Abstract
Algal biofuels represent one of the most promising means of sustainably replacing liquid fuels. However, significant challenges remain before alga-based fuels become competitive with fossil fuels. One of the largest challenges is the ability to harvest the algae in an economical and low-energy manner. In this article, we describe the isolation of a bacterial strain, Bacillus sp. strain RP1137, which can rapidly aggregate several algae that are candidates for biofuel production, including a Nannochloropsis sp. This bacterium aggregates algae in a pH-dependent and reversible manner and retains its aggregation ability after paraformaldehyde fixation, opening the possibility for reuse of the cells. The optimal ratio of bacteria to algae is described, as is the robustness of aggregation at different salinities and temperatures. Aggregation is dependent on the presence of calcium or magnesium ions. The efficiency of aggregation of Nannochloropsis oceanica IMET1 is between 70 and 95% and is comparable to that obtained by other means of harvest; however, the rate of harvest is fast, with aggregates forming in 30 s.
INTRODUCTION
Fossil fuels have provided the energy to drive development in the modern world. Most of the energy used in the United States comes from fossil fuels, including crude oil, coal, and natural gas (1). Our energy infrastructure, particularly transportation, is built around the availability of liquid fuels, meaning that at least in the near term, a suitable sustainable replacement to fossil oil is desirable. Biofuels have been proposed as a means to switch from the current extractive means of liquid fuel generation to a sustainable form of liquid fuel generation.
First-generation biofuels are derived from food crops by converting corn and sugarcane into ethanol. Production of biofuels has been spurred by the Renewable Fuel Standard, which requires the production of 136 billion liters of renewable fuel by 2022 (2). The act that established this standard also requires the fuels produced to have a lower carbon footprint than the petroleum fuels that they replace (2). Second-generation biofuels are derived from cellulose, which is converted to ethanol. The cellulose can come from a variety of sources, including corn stover, wood, and switchgrass. Fuels from algae are often listed as the third and perhaps most promising iteration of biofuels due to several advantages over other biofuels. The term alga-derived biofuels refers to the production of biodiesel from algal lipids, as opposed to ethanol derived from first- and second-generation approaches. Biodiesel can also be generated from oils extracted from canola, soy, and palm oils (3); however, chief among the advantages of algae is the productivity that is possible per unit area, with some algae being able to double their biomass in a few hours. Biofuels are based on converting biomass to fuel, so greater biomass productivity translates to greater fuel productivity. Other advantages of algae are the ability to grow in salt water or wastewater and to grow on land not suited for agriculture (4, 5). Algae can also be harvested multiple times per season, unlike many land-based crops (3).
Microalgae are promising candidates for producing renewable liquid fuels; however, several barriers must be reduced before large-scale production can be economically viable. It is important to note that these challenges are technical and economic in nature, as the process of creating biofuels from algae is proven. These barriers include supplying sufficient nutrients and CO2 to the algae as well as mixing and controlling the pH in the large facilities needed to produce significant amounts of biofuel (6). Other challenges come from a systems perspective, where the reliability of production must be improved (6). The risk associated with inconsistent production stems from the presence of algal grazers as well as infections by bacteria or viruses, which can lower productivity or even result in the rapid decline of algal cultures.
We have focused on the harvest step of algal biofuel production because this step must be improved for algal biofuels to become economically viable. In an analysis done by Richardson et al., harvesting was the number one operating expense and number two capital expense in an outdoor production system (6). Harvesting microalgae efficiently and cheaply is one of the biggest challenges for algal biofuels. Even in dense algal cultures, the biomass represents only a few grams per liter of water. This water must be removed before the biomass can be converted into fuel. Several strategies have been developed to concentrate algae, including filtration, centrifugation, sedimentation, dissolved air flotation, electrocoagulation, chemical flocculation, and bioaggregation (7). Each method has advantages and disadvantages with respect to cost, throughput, and posttreatment effects on both water and biomass, as reviewed in detail by Uduman et al. (7).
Bioaggregation can be achieved using biopolymers, such as chitosan (8–10), and extracellular polymeric substances, such as polysaccharides (11, 12). Bacterial bioaggregation is a natural process and is often observed in laboratory-grown algal cultures. Several bacteria have been identified as bioaggregation agents that can be used to aggregate algae (13–17). In this study, we isolated and identified a bacterium designated Bacillus sp. strain RP1137 that is capable of rapidly aggregating microalgae that are candidates for biofuel production. We characterize the conditions under which aggregation is effective and the range of algae which the bacterium can aggregate.
MATERIALS AND METHODS
Strains and culture conditions.
The bacterial strain Bacillus sp. RP1137 was grown in marine broth 2216 (BD, Franklin Lakes, NJ) at 30°C in 125-ml Erlenmeyer flasks with shaking at 180 rpm. Solid medium was prepared from marine broth 2216 by addition of 15 g/liter Difco technical agar (BD).
Nannochloropsis oceanica IMET1 was grown in f/2 medium (18) with a salinity of 20 ppt. This alga has been maintained for 10 years at the Institute of Marine and Environmental Technology by the Aquaculture Research Center in Baltimore, MD. The algae were grown in photobioreactors which consisted of 500-ml borosilicate glass bottles (Pyrex no. 1395; diameter, 13.6 cm; height, 26.2 cm) with three ports in the cap. Two ports were connected to tubes leading to the bottom of the bottle. Air was pumped into these two ports through 0.22-μm-pore-size syringe filters at a rate of 5 liters/min to provide constant mixing and to provide the carbon dioxide present in the air. The third port was used as a vent. The bottles were positioned 25 cm from 215-W Phillips white fluorescent lights with a light intensity of 275 μmol/m2/s at the front of the bottle and were grown on a light-dark photoperiod of 14 and 10 h, respectively, at 25°C. Algae were subcultured weekly using an inoculating volume of 10% and were used for the experiments at an optical density at 600 nm (OD600) of 1.0 and a pH of 10.2.
Tetraselmis chuii, Tetraselmis sucia, and a Phaeodactylum sp. were grown using the same setup described above and were also grown in f/2 medium. Cells were grown in 250-ml Erlenmeyer flasks at 180 rpm with shaking at 20°C and with a light-dark photoperiod of 14 and 10 h, respectively.
Neochloris oleoabundans MK8520 was grown in Neochloris medium. This medium was composed of 5.82 g/liter NaCl, 2.47 g/liter MgSO4·7H2O, 1 g/liter KNO3, 0.75 g/liter KCl, 66 mg/liter CaCl2, 1.24 g/liter H3BO3, 4.2 g/liter NaHCO3, 0.109 g/liter KH2PO4, 10 μg/liter thiamine HCl, 50 μg/liter biotin, 50 μg/liter vitamin B12, 0.24 mg/liter Na2MoO4·2H2O, 6.6 mg/liter FeCl3, 0.1 mg/liter CuSO4·5H2O, 0.08 mg/liter CoCl2·6H2O, 3.2 mg/liter MnCl2·4H2O, 0.22 mg/liter ZnCl2, and 40 mg/liter Na2EDTA. Cells were grown in 250-ml Erlenmeyer flasks at 180 rpm with shaking at 20°C and with a light-dark photoperiod of 14 and 10 h, respectively.
Nitzschia angularis MK8708 was grown in 100% Instant Ocean medium. This medium was composed of 35 g/liter Instant Ocean, 0.75 g/liter NaNO3, 0.3 g/liter Na2SiO3·9H2O, 15 mg/liter NaH2PO4, 20 mg/liter Fe2SO4·7H2O, 17 mg/liter Na2EDTA, 34.2 mg/liter H3BO3, 4.3 mg/liter MnCl2·4H2O, 0.3 mg/liter ZnCl2, 0.13 mg/liter CoCl2·6H2O, 0.025 mg/liter NaMoO4·2H2O, 0.01 mg/liter CuSO4·5H2O, 0.26 mg/liter NiSO4·6H2O, 0.3 mg/liter vitamin B12, 6 mg/liter thiamine, and 0.3 mg/liter biotin. Cells were grown in 250-ml Erlenmeyer flasks at 180 rpm with shaking at 20°C and with a light-dark photoperiod of 14 and 10 h, respectively.
