Abstract
Autoaggregation in lactic acid bacteria is directly related to the production of certain extracellular proteins, notably, aggregation-promoting factors (APFs). Production of aggregation-promoting factors confers beneficial traits to probiotic-producing strains, contributing to their fitness for the intestinal environment. Furthermore, coaggregation with pathogens has been proposed to be a beneficial mechanism in probiotic lactic acid bacteria. This mechanism would limit attachment of the pathogen to the gut mucosa, favoring its removal by the human immune system. In the present paper, we have characterized a novel aggregation-promoting factor in Lactobacillus plantarum. A mutant with a knockout of the D1 gene showed loss of its autoaggregative phenotype and a decreased ability to bind to mucin, indicating an adhesion role of this protein. In addition, heterologous production of the D1 protein or an internal fragment of the protein, characterized by its abundance in serine/threonine, strongly induced autoaggregation in Lactococcus lactis. This result strongly suggested that this internal fragment is responsible for the bioactivity of D1 as an APF. To our knowledge, this is the first report on a gene coding for an aggregation-promoting factor in Lb. plantarum.
INTRODUCTION
Lactobacillus plantarum is a lactic acid bacterium (LAB) with one of the largest genomes of LAB (1). This provides this species with a great capacity to adapt to changing environmental conditions, as well as a high degree of versatility to attach to different substrates (2). Lb. plantarum can be found in a wide array of fermented foods all over the world (3), and, in addition, some strains, such as 299v or WCFS1, have been proposed to confer benefits on human health, being considered potential probiotics (4, 5). Currently, we are lacking information about the molecular mechanisms of action by which beneficial bacteria interact with the human host and exert their benefits. In this way, extracellular proteins are being characterized as potential mediators between commensal bacteria and the cells of the gut mucosa (6, 7). These proteins may also have relevant roles in the interaction of the bacterium with its environment, notably, with the host immune system and in the maintenance of the mucosal barrier in the case of probiotic bacteria (8).
Certain strains exhibit a marked autoaggregative phenotype, with this trait being related to their capacity to coaggregate with other bacteria (9, 10). In fact, coaggregation with pathogens is among the proposed mechanisms by which probiotic LAB may exert their beneficial effects on the human host. This is thought to decrease the adhesion of the pathogen to the mucosa, thereby favoring the formation of a barrier that might prevent pathogen colonization and enhance the clearance of the pathogen (11). In some cases, the aggregation phenotype has been related to the production of certain extracellular proteins involved in the process of bacterial conjugation (12). In other cases, autoaggregation is dependent on the presence of small hydrophilic peptides, such as in the case of Lactobacillus gasseri 2459 (13). Finally, the autoaggregative phenotype is sometimes dependent on the presence of an extracellular surface-associated sex factor that mediates cell-to-cell contact (14) or on the presence of other types of cell wall-associated proteins (15, 16).
Particularly interesting is the case of the Lactobacillus species from the acidophilus group, which contain within their genomes one or more open reading frames coding for the so-called aggregation-promoting factors (APFs) (10). These are highly conserved genes that display their maximum expression rates at the stationary phase of growth (11) and that typically code for extracellular proteins that range in size from about 260 to 330 amino acids (17). The involvement of these APFs in the aggregation phenotype has been proved with the analysis of nonaggregative mutants obtained from aggregative wild-type strains; these nonaggregative mutants often display mutations in the apf locus (18, 19). In other microorganisms, APFs additionally act as binding proteins; for instance, the 35-kDa APF from Porphyromonas gingivalis is a hemin-binding protein (20). Production of APFs also has a great impact on cell shape maintenance; in this regard, overexpression of one of the two apf genes harbored by Lb. gasseri 4132 induced formation of elongated and irregular cells (21).
To date, we know the amino acid sequence of the major extracellular proteins secreted by Lb. plantarum, although their precise bioactivity has not been described, with the exception of that of a chitin-binding protein (22–25). In the present work, we have focused on protein D1 from Lb. plantarum NCIMB 8826 (GI|342240345). For this purpose, we developed a genetic system for knockout generation in Lb. plantarum, which allowed us to obtain a mutant strain lacking the D1 gene. Changes in the aggregation phenotype and adhesion capabilities of the mutant strain, together with the controlled heterologous production of the D1 protein in Lactococcus lactis, strongly suggest that D1 is a novel APF.
MATERIALS AND METHODS
Culture conditions.
The bacterial strains, plasmids, and primers used in this work are listed in Tables 1 and 2. Lb. plantarum NCIMB 8826, Lb. plantarum NCIMB 8826ΔD1, and the Lb. plantarum strains used for knockout construction were propagated on MRS agar (Becton, Dickinson France SAS, Le Pont-De-Claix, France) with or without erythromycin (Ery; 5 μg/ml) and/or chloramphenicol (Cm; 10 μg/ml) as selection markers. Isolated colonies were used to inoculate 10 ml of MRS broth and were used for total DNA extraction. Strains Lc. lactis NZ9000, Lc. lactis NZ9000(pNZ8110) (harboring the vector pNZ8110), Lc. lactis D1, and Lc. lactis ST were propagated on M17 medium supplemented with 1% (wt/vol) glucose (GM17 medium; BD) and incubated under aerobic conditions at 30°C. Cm (5 μg/ml) was added to GM17 medium as the selective agent when appropriate.
Table 1.
