Abstract
In biofilms, microbial activities form gradients of substrates and electron acceptors, creating a complex landscape of microhabitats, often resulting in structured localization of the microbial populations present. To understand the dynamic interplay between and within these populations, quantitative measurements and statistical analysis of their localization patterns within the biofilms are necessary, and adequate automated tools for such analyses are needed. We have designed and applied new methods for fluorescence in situ hybridization (FISH) and digital image analysis of directionally dependent (anisotropic) multispecies biofilms. A sequential-FISH approach allowed multiple populations to be detected in a biofilm sample. This was combined with an automated tool for vertical-distribution analysis by generating in silico biofilm slices and the recently developed Inflate algorithm for coaggregation analysis of microbial populations in anisotropic biofilms. As a proof of principle, we show distinct stratification patterns of the ammonia oxidizers Nitrosomonas oligotropha subclusters I and II and the nitrite oxidizer Nitrospira sublineage I in three different types of wastewater biofilms, suggesting niche differentiation between the N. oligotropha subclusters, which could explain their coexistence in the same biofilms. Coaggregation analysis showed that N. oligotropha subcluster II aggregated closer to Nitrospira than did N. oligotropha subcluster I in a pilot plant nitrifying trickling filter (NTF) and a moving-bed biofilm reactor (MBBR), but not in a full-scale NTF, indicating important ecophysiological differences between these phylogenetically closely related subclusters. By using high-resolution quantitative methods applicable to any multispecies biofilm in general, the ecological interactions of these complex ecosystems can be understood in more detail.
INTRODUCTION
A bacterial biofilm can be defined as a multicellular community attached to a surface and embedded in a matrix of extracellular material (1). The advantages of growing in biofilms are many. By growing attached to a surface, bacteria can stay indefinitely in a favorable environment under conditions where external forces, such as flowing water, would otherwise sweep them away. In biofilm communities, bacteria are more resistant to antibiotics (2) and may also avoid predation through microcolony formation (3). Microbial biofilms in general often contain intricate structures, such as channels and voids (4), affecting important processes, including nutrient delivery to the populations present (5). Biofilms containing multiple functional groups competing for resources or collaborating under mutualistic forms are therefore exceptionally complex, and even more so if each functional group contains several populations, each occupying its own ecological niche. The positions and spatial relationships of cells within biofilms or similar environments reveal information about important interactions. Different examples where such spatial information is important include oral biofilms involved in periodontal diseases (6), biofilms in streams (7), bacterial communities on leaves and root hairs (8), syntrophic propionate-oxidizing cells and methanogens in upflow anaerobic sludge blanket granules (9), methanogenic-sulfidogenic aggregates (10), and aerobic ammonia-oxidizing bacteria (AOB) and nitrite-oxidizing bacteria (NOB), which often grow in intricate heterogeneous multispecies biofilms (11–15).
Current techniques, such as confocal laser scanning microscopy (CLSM), allow the systematic collection of high-quality biofilm images suitable for digital image analysis. Especially when combined with specific molecular labeling methods, most notably fluorescence in situ hybridization (FISH) with rRNA-targeted probes (16), image analysis offers great possibilities for the use of spatial statistics and stereology for quantifying the spatial arrangement of microbial populations (8, 17, 18). Some of these approaches have already been implemented in computer software (8, 17). Nevertheless, certain research questions are still difficult to tackle. For instance, the directionally dependent (anisotropic) structure of stratified biofilms may cause biases when such samples are analyzed by stereological methods that assume isotropy (19). Stratification likely reflects the occurrence of important environmental or biological factors affecting biofilm growth, such as substrate concentration gradients. Therefore, adequate tools to analyze the distribution of the organisms in such biofilms would be highly useful.
Here, we developed improved methods for quantifying the vertical distribution and the coaggregation patterns of defined biofilm populations. Their use is demonstrated by analyses of stratified nitrifying biofilms inhabited by AOB from the phylogenetic Nitrosomonas oligotropha cluster 6a and NOB from the genus Nitrospira. Members of these lineages are frequently encountered in nitrifying biofilms in wastewater treatment plants (WWTP) and play major roles in the removal of nitrogen from sewage (12, 20–23). Previous investigations have demonstrated the formation of substrate gradients in such biofilms, which lead to pronounced stratification and distinct spatial distributions of AOB and NOB (12, 15, 24).
Different biofilms were obtained from a pilot size moving-bed biofilm reactor (MBBR), a pilot size nitrifying trickling filter (NTF) system, and a full-scale NTF of a domestic WWTP. A sequential FISH protocol was developed and applied to detect multiple nitrifying bacterial populations simultaneously and efficiently while preserving the possibility to quantify their abundance relative to all bacteria in the sample. This technique was combined with a new image analysis tool that automatically cuts biofilm images into sections that represent different depth zones, thus allowing the precise quantification of microorganisms in the layers of stratified biofilms. As AOB and NOB are partners in a mutualistic symbiosis (25), they frequently coaggregate in nitrifying biofilms and flocs (20, 21, 24). However, the overall strength of coaggregation and the preferred distances between the AOB and NOB microcolonies can vary among different phylogenetic clades (18). Therefore, the coaggregation patterns of the different nitrifiers were quantified by a recently developed algorithm that works with strongly anisotropic samples (19).
MATERIALS AND METHODS
Full-scale NTF and pilot plant design.
The Rya municipal WWTP in Göteborg, Sweden, applies a nitrification step in which approximately half of the effluent water from the WWTP is recirculated through six parallel NTFs, after which the water enters a series of anoxic activated-sludge basins for denitrification (for more details, see http://www.gryaab.se). In the NTFs, the nitrifying bacteria grow in biofilms on a fixed cross-flow material with a large surface-to-volume ratio (Munthers, Sweden; specific area, 230 m2 m−3).
The pilot plant consisted of two parts: aerated MBBRs (3.2 by 1.0 by 1.0 m), containing suspended plastic biofilm carriers (AnoxKaldnes, Sweden) and NTFs (height, 1.0 m; diameter, 0.5 m), filled with the same plastic cross-flow medium for biofilm growth as in the full-scale NTFs. The pilot plant received the same influent as the full-scale NTFs at the WWTP. The pilot plant performance is presented in detail elsewhere (23, 26). For about 2 months prior to sampling, the investigated full-scale NTF was fed with a mean ammonium concentration of 14.3 ± 3.2 mg N liter−1 (load, 17.42 ± 2.59 g N m−2 day−1). The corresponding concentrations during the period before sampling coming into the pilot plant NTF that was used in this investigation (NTF2) were 7.6 ± 1.1 mg N liter−1 (load, 6.5 ± 1.1 g N m−2 day−1) and 1.7 ± 0.8 mg N liter−1 (load, 0.85 ± 0.23 g N m−2 day−1) in the pilot plant MBBR tank 1 (T1).
Sampling and fixation of biofilm.
Biofilm samples for cryosectioning and DNA extraction were taken on 28 February 2006 from the pilot plant trickling filter NTF2 and MBBR T1. Full-scale NTF samples were taken on 21 May 2003, as previously described (14). Snails, worms, and other animals were removed before the biofilm was retrieved. For DNA extraction, the biofilm was brushed off with a toothbrush and suspended in 1× phosphate-buffered saline (PBS) in 1.5-ml Eppendorf tubes. The biofilm suspensions were centrifuged for 3 min (5,000 × g). Pellets for DNA extraction were kept at −20°C until use. Samples for cryosectioning were fixed by submerging the plastic pieces in 4% paraformaldehyde for 8 h at 4°C, followed by rinsing twice with PBS before application of the cryosectioning protocol.