Cyclotella cryptica MK89172 was grown in 50% Instant Ocean medium. This medium is the same as the 100% Instant Ocean medium listed above, except that 17.5 g/liter instead of 35 g/liter of Instant Ocean was used and the medium was supplemented with 8 g/liter glucose. Cells were grown in 250-ml Erlenmeyer flasks at 180 rpm with shaking at 20°C and with a light-dark photoperiod of 14 and 10 h, respectively.
16S rRNA gene-based strain identification.
To identify the aggregating strain, 16S rRNA gene PCR was performed using primers 27f and 1492r (19), and RP1137 cells were added directly to the PCR mixture as the DNA template. Cycling conditions were one cycle at 95°C for 3 min; 30 cycles of 95°C for 30 s, 46°C for 30 s, and 72°C for 90 s; and then one cycle of 72°C for 5 min. The ca. 1,465-bp product was purified using a Qiagen QIAquick PCR purification kit. The purified PCR product was sequenced using primers 27f, 1492r, 700f (GTGKAGCRGTGAAA), and 700r (CTACGCATTTCACY) to obtain full-length double-stranded sequence. The closest match for RP1137's 16S rRNA gene sequence was identified using NCBI's BLASTn algorithm.
Filtration aggregation assay.
Aggregation data were collected using a filtration aggregation 96-well assay. This assay was developed to allow testing of multiple parameters with replicates in a 96-well plate. Filtration rigs were built by sandwiching a 50-μm mesh between two pipette tip racks (Tip One; USA Scientific) and sealing this mesh in place with silicon. The resultant apparatus was designated a “filtration rig.” The filtration rig fit neatly in a 96-well plate and retained aggregates on the mesh, while nonaggregated algal cells (diameter, 2 to 3 μm) passed through into the wells of the microtiter plate. In a typical aggregation experiment, 150 μl of algae at 1 × 107 cells/ml was pipetted into a round-bottom 96-well plate. A 100-fold-concentrated bacterial suspension (1.5 μl; original density, 1 × 107 cells/ml) was added to the algae in the designated wells to give a 1:1 alga-to-bacterium ratio. Wells not receiving bacteria served as negative controls for aggregation induced by the addition of bacteria. The plate was sealed with Parafilm, and the contents were mixed by vortexing for 1 min. A filtration rig was placed on a round-bottom 96-well plate, and the 150 μl containing the aggregates was pipetted from the first microtiter plate into the top of the filtration rig using wide-bore pipette tips. The plate and rig were centrifuged at 100 × g for 10 s to ensure that the entire volume in the rig passed through the mesh into the second, lower 96-well plate. At this point, the aggregates were retained on the mesh and the nonaggregated cells had passed into the second plate. The algae in the second plate were suspended and diluted into the linear range for chlorophyll fluorescence measurement in a Spectromax M5 microplate reader. Chlorophyll fluorescence was measured with an excitation wavelength of 488 nm, a 515-nm-cutoff filter, and an emission wavelength of 685 nm. From these data, the percentage of algae that had aggregated and remained on the mesh was calculated from the fluorescence of the algae that were not aggregated using the formula P = [1 − (A/N)] · 100, where P is the percentage of algae aggregated, A is the fluorescence of the filtrate from the aggregated sample, and N is the fluorescence of the filtrate from the nonaggregated sample.
Fluorescence microscopy.
Microscopy was used to visualize the structure of the bacterial-algal aggregates. A 1-ml aliquot of Bacillus sp. RP1137 was stained with 2.5 μl of SYBR green I nucleic acid stain for 10 min in the dark. The bacteria were concentrated by centrifugation at 13,000 × g for 2 min, and the cell pellet was suspended in 10 μl of fresh medium, giving a 100-fold concentration of the bacteria. An 8-μl portion of this suspension was added to 800 μl of Nannochloropsis cells in f/2 medium at an OD600 of 1.0, and the components were mixed. Aggregates were pipetted onto a slide and visualized using laser scanning confocal microscopy with a Zeiss Axioskop microscope and a Bio-Rad Radiance 2100 laser light source. Both algal chlorophyll and SYBR green I-stained bacteria were excited with a 488-nm argon laser. The SYBR green I signal was visualized using a 415/30-nm band-pass filter, and the chlorophyll autofluorescence was visualized with a 600-nm long-pass filter. Controls comprising bacteria only and algae only were used to ensure that there was no cross bleeding of signals between the two channels.
Temperature dependence.
To determine the effect of temperature on aggregation, 150 μl of algae was transferred into PCR tubes. The PCR tubes were placed in a thermocycler to hold the algal suspensions at 10, 20, 30, or 40°C. These temperatures were chosen because they represent the range of temperatures likely to occur in an outdoor algal production pond. The algae at 1 × 107 cells/ml were incubated for 10 min to ensure that they reached the set temperatures. To half of the tubes at a given temperature, 1.5 μl of 100-fold-concentrated RP1137 cells (original density, 1 × 107 cells/ml) was added and the components were mixed by vortexing. The other half of the tubes served as no-aggregation controls. All samples were transferred to the top of the filtration aggregation rig, and the assay was continued as described above.
Salinity dependence.
The salinity dependence of aggregation was determined by harvesting a 5-ml aliquot of algae at 5,580 × g for each salinity tested. Half of the supernatant was removed (2.5 ml). Nannochloropsis was grown in 20-ppt salinity. To get 10-ppt salinity, 2.5 ml of distilled H2O (dH2O) was added; for 20-ppt salinity, 2.5 ml of 20-ppt NaCl was added; for 30-ppt salinity, 2.5 ml of 40-ppt NaCl was added; for 40-ppt salinity, 2.5 ml of 60-ppt NaCl was added; and for 156-ppt salinity, 2.5 ml of 292-ppt NaCl was added. For the 0-ppt-salinity sample, the cell pellet was suspended in dH2O and the pH was adjusted to match that of the other samples (pH 10.2). The last sample had an effective salinity of 0.005 ppt due to the NaOH used to adjust the pH. After salinity adjustment, the filtration aggregation assay was carried out to quantify the effect of salinity on aggregation.
pH dependence.
The pH dependence of aggregation was determined by adjusting the pH of a 200-ml aliquot of Nannochloropsis at 1 × 107 cells/ml in a beaker with constant stirring. The pH was adjusted with either 5 M NaOH or 5 M HCl; concentrated base or acid was used to limit changes in algal cell concentration. When pH stabilized at a desired value, 150 μl of the algal suspension was quickly pipetted into a 96-well plate. An overnight culture of Bacillus sp. RP1137 with a density 1 × 107 cells/ml was 100-fold concentrated, and 1.5 μl of the concentrate was added to half of the wells. The other half of the wells served as 0% aggregation negative controls. The filtration aggregation assay was carried out, and a final pH measurement was taken to ensure that the pH had not shifted significantly during the assay. This process was repeated for each pH tested.
Viability dependence.
Two 5-ml portions of algae and two 5-ml portions of bacteria were used. To one algal sample and one bacterial sample, 5 ml of 8% paraformaldehyde (PFA; pH 7) was added and the components were mixed. To the other algal and bacterial samples, 5 ml of dH2O was added. All samples were then incubated at room temperature for 1 h with mixing every 15 min. All samples were then harvested by centrifugation at 5,580 × g for 5 min. The supernatants were aspirated, and the algal samples were suspended in 5 ml of cell-free spent algal medium from the original algal culture. Bacterial samples were suspended in 5 ml of marine broth 2216. All samples were again concentrated by centrifugation, aspirated, and suspended. The algae were suspended in 5 ml to keep their original concentration, while the bacteria were suspended in 50 μl to give a 100-fold concentration. Aliquots (150 μl) of the algae at 1 × 107 cells/ml were transferred to a 96-well plate, and 1.5 μl of the concentrated bacteria per well (original density, 1 × 107 cells/ml) was used for aggregation experiments. The filtration aggregation assay was followed to complete quantitation of aggregation with the live and dead cells.