Strains and vectors used in this work
| Strain or vector | Relevant features | Reference or source |
|---|---|---|
| Strains | ||
| Lb. plantarum NCIMB 8826 | Human saliva isolate; reference genome sequence, Lb. plantarum WCFS1 (GenBank accession no. AL935263); Erys | NCIMBa |
| Lb. plantarum NCIMB 8826ΔD1 | Lb. plantarum NCIMB 8826 derivative strain in which the gene coding for protein D1 is deleted, Cmr Erys | This work |
| E. coli DH10B | F− mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara leu)7697 galU galK λ− rpsL nupG | Invitrogen |
| Lc. lactis NZ9000 | Strain MG1363 with nisRK genes integrated in the chromosome, host for plasmid pNZ8110 derivatives | 47 |
| Lc. lactis D1 | Strain harboring the D1 gene from Lb. plantarum NCIMB 8826 without the sequence coding for the signal peptide, cloned into pNZ8110, Cmr | This study |
| Lc. lactis ST | Strain producing the ST-rich domain encoded by the D1 gene from Lb. plantarum NCIMB 8826, cloned into pNZ8110, Cmr | This study |
| Vectors | ||
| pUC19E | pUC19 derivative | 48 |
| pBluescript II KS (+) | pUC ori, LacZ Apr | Agilent |
| pMN0 | pBluescript::Ery resistance cassette::P32cat | This work |
| pMN1 | pMN0::pepN terminator | This work |
| pMN1i | pMN1::1,171-bp Lb. plantarum region from residues 278917 to 280088 | This work |
| pMN18 | pMN1i::1,267-bp Lb. plantarum region from residues 280728 to 2819958 | This work |
| pNZ8110 | Vector for protein secretion using the signal sequence of protein Usp45 of Lc. lactis, chloramphenicol resistant | 35 |
| pNZ8110-D1 | D1 gene with genetic information coding for a C-terminal fused His tag, cloned in pNZ8110 | This work |
| pNZ8110-ST | ST-rich domain encoded in the D1 gene with genetic information coding for a C-terminal fused His tag, cloned in pNZ8110 | This work |
NCIMB, National Collections of Industrial and Marine Bacteria Ltd., Aberdeen, Scotland.
Table 2.
Primers used in this work
| Amplicon name | Primer | Directiona | Sequenceb (5′ to 3′) | Source or GenBank accession no. | Position |
|---|---|---|---|---|---|
| Terminator | TERM BamHI | Fw | GGGGATCCCCAAAATTAGAAAACCAAGGC | AM406671 | 306556–306576 |
| TERM PstIBglII | Rv | GGCTGCAGGAGATCTAAGCTCGCGTTATCGGTCC | AM406671 | 306866–306853 | |
| 1 | XbaI plantarum | Fw | GGGGCTCTAGAGCAATTGCTGAAACAGTATCG | AL935263 | 278917–278937 |
| BglII plantarum | Rv | GGGGCAGATCTAAAGCCACTCTAAATTAATAAACAAAT | AL935263 | 280088–280062 | |
| 2 | EcoRI plantarum | Fw | GGGGCGAATTCTTGTAATGTACACTCCTATTTTTTAGTCC | AL935263 | 280728–280756 |
| SalI plantarum | Rv | GGGGCGTCGACCTATTACCACAACACGCGGTCT | AL935263 | 281995–281974 | |
| Knockout verification | Plantarum KO-Fw | Fw | TGGATCGTGTCCGTATTAAAG | AL935263 | 279855- 279875 |
| Plantarum KO-Rv | Rv | ATTAACAATAAACAAACCGACAAC | AL935263 | 281182–281159 | |
| Heterologous expression | D1F | Fw | GGGGCGCCGGCACGGTAACTGTTAAAGCCGG | This study | |
| D1HTR | Rv | CGGGGGCCGGCCTAGTGATGGTGATGGTGATGGTACCAGCCATTAGCAAG | This study | ||
| STF | Fw | GGGGCGCCGGCGAAGTAAATGGTGATAGCACT | This study | ||
| STHTR | Rv | GGGGCGCCGGCCTAGTGATGGTGATGGTGATGATAGTTTGATGTTGAACT | This study |
Fw, forward; Rv, reverse.
Enzyme recognition sites are shown underlined, and the sequence coding for the histidine tag is double underlined.
Escherichia coli strains were grown at 37°C with constant shaking in Luria-Bertani broth with or without ampicillin (100 μg/ml) as the selection marker. Plasmid DNA from E. coli was extracted by a plasmid midikit (Qiagen Iberia S.L, Madrid, Spain), and genomic DNA from Lc. lactis and Lb. plantarum was extracted by a GenElute bacterial genomic DNA kit (Sigma-Aldrich, St. Louis, MO) following the manufacturer's instructions.
Construction of a D1 gene knockout mutant of Lb. plantarum NCIMB 8826.
