal isolates. Nonetheless, the strong rates of adhesion of several fecal isolates to 5637 urinary epithelial cells (17 isolates) and to PK-15 cells (1 isolate) indicates their uropathogenic potential and supports the fecal-perineal-urethral hypothesis (41). This hypothesis is also supported by the fact that there were no significant differences between the adhesion of human UPEC and human intestinal isolates to Caco-2 and 5637 cells and between the adhesion of porcine UPEC and porcine intestinal isolates to IPEC-J2 and PK-15 cells.
It is worth mentioning that species-specific adhesion was quite noticeable. Sixteen of the 17 isolates (94%) from the red squirrel and 7 of the 8 isolates (88%) from the European hare adhered to at least one cell line tested (mostly to Caco-2 cells). This was in contrast to the findings for many nonadherent isolates, like those from the house mouse, with 3 of the 9 isolates being adherent (33%), and from wild boars, with 10 of 22 isolates being adherent (45%). This indicates that one E. coli adhesin or one colonization strategy of commensal E. coli can be specific for one species.
Few studies have given any information on the role of exVAGs as intestinal colonization factors in vivo (16, 18, 47) and in vitro (2, 48). We can confirm here a link between the occurrence of the exVAGs afa-dra and sfa-foc and a high level of adhesion to epithelial cells. We can now demonstrate that adhesion via afa-dra and sfa-foc is restricted to one cell line (for afa-dra, Caco-2 cells; for sfa-foc, IPEC-J2 cells). tsh was previously defined to be a gene associated with adhesion to red blood cells, bovine mucin, turkey hemoglobin, mouse collagen and laminin, and human fibronectin (49). Our data implicate that tsh additionally supports adhesion to Caco-2 cells. mat was previously defined to be a gene associated with adhesion to HEp-2, HeLa, and HT29 cells (50, 51). We now show that mat can support adhesion to Caco-2 cells. The pap operon was previously associated with adhesion to Galα1-4Gal-containing glycolipids present on both human erythrocytes, especially of blood group P, and uroepithelial cells, such as human bladder carcinoma T24 cells (52, 53). We can now show that papC-positive isolates adhered specifically to IPEC-J2 cells.
Moreover, we discovered that the occurrence of other VAGs (pic, ibeA, vat, fyuA, malX, and irp2) was associated with increased adhesion to a specific cell line. While this increase was not drastic, it was still quite significant. These VAGs have a function in adhesion not known so far, or their presence is associated with other adhesion genes that have not been tested. The connection of these virulence factors to adhesion is supported by the high numbers of isolates which provide the basis for analysis: pic (n = 39 isolates with this exVAG), ibeA (n = 57), vat (n = 55), tsh (n = 31), mat (n = 257), fyuA (n = 94), malX (n = 62), irp2 (n = 94), and papC (n = 12). The association of each exVAG with adhesion to mostly one cell line indicates only that E. coli isolates with a broad variety of exVAGs might be better able to colonize different microhabitats. In addition, several exVAGs in parallel support adhesion in one microhabitat, which is obvious for Caco-2 cells, as most significant associations were observed between a VAG and Caco-2 cells. This might explain why human intestinal E. coli isolates carry such a large amount and such a great diversity of exVAGs compared with E. coli isolates from other animal groups. The better adhesion of the ECoR group B2 isolates can be explained by the higher prevalence of such adhesion-related genes in B2 isolates. The task for the future is to investigate and verify the direct role of each of these exVAGs as an adhesin.
In conclusion, we characterized 317 nonhemolytic E. coli isolates for the occurrence of VAGs and phylogenetic affiliation and then correlated these data with adhesion to four epithelial cell lines. We defined the reservoirs for potential pathogenic E. coli isolates and described the species-specific prevalence of exVAGs. We classified the adhesion of the E. coli isolates and identified a strong variation between isolates, cell lines, tissues, and species origin. Several known adhesins were associated with host cell-specific adhesion. Other new potential adhesion genes were described. Our study, which included the development of new tools, was initiated for the purpose of screening and comparing large numbers of isolates and is the basis for the subsequent analysis of single parameters, isolates, and observations.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by InnoProfile IP 03 IP 611, funded by the Bundesministerium für Bildung und Forschung (BMBF; Germany), and by Collaborative Research Group (SFB) 852 (Nutrition and intestinal microbiota-host interactions in the pig), funded by the Deutsche Forschungsgemeinschaft (DFG).
