Abstract
RjpA in Rhodococcus jostii is the ortholog of a channel-forming porin, MspA. Deletion of rjpA delayed growth of R. jostii on cholate but not on cholesterol. Eventual growth on cholate involved increased expression of other porins, namely, RjpB, RjpC, and RjpD. Porins appear essential for the uptake of bile acids by mycolic acid bacteria.
TEXT
Steroids are important biomolecules occurring in all domains of life. Steroids are ubiquitous in the environment as a result of excretion and biomass decomposition, as well as industrial and municipal waste discharges. These steroids can be detrimental to both humans (1) and ecosystems (2). Some bacteria degrade steroids, and a few pathways permitting catabolism of steroids have been partially elucidated (3–6). Recent studies have also indicated that steroid uptake and metabolism play important roles in the virulence of both human (7, 8) and animal (9) pathogens. Despite the importance of steroids and their transformation by microorganisms, there remain substantial gaps in our knowledge of microbial steroid uptake and metabolism.
Rhodococcus jostii is a soil-dwelling, metabolically versatile member of the mycolic acid-containing actinobacteria that grow on a range of steroids (10). BLAST analysis of the R. jostii genome revealed four proteins, RjpA (Ro04074), RjpB (Ro03127), RjpC (Ro03156), and RjpD (R08561), with 33, 33, 31, and 36% identities, respectively, to a channel-forming porin from Mycobacterium smegmatis known as MspA (Mycobacterium smegmatis porin A). Furthermore, the presence of signal peptide cleavage sites in RjpA, RjpB, and RjpD indicates that these proteins, like MspA, target the membrane. RjpA and MspA are reciprocal best BLAST hits, indicating that they are orthologous. A structural homology search using RjpA as a query also retrieved MspA, suggesting that RjpA forms a channel similar to that formed by MspA. The MspA monomers oligomerize to form a large (>100-kDa) homo-octameric, goblet-shaped protein with a central pore spanning the outer membrane (11). Loop regions of the protein lining the pore eyelet undergo conformational change, which may affect the uptake of ions and solutes by the porin (12). In M. smegmatis, MspA is involved primarily in the uptake of hydrophilic solutes, such as glucose (13) and phosphates (14). MspA has been shown to be the main conduit for hydrophilic antibiotics, such as fluoroquinolones and chloramphenicol (15), in addition to nutrients. Deletion of MspA also caused a marked increase in resistance to hydrophobic antibiotics, such as rifampin and erythromycin (16). Furthermore, deletion of MspA resulted in a 3-fold reduction in the uptake of the bile acid steroid chenodeoxycholate (16).
The similarity of RjpA to the MspA porin and the effect of MspA deletion on the uptake of hydrophobic antibiotics and a steroid led to the hypothesis that RjpA is involved in the uptake of steroids by R. jostii, which this study investigated.
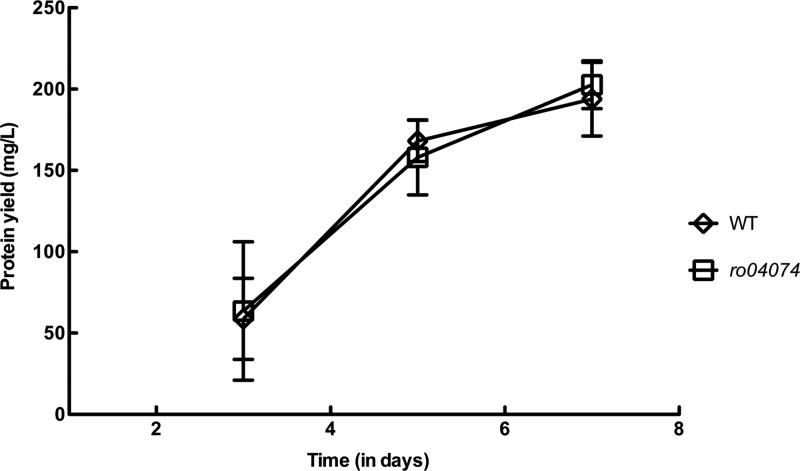