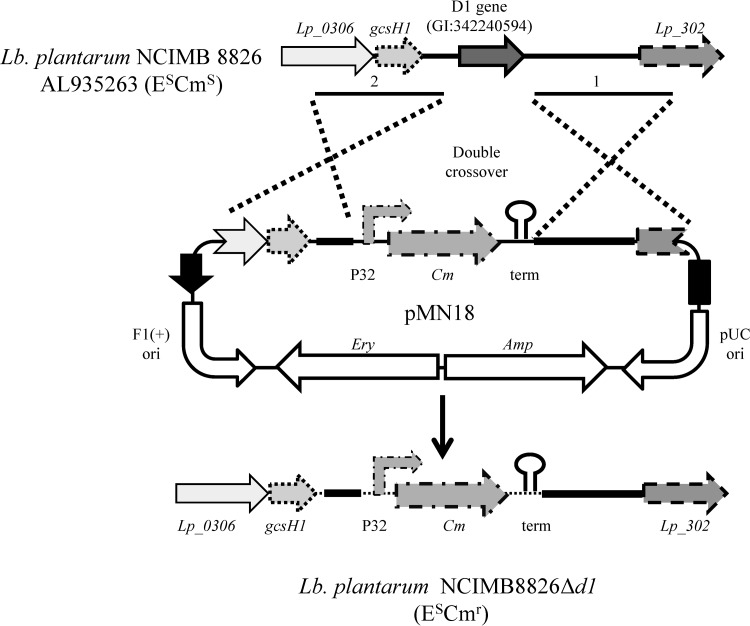
E. coli DH10B (Invitrogen, Carlsbad, CA) was used in all the cloning steps for plasmid pMN18 construction. Suicide vector pMN18 is a derivative of E. coli plasmid pMN1 (see Fig. S1 in the supplemental material for the construction of pMN1) that possesses a pUC replication origin (pUC ori), an ampicillin resistance gene, and an erythromycin resistance gene. In addition, pMN1 has two multiple-cloning sites (MCSs) separated by a chloramphenicol resistance region. On the basis of the complete genome sequence of Lb. plantarum NCIMB 8826 (GenBank accession number AL935263), primers to amplify regions 1,000 bp upstream and 1,000 bp downstream of the gene coding for the D1 protein (GI|342240594) were designed. In order to clone each fragment in the right orientation into the MCS flanking the chloramphenicol resistance gene of pMN1, specific restriction sites were added (Table 2). Both fragments were amplified from total DNA of Lb. plantarum NCIMB 8826 as the template using Pwo DNA polymerase (Roche Farma, Madrid, Spain). Amplicon 1 was cut with XbaI and BglII (New England BioLabs, Ipswich, MA) and ligated with pMN1 digested with the same enzymes using T4 DNA ligase (Invitrogen), resulting in an intermediate plasmid (pMN1i). Once checked, it was cut with EcoRI and SalI (New England BioLabs) and ligated with amplicon 2, which had previously been cut with the same enzymes, resulting in plasmid pMN18. Plasmid DNA from E. coli was sequenced (Macrogen Inc., Seoul, South Korea), and Lb. plantarum NCIMB 8826 competent cells, prepared as described above, were transformed with the plasmid (26). Cells were transformed with 1 μg of pMN18, and direct selection was achieved in plates of MRS with Cm (10 μg/ml). Single resistant isolates were grown in MRS broth with Cm (10 μg/ml). In order to improve the success of the double-crossover event, three independent subcultures in MRS broth (with 10 μg/ml Cm) were made; single isolates from each subculture were analyzed for erythromycin sensitivity by replica plating. Colonies displaying chloramphenicol resistance and erythromycin sensitivity were the result of a double-crossover event resulting in the replacement of the D1 protein-encoding gene by the Cm resistance region (Fig. 1). The genetic organization of mutant candidates was verified by PCR using primers Plantarum KO-Fw and Plantarum KO-Rv (Table 2). Amplicons of the correct size were sequenced (Macrogen) to verify the genotype of the mutant strain, which was designated Lb. plantarum NCIMB 8826ΔD1. In this genetic system, insertion of a chimeric chloramphenicol resistance gene with a strong promoter (P32) (27) and inclusion of a strong transcriptional terminator (PepN) (28) allow plasmid selection in a single copy and allow read-through to be avoided.
Fig 1.
Schematic representation of D1 gene deletion by homologous recombination using plasmid pMN18. E, erythromycin; Cm, Ery, and Amp, chloramphenicol, erythromycin, and ampicillin resistance cassettes, respectively.
Cloning of the sequence coding for the D1 gene and for the ST fragment in Lactococcus lactis.
Total DNA of Lb. plantarum NCIMB 8826 was extracted and purified from overnight cultures using a DNeasy blood and tissue kit (Qiagen), following the manufacturer's instructions. The coding sequence of the D1 gene was PCR amplified using the primer pair D1F (forward) and D1R (reverse) (Table 1), without the native signal peptide of Lb. plantarum D1. The internal gene sequence coding for the serine/threonine-rich zone (ST fragment) of the D1 protein was amplified using primers STF (forward) and STHTR (reverse). In both cases, genetic information coding for a C-terminal histidine tag was included.
Plasmid pNZ8110, containing the lactococcal Usp45 signal peptide, was extracted from the Lc. lactis NZ9000(pNZ8110) strain using the Qiagen plasmid midikit, following the manufacturer's instructions. PCR products and plasmid pNZ8110 were digested with NaeI (Promega Biotech Ibérica, Alcobendas, Spain), and the digestion product was further dephosphorylated using alkaline phosphatase (Promega). Digestion products were ligated using T4 DNA ligase (Promega) and then transformed into Lc. lactis NZ9000. Two single clones, named Lc. lactis D1 and Lc. lactis ST, were selected for further studies using chloramphenicol as the selective marker. Sequencing of the resulting plasmid was carried out in order to ensure that undesirable mutations were not generated. These strains produced a recombinant D1 protein or its ST domain. Both carried one extra glycine at the N terminus of the mature protein after cleavage by sortase (the extra glycine came from codon GGC, which originated in the NaeI restriction site [5′-GCCGGC-3′] reconstituted after ligation).
Protein manipulations.
Production of recombinant D1 and the ST fragment was induced by adding 40 ng/ml nisin to cultures of strain Lc. lactis D1 or Lc. lactis ST in the exponential phase of growth, usually at an A600 of 0.3. Cytoplasmic, surface-associated, and extracellular proteins were isolated after different induction times (0, 3, 6, and 16 h) in order to assess the phenotype of the knockout strain and the subcellular localization of D1, as already described (24). Proteins were resolved as described by Laemmli (29) in 12% (wt/vol) polyacrylamide gels and visualized with GelCode blue safe protein stain (Thermo Scientific, Rockford, IL). D1 was identified by band excision and in-gel digestion with trypsin using standard protocols, and the resulting peptide mixture was analyzed by tandem mass spectrometry (MS/MS) (24).
Characterization of the aggregation phenotype.
Lb. plantarum NCIMB 8826 (wild type) and Lb. plantarum NCIMB 8826ΔD1 (a mutant not producing the D1 protein) were grown in 50 ml MRS for 24 h. Cultures were harvested by centrifugation, washed with phosphate-buffered saline (PBS) buffer (Oxoid), and resuspended in 60 ml of PBS buffer. Cell suspensions were divided into aliquots of 10 ml and left to precipitate overnight (ON) at room temperature. In the case of Lactococcus strains expressing the gene coding for the D1 protein or for the ST fragment, gene expression was induced by adding 40 ng/ml nisin to Lc. lactis(pNZ8110), Lc. lactis D1, or Lc. lactis ST cultures in the exponential phase of growth (usually at an A600 of 0.3). Aggregation was measured by following the decreases in the A600 of the culture supernatants for 70 h.