We thank Hugo Mildenberger for preparing the database and for discussions regarding statistical analysis and Ingo Berger for designing parts of the schemata of Fig. 1 and technical support.
D. Roggenbuck is a shareholder of GA Generic Assays GmbH and Medipan GmbH. W. Lehmann is a shareholder of the company Attomol GmbH. The remaining authors declare that we have no competing financial interest.
Footnotes
Published ahead of print 19 July 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01384-13.
REFERENCES
- 1.Pizarro-Cerdá J, Cossart P. 2006. Bacterial adhesion and entry into host cells. Cell 124:715–727 [DOI] [PubMed] [Google Scholar]
- 2.Schierack P, Kleta S, Tedin K, Babila JT, Oswald S, Oelschlaeger TA, Hiemann R, Paetzold S, Wieler LH. 2011. E. coli Nissle 1917 affects Salmonella adhesion to porcine intestinal epithelial cells. PLoS One 6:e14712. 10.1371/journal.pone.0014712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Russo TA, Johnson JR. 2000. Proposal for a new inclusive designation for extraintestinal pathogenic isolates of Escherichia coli: ExPEC. J. Infect. Dis. 181:1753–1754 [DOI] [PubMed] [Google Scholar]
- 4.Pallen MJ, Wren BW. 2007. Bacterial pathogenomics. Nature 449:835–842 [DOI] [PubMed] [Google Scholar]
- 5.Ahmed N, Dobrindt U, Hacker J, Hasnain SE. 2008. Genomic fluidity and pathogenic bacteria: applications in diagnostics, epidemiology and intervention. Nat. Rev. Microbiol. 6:387–394 [DOI] [PubMed] [Google Scholar]
- 6.Johnson JR. 1991. Virulence factors in Escherichia coli urinary tract infection, vol 4 American Society for Microbiology, Washington, DC: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaper JB, Nataro JP, Mobley HL. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123–140 [DOI] [PubMed] [Google Scholar]
- 8.Dobrindt U, Hochhut B, Hentschel U, Hacker J. 2004. Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2:414–424 [DOI] [PubMed] [Google Scholar]
- 9.Hacker J, Blum-Oehler G, Mühldorfer I, Tschäpe H. 1997. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol. Microbiol. 23:1089–1097 [DOI] [PubMed] [Google Scholar]
- 10.Hultgren S, Normark JS, Abraham SN. 1991. Chaperone-assisted assembly and molecular architecture of adhesive pili. Annu. Rev. Microbiol. 45:383–415 [DOI] [PubMed] [Google Scholar]
- 11.Mundy R, Schuller S, Girard F, Fairbrother JM, Phillips AD, Frankel G. 2007. Functional studies of intimin in vivo and ex vivo: implications for host specificity and tissue tropism. Microbiology 153:959–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Korhonen TK, Vaisanen-Rhen V, Rhen M, Pere A, Parkkinen J, Finne J. 1984. Escherichia coli fimbriae recognizing sialyl galactosides. J. Bacteriol. 159:762–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ott M, Hoschutzky H, Jann K, Van Die I, Hacker J. 1988. Gene clusters for S fimbrial adhesin (sfa) and F1C fimbriae (foc) of Escherichia coli: comparative aspects of structure and function. J. Bacteriol. 170:3983–3990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lasaro MA, Salinger N, Zhang J, Wang Y, Zhong Z, Goulian M, Zhu J. 2009. F1C fimbriae play an important role in biofilm formation and intestinal colonization by the Escherichia coli commensal strain Nissle 1917. Appl. Environ. Microbiol. 75:246–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schierack P, Rödiger S, Kuhl C, Hiemann R, Roggenbuck D, Li G, Weinreich J, Berger E, Nolan LK, Nicholson B, Römer A, Frömmel U, Wieler LH, Schröder C. 2013. Porcine E. coli: virulence-associated genes, resistance genes and adhesion and probiotic activity tested by a new screening method. PLoS One 8:e59242. 10.1371/journal.pone.0059242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schierack P, Walk N, Ewers C, Wilking H, Steinrück H, Filter M, Wieler LH. 2008. ExPEC—typical virulence-associated genes correlate with successful colonization by intestinal E. coli in a small piglet group. Environ. Microbiol. 10:1742–1751 [DOI] [PubMed] [Google Scholar]