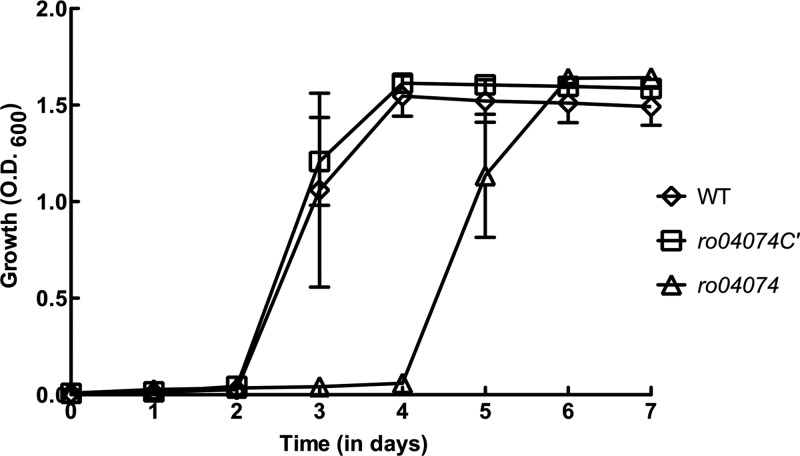
The rjpA gene was deleted from R. jostii, and the mutant was tested for its ability to grow on cholesterol or cholate. The rjpA gene of R. jostii was completely deleted, in frame and unmarked, using the sacB counterselection system described previously (17) and primers in Table 1. Deletion of rjpA did not affect the growth of R. jostii on cholesterol (Fig. 1). This result suggests that RjpA is not involved in the uptake of highly hydrophobic steroids, such as cholesterol. Both R. jostii and Mycobacterium tuberculosis take up cholesterol and, in the case of R. jostii, also β-sitosterol, via the Mce4 system, an unusually complex ATP-binding cassette transporter (7, 10). Functionally essential components of the Mce4 system include putatively extracytoplasmic Mce proteins, which we speculate may facilitate movement of hydrophobic steroids across the outer membrane and periplasm. The Mce4 system was not required for the uptake of, and growth on, cholate. In contrast to results with cholesterol, growth of the rjpA mutant on the more hydrophilic bile acid cholate was impaired. Initiation of growth of the mutant on cholate was 2 days later than that of the wild type (Fig. 2). Subsequently, the mutant grew at the same rate and to the same final cell density as the wild type. The 2-day difference in growth initiation was highly reproducible in independent experiments.
Table 1.
Primers and probes used in this study
| No. | Primer | Sense or probe | 5′–3′ sequencea | Function |
|---|---|---|---|---|
| 1 | ro04074 | Forward | TGG TCA GGA TGC AGG GAG TT | Primers and probes for quantification of porin genes by RT-QPCR |
| 2 | ro04074 | Probe rpJA | TGG CTC CAT CTC C | |
| 3 | ro04074 | Reverse | GTG ACC GGC CCG ATA GC | |
| 4 | ro03127 | Forward | GAG GCA ACC GGG TCG AA | |
| 5 | ro03127 | Probe rpJB | TCC TGC AAG GTG ATA CC | |
| 6 | ro03127 | Reverse | CGG CGG GAC CAC TTG ATA C | |
| 7 | ro03156 | Forward | CAG TGG CTC AAC GAC GTG AT | |
| 8 | ro03156 | Probe rpJC | CTC AAC GGC ACC CC | |
| 9 | ro03156 | Reverse | TGG ACT TGA TCG CGG TGT T | |
| 10 | ro08561 | Forward | CCG CCG CTG GAT GGT | |
| 11 | ro08561 | Probe rpJD | TTC CCA CCA GTG TCG AG | |
| 12 | ro08561 | Reverse | GGC GTA ACC GTT GTG GAA GA | |
| 13 | ro04074_P1 | Forward | CCCAAGCTTAGGCGATCAGCAAGCCGAGGACG | Upstream flanking region amplification primers |
| 14 | ro04074_P2 | Reverse | TGCTCTAGACTGCGACTGCCACCGAGCGTG | |
| 15 | ro04074_P3 | Forward | TGCTCTAGAGGCCGTGACGGAGAACGGTG | Downstream flanking region amplification primers |
| 16 | ro04074_P4 | Reverse | CGCGGATCCCGGTGTGCTGGGCGCG | |
| 17 | ro04074_P7 | Forward | CGGATTCGAGGAGCGGCAGGTG | rjpA deletion confirmation primers |
| 18 | ro04074_P8 | Reverse | GGCGGTGAGGTGGTG GTGCATG | |
| 19 | ro04074_P9 | Forward | GGAATTCCATATGGGTCGGCATGACGGACATCAGC | Primers to amplify rjpA for complementation |
| 20 | ro04074_P10 | Reverse | CCCAAGCTTCTAGTTGAGCTTCTGCGGCACACCG |
Restriction sites are underlined.