Cryosectioning.
Biofilm cryosections were produced as previously described (14). After fixation, the plastic pieces were covered with a thick layer (several millimeters) of Tissue-Tek O.C.T. Compound (Sakura Finetek Europe B.V., The Netherlands) on the biofilm side, placed in a closed and parafilm-sealed petri dish, and incubated overnight at 4°C. Excess O.C.T. compound was removed from the back side of the plastic pieces, and a second layer of O.C.T. compound was added at the biofilm side to an approximate thickness of 2 mm. The embedded pieces were placed in a liquid nitrogen fume chamber until the O.C.T. compound was completely frozen. The plastic edge of such a piece was then gently bent, by using forceps, to detach the frozen block containing the biofilm. Subsequently, the detached biofilm blocks were again embedded in O.C.T. and placed in the liquid nitrogen fume chamber until frozen solid. The blocks were stored at −70°C until use. They were sectioned in 10-μm-thick vertical slices with a HM550 microtome cryostat (Microm International GmbH, Walldorf, Germany) at −20°C. The slices were collected on SuperFrost Plus Gold microscope slides (Menzel GmbH, Braunschweig, Germany). After dehydration in an ethanol series (50%, 80%, and 96% [vol/vol]), the microscope slides were stored at −20°C until use.
Fluorescence in situ hybridization.
To ensure that all cells in the sliced biofilm would be fixed, the cryosectioned biofilm samples were fixed again directly on the microscope slides at room temperature for 20 min with 4% paraformaldehyde, followed by submersion in PBS for 30 min. FISH was performed at 46°C for 4 h (27). To facilitate FISH, a hydrophobic barrier frame was applied to the glass slides around the regions containing biofilm sections by using a Liquid Blocker Mini Pap Pen (Daido Sangyo, Tokyo, Japan). Probes specific for nitrifiers were hybridized together with the EUB338 probe mixture (labeled with a different fluorochrome) to biofilm samples. When probes with different hybridization stringency optima were applied to the same sample, several hybridizations were performed, beginning with the probe(s) requiring the most stringent conditions (27). The probe sequences and hybridization conditions are listed in Table S1 in the supplemental material.
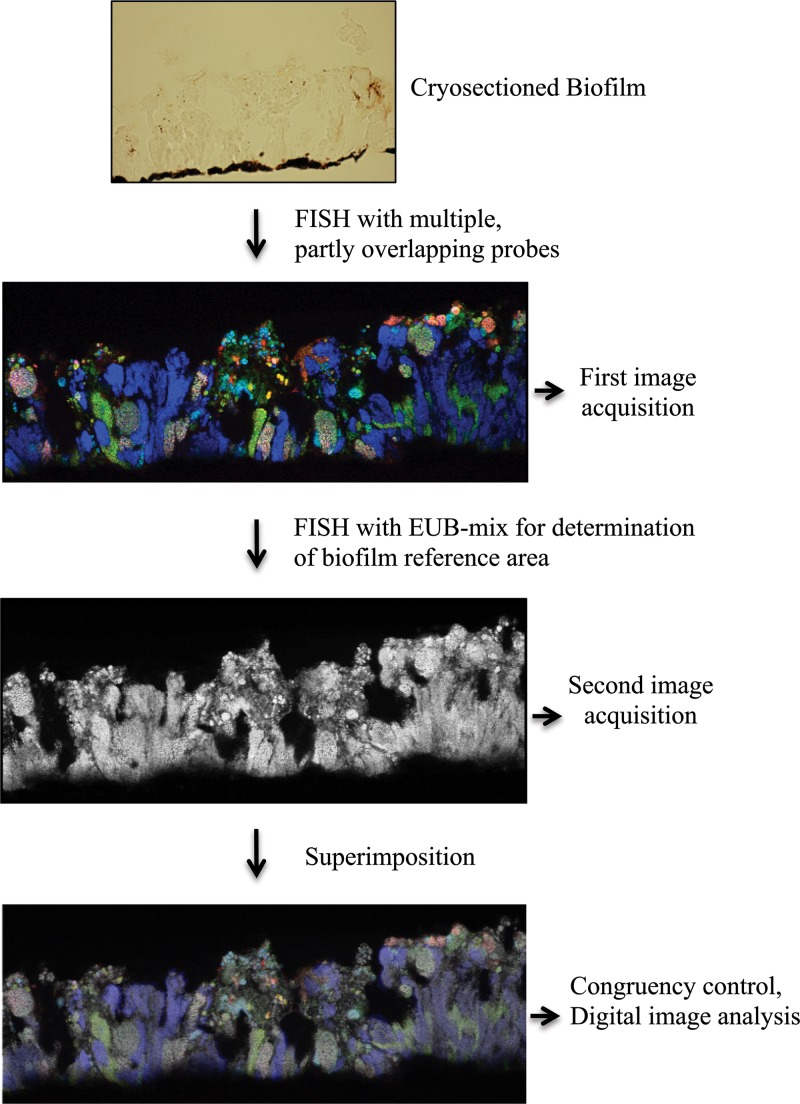
For sequential FISH of the cryosectioned biofilms, probes specific for AOB and NOB were used in the first round of in situ hybridization and microscopy. While recording the images of these probe signals by confocal laser scanning microscopy, the field of view (FOV) position on the glass slide, indicated as x-y coordinates provided by the motorized microscope stage control unit, was recorded and later used for positioning during image acquisition of the bacterial reference area. After carefully removing the coverslip and rinsing the glass slide in double-distilled water, a second hybridization step was performed with the EUB338 probe mixture only (with 10% formamide in the hybridization buffer). New images containing the EUB probe signal were recorded at the same locations in the biofilm where images had already been taken after the first FISH step. Minor adjustments of confocal microscope settings were made to obtain optimal congruency between the micrographs of the nitrifying community and the bacterial biofilm reference. Corresponding images showing the same FOV were aligned and superimposed using Photoshop CS4 Extended (Adobe Systems, San Jose, CA, USA). Pairs of corresponding images were carefully compared to ensure that the shape and size of identical biomass objects had not been altered during the repeated hybridization and image acquisition procedures. In the rare cases where biofilm pieces had been lost or partly destroyed, the affected images were excluded from further analysis. All fluorescent probes and unlabeled competitors were obtained from Thermo Electron (Interactiva Division, Ulm, Germany) or MWG Biotech (Ebersberg, Germany). Fluorescent probes were 5′ labeled with the sulfoindocyanine dye indocarbocyanine (Cy3) or indodicarbocyanine (Cy5) or with fluorescein or Alexa 488 dye. In addition, probe Nse1472 was used with double labeling of oligonucleotide probes (DOPE) (5′ and 3′ labeled with Cy3) (28). All slides were mounted in Citifluor AF1 (Citifluor Ltd., London, United Kingdom) prior to microscopy.
Microscopy and digital image analysis.