Cell ratio.
For cell ratio experiments, the concentrations of bacteria and algae were determined using an Accuri C6 flow cytometer. Medium that had been filtered through a 0.22-μm-pore-size filter was used to grow both the algae and the bacteria. The same medium was used as the blank for determining the correct parameters for detecting cells. Chlorophyll fluorescence was used as the cutoff for gating algal cells. A forward-scatter-area cutoff of greater than 5,000 was used for gating RP1137 cells and excluded noncellular debris found in both sterile-filtered (pore size, 0.22 μm) marine broth 2216 and the RP1137 cultures. The algal cell concentration and volume (150 μl) were held constant in all wells in the 96 well-plate, while the concentration of the bacteria added was adjusted. The bacterial volume added was held constant. The bacterial culture was either concentrated by centrifugation or diluted to achieve the bacterium-alga ratios tested of 25:1, 5:1, 1:1, 1:5, 1:25, and 1:125. As before, 1.5 μl of concentrated bacteria was added to each well, and the filtration aggregation assay was used to quantify the percentage of algae that were aggregated.
Aggregation of other algae.
T. chuii, T. sucia, a Phaeodactylum sp., N. oleoabundans. MK8520, C. cryptica MK89172, and N. angularis MK8708 were grown until the pH of cultures was between 9.5 and 10. The strains were tested using the filtration aggregation assay as described above.
Filament shearing assay.
RP1137 cells were split into two portions. One portion served as the control, and the other portion was sheared by passing the cells through a 28.5-gauge needle 80 times. The cells were then tested using the filtration aggregation assay as described above.
Proteinase K digestion assay.
RP1137 cells were digested with 100 μg/ml proteinase K in 1× phosphate-buffered saline (PBS) buffer at pH 7.4. Nondigested samples were incubated in 1× PBS. Samples were incubated for 2 h at room temperature or 50°C. The bacteria were then washed twice with 1× PBS to remove the proteinase K and then tested using the filtration aggregation assay.
Carbohydrate inhibition assay.
Glucose, galactose, mannose, and lactose were individually added to aliquots of Nannochloropsis cells at concentrations ranging from 200 to 800 mM. RP1137 cells were added to these algal aliquots to assay the effectiveness of each carbohydrate at inhibiting aggregation.
Ca2+ and Mg2+ dependence assay.
Nannochloropsis and RP1137 cells were washed three times in pH 10.3 deionized water. The washing procedure involved concentrating the cells via centrifugation, followed by aspiration of the supernatant and resuspension of the cells in pH 10.3 deionized water. A 0.5 M stock of CaCl2 or MgSO4 was added to the algae to obtain the final Ca2+ or Mg2+ concentrations of 0.125, 0.25, 0.5, 1, 2, 4, 8, and 16 mM. The algal solution containing the metal ions was then tested using the filtration aggregation assay with RP1137 cells.
Nucleotide sequence accession number.
The GenBank accession number for the 16S rRNA gene sequence from Bacillus sp. RP1137 is KF015297.
RESULTS
Isolation of an alga-aggregating bacterium.
An environmental bacterium that was able to rapidly aggregate algae was isolated from a Nannochloropsis oceanica IMET1 aggregation experiment. When this bacterium was added to Tetraselmis cultures and mixed, it quickly aggregated the algae, with most of the algae settling out of solution in large aggregates in 30 s (Fig. 1). This new strain was serendipitously found as a contaminant in a broth culture of HW001, a bacterium previously reported to have the ability to aggregate algae (16). The contaminant was isolated into pure culture and was shown to have an ability to aggregate algae superior to that of HW001. The 16S rRNA gene of this new isolate was PCR amplified and sequenced. The sequence was identified using BLASTn and was found to have a 99% sequence identity to Bacillus megaterium strain PPB7. This aggregating strain was designated Bacillus sp. RP1137. Previous work by Wang et al. showed that bacteria whose 16S rRNA sequence matches the 16S rRNA sequence of Bacillus sp. RP1137 are not present in Nannochloropsis cultures, and indeed, no bacteria within the phylum Firmicutes were found (16), indicating that it is unlikely that RP1137 is a major component of the natural algal bacterial community. The original cryopreserved stock of HW001 was examined and found to be pure. Comparison of the 16S rRNA gene sequences to all sequences derived from culture-based and culture-independent analyses in the laboratory revealed no matches. Bacillus megaterium species can be found in seawater, freshwater, and soil (20), suggesting that the environment where the strain originated will likely remain unknown.
Fig 1.
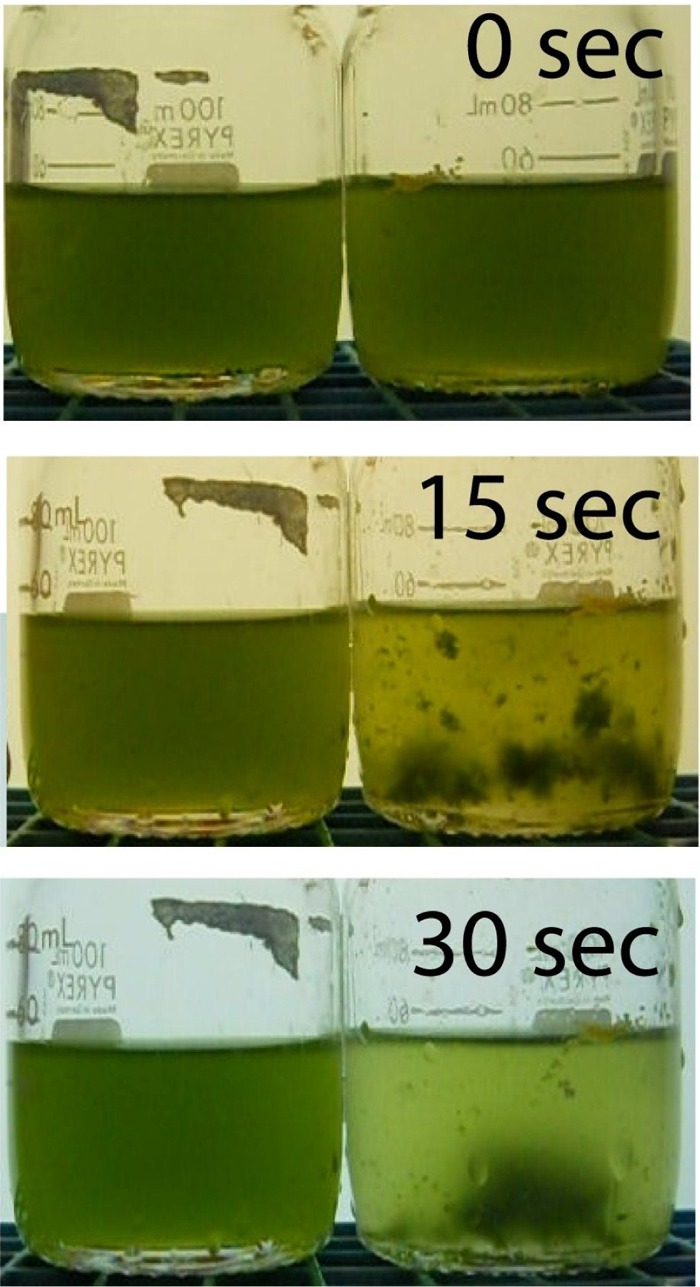
Aggregation of algae is rapid upon addition of the bacterium Bacillus sp. RP1137. Aggregates form within 15 s, and by 30 s the aggregates have settled out of solution. RP1137 cells were added to the bottle of a Tetraselmis sp. on the right, and the bottle on the left served as a control.