- 17.Wold AE, Caugant DA, Lidin-Janson G, de Man P, Svanborg C. 1992. Resident colonic Escherichia coli strains frequently display uropathogenic characteristics. J. Infect. Dis. 165:46–52 [DOI] [PubMed] [Google Scholar]
- 18.Nowrouzian FL, Adlerberth I, Wold AE. 2006. Enhanced persistence in the colonic microbiota of Escherichia coli strains belonging to phylogenetic group B2: role of virulence factors and adherence to colonic cells. Microbes Infect. 8:834–840 [DOI] [PubMed] [Google Scholar]
- 19.Moreno E, Johnson JR, Perez T, Prats G, Kuskowski MA, Andreu A. 2009. Structure and urovirulence characteristics of the fecal Escherichia coli population among healthy women. Microbes Infect. 11:274–280 [DOI] [PubMed] [Google Scholar]
- 20.Johnson AM, Kaushik RS, Francis DH, Fleckenstein JM, Hardwidge PR. 2009. Heat-labile enterotoxin promotes Escherichia coli adherence to intestinal epithelial cells. J. Bacteriol. 191:178–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ewers C, Antao EM, Diehl I, Philipp HC, Wieler LH. 2009. Intestine and environment of the chicken as reservoirs for extraintestinal pathogenic Escherichia coli strains with zoonotic potential. Appl. Environ. Microbiol. 75:184–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tenaillon O, Skurnik D, Picard B, Denamur E. 2010. The population genetics of commensal Escherichia coli. Nat. Rev. Microbiol. 8:207–217 [DOI] [PubMed] [Google Scholar]
- 23.Merlino J, Siarakas S, Robertson GJ, Funnell GR, Gottlieb T, Bradbury R. 1996. Evaluation of CHROMagar Orientation for differentiation and presumptive identification of gram-negative bacilli and Enterococcus species. J. Clin. Microbiol. 34:1788–1793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schierack P, Römer A, Jores J, Kaspar H, Guenther S, Filter M, Eichberg J, Wieler LH. 2009. Isolation and characterization of intestinal Escherichia coli clones from wild boars in Germany. Appl. Environ. Microbiol. 75:695–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guenther S, Filter M, Tedin K, Szabo I, Wieler LH, Nöckler K, Walk N, Schierack P. 2010. Enterobacteriaceae populations during experimental Salmonella infection in pigs. Vet. Microbiol. 142:352–360 [DOI] [PubMed] [Google Scholar]
- 26.Ewers C, Guenther S, Wieler LH, Schierack P. 2009. Mallard ducks—a waterfowl species with high risk of distributing Escherichia coli pathogenic for humans. Environ. Microbiol. Rep. 1:510–517 [DOI] [PubMed] [Google Scholar]
- 27.Rödiger S, Schierack P, Böhm A, Nitschke J, Berger I, Frömmel U, Schmidt C, Ruhland M, Schimke I, Roggenbuck D, Lehmann W, Schröder C. 2013. A highly versatile microscope imaging technology platform for the multiplex real-time detection of biomolecules and autoimmune antibodies. Adv. Biochem. Eng. Biotechnol. 133:35–74 [DOI] [PubMed] [Google Scholar]
- 28.Schierack P, Nordhoff M, Pollmann M, Weyrauch KD, Amasheh S, Lodemann U, Jores J, Tachu B, Kleta S, Blikslager A, Tedin K, Wieler LH. 2006. Characterization of a porcine intestinal epithelial cell line for in vitro studies of microbial pathogenesis in swine. Histochem. Cell Biol. 125:293–305 [DOI] [PubMed] [Google Scholar]