Images were collected using a Bio-Rad Radiance 2000 MP confocal laser scanning microscope (Bio-Rad, Hemel Hempstead, United Kingdom) equipped with a red diode laser (638 nm), a He/Ne laser (543 nm), and an argon laser (457, 476, 488, and 514 nm). Images for quantification were collected, using a Nikon Plan Fluor 40×/1.40 oil objective and the bundled software LaserSharp 2000, as 8-bit/pixel greyscale or 24-bit/pixel RGB images of 512 by 512 pixels (resolution, 1.65 pixels/μm). A Kalman filter (n = 2) was applied for noise reduction during image recording. Multicolor images of cryosectioned biofilm (Fig. 1), as obtained by sequential FISH, were used for measuring the spatial-localization patterns of nitrifiers and their population abundances at different depths of the stratified biofilms. These images were imported in the RGB TIFF format into the digital image analysis program daime (version 2.0) (17), and all subsequent steps were performed using this software. Image segmentation, which defined the biomasses of the different probe-labeled populations, was based on the colors of the different probe-labeled organisms, but not on the intensities of the fluorescence signals (for details, see the supplemental material).
Fig 1.
The sequential-FISH procedure. The first FISH was performed with multiple partly overlapping probes targeting NOB, AOB, or Betaproteobacteria. In a second iteration, the same biofilm was subjected to FISH with the EUB338 probe mixture. After each round of FISH, CLSM images of the same microscope FOVs were acquired at the same positions. The images were aligned and superimposed in silico. For illustrative purposes, the brightness and contrast of the multicolor FISH images were increased. The pixel intensities and contrast in the original images were lower but sufficient for image segmentation and analysis.
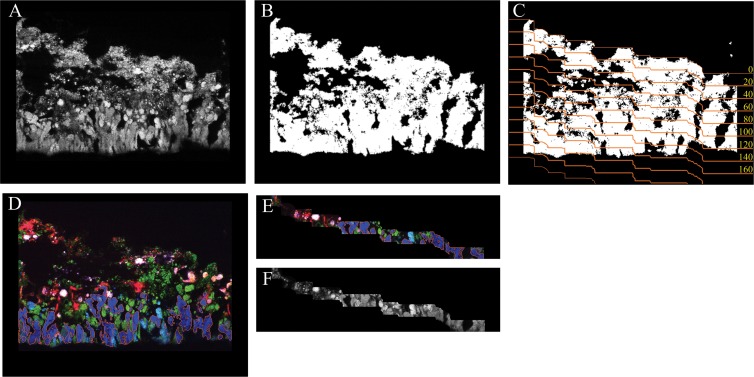
The newly developed automated virtual slicing of biofilm images (the Slicer tool) works as follows. First, an image that shows all the biomass of the biofilm is required. This usually is an image of the EUB probe mixture signal, as in this case, or a general nucleic acid stain. Prior to segmentation (i.e., detection of biomass versus background), a copy (I′) was made of each FISH image (I) to be segmented. All pixel intensities in I′ were multiplied by a factor of 3 to brighten the image and facilitate the following steps. Subsequently, dark background signals were removed and FISH signals were further brightened by histogram stretching, which set all pixels in I′ below the first pronounced peak of the image histogram to zero and all pixels above the last histogram peak to maximal intensity. The image I′ was then subjected to two iterations of median filtering with a kernel of 3 by 3 pixels. These steps decreased small color variations among pixels belonging to the biomass of the same probe-target population. The different populations were then segmented by using the color-based “magic wand” tool of daime, with a tolerance setting of 20%. Very small objects, which most likely represented remaining noise, were excluded from further analysis. Finally, the segmentation data (i.e., object definitions) were transferred from I′ to the unchanged original image I (by using the respective tool of daime), and I′ was discarded. In some cases, objects of different probe-target populations overlapped due to ambiguous colors at the borders of the respective cell clusters. These overlapping regions would cause biases and thus had to be excluded from further analysis. For this purpose, the object definitions were extracted from all segmented images and were converted to binary images that contained only white (biomass) and black (background) pixels (called “object masks” in daime). Overlapping regions were then removed by a sequence of Boolean AND and XOR operations, the binary images were resegmented, and these segmentation data (called “object layers” in daime) were transferred to the original FISH images.
After segmentation, a binary image is created from this general biomass image, in which biomass is depicted by white pixels of value 1 and background by black pixels of value 0 (Fig. 2A and B). Small biomass particles and noise, which may interfere with the subsequent steps, are automatically removed from this image. Subsequently, the binary image is automatically sliced (Fig. 2C) with a user-defined thickness by the algorithm described in the supplemental material. Here, images containing the signals of nitrifier-specific FISH probes (Fig. 2D) or the EUB probe mixture (Fig. 2A) were virtually sliced (Fig. 2E and F). The direction of slicing was from the top to the bottom of the biofilm, with a slice thickness of 20 μm, surface line smoothing of 95%, and a size limit of up to 5,000 pixels for biomass particles or noise to be ignored during the definition of the baseline for slicing. The resulting slices were analyzed as described elsewhere (17, 29) to quantify the abundances (biovolume fractions) of AOB and NOB in the respective biofilm layers. These analyses were performed for the reactors NTF2 (based on 33 FOVs of cryosectioned biofilm), MBBR T1 (14 FOVs), and the full-scale NTF (46 FOVs). Differences between nitrifier abundances in the biofilm layers were tested for significance by Kruskal-Wallis one-way analysis of variance (ANOVA) on ranks, with pairwise multiple comparisons by Dunn's method, using SigmaPlot 11.0 (Systat Software).
Fig 2.
Automated slicing of biofilm images. (A) Greyscale image of cryosectioned biofilm labeled by FISH with the EUB338 probe mixture. (B) The same image as in panel A after binarization and the removal of noise and small particles. (C) The same image as in panel A after binarization and with the edges of the automatically defined slices (orange lines), which have a thickness of 20 μm in this example. The yellow numbers indicate the depth (in μm) starting at the top surface line of the biofilm. (D) The same biofilm region with different nitrifiers labeled by FISH with specific probes. The orange borders around the biomass of Nitrospira-like NOB (dark blue) indicate that this population has been identified by image segmentation. (E) A subimage of panel D after automatic slicing as defined for panel C. The slice shown represents the zone at 100 to 120 μm. Note that the segmentation of the Nitrospira biomass has been preserved (the orange borders are still present). (F) A subimage of panel A after automatic slicing as defined for panel C. The slice shown represents the zone at 100 to 120 μm. Together, the images in panels E and F can be used for quantifying the biovolume fraction of Nitrospira in this region of the biofilm.