To investigate the nature of the aggregates formed when Bacillus sp. RP1137 was added to Nannochloropsis cultures, the aggregates were visualized by laser scanning confocal microscopy. Visualization of the microstructure of the aggregates was done to help elucidate the mechanism of aggregation. The bacteria in these images are filamentous and are intercalated between algal cells (Fig. 2). In general, the bacteria are mostly in contact with algae, with little bacterium-to-bacterium contact along the length of the cell. The algae are in contact with both the bacterial cells and other algal cells. Packing of the cells in the aggregates is tight, with apparent cell-to-cell contact, which suggests that the mechanism of aggregation involves interactions at the cell surface.
Fig 2.
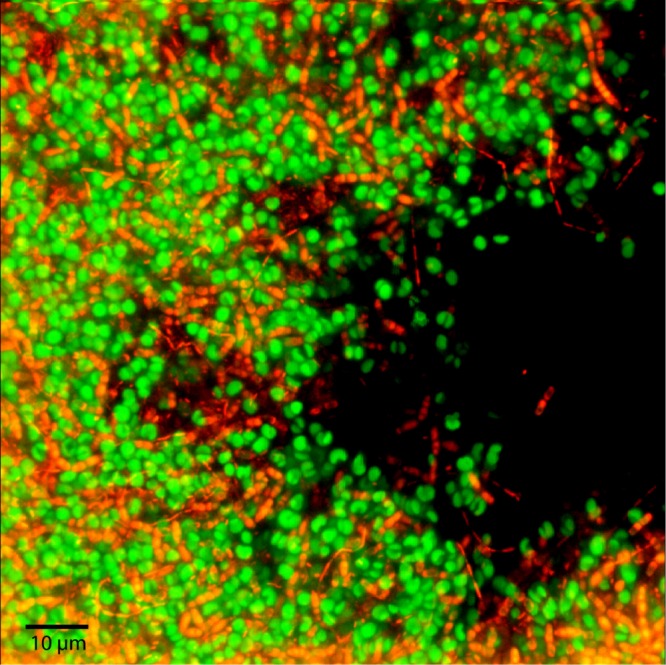
Laser scanning confocal micrograph of an N. oceanica IMET1-Bacillus sp. RP1137 aggregate. Algae (green) were visualized by chlorophyll autofluorescence, and the bacteria (orange) were visualized using SYBR green I staining.
Determination of the optimal bacterium-alga ratio.
Microscopy showed that the bacterial filaments within an aggregate are intertwined throughout the structure and interact with multiple algal cells. This raised the question of how many bacteria are needed per algal cell to efficiently form these aggregates. The optimal bacterium-to-alga ratio is also important for practical use of this strain in harvesting algae, to maximize aggregation efficiency with a minimum of bacteria. To test multiple ratios of bacteria to algae and get statistically meaningful data, a 96-well plate aggregation assay was developed and is discussed in detail in the Materials and Methods section. Briefly, the bacteria and algae are combined in a 96-well plate and mixed to form aggregates. The entire volume of each well (aggregated and nonaggregated cells) is passed through a mesh with 50-μm openings. Nannochloropsis cells in culture are mostly found as single, spherical cells with a diameter of 2 to 3 μm. The few natural aggregates are small and have an average cross-sectional area of 71.6 μm2 (16). The aggregates easily pass through the mesh, which has 2,500-μm2 openings. The algal-bacterial aggregates are retained on this mesh, while nonaggregated cells pass through into a second 96-well plate. The algal cells that pass through into the second plate are then quantitated using chlorophyll fluorescence, and the quantity is compared to the quantity for a control where only algae (no bacteria were added to induce aggregation) were passed through the mesh. From these data, the percentage of algae that have been aggregated by the bacteria can be calculated. This process is referred to here as the filtration aggregation assay.
To determine the optimal bacterium-to alga ratio, the volume and concentration of algae were held constant in each well and different concentrations of bacteria were added. Ratios of between 25 bacteria to 1 algal cell and 1 bacterium to 125 algal cells were tested. The ratio of bacteria to algae for optimal aggregation was 1:1, with 1:5 bacteria to algae being only slightly less efficient than 1:1 bacteria to algae at aggregating Nannochloropsis (Fig. 3). These data are consistent with the visual results seen in fluorescence micrographs, where one bacterial cell interacts with several algal cells. At less than one bacterium per five algal cells, the efficiency of aggregation decreases. The efficiency of aggregation also decreases when the number of bacteria used becomes higher than the number of algae. This is likely due to increased self-aggregation between bacterial cells, leaving fewer bacteria to aggregate algal cells. Self-aggregation does occur when the bacteria are present in pure culture if the pH of the culture is increased to >10, which is similar to the pH in dense Nannochloropsis cultures. The data presented above indicate that there is an optimal ratio of bacteria to algae cell for bioaggregation and at higher or lower ratios, the efficiency of aggregation decreases.
Fig 3.
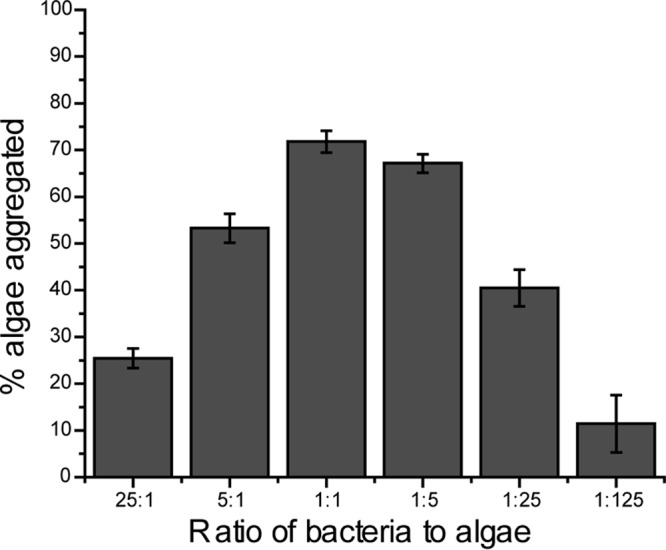
Aggregation efficiency is the highest when one bacterium is added for every one to five algal cells present. At ratios above and below these ratios, aggregation efficiency is reduced. The result for the ratio of 5:1 is statistically significantly different from that for the ratio of 1:1 (P = 8.0E−5). The result for the ratio of 1:1 is statistically significantly different from that for the ratio of 1:5 (P = 0.009). Bars and error bars represent the means and standard errors, respectively, of five independent aggregation reactions. A value of 100% is aggregation of all algal cells.
Characterization of the physical conditions that affect algal aggregation.
Next we were interested in determining which physical factors affect aggregation of the algae. From the microscopy data, it appears the mechanism of aggregation likely involves cell-to-cell contact, as opposed to aggregation of the algae by unassociated cellular exudates. To begin unraveling the nature of this cell-to-cell adhesion, we decided to test if aggregation is pH dependent. Solution pH dictates the surface charge of exposed proteins and will affect both specific and nonspecific protein-driven interactions. From a practical standpoint, the effect of pH on aggregation is also important because dense algal cultures can quickly increase pH through carbon fixation and also rapidly decrease pH through respiration. The effect of pH on aggregation was tested by adjusting the pH of the aliquots of algal cultures with either NaOH or HCl. The pH values tested ranged from 6 to 10. In this experiment, the pH of the algae was set, and then the filtration aggregation assay was run and a final pH reading was taken to ensure that the pH did not change significantly during the experiment. The pH value did have a significant effect on aggregation, as shown in Fig. 4. At pH 8 and below there was no significant aggregation, and at pH 9 and above aggregation was the highest. These data show that the aggregation of Nannochloropsis oceanica IMET1 by Bacillus sp. RP1137 is pH dependent, with aggregation occurring only when the pH is above 9. When the pH of the solution was decreased, disaggregation was observed, showing that aggregation was also reversible (data not shown).