Adhesion assays.
Adhesion of Lb. plantarum NCIMB 8826, Lb. plantarum NCIMB 8826ΔD1, Lc. lactis NZ9000(pNZ8110) Lc. lactis D1, and Lc. lactis ST to different matrices was performed as described before (30). Briefly, mucin (type II or type III; Sigma) or fibronectin (Sigma) at 20 mg/ml in PBS was coated onto Immuno MicroWell 96-well plates (Nunc, Roskilde, Denmark) at 4°C for 16 h. Just before their use, wells were blocked for 1 h with 300 μl of 1% (wt/vol) bovine serum albumin and washed twice with PBS. ON cultures were used to inoculate fresh and prewarmed growth medium (1%, vol/vol). Cultures were grown to an A600 of 0.3 and then induced or not with 40 ng/ml nisin, in the case of Lactococcus cultures. After an induction period of 3 h, cells were collected by centrifugation (10,000 × g for 5 min at 4°C), washed twice, and resuspended in PBS. For L. plantarum cultures, cells in the mid-exponential phase of growth were used for the adhesion assays. The A600 of the bacterial suspensions was adjusted to 0.7, and the numbers of CFU/ml were determined by plate counting. Cellular suspensions containing 107 CFU/ml were incubated with 100 μmol/liter carboxyfluorescein diacetate (CFDA; Molecular Probes, OR) at 37°C for 30 min, as already described (31). Suspensions were washed twice and resuspended in the same volume of PBS. Volumes of 200 μl of CFDA-labeled suspensions were loaded onto coated 96-well plates and incubated at 37°C for 1 h. After the incubation period, the medium was removed with a micropipette and the wells were washed three times with 300 μl PBS. Then, 300 μl of a solution containing 1% (wt/vol) sodium dodecyl sulfate in 0.1 N NaOH was added to the wells and the plates were incubated at 37°C for 1 h. Finally, the well contents were homogenized and transferred to black 96-well plates (Nunc), suitable for fluorescence scanning. The fluorescence (excitation λ, 485 nm; emission λ, 538 nm) was read in a Cary Eclipse fluorescence spectrophotometer (Varian, Palo Alto, CA). Negative controls without bacteria were used to calculate the unspecific CFDA absorption to the wells.
Adhesion was expressed as the percentage of fluorescence recovered after binding corrected by the fluorescence of the bacterial suspension added to the wells. Each assay was performed in three independent experiments with two technical replicates.
Statistical methods.
Adhesion rates were calculated as the mean ± standard deviation of three independent experiments with two technical replicates. Data on adhesion rates were analyzed statistically with independent Student's t tests.
Nucleotide sequence accession numbers.
The DNA sequence of the D1 gene was deposited in the GenBank database under accession numbers HQ262413 and HQ262414.
RESULTS AND DISCUSSION
Extracellular proteins are currently being evaluated as potential mediators between probiotic LAB and the human host. Nowadays, many extracellular proteins secreted by this bacterial group are not properly annotated in public databases because their functions are not well understood, as in the case of Lb. plantarum (24). In a previous work, we showed that an uncharacterized extracellular protein from Lb. plantarum, denominated D1, displayed affinity to mucin (mucin type III, from porcine stomach) and fibronectin (24). In the present study, we constructed a mutant strain lacking the gene coding for the D1 protein in order to further elucidate its possible function.
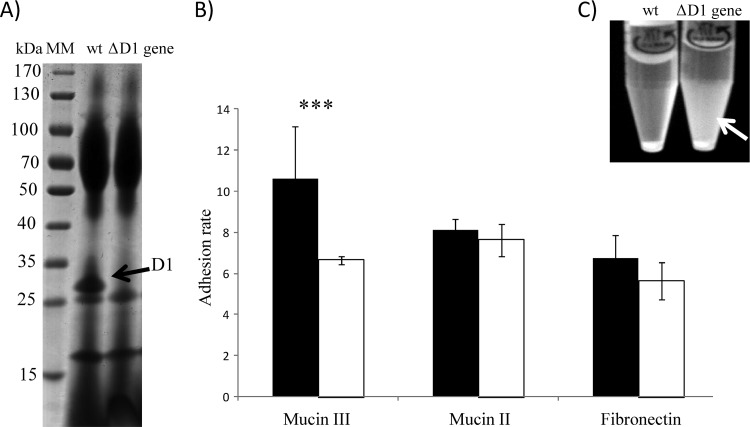
The gene coding for the D1 protein was knocked out from the wild-type strain (Lb. plantarum NCIMB 8826) (Fig. 1), resulting in a mutant strain (Lb. plantarum NCIMB 8826ΔD1). PCR amplification of the corresponding fragment confirmed the absence of the D1 gene and the insertion of the Cm resistance gene. Furthermore, SDS-PAGE of the proteins secreted in the supernatants of both strains confirmed that NCIMB 8826ΔD1 was not able to secrete the D1 protein to the extracellular milieu (Fig. 2A). Further characterization of the phenotype displayed by the NCIMB 8826ΔD1 strain confirmed the loss of affinity to mucin III and a drastic decrease in its autoaggregative capacity (Fig. 2B and C), suggesting a potential role for D1 as an adhesin (affinity to mucin and fibronectin) and aggregation-promoting factor (APF). Since mucins differ in the glycan moieties that decorate the protein backbone, including the amount of bound sialic acid, it can be hypothesized that the D1 protein may have a greater affinity for those present in mucin type III (32). However, this is a very speculative statement that deserves further research, including moving from the porcine mucins used in this study to the mucins present in the human gastrointestinal tract.