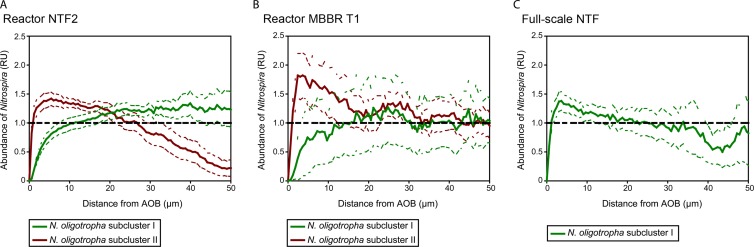
The coaggregation patterns of AOB and NOB were quantified in multicolor sequential FISH images of cryosections that were not virtually sliced but showed the whole biofilm (e.g., the image in Fig. 2D). For this purpose the Inflate algorithm was used (19). This approach examines two FISH probe-defined microbial populations, the “analyzed” and the “reference” populations. It reveals whether the spatial arrangement of the analyzed population relative to the reference population is coaggregation, random, or mutual avoidance. The Inflate algorithm creates its own random-distribution control: it makes artificial images in which one of the populations becomes randomly distributed without changing the population density in the image. These images are analyzed and used to define the “random-distribution threshold” (see Fig. 6, horizontal dashed lines in the plots). The coaggregation is significant at distances where the 95% confidence intervals (see Fig. 6, dashed green or red curves) are completely above the horizontal dashed line (y = 1.0). For a detailed description of this method, see reference 19.
Fig 6.
Coaggregation analysis of Nitrospira and AOB in biofilms from three systems. The mean abundance of Nitrospira relative to the abundance that would be expected if the nitrifiers were randomly distributed is plotted against the distance from the biomass of AOB. A value of 1.0 (horizontal dashed lines) indicates random distribution at the respective distance, whereas larger values indicate coaggregation of Nitrospira with AOB. Values smaller than 1.0 indicate that the abundance of Nitrospira was below the abundance of a randomly distributed organism. The colored dashed lines indicate 95% confidence intervals. RU, relative units. According to the terminology (20), the y axis shows the normalized positional pixel fraction of Nitrospira. A 95% confidence interval above or below the normalized y axis indicates coaggregation or repulsion, respectively. (A) Results obtained for biofilm from reactor NTF2. (B) The same analysis for reactor MBBR T1. (C) Results for the full-scale NTF. Here, AOB of subcluster II were not analyzed due to their low abundance.
Images of the EUB338 probe mixture signal (recorded in the second round of sequential FISH) were used as “biomass masks” for the Inflate algorithm, which delimited the biomass-containing image regions (Fig. 2A). The spatial-arrangement patterns of nitrifiers were analyzed for the reactors NTF2, MBBR T1, and the full-scale NTF based on the same images that were used to quantify the abundances of AOB and NOB at different depths of the biofilm (see above). The analyzed spatial range was 0 to 100 μm. NOB of the genus Nitrospira were always the analyzed population, whereas N. oligotropha-like AOB of either subcluster 1 or 2 were the reference population.
RESULTS
Sequential FISH on cryosectioned biofilms.
To enable the simultaneous in situ detection and quantification of several nitrifying populations, we applied a “sequential-FISH” protocol to cryosections of the biofilms. In silico alignment and superimposition of the acquired images resulted in mixed color patterns, which were more complex than standard FISH images. In this study, six specifically targeted populations were simultaneously detected in the examined biofilm (Fig. 1), with colors as shown in Table 1. The phylogenetic affiliation of these probe-defined populations was determined by 16S rRNA comparative sequence analysis (see the supplemental material).
Table 1.
Probe combinations, cell targets, and final colors (as shown in Fig. 1) of detected cells in the analyzed biofilmsa
| Probe combination | Probe target | Color | Detection of probe targetb |
||
|---|---|---|---|---|---|
| Full-scale NTF | MBBR T1 | NTF2 | |||
| 1st image acquisition | |||||
| 6a192 (Cy5) + Nmo218 (Cy3) + Bet42a (Alexa 488) | N. oligotropha subcluster I | Gray/pink | + | + | + |
| 6a192 (Cy5)-Nmo218 (Cy3) + Bet42a (Alexa 488) | N. oligotropha subcluster II | Cyan | + | + | + |
| Ntspa662 (Cy5)-Ntspa 1151 (Cy3) | Nitrospira sublineage I | Blue | + | + | + |
| Ntspa662 (Cy5) + Ntspa 1151 (Cy3) | Nitrospira sublineage II | Purple | ND | + | NA |
| Nse1472 (Cy3) + Bet42a (Alexa 488) | N. europaea | Yellow | NA | + | + |
| Bet42a (Alexa 488) | Betaproteobacteria | Green | + | + | + |
| 2nd image acquisition | |||||
| EUB mixture (Alexa 488) | Eubacteria | White | + | + | + |
All probes in the first round of FISH were visualized simultaneously.
+, target detected; ND, target not detected or properly distinguished; NA, probe combination not applied (the population was previously confirmed to be absent or present in very low numbers in the system).
Automated virtual slicing of biofilm images.
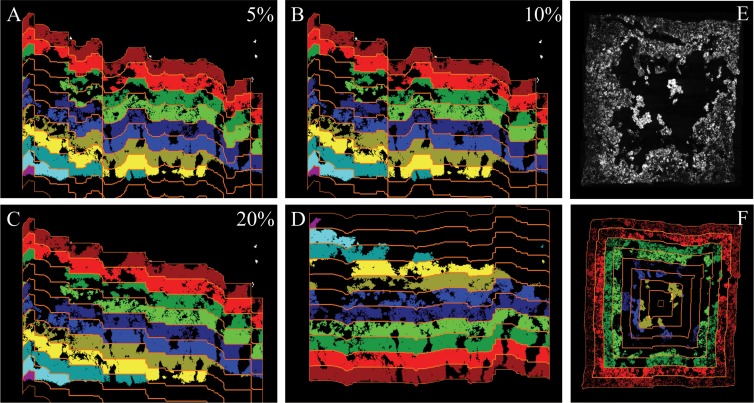
The abundances of AOB and NOB were quantified at different biofilm depths to analyze and compare the vertical distributions of the nitrifiers in the biofilms from different reactors. For this purpose, we developed a new image-processing tool, the Slicer, which automatically cuts biofilm images into slices of a user-defined thickness. The algorithm detects the surface lines of the biofilm in the images and produces virtual slices whose contours follow that of the surface (Fig. 2 and 3). Prior to slicing, the user can adjust a smoothing parameter, which determines whether (small) irregularities of the biofilm surface are ignored during slicing (Fig. 3). Moreover, the Slicer can cut biofilm images in any user-defined combination of four possible directions (from top to bottom, bottom to top, left to right, or right to left). This enables detailed analyses of biofilms grown on a flat, L-shaped, U-shaped, or round substratum (Fig. 3E and F). The Slicer has been implemented in version 2.0 of the digital image analysis software daime (30), which can be downloaded at www.microbial-ecology.net/daime. The Slicer supports batch processing and slices in one run many biofilm images that show different microscope FOVs. Analyzing larger batches of images improves the statistical reliability of the results. For details of the algorithm, see Materials and Methods and the supplemental material.
Fig 3.
Effects of different algorithm parameters on the automatic slicing of biofilm images. (A to C) The same biofilm image sliced with 5, 10, or 20% smoothing of the surface line. The slicing direction is from top to bottom. The slices are colorized for illustrational purposes. (D) The same biofilm image as in panels A to C but sliced from bottom to top (with 20% surface line smoothing). (E) Image of biofilm grown on all four walls of a hollow plastic carrier. (F) Application of the multidirectional-slicing algorithm on the biofilm in panel E. The biofilm image is sliced simultaneously in four directions.
Vertical-distribution analysis.