Fig 4.
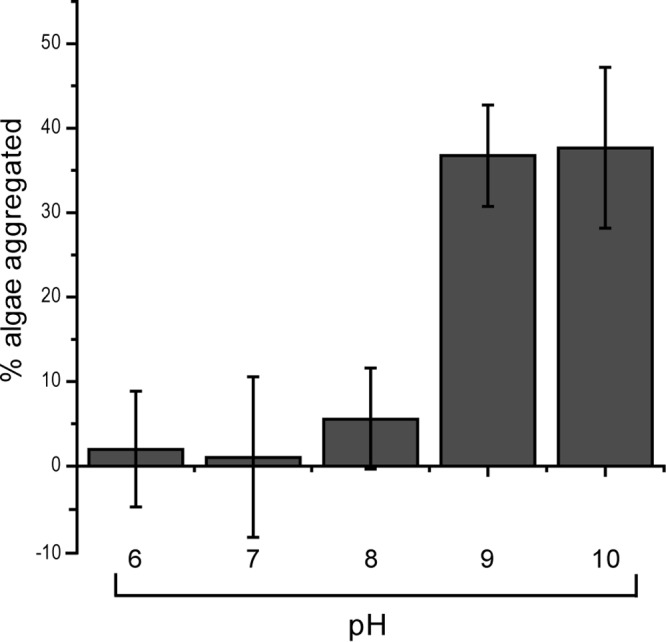
Aggregation efficiency is pH dependent, with optimal aggregation occurring at pH 9 and above. Aggregation at pH 9 is significantly higher than that at pH 8 (P = 5.9E−8). Aggregation above pH 9 is not significantly increased (P > 0.05). There was no significant change in aggregation below pH 8 (P > 0.05). Bars and error bars represent the means and standard errors, respectively, of eight independent aggregation reactions. A value of 100% is aggregation of all algal cells.
In addition to pH, the salinity and temperature of algal production ponds can vary. The effect of salinity on aggregation was determined by adjusting the salinity of aliquots of Nannochloropsis cultures from 0 to 156 ppt. Optimal aggregation occurred at 20-ppt salinity, with minor but significant decreases in aggregation occurring at between 20 ppt and 156 ppt (see Fig. S1 in the supplemental material). Aggregation was also significantly decreased at 0-ppt salinity compared to 20-ppt salinity. The aggregates at 0 ppt were also noticeably smaller (data not shown).
Outdoor algal production ponds can experience changes in temperature, which could result in different potential harvesting conditions. We tested the effect of temperature by performing the aggregation process at between 10 and 40°C. Large differences that would be significant in practical applications at different temperatures were not observed; however, minor statistically significant changes were observed (see Fig. S2 in the supplemental material). Optimal aggregation occurred at 20°C.
One concern in using bacterial bioaggregation as a means of harvesting microalgae is that the production of the bacteria must be scaled with the production of the microalgae. One way to dramatically reduce the number of bacteria needed is to recycle the bacterial cells, effectively using the bacteria as aggregating microparticles. This would be made easier if dead cells retained their aggregation phenotype. To test if dead cells still aggregated algae, Bacillus sp. RP1137 cells were killed with 4% paraformaldehyde and the aggregation potential of these dead cells was compared to that of the same number of live cells. The efficiency of aggregation by the PFA-killed cells had a minor but significant increase relative to that by live cells (Fig. 5A). These data show that the aggregation phenotype of RP1137 is not due to a response by the cell and is instead a passive characteristic that is likely part of the cell surface. As a further step, aggregation of live and PFA-killed Nannochloropsis algae was attempted with both live and dead Bacillus sp. RP1137 cells. The aggregation efficiency with PFA-killed algal cells had a minor but significant increase relative to that with live algal cells (P = 0.009). As with the live algae, the dead bacteria had a minor but significant increase in aggregation efficiency relative to that of the live RP1137 cells (Fig. 5B).
Fig 5.
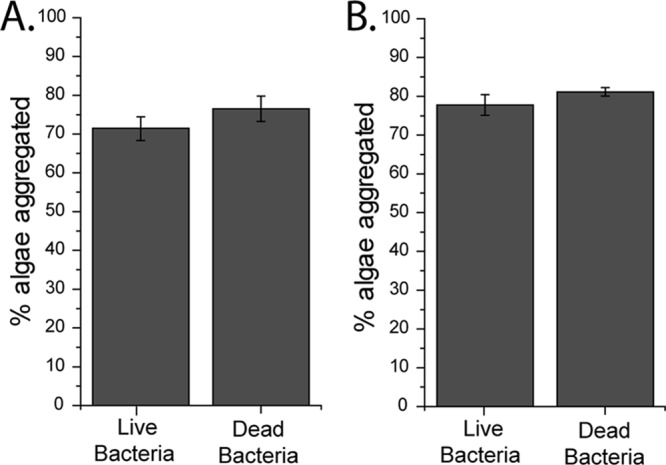
The mechanism of aggregation does not depend on live bacteria or live algae. (A) Live algae were aggregated with both live and dead bacteria. Dead bacteria had a minor but significant increase in aggregation relative to live bacteria (P = 0.06). (B) Dead algae were aggregated with both live and dead bacteria, with dead bacteria aggregating the dead bacteria significantly better (P = 0.01). Bars and error bars represent the means and standard errors, respectively, of eight independent aggregation reactions. A value of 100% is aggregation of all algal cells.
Aggregation of candidate biofuel-producing algae.
Bacillus sp. RP1137 effectively aggregates Nannochloropsis oceanica IMET1 in a pH-dependent and reversible manner. The Bacillus sp. RP1137 cells can also be used effectively after the cells have been killed by fixation. Next, we were interested in seeing if this bacterium can aggregate algae other than Nannochloropsis. A panel of algal strains that are of interest for their potential as biofuel production strains was tested. The strains tested were T. chuii, T. sucia, a Phaeodactylum sp., N. oleoabundans MK8520, C. cryptica MK89172, and N. angularis MK8708. The bacterium Bacillus sp. RP1137 was able to aggregate both of the Tetraselmis species, with a measured aggregation efficiency of 78.3% ± 4.3% for T. sucia and an efficiency of 13.1% ± 3.1% for T. chuii. The bacterium was also able to aggregate the Phaeodactylum sp. with a 17.3% ± 3% efficiency. N. oleoabundans, C. cryptica, and N. angularis had little or no aggregation, with efficiencies of 6% ± 4.4%, 1.3% ± 9%, and 0.02% ± 5.7%, respectively. However, these three nonaggregating algae could be aggregated in a bacterium-dependent manner if concentrated base was added to the solution. The concentrated base increased the pH to ca. 11, which is above the pH that the algae can achieve naturally (data not shown).
Characterization of mechanism of aggregation.
Next, we were interested in exploring the mechanism of algal aggregation by the bacterium RP1137. Nannochloropsis oceanica IMET1 was used as the model alga for these investigations. From the confocal microscopy images, we hypothesized that RP1137's filamentous morphology was important for aggregation of algae. To test this, we sheared the filaments by passing the cells through a 28.5-gauge needle repeatedly. Shearing reduced the RP1137 population to single cells and doublets (Fig. 6A). Sheared and unsheared cells were used for aggregation assays. The results showed that the morphology of the cells did have a significant effect on aggregation (P = 2.71E−7), with sheared cells having a 2.6% decrease in aggregation efficiency compared to that of the unsheared cells (Fig. 6B). While the difference is statistically significant, this minor difference between filaments and single cells or doublets indicates that the filamentous form of RP1137 is not a major factor in the process of aggregation.
Fig 6.