Fig 1.
Growth of R. jostii (wild type [WT]) and the rjpA (Δro04074) mutant on cholesterol (n = 3; bars indicate standard errors). Cultures were grown on defined medium as previously described (22), with 2.0 mM cholesterol as the sole organic substrate. Because precipitated cholesterol interfered with optical density measurement, growth was measured as protein using the bicinchoninic acid (BCA) assay (Pierce) after hot alkaline lysis.
Fig 2.
Growth of R. jostii (WT), the rjpA mutant (Δro04074), and the rjpA complementation strain (Δro04074C) on cholate (n = 3; bars indicate standard errors). Growth conditions were as described for Fig. 1, except that 2.0 mM cholate was the sole organic substrate.
To verify that deletion of rjpA caused delayed growth on cholate, the mutation was complemented. The rjpA gene was cloned into the pTip-QC2 vector and expressed in the rjpA mutant strain, using previously described methods (10) and primers in Table 1. This complementation completely restored the wild-type growth phenotype on cholate (Fig. 2).
The ability of the rjpA mutant, following the 2-day delay, to grow on cholate at the same rate and to the same final density as the wild type suggested that R. jostii may compensate for the rjpA deletion by upregulating other porins. Three paralogs of rjpA, ro03127 (rjpB), ro03156 (rjpC), and ro08561 (rjpD), were identified by BLAST search. To determine whether the other rjp porins might compensate for the rjpA deletion, levels of expression of all four porin genes during exponential growth (optical density at 600 nm [OD600] = 0.8) were compared using reverse transcriptase quantitative-PCR (RT-QPCR). The wild type expressed rjpA at much higher levels than the other porin genes on both pyruvate and cholate (Table 2), indicating that under our growth conditions, RjpA is the major porin in R. jostii. Compared to the wild type, the mutant greatly increased expression of rjpB, rjpC, and rjpD during growth on cholate, by 45-, 65-, and 26-fold, respectively. In contrast, on pyruvate, the rjpA mutant grew normally and did not increase expression of rjpB, rjpC, and rjpD, relative to that of the wild type. Thus, despite its high level of expression, RjpA does not play a critical role in growth on pyruvate. Expression of sigA is expected to be correlated with the cellular growth rate, and it did not vary by more than 2-fold among the assays. This indicates that the large differences in expression of rjp genes observed are not attributable to major differences in growth rates among the two strains on the two substrates.
Table 2.
Abundance of porin transcripts during exponential growth of R. jostii or the rjpA mutant strain on either pyruvate or cholate
| Gene | No. of gene transcripts (no. of copies/ng of DNA) after growth ona: |
|||
|---|---|---|---|---|
| Pyruvate (20 mM) |
Cholate (2 mM) |
|||
| R. jostii | Mutant | R. jostii | Mutant | |
| rjpA | 445,800 ± 195, 800 | 250 ± 160 | 28,300 ± 1,860 | 4 ± 4 |
| rjpB | 800 ± 320 | 180 ± 9 | 35 ± 4 | 1,500 ± 440 |
| rjpC | 50 ± 5 | 30 ± 1 | 22 ± 1 | 1,400 ± 140 |
| rjpD | 1,330 ± 240 | 350 ± 32 | 144 ± 39 | 3,800 ± 290 |
| sigA | 15,800 ± 330 | 30,800 ± 7,200 | 21,000 ± 720 | 27,300 ± 2,500 |
Overall, the results indicate that porins are essential for efficient cholate uptake. Of the porins, RjpA plays the main role in cholate uptake, but at least one of the other three porins can also serve that function and compensate for the loss of RjpA. A similar phenomenon was observed in M. smegmatis growing on glycerol (18). The mspA gene codes for the most highly expressed porin in M. smegmatis. Only mspA and, to a much lesser extent, mspC were expressed in wild-type M. smegmatis (18). In response to the deletion of mspA, expression of mspB and mspD was increased. Moreover, the deletion of mspA resulted in a significant decrease of nutrient permeability across the outer membrane, while the deletion of other porin genes caused no significant reduction in nutrient uptake (18).