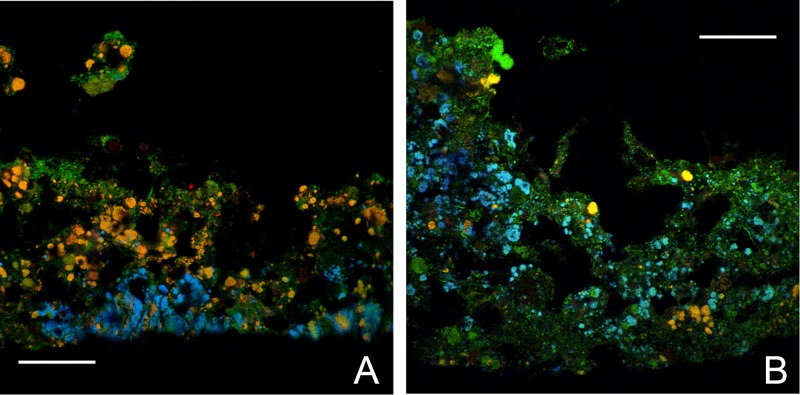
The biofilms in the full-scale NTF, the pilot NTF2, and the pilot MBBR T1 were different for reasons such as reactor design, shear forces, temperature, and ammonium concentrations during the period before the sampling. The biofilm in the full-scale NTF was thin and compact, with a mean thickness of 127 ± 10 μm (standard error [SE]). Similarly, the thicker (193- ± 10-μm) NTF2 biofilm also had a compact architecture, whereas the MBBR biofilm was less dense, with a mean thickness of 387 ± 33 μm (Fig. 4).
Fig 4.
Images exemplifying biofilm structure and stratification. Cryosectioned biofilms from the pilot plant NTF2 (A) and MBBR T1 (B) show cells hybridized with the AOB probe mixture and the EUB338 probe mixtures (yellow), Nitrospira cells hybridized with probe Ntspa662 and the EUB338 probe mixture (cyan), and other bacteria hybridized with the EUB338 probe mixture only (green). Scale bars = 60 μm.
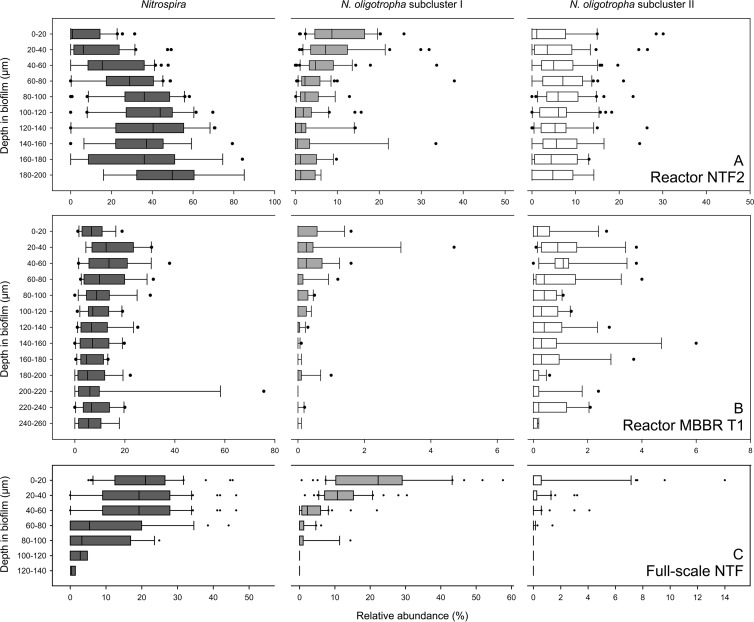
The vertical distribution of the biovolume fractions of the probe-labeled nitrifiers was quantified in cryosectioned biofilms from the three reactors using the new Slicer algorithm. In biofilm from reactor NTF2 (Fig. 5A), the biovolume fractions of NOB from the genus Nitrospira increased significantly (P < 0.001) with biofilm depth and the abundance of Nitrospira in the uppermost region of the biofilm (0 to 40 μm) was significantly (P < 0.05) lower than the abundance in the deeper regions below 80 μm. Especially in the zone closest to the substratum (180 to 200 μm), Nitrospira was highly abundant, with a median biovolume fraction of 49.8% relative to all bacteria. A stratified distribution was also observed for the N. oligotropha-like AOB of subcluster I (P < 0.001), but in contrast to Nitrospira, these AOB were more abundant in the top 0 to 40 μm of the biofilm than in the zone below 60 μm (P < 0.05). These AOB reached their highest density at a depth of 0 to 20 μm, where their median biovolume fraction was 8.7%. N. oligotropha-like AOB of subcluster II had their highest biovolume fraction (median, 7.1%) at a depth of 60 to 80 μm and did not show significant variability in their vertical distribution (P = 0.31).
Fig 5.
Vertical-distribution analysis of Nitrospira (dark-gray boxes), N. oligotropha subcluster I (light-gray boxes), and N. oligotropha subcluster II (white boxes) from NTF2 (A), MBBR T1 (B), and the full-scale NTF (C). Relative proportions as percentages of the total bacterial community, as determined by the EUB338 probe mixture, are shown as box plots for each population at the different depths in the corresponding biofilms. Note the differences in scales on the x and y axes. The boundary of the box closest to zero indicates the 25th percentile, a line within the box marks the median, and the boundary of the box farthest from zero indicates the 75th percentile. The whiskers left and right of the box indicate the 90th and 10th percentiles. The solid circles are outlying points.
The vertical distribution of the nitrifiers in the biofilm from the MBBR T1 was more homogeneous than in the NTF2. Slight stratification was observed only for the two groups of N. oligotropha-like AOB, which were less frequent in deep biofilm zones below 180 μm than in the zone from 20 to 60 μm (P < 0.05). However, the MBBR T1 biofilm was distinctly different from the other two analyzed biofilms. Its structure was less dense and included an upper, loosely attached, and highly irregular fraction resembling a floccular structure (Fig. 4). As we could not apply the automated Slicer tool to this upper fraction, Fig. 5B shows the distribution of nitrifiers only in the lower, more compact regions. However, we quantified the AOB and NOB separately in the loose upper part of the biofilm (without slicing) and obtained values similar to the abundances at depths of 0 to 20 μm in the compact portion (Nitrospira, 7.0% ± 4.7% in the loose region versus 7.7% ± 5.2% at 0 to 20 μm; N. oligotropha subcluster I, 0.2% ± 0.2% versus 0.3% ± 0.5%; N. oligotropha subcluster II, 0.5% ± 0.9% versus 1.1% ± 1.8%). All analyzed nitrifiers were much less abundant in MBBR T1 than in NTF2 (Fig. 5).
The nitrifying biofilm from the full-scale NTF was strongly stratified (Fig. 5C). Nitrospira clearly was more abundant from 0 to 60 μm than below 60 μm (P < 0.05), with the highest median biovolume fraction of 21% at 0 to 20 μm. Interestingly, this distribution pattern contrasted with the trend observed in NTF2, where Nitrospira became more abundant toward the base of the biofilm (Fig. 5A and C). N. oligotropha-like AOB of subcluster I occurred much more frequently at 0 to 40 μm than deeper in the biofilm (P < 0.05) and reached their highest density (median biovolume fraction, 22%) in the uppermost layer (0 to 20 μm). The very low density of the AOB of subcluster II in this biofilm hampered vertical-distribution analysis of this population. Indeed, in the zones below 40 μm, these AOB were hardly detectable by FISH (Fig. 5C). Although detected by FISH, cells belonging to the Nitrosomonas europaea-Nitrosococcus mobilis cluster 7 or sublineage II Nitrospira were not encountered in high enough numbers to allow a reliable spatial distribution analysis in any of the investigated biofilms.