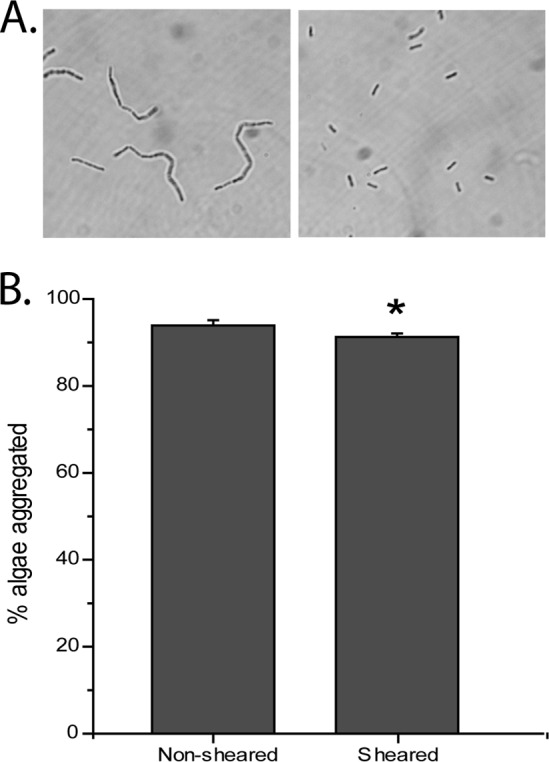
Bacterial filament length has a minor effect on aggregation efficiency. (A) Phase-contrast image of RP1137 before shearing (left) and the same population of cells after shearing (right). (B) Aggregation of Nannochloropsis with nonsheared cells and sheared cells showing a statistically significant (P = 2.71E−7) but minor effect due to filament length. Bars and error bars represent the means and standard errors, respectively, of 16 independent aggregation reactions. A value of 100% is aggregation of all algal cells.
Fixed RP1137 cells are still able to aggregate algae, suggesting that the cell surface rather than active metabolic processes are important. To determine if the aggregation potential of the cells is dependent on exposed surface proteins, we digested RP1137 cells with proteinase K. This approach has been used with Helicobacter pylori to digest off and identify surface proteins (21). When cells were incubated at room temperature with proteinase K, there was no significant difference in aggregation efficiency between digested and nondigested RP1137 cells (see Fig. S3 in the supplemental material). Cells incubated with proteinase K at 50°C displayed a significant (P = 0.002) yet small decrease in aggregation efficiency of 4% compared to that for the nondigested controls, indicating that proteinase K-cleavable surface proteins are not a major factor in the aggregation process.
Adhesion between different cells is often mediated by lectin-carbohydrate-type interactions. These interactions can be disrupted by blocking the lectin with the addition of other carbohydrates. To determine whether lectin-carbohydrate interactions are important in aggregation, glucose, galactose, lactose, and mannose were added to the aggregation reaction mixtures at concentrations ranging from 200 mM to 800 mM. No significant inhibition of aggregation was observed during any of these experiments (data not shown).
We next hypothesized that divalent cations are required for aggregation. As an initial test of this hypothesis, we added 50 mM EDTA and 50 mM ethylene glycol tetraacetic acid (EGTA) to Nannochloropsis in f/2 medium while maintaining the pH at 10.3. Both EDTA and EGTA chelate divalent cations, with EDTA having a higher affinity for magnesium and EGTA having a higher affinity for calcium. EDTA and EGTA at 50 mM were sufficient to inhibit aggregation by RP1137 cells (Fig. 7A). To confirm the importance of divalent cations, we next performed dose-dependent aggregation assays in deionized water with different concentrations of magnesium or calcium ions. The cells were washed several times with deionized water (pH 10.3) to remove trace ions. Fixed algae and fixed bacterial cells were used. Aggregation was found to be highly dependent on the concentration of magnesium and calcium present in solution with the algae (Fig. 7B). At micromolar concentrations of either ion, little or no aggregation was observed. At 8 mM calcium, maximum aggregation efficiency was observed, while maximum aggregation efficiency with magnesium occurred at 16 mM. Addition of calcium or magnesium to Nannochloropsis alone did not precipitate aggregation in the absence of RP1137 cells. These data show that aggregation of Nannochloropsis by RP1137 is highly dependent on the magnesium and calcium concentrations.
Fig 7.
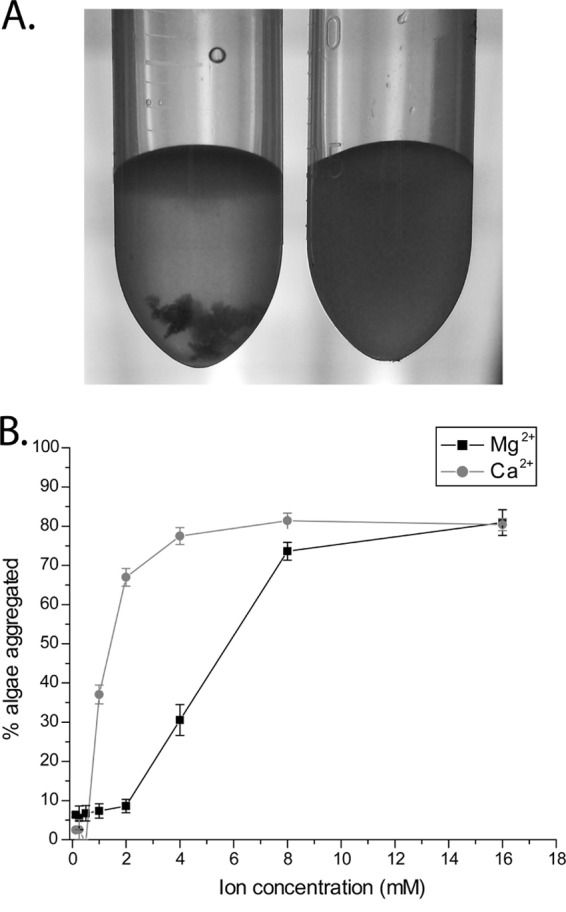
Aggregation is dependent on divalent cations. (A) Aggregation in artificial seawater is inhibited by the addition of EDTA and EGTA (right) compared to that for the untreated control (left). (B) Dose-dependent effect of calcium and magnesium on aggregation, showing increasing aggregation with increasing concentrations of calcium or magnesium. Points and error bars represent the means and standard errors, respectively, of four independent aggregation reactions. A value of 100% is aggregation of all algal cells.
DISCUSSION
We show that live and dead Bacillus sp. RP1137 cells can rapidly aggregate Nannochloropsis oceanica IMET1 in a pH-dependent manner. We previously reported a novel bacterial isolate from Permian groundwater that can aggregate Nannochloropsis oceanica IMET1, but this process occurred over hours to days (16). In contrast, aggregation of Nannochloropsis oceanica IMET1 by Bacillus sp. RP1137 occurred extremely rapidly (in seconds). We also showed that Bacillus sp. RP1137 can aggregate other biofuel-producing algae. The purpose of this study was to identify and evaluate the aggregation ability of this bacterial isolate and specifically to test the robustness of its aggregation phenotype under different conditions. Due to the practical implications of the work, experiments were generally carried out in algal growth medium so that the results could be directly interpreted. It is also worth noting that a simple assay was developed to quantify aggregation potential, to avoid some pitfalls that befall other methods when trying to measure algae aggregated upon addition of bacteria. For example, using the dry weight of the aggregated algae is complicated by the unknown biomass that is contributed by the bacteria to the aggregates. The other common method to quantitate aggregation is to measure sedimentation rates, usually via determination of the absorbance of the supernatant (15). The bacteria added during aggregation assays contribute to the absorbance, occluding the removal of algae from the water column. To deal with these complications, the filtration aggregation assay measures chlorophyll autofluorescence to exclude any signal from the bacteria. Additionally, our method measures the uniform, nonaggregated algal fraction to get an accurate signal of the amount of algae remaining after aggregation. By comparing the remaining fluorescence after aggregation to the fluorescence of the same algal stock that received no treatment, the percentage of algae aggregated can be calculated.