Fig 2.
(A) SDS-PAGE analysis of extracellular proteins from wild-type strain Lb. plantarum NCIMB 8826 and the mutant strain Lb. plantarum NCIMB 8826ΔD1, showing the absence of protein D1 in the latter. Lane MM, molecular marker; lane wt, wild type. (B) Rates of Lb. plantarum NCIMB 8826 (black boxes) and Lb. plantarum NCIMB 8826ΔD1 (white boxes) adhesion to mucin type III, mucin type II, and fibronectin. ***, P < 0.001. (C) Loss of the autoaggregative phenotype in the NCIMB 8826ΔD1 strain after 24 h in PBS buffer. White arrow, unaggregated bacteria that remained in suspension.
APFs and their genes have been characterized in many Lactobacillus species, but to our knowledge, there is no genetic information regarding Lb. plantarum, with the exception of reports on the autoaggregative phenotype of some strains (12). APFs confer certain properties to the producing strains that contribute to their adaptation to the environment (11).
One of the effects that has been directly linked to APF production is the ability of the producing bacterium to coaggregate with other bacteria (13). In this sense, the results of much research involving the coaggregation of autoaggregative lactobacilli with pathogens can be found in the scientific literature (11, 33, 34). Coaggregation with other microorganisms has been proposed to be a mechanism by which probiotic bacteria decrease the ability of pathogens to bind to human mucosae and favors the action of potential antimicrobials produced by probiotic strains, such as bacteriocins or short-chain fatty acids (9, 10).
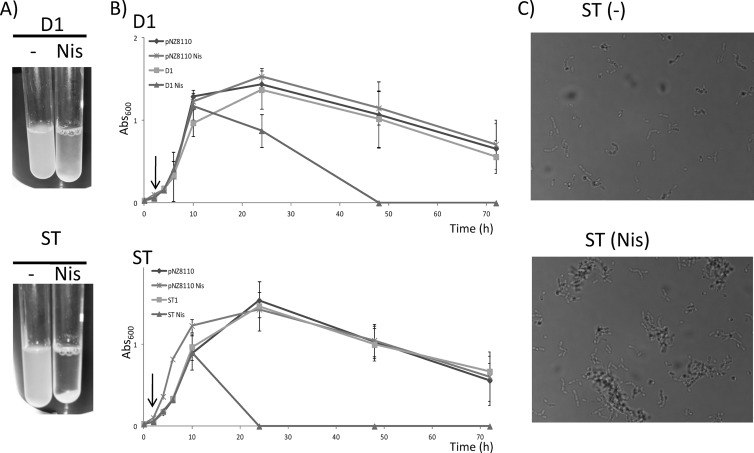
Analysis of the amino acid sequence of protein D1 showed an internal region characterized by an abundance of uncharged polar amino acids, notably, serine and threonine. To better characterize the involvement of this domain in the aggregation phenotype, we cloned both the sequence coding for the whole protein and the sequence coding for the serine/threonine-rich domain (ST) (24). In the present study, both DNA sequences (in the case of the D1 gene without the sequence coding for its own signal peptide) were cloned into pNZ8110 from Lactococcus lactis, a plasmid harboring a promoter inducible by nisin and a signal peptide that allows secretion of the synthesized protein (35). Heterologous production of the D1 protein caused bacterial aggregation in ON cultures, but autoaggregation was significantly magnified in the case of strain ST, in which a complete bacterial precipitation was observed after 24 h of culture (Fig. 3A). Heterologous production of other Lactobacillus proteins with a fused C-terminal His tag in Lc. lactis, using the same genetic system, did not induce culture aggregation (25). For this reason, we studied the autoaggregation kinetics of induced and uninduced D1 and ST cultures over time. As can be observed in Fig. 3B, the A600 of induced D1 and ST cultures decreased more quickly than that of the corresponding uninduced cultures. Additionally, autoaggregation was evidenced in the case of induced ST cultures by the formation of cell clumps, as observed under a phase-contrast microscope (Fig. 3C). This supports the suggestion that this ST domain, encoded by the amino acidic sequence of protein D1, is responsible for the autoaggregation phenotype promoted by protein D1.
Fig 3.
(A) Photographs of culture turbidity of induced (with nisin [Nis]) or uninduced (−) 24-h cultures of Lc. lactis strains D1 and ST, showing the strong aggregative phenotype of the latter when the peptide ST is produced. (B) Growth curves of strain NZ9000 harboring the empty plasmid pNZ8110, strain D1, and strain ST induced or not with nisin (the induction point is marked with an arrow). The A600 of the cultures was measured over time without agitation and was used as a measure of the rate of aggregation of the bacteria. (C) Photographs of culture turbidity of induced (with nisin [Nis]) and uninduced (−) 24-h cultures of strain ST showing the clumps that originated after bacterial aggregation.