Coaggregation analysis.
In addition to the vertical distribution of the nitrifiers, we quantified the spatial coaggregation patterns of AOB and NOB in the biofilms from the pilot NTF2, the pilot MBBR T1, and the full-scale NTF. The aim was to determine whether the coaggregation patterns with NOB differ for the two analyzed N. oligotropha-like AOB subclusters. For this purpose, the Inflate algorithm (19) was applied. This method performs a pairwise analysis of two FISH probe-defined microbial populations and measures whether these organisms coaggregate, are randomly distributed, or avoid each other in situ. In the biofilm from reactor NTF2, the NOB (Nitrospira) strongly coaggregated with N. oligotropha-like AOB of subcluster II within short spatial distances from 1 to 20 μm (Fig. 6A). At distances beyond 33 μm from these AOB, Nitrospira abundance was lower than expected for randomly distributed populations (Fig. 6A), demonstrating a strong tendency of Nitrospira to coaggregate with subcluster II AOB. In contrast, no coaggregation, but rather repulsion, was observed between Nitrospira and N. oligotropha-like AOB of subcluster I at distances below about 8 μm, whereas these organisms clearly coaggregated at distances between 18 and 43 μm (Fig. 6A). A similar localization pattern was observed in reactor MBBR T1, although the difference between AOB subclusters I and II was less pronounced than in NTF2 and the nitrifiers were randomly distributed relative to each other at distances beyond 12 μm (Fig. 6B). In the full-scale NTF, Nitrospira coaggregated with AOB subcluster I over a distance ranging from 2 to 12 μm and showed random distribution or repulsion at longer distances (Fig. 6C). The localization pattern of AOB of subcluster II was not quantified due to their low density in the biofilm (Fig. 5C).
DISCUSSION
Sequential FISH on cryosectioned biofilms.
Sequential FISH enabled the simultaneous visualization, quantification, and spatial-distribution analysis of more populations than would be possible in one regular FISH experiment (Fig. 1 and Table 1). Without the sequential approach, one CLSM channel would always have been occupied by the EUB338 probe mixture signal, which was required for abundance quantification and as a “biomass mask” for the coaggregation analysis. However, since the EUB338 probe mixture was applied in a second FISH iteration, all CLSM channels were available for detecting six specific populations in the first round of FISH. A proper combination of hierarchically nested probes ensured color patterns that could be interpreted and were not ambiguous (Table 1). Sequential FISH required image acquisition of the same FOV after each iteration, which was feasible with the motorized stage of the CLSM. The characteristic shape of the cryosections enabled precise alignment of the images of each FOV from the first and second iteration. The technique could also be a time-efficient alternative for analyzing other sample types with a less pronounced spatial architecture, as long as the previously recorded FOVs can be recognized in the second round of image acquisition. Recently, a powerful new method combining FISH with multispectral imaging (combinatorial labeling and spectral imaging [CLASI]-FISH) was published (31), enabling visualization of 28 (and potentially more) different combinatorial labels simultaneously. The potential of this method, in combination with the image analysis techniques presented here, is undoubtedly promising. However, the independent requirements for multispectral CLASI-FISH are beyond the capacity of standard CLSMs, where the sequential-FISH approach would be a useful alternative. Another interesting alternative is FISH using multicolored, double-labeled oligonucleotide probes (32). This method allows the simultaneous detection of up to six independent populations with higher sensitivity and less bias than offered by CLASI-FISH (32). However, as noted above, if one of the three dyes in multicolor DOPE-FISH is used for the EUB mixture, which is needed for quantitative analyses, the number of independent bacterial populations that can be identified simultaneously is significantly reduced. Thus, the sequential approach used in this study would also be of use in combination with multicolor DOPE-FISH.
Spatial distribution analysis of biofilm microbial populations.
Biofilm spatial distribution is one of the few means whereby possible differences in ecophysiology and niche differentiation of so far uncultured microorganisms can be deduced (18). In order to quantify population abundances at different depths, one could simply use image-editing software to manually cut the images of FISH-stained cryosectioned biofilm into vertically stacked pieces (slices). Image analysis could then be used to quantify the microbial populations in each slice. Such manual editing, however, would become very tedious with the many biofilm images needed for statistically reliable results. Moreover, one could not simply cut the images in straight horizontal stripes, because the surfaces of most biofilms have a complex irregular contour and are not parallel to the borders of the image. Therefore, we developed and applied the automated Slicer tool, which solves most of these problems. By using this approach, we found that the vertical distribution of Nitrospira sublineage I and two clusters of N. oligotropha-like cells were distinctly different and varied between the analyzed systems (Fig. 5).
In the MBBR T1 biofilm, the relative abundance of all nitrifiers was lower than in NTF2, probably due to the lower NH4+ levels. Nitrospira and N. oligotropha subclusters I and II showed less stratified distributions in the MBBR biofilm. This lack of stratification might be attributed to the distinct physical structure of the biofilm. It was relatively thick, with a comparably low cell density, which may reflect differences in important factors, such as substrate availability and shear forces. A similar biofilm architecture in an MBBR system was reported earlier, and it was suggested that the properties of the MBBR carriers influenced the biofilm structure (12). N. oligotropha subcluster II reached higher densities than N. oligotropha subcluster I in deeper biofilm layers in both NTF2 and MBBR T1. A possible explanation would be that subcluster II is more competitive at lower O2 and/or ammonium concentrations. A more K-like strategy (i.e., slow growth rate and high substrate affinity) for subcluster II, relative to subcluster I, would also explain its very low abundance in the full-scale NTF, where the ambient NH4+ level was the highest, while subcluster I had a high relative abundance and showed a highly stratified depth distribution. Indeed, a similar ammonium-dependent niche differentiation between the two N. oligotropha subclusters was reported in earlier investigations of the MBBR system (23, 33) and may now also be extended to their distinct biofilm vertical-distribution patterns (Fig. 5A and B) and coaggregation patterns with Nitrospira (see below) (Fig. 6). Further evidence for niche partitioning of N. oligotropha-related strains has been provided by Limpiyakorn and colleagues (22), who observed differences in the autoecology of N. oligotropha and connected them to different ammonium concentrations. They also suggested that ammonium sensitivity at concentrations in the millimolar range may inhibit some members of N. oligotropha (34).