- 29.Egerer K, Roggenbuck D, Hiemann R, Weyer MG, Buttner T, Radau B, Krause R, Lehmann B, Feist E, Burmester GR. 2010. Automated evaluation of autoantibodies on human epithelial-2 cells as an approach to standardize cell-based immunofluorescence tests. Arthritis Res. Ther. 12:R40. 10.1186/ar2949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grossmann K, Roggenbuck D, Schröder C, Conrad K, Schierack P, Sack U. 2011. Multiplex assessment of non-organ-specific autoantibodies with a novel microbead-based immunoassay. Cytometry A 79:118–125 [DOI] [PubMed] [Google Scholar]
- 31.Roggenbuck D, Reinhold D, Hiemann R, Anderer U, Conrad K. 2011. Standardized detection of anti-ds DNA antibodies by indirect immunofluorescence—a new age for confirmatory tests in SLE diagnostics. Clin. Chim. Acta 412:2011–2012 [DOI] [PubMed] [Google Scholar]
- 32.Frömmel U, Berger I, Rödiger S, Schierack P, Schröder C. 2011. Multiplex-PCR-Mikropartikel-Assay zum Nachweis bakterieller Gene. Pabst Science Publishers, Lengerich, Germany [Google Scholar]
- 33.Rödiger S, Friedrichsmeier T, Kapat P, Michalke M. 2012. RKWard: a comprehensive graphical user interface and integrated development environment for statistical analysis with R. J. Stat. Softw. 49:1–34 [Google Scholar]
- 34.Derveaux S, Stubbe BG, Braeckmans K, Roelant C, Sato K, Demeester J, De Smedt SC. 2008. Synergism between particle-based multiplexing and microfluidics technologies may bring diagnostics closer to the patient. Anal. Bioanal. Chem. 391:2453–2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fulton RJ, McDade RL, Smith PL, Kienker LJ, Kettman JR., Jr 1997. Advanced multiplexed analysis with the FlowMetrix system. Clin. Chem. 43:1749–1756 [PubMed] [Google Scholar]
- 36.Schmitt M, Bravo IG, Snijders PJF, Gissmann L, Pawlita M, Waterboer T. 2006. Bead-based multiplex genotyping of human papillomaviruses. J. Clin. Microbiol. 44:504–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanchez S, Garcia-Sanchez A, Martinez R, Blanco J, Blanco JE, Blanco M, Dahbi G, Mora A, Hermoso de Mendoza J, Alonso JM, Rey J. 2009. Detection and characterisation of Shiga toxin-producing Escherichia coli other than Escherichia coli O157:H7 in wild ruminants. Vet. J. 180:384–388 [DOI] [PubMed] [Google Scholar]
- 38.Oelschlaeger TA, Dobrindt U, Hacker J. 2002. Virulence factors of uropathogens. Curr. Opin. Urol. 12:33–38 [DOI] [PubMed] [Google Scholar]
- 39.Katouli M. 2010. Population structure of gut Escherichia coli and its role in development of extra-intestinal infections. Iran J. Microbiol. 2:59–72 [PMC free article] [PubMed] [Google Scholar]
- 40.Escobar-Paramo P, Le Menac'h A, Le Gall T, Amorin C, Gouriou S, Picard B, Skurnik D, Denamur E. 2006. Identification of forces shaping the commensal Escherichia coli genetic structure by comparing animal and human isolates. Environ. Microbiol. 8:1975–1984 [DOI] [PubMed] [Google Scholar]
- 41.Yamamoto S, Tsukamoto T, Terai A, Kurazono H, Takeda Y, Yoshida O. 1997. Genetic evidence supporting the fecal-perineal-urethral hypothesis in cystitis caused by Escherichia coli. J. Urol. 157:1127–1129 [PubMed] [Google Scholar]
- 42.Hancock V, Nielsen EM, Krag L, Engberg J, Klemm P. 2009. Comparative analysis of antibiotic resistance and phylogenetic group patterns in human and porcine urinary tract infectious Escherichia coli. APMIS 117:786–790 [DOI] [PubMed] [Google Scholar]
- 43.Moreno E, Andreu A, Pigrau C, Kuskowski MA, Johnson JR, Prats G. 2008. Relationship between Escherichia coli strains causing acute cystitis in women and the fecal E. coli population of the host. J. Clin. Microbiol. 46:2529–2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ewers C, Li G, Wilking H, Kieling S, Alt K, Antáo E-M, Laturnus C, Diehl I, Glodde S, Homeier T, Böhnke U, Steinrück H, Philipp H-C, Wieler LH. 2007. Avian pathogenic, uropathogenic, and newborn meningitis-causing Escherichia coli: how closely related are they? Int. J. Med. Microbiol. 297:163–176 [DOI] [PubMed] [Google Scholar]