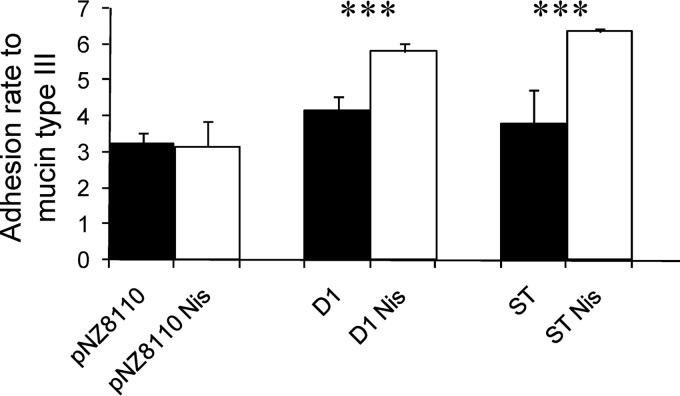
In our study, the 8826 strain and the Lc. lactis strains producing either D1 protein or the ST fragment adhered better to mucin (Fig. 4). Several scientific works have reported on the relationship between autoaggregation and adhesion to cellular and noncellular components of the human mucosa, although the precise mechanisms are not yet well understood. For instance, enteroaggregative Escherichia coli (EAEC) bases its pathogenic potential on flagellin-promoted aggregation, which enhances bacterial adhesion to epithelial cells (36). This allows a physical closeness of EAEC strains to epithelial cells, enhancing the bioactivity of their toxins and leading to several forms of diarrhea (36). Glucosyltransferase (GtfA) from Lb. reuteri has been shown to mediate sucrose-dependent autoaggregation at acid pH in this species. In addition, an isogenic gtfA-defective knockout mutant was less efficient when submitted to a colonization experiment in the gastrointestinal tract of a mouse model (37). Regarding APFs, it is known that they are also involved in the adhesion to human intestinal mucus (38). In addition to adhesion, APFs can enhance the ecological competitiveness of a given probiotic bacterium in the human gastrointestinal tract by increasing its resistance to several stresses, such as resistance to bile salts (11).
Fig 4.
Adhesion of recombinant Lc. lactis strains to mucin type III. White bars, adhesion in nisin (Nis)-induced cultures; black bars, adhesion in uninduced cultures. D1 and ST refer to the corresponding Lactococcus strains expressing the D1 gene or the ST-rich fragment, as indicated in Table 1. Lc. lactis harboring the empty plasmid pNZ8110 was used as a negative control. ***, P < 0.001.
Proteins with serine-rich domains have been shown to act as adhesins for binding to eukaryotic components and as virulence mechanisms. This is the case of SraP, a serine-rich extracellular protein of Staphylococcus aureus able to bind to human platelets (39). Also, GspB, a cell wall-attached protein from Streptococcus gordonii, mediates binding to sialylated carbohydrate moieties present on human platelets (40). Other adhesins belonging to this serine-rich family include Fap1 of Streptococcus parasanguis (41), Hsa of Streptococcus gordonii (42), the Srr proteins of Streptococcus agalactiae (43), and the SrpA proteins of Streptococcus sanguis (44) and Streptococcus cristatus (45).
In the case of Lb. plantarum, this serine/threonine-rich protein is related to the homeostasis in the gastrointestinal environment, not only in the attachment to host surfaces and in the autoaggregative phenotype but also in the molecular cross talk with the human host. Our research group has evidenced that purified ST peptide is able to interact with human dendritic cells isolated from the gut mucosa, promoting a tolerogenic and anti-inflammatory phenotype (46). In this context, work is ongoing to support our findings with in vivo research.
Concluding remarks.
To sum up, in the present work we have described a gene coding for an APF produced by Lb. plantarum. The results obtained with a knockout strain for the D1 gene support the role of this protein as an APF. In addition, heterologous production of the D1 protein in Lc. lactis strongly suggested that its activity is located in an internal domain rich in serine and threonine (the ST fragment). APFs confer onto the producing strains certain properties that, in some cases, contribute to adaptation/interaction with the environment. Results of our previous research revealed that it is precisely this peptide, encrypted in the D1 protein, that is the soluble factor mediating the molecular cross talk between Lb. plantarum and intestinal dendritic cells (46). Further research using high-throughput techniques and in vivo experiments will shed light on the signalization pathways on mucosal cells triggered by this peptide and will help to elucidate the precise molecular mechanisms of action of probiotic bacteria.
Supplementary Material
ACKNOWLEDGMENTS
Arancha Hevia and Borja Sánchez were recipients of an FPI grant and a Juan de la Cierva postdoctoral contract, respectively, from the Spanish Ministry of Economy and Competitiveness. This research was supported by grants AGL2010-14952 and RM2010-00012-00-00 from the Spanish Ministry of Economy and Competitiveness.
Footnotes
Published ahead of print 26 July 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01657-13.
REFERENCES
- 1.Chevallier B, Hubert JC, Kammerer B. 1994. Determination of chromosome size and number of rrn loci in Lactobacillus plantarum by pulsed-field gel-electrophoresis. FEMS Microbiol. Lett. 120:51–56 [DOI] [PubMed] [Google Scholar]
- 2.Kleerebezem M, Boekhorst J, van Kranenburg R, Molenaar D, Kuipers OP, Leer R, Tarchini R, Peters SA, Sandbrink HM, Fiers MW, Stiekema W, Lankhorst RMK, Bron PA, Hoffer SM, Groot MN, Kerkhoven R, de Vries M, Ursing B, de Vos WM, Siezen RJ. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. U. S. A. 100:1990–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tallon R, Arias S, Bressollier P, Urdaci MC. 2007. Strain- and matrix-dependent adhesion of Lactobacillus plantarum is mediated by proteinaceous bacterial compounds. J. Appl. Microbiol. 102:442–451 [DOI] [PubMed] [Google Scholar]
- 4.de Vries MC, Vaughan EE, Kleerebezem M, de Vos WM. 2006. Lactobacillus plantarum—survival, functional and potential probiotic properties in the human intestinal tract. Int. Dairy J. 16:1018–1028 [Google Scholar]