Genomic data and incubation experiments have suggested that Nitrospira is microaerophilic (35, 36) and thus prefers hypoxic conditions, which are usually found in deeper biofilm layers. This could explain why Nitrospira densities were higher in the deeper layers of the biofilm in the pilot plant trickling filter NTF2 (Fig. 5A). This observation is in agreement with earlier investigations of nitrifying biofilms (15, 37). However, in the full-scale NTF biofilm, Nitrospira cells were most abundant in the upper biofilm region (Fig. 5C). An earlier study could not detect this Nitrospira stratification in the full-scale NTF (14), but reanalyzing the older CLSM images (see Fig S3 in the supplemental material) and biofilm samples with the new automated Slicer method showed the same stratification presented here (Fig. 5C). The previous, less conclusive results were due to the use of a manual slicing approach with lower resolution (Pär Lydmark, personal communication). The high ammonium load in the full-scale NTF likely caused the high activity of the N. oligotropha subcluster I AOB, which also were most abundant in the upper biofilm layers (Fig. 5C). These AOB not only produced nitrite, but also may have consumed a significant fraction of the available oxygen, creating suitable conditions for Nitrospira in the upper region of the biofilm.
The measured coaggregation patterns showed the distribution of the NOB population in relation to the two AOB populations. In NTF2 and MBBR T1, Nitrospira specifically coaggregated with microcolonies of N. oligotropha subcluster II, which was more abundant than subcluster I in the deeper layers (Fig. 6A and B). Nitrospira might have relied on local NO2− production by the AOB at these biofilm depths, but NO2− was also produced at other depths, and other interactions between specific NOB-AOB are possible, such as in the full-scale NTF, where almost no N. oligotropha subcluster II cells were present and where Nitrospira coaggregated with N. oligotropha subcluster I (Fig. 6C).
Another biological interaction that might drive specific AOB-NOB localization patterns is competition for electron acceptors (repulsion), but this might be balanced by cooperation, as AOB produce the electron donor for NOB and NOB remove toxic NO2−. Other possible but still unknown interactions might be exchange of organic exudates (38) or siderophore parasitism, as suggested for N. europaea (39). Interestingly, the spatial distribution of Nitrobacter in biofilms was recently reported to depend on the community composition of AOB (40). Distinct spatial-distribution patterns of Nitrospira relative to AOB were previously found to reflect niche differentiation in terms of different nitrite requirements (18).
Although the results do not necessarily explain overall reactor function, the observed spatial distribution patterns are relevant for describing the niche differentiation of nitrifying bacteria, which would have been overlooked without the relatively high phylogenetic resolution that was offered by the applied combinations of FISH probes and sequential FISH (Table 1). Reaching adequately high levels of phylogenetic and spatial resolution when analyzing nitrifying community structures may thus be pivotal for avoiding simplified descriptions of these functional guilds. We suggest that functional diversity within certain AOB lineages is substantial and that the new methods presented here will contribute to the elucidation of this diversity.
In summary, by applying the new methodology described here, we have gained knowledge about the interactions in nitrifying biofilms. The niche partitioning of N. oligotropha lineages that has been suggested by us and others was supported. We also showed for the first time that these N. oligotropha lineages had different coaggregation patterns with the dominant NOB, indicating important sublineage ecophysiological differences, and that both AOB and NOB had distinct vertical-distribution patterns, different from what was reported when a less precise approach was used for analysis (14). This investigation is a proof of concept and shows that the described methods can be used for analyzing distributional patterns of microbial populations in practically any multispecies biofilm.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge financial support from the Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (Formas; contracts no. 214-2004-1678, 243-2008-131, and 2010-2259), the Royal Society of Sciences and Letters in Gothenburg (Kungliga Vetenskaps-och Vitterhetssamhället i Göteborg), Göteborgs Universitets Donationsfonder Adlerbertska Forskningsfonden, and the Austrian Science Fund (FWF; grant I44-B06).
We acknowledge Pär Lydmark and Magnus Lind for providing cryosectioned biofilm samples and previously acquired CLSM micrographs from the full-scale NTF. We also acknowledge the technical staff at Gryaab, the Rya WWTP in Gothenburg, for sampling assistance and pilot plant maintenance.
Footnotes
Published ahead of print 26 July 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01727-13.
REFERENCES
- 1.Branda SS, Vik S, Friedman L, Kolter R. 2005. Biofilms: the matrix revisited. Trends Microbiol. 13:20–26 [DOI] [PubMed] [Google Scholar]
- 2.Nickel JC, Ruseska I, Wright JB, Costerton JW. 1985. Tobramycin resistance of Pseudomonas aeruginosa cells growing as biofilm on urinary catheter material. Antimicrob. Agents Chemother. 27:619–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matz C, Kjelleberg S. 2005. Off the hook: how bacteria survive protozoan grazing. Trends Microbiol. 13:302–307 [DOI] [PubMed] [Google Scholar]
- 4.Lawrence JR, Korber DR, Hoyle BD, Costerton JW, Caldwell DE. 1991. Optical sectioning of microbial biofilms. J. Bacteriol. 173:6558–6567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Beer D, Stoodley P, Roe F, Lewandowski Z. 1994. Effects of biofilm structures on oxygen distribution and mass transport. Biotechnol. Bioeng. 43:1131–1138 [DOI] [PubMed] [Google Scholar]
- 6.Schillinger C, Petrich A, Lux R, Riep B, Kikhney J, Friedmann A, Wolinsky LE, Göbel UB, Daims H, Moter A. 2012. Co-localized or randomly distributed? Pair cross correlation of in vivo grown subgingival biofilm bacteria quantified by digital image analysis. PLoS One 7:e37583. 10.1371/journal.pone.0037583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Augspurger C, Karwautz C, Mußmann M, Daims H, Battin TJ. 2010. Drivers of bacterial colonization patterns in stream biofilms. FEMS Microbiol. Ecol. 72:47–57 [DOI] [PubMed] [Google Scholar]
- 8.Gantner S, Schmid M, Durr C, Schuhegger R, Steidle A, Hutzler P, Langebartels C, Eberl L, Hartmann A, Dazzo F. 2006. In situ quantitation of the spatial scale of calling distances and population density-independent N-acylhomoserine lactone-mediated communication by rhizobacteria colonized on plant roots. FEMS Microbiol. Ecol. 56:188–194 [DOI] [PubMed] [Google Scholar]
- 9.Harmsen HJ, Kengen HM, Akkermans AD, Stams AJ, de Vos WM. 1996. Detection and localization of syntrophic propionate-oxidizing bacteria in granular sludge by in situ hybridization using 16S rRNA-based oligonucleotide probes. Appl. Environ. Microbiol. 62:1656–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Santegoeds CM, Damgaard LR, Hesselink G, Zopfi J, Lens P, Muyzer G, de Beer D. 1999. Distribution of sulfate-reducing and methanogenic bacteria in anaerobic aggregates determined by microsensor and molecular analyses. Appl. Environ. Microbiol. 65:4618–4629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foesel BU, Gieseke A, Schwermer C, Stief P, Koch L, Cytryn E, de la Torré JR, van Rijn J, Minz D, Drake HL, Schramm A. 2008. Nitrosomonas Nm143-like ammonia oxidizers and Nitrospira marina-like nitrite oxidizers dominate the nitrifier community in a marine aquaculture biofilm. FEMS Microbiol. Ecol. 63:192–204 [DOI] [PubMed] [Google Scholar]