- 45.Johnson JR, Oswald E, O'Bryan TT, Kuskowski MA, Spanjaard L. 2002. Phylogenetic distribution of virulence-associated genes among Escherichia coli isolates associated with neonatal bacterial meningitis in the Netherlands. J. Infect. Dis. 185:774–784 [DOI] [PubMed] [Google Scholar]
- 46.Römer A, Wieler LH, Schierack P. 2012. Analyses of intestinal commensal Escherichia coli strains from wild boars suggest adaptation to conventional pig production conditions. Vet. Microbiol. 161:122–129 [DOI] [PubMed] [Google Scholar]
- 47.Hudault S, Spiller OB, Morgan BP, Servin AL. 2004. Human diffusely adhering Escherichia coli expressing Afa/Dr adhesins that use human CD55 (decay-accelerating factor) as a receptor does not bind the rodent and pig analogues of CD55. Infect. Immun. 72:4859–4863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Servin AL. 2005. Pathogenesis of Afa/Dr diffusely adhering Escherichia coli. Clin. Microbiol. Rev. 18:264–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kostakioti M, Stathopoulos C. 2004. Functional analysis of the Tsh autotransporter from an avian pathogenic Escherichia coli strain. Infect. Immun. 72:5548–5554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rendón MA, Saldaña Z, Erdem AL, Monteiro-Neto V, Vázquez A, Kaper JB, Puente JL, Girón JA. 2007. Commensal and pathogenic Escherichia coli use a common pilus adherence factor for epithelial cell colonization. Proc. Natl. Acad. Sci. U. S. A. 104:10637–10642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saldaña Z, Xicohtencatl-Cortes J, Avelino F, Phillips AD, Kaper JB, Puente JL, Giron JA. 2009. Synergistic role of curli and cellulose in cell adherence and biofilm formation of attaching and effacing Escherichia coli and identification of Fis as a negative regulator of curli. Environ. Microbiol. 11:992–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lindberg F, Lund B, Johansson L, Normark S. 1987. Localization of the receptor-binding protein adhesin at the tip of the bacterial pilus. Nature 328:84–87 [DOI] [PubMed] [Google Scholar]
- 53.Strömberg N, Marklund BI, Lund B, Ilver D, Hamers A, Gaastra W, Karlsson KA, Normark S. 1990. Host-specificity of uropathogenic Escherichia coli depends on differences in binding specificity to Galα1-4Gal-containing isoreceptors. EMBO J. 9:2001–2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chapman TA, Wu X-Y, Barchia I, Bettelheim KA, Driesen S, Trott D, Wilson M, Chin JJC. 2006. Comparison of virulence gene profiles of Escherichia coli strains isolated from healthy and diarrheic swine. Appl. Environ. Microbiol. 72:4782–4795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Casey T, Bosworth BT. 2009. Design and evaluation of a multiplex PCR assay for the simultaneous identification of genes for nine different virulence factors associated with Escherichia coli that cause diarrhea and edema disease in swine. J. Vet. Diagn. Invest. 21:25–30 [DOI] [PubMed] [Google Scholar]
- 56.Guion CE, Ochoa TJ, Walker CM, Barletta F, Cleary TG. 2008. Detection of diarrheagenic Escherichia coli by use of melting-curve analysis and real-time multiplex PCR. J. Clin. Microbiol. 46:1752–1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Clermont O, Bonacorsi S, Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555–4558 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.