- 5.Molin G. 2001. Probiotics in foods not containing milk or milk constituents, with special reference to Lactobacillus plantarum 299v. Am. J. Clin. Nutr. 73:380S–385S [DOI] [PubMed] [Google Scholar]
- 6.Lebeer S, Vanderleyden J, De Keersmaecker SC. 2010. Host interactions of probiotic bacterial surface molecules: comparison with commensals and pathogens. Nat. Rev. Microbiol. 8:171–184 [DOI] [PubMed] [Google Scholar]
- 7.Turroni F, Ventura M, Buttó LF, Duranti S, O'Toole PW, Motherway MO, van Sinderen D. 21 March 2013. Molecular dialogue between the human gut microbiota and the host: a Lactobacillus and Bifidobacterium perspective. Cell. Mol. Life Sci. [Epub ahead of print.] 10.1007/s00018-013-1318-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanchez B, Bressollier P, Urdaci MC. 2008. Exported proteins in probiotic bacteria: adhesion to intestinal surfaces, host immunomodulation and molecular cross-talking with the host. FEMS Immunol. Med. Microbiol. 54:1–17 [DOI] [PubMed] [Google Scholar]
- 9.Kmet V, Callegari ML, Bottazzi V, Morelli L. 1995. Aggregation-promoting factor in pig intestinal Lactobacillus strains. Lett. Appl. Microbiol. 21:351–353 [DOI] [PubMed] [Google Scholar]
- 10.Kmet V, Lucchini R. 1997. Aggregation-promoting factor in human vaginal Lactobacillus strains. FEMS Immunol. Med. Microbiol. 19:111–114 [DOI] [PubMed] [Google Scholar]
- 11.Goh YJ, Klaenhammer TR. 2010. Functional roles of aggregation-promoting-like factor in stress tolerance and adherence of Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 76:5005–5012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reniero R, Cocconcelli P, Bottazzi V, Morelli L. 1992. High-frequency of conjugation in Lactobacillus mediated by an aggregation-promoting factor. J. Gen. Microbiol. 138:763–768 [Google Scholar]
- 13.Boris S, Suarez JE, Barbes C. 1997. Characterization of the aggregation promoting factor from Lactobacillus gasseri, a vaginal isolate. J. Appl. Microbiol. 83:413–420 [DOI] [PubMed] [Google Scholar]
- 14.Stentz R, Jury K, Eaton T, Parker M, Narbad A, Gasson M, Shearman C. 2004. Controlled expression of CluA in Lactococcus lactis and its role in conjugation. Microbiology 150(Pt 8):2503–2512 [DOI] [PubMed] [Google Scholar]
- 15.Lozo J, Jovcic B, Kojic M, Dalgalarrondo M, Chobert JM, Haertle T, Topisirovic L. 2007. Molecular characterization of a novel bacteriocin and an unusually large aggregation factor of Lactobacillus paracasei subsp. paracasei BGSJ2-8, a natural isolate from homemade cheese. Curr. Microbiol. 55:266–271 [DOI] [PubMed] [Google Scholar]
- 16.Schroeder K, Jularic M, Horsburgh SM, Hirschhausen N, Neumann C, Bertling A, Schulte A, Foster S, Kehrel BE, Peters G, Heilmann C. 2009. Molecular characterization of a novel Staphylococcus aureus surface protein (SasC) involved in cell aggregation and biofilm accumulation. PLoS One 4:e7567. 10.1371/journal.pone.0007567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ventura M, Jankovic I, Walker DC, Pridmore RD, Zink R. 2002. Identification and characterization of novel surface proteins in Lactobacillus johnsonii and Lactobacillus gasseri. Appl. Environ. Microbiol. 68:6172–6181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marcotte H, Ferrari S, Cesena C, Hammarstrom L, Morelli L, Pozzi G, Oggioni MR. 2004. The aggregation-promoting factor of Lactobacillus crispatus M247 and its genetic locus. J. Appl. Microbiol. 97:749–756 [DOI] [PubMed] [Google Scholar]
- 19.Siciliano RA, Cacace G, Mazzeo MF, Morelli L, Elli M, Rossi M, Malorni A. 2008. Proteomic investigation of the aggregation phenomenon in Lactobacillus crispatus. Biochim. Biophys. Acta 1784:335–342 [DOI] [PubMed] [Google Scholar]
- 20.Shibata Y, Hiratsuka K, Hayakawa M, Shiroza T, Takiguchi H, Nagatsuka Y, Abiko Y. 2003. A 35-kDa co-aggregation factor is a hemin binding protein in Porphyromonas gingivalis. Biochem. Biophys. Res. Commun. 300:351–356 [DOI] [PubMed] [Google Scholar]
- 21.Jankovic I, Ventura M, Meylan V, Rouvet M, Elli M, Zink R. 2003. Contribution of aggregation-promoting factor to maintenance of cell shape in Lactobacillus gasseri 4B2. J. Bacteriol. 185:3288–3296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beck HC, Madsen SM, Glenting J, Petersen J, Israelsen H, Norrelykke MR, Antonsson M, Hansen AM. 2009. Proteomic analysis of cell surface-associated proteins from probiotic Lactobacillus plantarum. FEMS Microbiol. Lett. 297:61–66 [DOI] [PubMed] [Google Scholar]
- 23.Madsen SM, Glenting J, Vrang A, Ravn P, Riemann HK, Israelsen H, Norrelykke MR, Hansen AM, Antonsson M, Ahrne S, Beck HC. 2005. Cell surface-associated glycolytic enzymes from Lactobacillus plantarum 299v mediate adhesion to human epithelial cells and extracellular matrix proteins. Proceedings of the 12th European Congress of Biotechnology, Copenhagen, Denmark, 21 to 24 August 2005 [Google Scholar]
- 24.Sanchez B, Schmitter JM, Urdaci MC. 2009. Identification of novel proteins secreted by Lactobacillus plantarum that bind to mucin and fibronectin. J. Mol. Microbiol. Biotechnol. 17:158–162 [DOI] [PubMed] [Google Scholar]
- 25.Sanchez B, Gonzalez-Tejedo C, Ruas-Madiedo P, Urdaci MC, Margolles A. 2011. Lactobacillus plantarum extracellular chitin-binding protein and its role in the interaction between chitin, Caco-2 cells, and mucin. Appl. Environ. Microbiol. 77:1123–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aukrust T, Blom H. 1992. Transformation of Lactobacillus strains used in meat and vegetable fermentations. Food Res. Int. 25:253–261 [Google Scholar]