We are beginning to understand steroid uptake by bacteria. This and other studies suggest that different uptake mechanisms are employed for the most hydrophobic steroids versus the more hydrophilic bile acids. So far, three systems that transport bile acids across the cytoplasmic membrane have been characterized. One is the well-characterized BaiG protein, a 50-kDa integral membrane protein from Eubacterium sp. strain VP1 12708 (19). BaiG cloned into Escherichia coli has been shown to transport unconjugated cholate and chenodeoxycholate (19). The second bile acid transporter was identified in Lactobacillus johnsonii and cloned into E. coli to demonstrate its ability to transport cholate (20). Recently, a transporter from Neisseria meningitidis homologous to the human apical sodium-dependent bile acid transporter (ASBT) was structurally characterized (21). The current study advances our understanding of the role of porins in transport of a bile acid across an outer membrane. Stephan et al. (16) proposed that the presence of porins might affect rates of diffusion of chenodeoxycholate through the lipids of the outer membrane of M. smegmatis. However, based on the essentiality of porins for growth of RHA1 on cholate, a more parsimonious conclusion may be that bile acids diffuse through porin channels. Clearly, further biochemical and structural studies are needed to elucidate the mechanism by which RjpA and other porins facilitate the uptake of bile acids. It is likely that bile acid uptake by mycolic acid bacteria, in general, requires porins. It is further possible that bile acid uptake by Gram-negative bacteria also requires porins, as their outer membranes are structurally analogous to those of mycolic acid bacteria.
ACKNOWLEDGMENTS
We thank Jie Liu for providing RHA1 genomic DNA and Gordon Steward for technical assistance.
This research was funded by a CIHR operating grant.
Footnotes
Published ahead of print 26 July 2013
REFERENCES
- 1.Caserta D, Maranghi L, Mantovani A, Marci R, Maranghi F, Moscarini M. 2008. Impact of endocrine disruptor chemicals in gynaecology. Hum. Reprod. Update 14:59–72 [DOI] [PubMed] [Google Scholar]
- 2.Sumpter JP. 1998. Xenoendorine disrupters—environmental impacts. Toxicol. Lett. 102–103:337–342 [DOI] [PubMed] [Google Scholar]
- 3.Horinouchi M, Hayashi T, Kudo T. 2012. Steroid degradation in Comamonas testosteroni. J. Steroid Biochem. Mol. Biol. 129:4–14 [DOI] [PubMed] [Google Scholar]
- 4.Van der Geize R, Yam K, Heuser T, Wilbrink MH, Hara H, Anderton MC, Sim E, Dijkhuizen L, Davies JE, Mohn WW, Eltis LD. 2007. A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into Mycobacterium tuberculosis survival in macrophages. Proc. Natl. Acad. Sci. U. S. A. 104:1947–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Philipp B. 2011. Bacterial degradation of bile salts. Appl. Microbiol. Biotechnol. 89:903–915 [DOI] [PubMed] [Google Scholar]
- 6.Mohn WW, Wilbrink MH, Casabon I, Stewart GR, Liu J, van der Geize R, Eltis LD. 2012. Gene cluster encoding cholate catabolism in Rhodococcus spp. J. Bacteriol. 194:6712–6719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pandey AK, Sassetti CM. 2008. Mycobacterial persistence requires the utilization of host cholesterol. Proc. Natl. Acad. Sci. U. S. A. 105:4376–4380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yam KC, D'Angelo I, Kalscheuer R, Zhu H, Wang JX, Snieckus V, Ly LH, Converse PJ, Jacobs WR, Jr, Strynadka N, Eltis LD. 2009. Studies of a ring-cleaving dioxygenase illuminate the role of cholesterol metabolism in the pathogenesis of Mycobacterium tuberculosis. PLoS Pathog. 