One of the most important questions for the practicality of using bacteria as a means to harvest algae is how many bacteria are needed to harvest a given number of algae. Here the data show that a ratio of one bacterium to five algal cells is sufficient to efficiently harvest the algae. At ratios below this ratio, the efficiency of aggregation decreased; this decrease was likely due to individual bacterial cells being coated by algae and preventing the formation of larger flocs. When the number of bacteria increased above one to one, the efficiency of aggregation also decreased. This may be due to increased bacterium-to-bacterium interactions, which again occlude the formation of larger aggregates. These hypotheses will require further investigation. The bacteria can grow to densities 10 times higher than those of the algae, meaning that if the bacteria are used once, then 1 unit volume of bacteria is needed to aggregate 50 volumes of algae. This represents a substantial amount of bacteria to be cultured for a large-scale algal production facility. As a means to decrease the amount of bacteria needed and gain information about the mechanism of aggregation, we tested if bacteria that had been killed with paraformaldehyde could still effectively aggregate the algae. These killed cells exhibited an aggregation potential equivalent to that of their live counterparts. Mechanistically, this rules out any response by the cell to initiate aggregation and suggests that the mechanism of aggregation is associated with factors located at the cell surface. When cells were fixed by paraformaldehyde, their structure was preserved, which opens the possibility of incorporating the fixed cells onto a solid surface that would allow them to be reused after the algae have been removed from the water. This type of reuse of the bacterial cells could be facilitated by the finding that the aggregation process can be reversed by lowering the pH. An additional advantage of using killed cells is that there is no concern of contamination of the main algal culture or the environment, because the cells will not propagate.
The physical conditions under which harvesting of algae by Bacillus sp. RP1137 is most effective were investigated. Of the conditions tested, pH was the most critical, with aggregation occurring when the pH was at or above 9. In general, a lower percentage of algae was aggregated in experiments testing the effects of pH on aggregation than in other experiments. This may be because these earlier pH experiments were done with RP1137 cells at a different phase of growth than cells in later experiments. Preliminary data suggest that the aggregation potential of RP1137 changes over the growth period of the culture (results not shown). Obtaining pH values in this range does not require addition of base to the medium because algal cultures naturally increase the pH via the consumption of CO2 during photosynthesis. Maximum values of pH of 10.2 to 10.4 were routinely observed in the Nannochloropsis cultures used in this study, with the pH varying from 7.5 in newly inoculated cultures to nearly 10.3 to 10.4 in dense cultures. In naturally occurring systems and algal production ponds, the pH varies throughout the day (22). Water temperature also varies during a day and throughout the year. We tested the effect of temperature and found only minor differences among the temperatures tested. There was also no large effect of salinity on aggregation, except at 0-ppt salinity, where aggregation was 15% lower than that for the samples tested with salinity at 20 ppt. It should be noted that in the pH, temperature, and salinity experiments, data were collected on aliquots of algae that were first grown under standard conditions and for which, after harvesting of a sample, the condition being tested was then varied so that the direct effect of these conditions on the aggregation ability of these cells could be measured. These measurements are likely dependent on how the particular condition tested impacts the surface chemistry of the cells. However, cells grown for extended periods of time under differing conditions of salinity, temperature, or pH may have different surface characteristics than the cells tested in these experiments.
The aggregation of several different algae by RP1137 was tested to determine if this bacterium could be used for harvesting other algae besides Nannochloropsis. The results showed that this bacterium could aggregate two strains of Tetraselmis and a Phaeodactylum species but was not able to aggregate N. oleoabundans, C. cryptica, and N. angularis effectively without first increasing the pH by the addition of base. These data show that Bacillus sp. RP1137 has the potential to aggregate other algae and may be useful for the harvesting of other algae besides the potential biofuel producer N. oceanica IMET1, although optimization would be needed to achieve the efficiencies that we achieved with Nannochloropsis aggregation.
The mechanism by which RP1137 interacts with and aggregates algae was investigated. RP1137 forms long filaments which were hypothesized to aid aggregation by increasing bridging between separate algal cells. We tested this hypothesis by shearing the filaments to break them into single cells or doublets and then using the sheared cells in aggregation assays. We found a significant, though minor, decrease in the percentage of algae aggregated in the sheared population. This suggests that the filaments are not the major phenotype responsible for aggregation. The minor effect of filament length and the fact that killed cells retain the ability to aggregate microalgae suggest that the cell surface characteristics may be important in aggregation. There are different types of potential cell surface interactions that could lead to aggregation, including specific and nonspecific protein interactions. To look for an effect due to surface proteins, we digested RP1137 cells with proteinase K to digest exposed and cleavable proteins. Digestion at room temperature yielded no significant difference in the aggregation potential of digested cells relative to that of a nondigested control. A significant, though minor, decrease in the percentage of algae aggregated was observed when the cells were digested at 50°C, at which proteinase K has higher activity. The lack of a large effect upon digestion with proteinase K does not exclude the possibility that surface-localized proteins play a role in aggregation. The strong effect on aggregation of higher pH values, when the bacterial cell surface would likely become increasingly negatively charged, suggested that the aggregation mechanism may be charge related. The finding that aggregation was completely abolished by washing the algae and bacteria several times in deionized water indicated that ionic species may be important for aggregation. Addition of EDTA and EGTA completely abolished aggregation in f/2 medium, suggesting that divalent cations are important for aggregation. The calculated concentrations of the major divalent cations in the f/2 medium used were 5.9 mM and 30.5 mM for the calcium and magnesium ions, respectively. To confirm the dependence of aggregation on these divalent cations, experiments in which magnesium or calcium concentrations were varied were performed. In these experiments, a strong dose-dependent increase in aggregation was observed with the addition of calcium or magnesium. Our preliminary investigation of the mechanism of aggregation therefore points to a charge-based mechanism that is dependent on divalent cations. In Gram-positive organisms like RP1137, it is known that negatively charged teichoic acids and peptidoglycan can bind magnesium and calcium ions (21). Therefore, we hypothesize that these divalent cations bind to these components in the cell wall of RP1137 and reduce the negative surface charge. Calcium ions may also neutralize the surface charge on Nannochloropsis cells, allowing both cell types to overcome electrostatic repulsion and form aggregates. Little is known about the cell wall of Nannochloropsis, though it is likely decorated with polysaccharides. Aggregation via charge neutralization with divalent cations has been hypothesized in other studies in which bacteria were found to be involved in aggregation (20) and in which algal autoflocculation in response to calcium was studied (23). The underlying mechanism of aggregation by RP1137 is likely similar to that of another bacillus, Paenibacillus kribbensis, the aggregation potential of which was also correlated with higher pH and the presence of calcium (15, 17). Investigations into the detailed mechanism of aggregation of algae by RP1137 are ongoing and will clarify the mechanism of action.
Choosing the best method of harvesting algae will differ on the basis of the species used and the culturing system; however, some common measures can be used to compare different methods of harvest. In particular, efficiency of harvest, harvest rate, and energy input are important parameters. Efficiency of harvest refers to the percentage of algae that are removed from the water. In this study, the bacterium Bacillus sp. RP1137 has a typical harvest efficiency with Nannochloropsis, where 70 to 95% of the chlorophyll is removed from the water. Preliminary work suggests that the difference in efficiency is due to changes in bacterial growth stage and warrants further investigation. These values are similar to the harvest efficiency of 83% reported for the aggregation of Chlorella vulgaris by the bacterium Paenibacillus kribbensis (15, 17); however, Bacillus sp. RP1137 is capable of aggregating multiple algae. Other flocculating agents, such as aluminum sulfate and polyacrylamide, resulted in harvest efficiencies of 72% and 78%, respectively (15). Harvest can also be done using the following other physical means with the indicated harvest efficiencies: tangential flow filtration (70 to 89%), dissolved air flotation (80 to 90%), electrocoagulation (95%), and centrifugation, which has a greater than 95% harvest efficiency (7). It should be pointed out that these methods were tested with different algal strains, but they do provide a baseline of what can typically be achieved by the different harvest methods available. The Bacillus sp. RP1137-based harvest efficiency compares favorably with the harvest efficiencies of these known methods of algal harvest. Another important measure of a harvesting system is the rate at which the algae can be removed from the water, sometimes known as harvest productivity. The rate of aggregation by Bacillus sp. RP1137 is fast, with large aggregates forming in seconds, which is similar to the times for other flocculation-type harvest methods. In comparison, centrifuges, while having excellent harvesting efficiencies, have low harvesting rates.