- 27.Goffin P, Lorquet F, Kleerebezem M, Hols P. 2004. Major role of NAD-dependent lactate dehydrogenases in aerobic lactate utilization in Lactobacillus plantarum during early stationary phase. J. Bacteriol. 186:6661–6666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan PS, van Alen-Boerrigter IJ, Poolman B, Siezen RJ, de Vos WM, Konings WN. 1992. Characterization of the Lactococcus lactis pepN gene encoding an aminopeptidase homologous to mammalian aminopeptidase N. FEBS Lett. 306:9–16 [DOI] [PubMed] [Google Scholar]
- 29.Laemmli UK. 1970. Cleavage of structural proteins during assembly of head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 30.Sanchez B, Arias S, Chaignepain S, Denayrolles M, Schmitter JM, Bressollier P, Urdaci MC. 2009. Identification of surface proteins involved in the adhesion of a probiotic Bacillus cereus strain to mucin and fibronectin. Microbiology 155(Pt 5):1708–1716 [DOI] [PubMed] [Google Scholar]
- 31.Laparra JM, Sanz Y. 2009. Comparison of in vitro models to study bacterial adhesion to the intestinal epithelium. Lett. Appl. Microbiol. 49:695–701 [DOI] [PubMed] [Google Scholar]
- 32.Fogg FJ, Hutton DA, Jumel K, Pearson JP, Harding SE, Allen A. 1996. Characterization of pig colonic mucins. Biochem. J. 316:937–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lang C, Bottner M, Holz C, Veen M, Ryser M, Reindl A, Pompejus M, Tanzer JM. 2010. Specific Lactobacillus/mutans Streptococcus co-aggregation. J. Dent. Res. 89:175–179 [DOI] [PubMed] [Google Scholar]
- 34.Schachtsiek M, Hammes WP, Hertel C. 2004. Characterization of Lactobacillus coryniformis DSM 20001(T) surface protein Cpf mediating coaggregation with and aggregation among pathogens. Appl. Environ. Microbiol. 70:7078–7085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mierau I, Kleerebezem M. 2005. 10 years of the nisin-controlled gene expression system (NICE) in Lactococcus lactis. Appl. Microbiol. Biotechnol. 68:705–717 [DOI] [PubMed] [Google Scholar]
- 36.Khan MA, Kang J, Steiner TS. 2004. Enteroaggregative Escherichia coli flagellin-induced interleukin-8 secretion requires Toll-like receptor 5-dependent p38 MAP kinase activation. Immunology 112:651–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walter J, Schwab C, Loach DM, Ganzle MG, Tannock GW. 2008. Glucosyltransferase A (GtfA) and inulosucrase (Inu) of Lactobacillus reuteri TMW1.106 contribute to cell aggregation, in vitro biofilm formation, and colonization of the mouse gastrointestinal tract. Microbiology 154(Pt 1):72–80 [DOI] [PubMed] [Google Scholar]
- 38.Castagliuolo I, Galeazzi F, Ferrari S, Elli M, Brun P, Cavaggioni A, Tormen D, Sturniolo GC, Morelli L, Palu G. 2005. Beneficial effect of auto-aggregating Lactobacillus crispatus on experimentally induced colitis in mice. FEMS Immunol. Med. Microbiol. 43:197–204 [DOI] [PubMed] [Google Scholar]
- 39.Siboo IR, Chambers HF, Sullam PM. 2005. Role of SraP, a serine-rich surface protein of Staphylococcus aureus, in binding to human platelets. Infect. Immun. 73:2273–2280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bensing BA, Siboo IR, Sullam PM. 2007. Glycine residues in the hydrophobic core of the GspB signal sequence route export toward the accessory Sec pathway. J. Bacteriol. 189:3846–3854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu H, Fives-Taylor PM. 1999. Identification of dipeptide repeats and a cell wall sorting signal in the fimbriae-associated adhesin, Fap1, of Streptococcus parasanguis. Mol. Microbiol. 34:1070–1081 [DOI] [PubMed] [Google Scholar]
- 42.Takahashi Y, Konishi K, Cisar JO, Yoshikawa M. 2002. Identification and characterization of hsa, the gene encoding the sialic acid-binding adhesin of Streptococcus gordonii DL1. Infect. Immun. 70:1209–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seifert KN, Adderson EE, Whiting AA, Bohnsack JF, Crowley PJ, Brady LJ. 2006. A unique serine-rich repeat protein (Srr-2) and novel surface antigen (epsilon) associated with a virulent lineage of serotype III Streptococcus agalactiae. Microbiology 152(Pt 4):1029–1040 [DOI] [PubMed] [Google Scholar]
- 44.Plummer C, Wu H, Kerrigan SW, Meade G, Cox D, Douglas CWI. 2005. A serine-rich glycoprotein of Streptococcus sanguis mediates adhesion to platelets via GPIb. Br. J. Haematol. 129:101–109 [DOI] [PubMed] [Google Scholar]
- 45.Handley PS, Correia FF, Russell K, Rosan B, DiRienzo JM. 2005. Association of a novel high molecular weight, serine-rich protein (SrpA) with fibril-mediated adhesion of the oral biofilm bacterium Streptococcus cristatus. Oral Microbiol. Immunol. 20:131–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bernardo D, Sanchez B, Al-Hassi HO, Mann ER, Urdaci MC, Knight SC, Margolles A. 2012. Microbiota/host crosstalk biomarkers: regulatory response of human intestinal dendritic cells exposed to Lactobacillus extracellular encrypted peptide. PLoS One 7:e36262. 10.1371/journal.pone.0036262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuipers OP, de Ruyter PGGA, Kleerebezem M, de Vos WM. 1998. Quorum sensing-controlled gene expression in lactic acid bacteria. J. Biotechnol. 64:15–21 [Google Scholar]
- 48.Leenhouts KJ, Kok J, Venema G. 1990. Stability of integrated plasmids in the chromosome of Lactococcus lactis. Appl. Environ. Microbiol. 56:2726–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.