5:e1000344. 10.1371/journal.ppat.1000344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van der Geize R, Grommen AW, Hessels GI, Jacobs AA, Dijkhuizen L. 2011. The steroid catabolic pathway of the intracellular pathogen Rhodococcus equi is important for pathogenesis and a target for vaccine development. PLoS Pathog. 7:e1002181. 10.1371/journal.ppat.1002181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mohn WW, van der Geize R, Stewart GR, Okamoto S, Liu J, Dijkhuizen L, Eltis LD. 2008. The actinobacterial mce4 locus encodes a steroid transporter. J. Biol. Chem. 283:35368–35374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faller M, Niederweis M, Schulz GE. 2004. The structure of a mycobacterial outer-membrane channel. Science 303:1189–1192 [DOI] [PubMed] [Google Scholar]
- 12.Huff J, Pavlenok M, Sukumaran S, Niederweis M. 2009. Functions of the periplasmic loop of the porin MspA from Mycobacterium smegmatis. J. Biol. Chem. 284:10223–10231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stahl C, Kubetzko S, Kaps I, Seeber S, Engelhardt H, Niederweis M. 2001. MspA provides the main hydrophilic pathway through the cell wall of Mycobacterium smegmatis. Mol. Microbiol. 40:451–464 [DOI] [PubMed] [Google Scholar]
- 14.Wolschendorf F, Mahfoud M, Niederweis M. 2007. Porins are required for uptake of phosphates by Mycobacterium smegmatis. J. Bacteriol. 189:2435–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Danilchanka O, Pavlenok M, Niederweis M. 2008. Role of porins for uptake of antibiotics by Mycobacterium smegmatis. Antimicrob. Agents Chemother. 52:3127–3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stephan J, Mailaender C, Etienne G, Daffe M, Niederweis M. 2004. Multidrug resistance of a porin deletion mutant of Mycobacterium smegmatis. Antimicrob. Agents Chemother. 48:4163–4170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van der Geize R, Hessels GI, van Gerwen R, van der Meijden P, Dijkhuizen L. 2001. Unmarked gene deletion mutagenesis of kstD, encoding 3-ketosteroid Delta1-dehydrogenase, in Rhodococcus erythropolis SQ1 using sacB as counter-selectable marker. FEMS Microbiol. Lett. 205:197–202 [DOI] [PubMed] [Google Scholar]
- 18.Stephan J, Bender J, Wolschendorf F, Hoffmann C, Roth E, Mailander C, Engelhardt H, Niederweis M. 2005. The growth rate of Mycobacterium smegmatis depends on sufficient porin-mediated influx of nutrients. Mol. Microbiol. 58:714–730 [DOI] [PubMed] [Google Scholar]
- 19.Mallonee DH, Hylemon PB. 1996. Sequencing and expression of a gene encoding a bile acid transporter from Eubacterium sp. strain VPI 12708. J. Bacteriol. 178:7053–7058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elkins CA, Savage DC. 1998. Identification of genes encoding conjugated bile salt hydrolase and transport in Lactobacillus johnsonii 100-100. J. Bacteriol. 180:4344–4349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu NJ, Iwata S, Cameron AD, Drew D. 2011. Crystal structure of a bacterial homologue of the bile acid sodium symporter ASBT. Nature 478:408–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swain K, Casabon I, Eltis LD, Mohn WW. 2012. Two transporters essential for reassimilation of novel cholate metabolites by Rhodococcus jostii RHA1. J. Bacteriol. 194:6720–6727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goncalves ER, Hara H, Miyazawa D, Davies JE, Eltis LD, Mohn WW. 2006. Transcriptomic assessment of isozymes in the biphenyl pathway of Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 72:6183–6193 [DOI] [PMC free article] [PubMed] [Google Scholar]