Aggregation is typically a low-energy method to remove algae from water (7). We do not have data on the energy associated with harvesting using Bacillus sp. RP1137; however, it could be estimated that the energy needed would involve the energy needed to grow the bacterium and the energy needed to apply the bacteria to the algal culture. Other systems of harvest are often energy intensive, with centrifugation requiring 8 kWh/m3 of water, which contributes to the operating cost (7). Energy efficiency must be considered because the final product of algal biofuels is energy. Any system that costs more energy than is recovered from the algae is not sustainable.
Creating the next generation of liquid fuel infrastructure on the basis of the sustainable conversion of algal biomass to fuels will require the development of new technologies to reduce the barriers to large-scale production. One of the largest barriers to economically viable scale-up remains the harvesting step. Here we presented a new bacterial isolate that rapidly aggregates algae in a reversible manner. After fixation, the aggregation phenotype remains, a situation which effectively renders the cells specialized aggregating microparticles that may permit reuse of the cells with further research. Our results indicate that Bacillus sp. RP1137 may be useful in reducing one of the barriers to large-scale algal biofuel production.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a Maryland Industrial Partnerships grant.
We thank Paul Behrens for the gift of the algal strains. We also thank H. Wang for discussions and L. Blasiak for comments on the manuscript. The manuscript was improved by the constructive comments in seven reviews. The Aquaculture Research Center at IMET is thanked for providing algal cultures.
Footnotes
Published ahead of print 26 July 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01496-13.
REFERENCES
- 1.U.S. Energy Information Administration 13 January 2013, posting date Your guide to understanding energy. U.S. Energy Information Administration, Washington, DC [Google Scholar]
- 2.U.S. Environmental Protection Agency 13 January 2013, posting date Renewable fuel standard (RFS). U.S. Environmental Protection Agency, Washington, DC [Google Scholar]
- 3.Schenk PM, Thomas-Hall SR, Stephens E, Marx UC, Mussgnug JH, Posten C, Kruse O, Hankamer B. 2008. Second generation biofuels: high-efficiency microalgae for biodiesel production. Bioenerg. Res. 1:20–43 [Google Scholar]
- 4.Waltz E. 2009. Biotech's green gold? Nat. Biotechnol. 27:15–18 [DOI] [PubMed] [Google Scholar]
- 5.Chisti Y. 2008. Biodiesel from microalgae beats bioethanol. Trends Biotechnol. 26:126–131 [DOI] [PubMed] [Google Scholar]
- 6.Richardson JW, Johnson MD, Outlaw JL. 2012. Economic comparison of open pond raceways to photo bio-reactors for profitable production of algae for transportation fuels in the Southwest. Algal Res. 1:93–100 [Google Scholar]
- 7.Uduman N, Qi Y, Danquah MK, Forde GM, Hoadley A. 2010. Dewatering of microalgal cultures: a major bottleneck to algae-based fuels. J. Renewable Sustainable Energy 2:012701 http://dx.doi.org/10.1063/1.3294480 [Google Scholar]
- 8.Sirin S, Trobajo R, Ibanez C, Salvado J. 2012. Harvesting the microalgae Phaeodactylum tricornutum with polyaluminum chloride, aluminium sulphate, chitosan and alkalinity-induced flocculation. J. Appl. Phycol. 24:1067–1080 [Google Scholar]
- 9.Lavoie A, Noüe J. 1983. Harvesting microalgae with chitosan. J. World Maricult. Soc. 14:685–694 [Google Scholar]
- 10.Divakaran R, Sivasankara Pillai V. 2002. Flocculation of algae using chitosan. J. Appl. Phycol. 14:419–422 [Google Scholar]
- 11.Pavoni JL, Tenney MW, Echelberger WF., Jr 1972. Bacterial exocellular polymers and biological flocculation. J. Water Pollut. Control Fed. 44:414–429 [PubMed] [Google Scholar]
- 12.Yuan SJ, Sun M, Sheng GP, Li Y, Li WW, Yao RS, Yu HQ. 2011. Identification of key constituents and structure of the extracellular polymeric substances excreted by Bacillus megaterium TF10 for their flocculation capacity. Environ. Sci. Technol. 45:1152–1157 [DOI] [PubMed] [Google Scholar]
- 13.Nontembiso P, Sekelwa C, Leonard MV, Anthony OI. 2011. Assessment of bioflocculant production by Bacillus sp. Gilbert, a marine bacterium isolated from the bottom sediment of Algoa Bay. Mar. Drugs 9:1232–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gardes A, Iversen MH, Grossart HP, Passow U, Ullrich MS. 2011. Diatom-associated bacteria are required for aggregation of Thalassiosira weissflogii. ISME J. 5:436–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oh HM, Lee SJ, Park MH, Kim HS, Kim HC, Yoon JH, Kwon GS, Yoon BD. 2001. Harvesting of Chlorella vulgaris using a bioflocculant from Paenibacillus sp AM49. Biotechnol. Lett. 23:1229–1234 [Google Scholar]
- 16.Wang H, Laughinghouse HDT, Anderson MA, Chen F, Willliams E, Place AR, Zmora O, Zohar Y, Zheng T, Hill RT. 2012. Novel bacterial isolate from Permian groundwater, capable of aggregating potential biofuel-producing microalga Nannochloropsis oceanica IMET1. Appl. Environ. Microbiol. 78:1445–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoon JH, Oh HM, Yoon BD, Kang KH, Park YH. 2003. Paenibacillus kribbensis sp. nov. and Paenibacillus terrae sp. nov., bioflocculants for efficient harvesting of algal cells. Int. J. Syst. Evol. Microbiol. 53:295–301 [DOI] [PubMed] [Google Scholar]
- 18.Guillard RRL. 1975. Culture of phytoplankton for feeding marine invertebrates, p 29–60 In Culture of marine invertebrate animals. Plenum, New York, NY [Google Scholar]
- 19.Lane D. 1991. 16S/23S rRNA sequencing, p 115–147 In Stackebrandt E, Goodfellow M. (ed), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Inc., New York, NY [Google Scholar]
- 20.Lee J, Cho DH, Ramanan R, Kim BH, Oh HM, Kim HS. 2013. Microalgae-associated bacteria play a key role in the flocculation of Chlorella vulgaris. Bioresour. Technol. 131:195–201 [DOI] [PubMed] [Google Scholar]
- 21.Marquis RE, Mayzel K, Carstensen EL. 1976. Cation exchange in cell walls of gram-positive bacteria. Can. J. Microbiol. 22:975–982 [DOI] [PubMed] [Google Scholar]
- 22.Dubinsky Z, Rotem J. 1974. Relations between algal populations and the pH of their media. Oecologia 16:53–60 [DOI] [PubMed] [Google Scholar]
- 23.Schlesinger A, Eisenstadt D, Bar-Gil A, Carmely H, Einbinder S, Gressel J. 2012. Inexpensive non-toxic flocculation of microalgae contradicts theories; overcoming a major hurdle to bulk algal production. Biotechnol. Adv. 30:1023–1030 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