- 12.Gieseke A, Bjerrum L, Wagner M, Amann R. 2003. Structure and activity of multiple nitrifying bacterial populations co-existing in a biofilm. Environ. Microbiol. 5:355–369 [DOI] [PubMed] [Google Scholar]
- 13.Gieseke A, Purkhold U, Wagner M, Amann R, Schramm A. 2001. Community structure and activity dynamics of nitrifying bacteria in a phosphate-removing biofilm. Appl. Environ. Microbiol. 67:1351–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lydmark P, Lind M, Sorensson F, Hermansson M. 2006. Vertical distribution of nitrifying populations in bacterial biofilms from a full-scale nitrifying trickling filter. Environ. Microbiol. 8:2036–2049 [DOI] [PubMed] [Google Scholar]
- 15.Okabe S, Itoh T, Satoh H, Watanabe Y. 1999. Analyses of spatial distributions of sulfate-reducing bacteria and their activity in aerobic wastewater biofilms. Appl. Environ. Microbiol. 65:5107–5116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amann RI, Ludwig W, Schleifer K-H. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daims H. 2009. Use of fluorescence in situ hybridization and the daime image analysis program for the cultivation-independent quantification of microorganisms in environmental and medical samples. Cold Spring Harb. Protoc. 2009:pdb.prot5253. 10.1101/pdb.prot5253 [DOI] [PubMed] [Google Scholar]
- 18.Maixner F, Noguera DR, Anneser B, Stoecker K, Wegl G, Wagner M, Daims H. 2006. Nitrite concentration influences the population structure of Nitrospira-like bacteria. Environ. Microbiol. 8:1487–1495 [DOI] [PubMed] [Google Scholar]
- 19.Daims H, Wagner M. 2011. In situ techniques and digital image analysis methods for quantifying spatial localization patterns of nitrifiers and other microorganisms in biofilm and flocs. Methods Enzymol. 496:185–215 [DOI] [PubMed] [Google Scholar]
- 20.Daims H, Nielsen JL, Nielsen PH, Schleifer K-H, Wagner M. 2001. In situ characterization of Nitrospira-like nitrite-oxidizing bacteria active in wastewater treatment plants. Appl. Environ. Microbiol. 67:5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Juretschko S, Timmermann G, Schmid M, Schleifer K-H, Pommerening-Roser A, Koops HP, Wagner M. 1998. Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira-like bacteria as dominant populations. Appl. Environ. Microbiol. 64:3042–3051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Limpiyakorn T, Kurisu F, Sakamoto Y, Yagi O. 2007. Effects of ammonium and nitrite on communities and populations of ammonia-oxidizing bacteria in laboratory-scale continuous-flow reactors. FEMS Microbiol. Ecol. 60:501–512 [DOI] [PubMed] [Google Scholar]
- 23.Lydmark P, Almstrand R, Samuelsson K, Mattsson A, Sorensson F, Lindgren P-E, Hermansson M. 2007. Effects of environmental conditions on the nitrifying population dynamics in a pilot wastewater treatment plant. Environ. Microbiol. 9:2220–2233 [DOI] [PubMed] [Google Scholar]
- 24.Schramm A, de Beer D, Wagner M, Amann R. 1998. Identification and activities in situ of Nitrosospira and Nitrospira spp. as dominant populations in a nitrifying fluidized bed reactor. Appl. Environ. Microbiol. 64:3480–3485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stein LY, Arp DJ. 1998. Loss of ammonia monooxygenase activity in Nitrosomonas europaea upon exposure to nitrite. Appl. Environ. Microbiol. 64:4098–4102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Almstrand R, Lydmark P, Sorensson F, Hermansson M. 2011. Nitrification potential and population dynamics of nitrifying bacterial biofilms in response to controlled shifts of ammonium concentrations in wastewater trickling filters. Bioresour. Technol. 102:7685–7691 [DOI] [PubMed] [Google Scholar]
- 27.Manz W, Amann R, Ludwig W, Wagner M. 1992. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15:593–600 [Google Scholar]
- 28.Stoecker K, Dorninger C, Daims H, Wagner M. 2010. Double labeling of oligonucleotide probes for fluorescence in situ hybridization (DOPE-FISH) improves signal intensity and increases rRNA accessibility. Appl. Environ. Microbiol. 76:922–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Daims H, Wagner M. 2007. Quantification of uncultured microorganisms by fluorescence microscopy and digital image analysis. Appl. Microbiol. Biotechnol. 75:237–248 [DOI] [PubMed] [Google Scholar]
- 30.Daims H, Lücker S, Wagner M. 2006. daime, a novel image analysis program for microbial ecology and biofilm research. Environ. Microbiol. 8:200–213 [DOI] [PubMed] [Google Scholar]
- 31.Valm AM, Welch JLM, Rieken CW, Hasegawa Y, Sogin ML, Oldenbourg R, Dewhirst FE, Borisy GG. 2011. Systems-level analysis of microbial community organization through combinatorial labeling and spectral imaging. Proc. Natl. Acad. Sci. U. S. A. 108:4152–4157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Behnam F, Vilcinskas A, Wagner M, Stoecker K. 2012. A straightforward DOPE (double labeling of oligonucleotide probes)-FISH (fluorescence in situ hybridization) method for simultaneous multicolor detection of six microbial populations. Appl. Environ. Microbiol. 78:5138–5142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Almstrand R, Lydmark P, Lindgren PE, Sorensson F, Hermansson M. 2013. Dynamics of specific ammonia-oxidizing bacterial populations and nitrification in response to controlled shifts of ammonium concentrations in wastewater. Appl. Microbiol. Biotechnol. 97:2183–2191 [DOI] [PubMed] [Google Scholar]
- 34.Limpiyakorn T, Kurisu F, Yagi O. 2006. Quantification of ammonia-oxidizing bacteria populations in full-scale sewage activated sludge systems and assessment of system variables affecting their performance. Water Sci. Technol. 54:91–99 [DOI] [PubMed] [Google Scholar]
- 35.Park H-D, Noguera DR. 2008. Nitrospira community composition in nitrifying reactors operated with two different dissolved oxygen levels. J. Microbiol. Biotechnol. 18:1470–1474 [PubMed] [Google Scholar]
- 36.Lücker S, Wagner M, Maixner F, Pelletier E, Koch H, Vacherie B, Rattei T, Sinninghe Damsté JS, Spieck E, Le Paslier D, Daims H. 2010. A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc. Natl. Acad. Sci. U. S. A. 107:13479–13484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schramm A, de Beer D, Gieseke A, Amann R. 2000. Microenvironments and distribution of nitrifying bacteria in a membrane-bound biofilm. Environ. Microbiol. 2:680–686 [DOI] [PubMed] [Google Scholar]
- 38.Rittmann BE, Regan JM, Stahl DA. 1994. Nitrification as a source of soluble organic substrate in biological treatment. Water Sci. Technol. 30:1–8 [Google Scholar]
- 39.Chain P, Lamerdin J, Larimer F, Regala W, Lao V, Land M, Hauser L, Hooper A, Klotz M, Norton J, Sayavedra-Soto L, Arciero D, Hommes N, Whittaker M, Arp D. 2003. Complete genome sequence of the ammonia-oxidizing bacterium and obligate chemolithoautotroph Nitrosomonas europaea. J. Bacteriol. 185:2759–2773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Terada A, Lackner S, Kristensen K, Smets BF. 2010. Inoculum effects on community composition and nitritation performance of autotrophic nitrifying biofilm reactors with counter-diffusion geometry. Environ. Microbiol. 12:2858–2872 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.