Abstract
The occurrence of waterborne pathogens was investigated at three drinking water intakes located about 2 km offshore in Lake Ontario. Water sampling was conducted over 3 years for Campylobacter spp., Cryptosporidium spp., Giardia spp., cultivable enteric viruses, and water quality parameters. All pathogens were detected in the offshore source water for each water treatment plant (WTP1 to WTP3), although at relatively low frequencies and concentrations. Giardia was the most common pathogen, occurring in 36% of water samples from the influent of WTP1 (n = 46), and with a maximum concentration of 0.70 cysts/liter in this influent. Cryptosporidium occurred as frequently as 15% in the WTP2 influent (n = 35), with a maximum concentration of 0.40 oocysts/liter in the WTP1 influent. The human Bacteroidales HF183 DNA marker was most common in the WTP1 influent (19%), and this was the only WTP where the Cryptosporidium hominis genotype was detected. No water quality parameter was predictive of pathogen occurrence across all three WTP influents. Escherichia coli was often below detection when pathogens were detected, and spikes in E. coli concentrations often did not coincide with pathogen occurrence. After summer rain events, river plumes had E. coli concentrations as high as 222 CFU/100 ml in surface waters 2 km offshore, without impacting drinking water intakes below the thermocline on the lake bottom. At times, prechlorination to control mussels at offshore intake cribs compromised the use of E. coli for “raw” water quality assessment, particularly for chlorine-resistant Cryptosporidium. E. coli measured by standard methods did not reliably predict pathogen occurrence at drinking water intakes in offshore ecosystems.
INTRODUCTION
The Great Lakes are the source of drinking water for over 24 million people in Canada and the United States (1). More than 100 communities around the Lakes obtain their drinking water from offshore intakes on the lake bottom within ∼2 km of the shoreline. Despite the importance of water quality at offshore drinking water intakes, there has been little systematic investigation of the occurrence of waterborne pathogens in these offshore ecosystems.
While the Great Lakes are widely regarded as a good source of drinking water (2), this was not always the case. In the 1800s, thousands of illnesses and deaths occurred in some years from typhoid and cholera outbreaks in communities that obtained their drinking water from the Lakes. Outbreaks of cholera claimed over 200 lives in Toronto in 1832 (3), and over 1,400 lives in Chicago in 1854 (4). An extensive study conducted by the International Joint Commission (IJC) in 1913 found significant sewage contamination in boundary waters between Canada and the United States (5, 6). More than 1,500 sampling sites were investigated by 17 collaborating laboratories, and more than 18,000 water samples were analyzed for indicator bacteria. This study has been considered among the world's most extensive bacteriological investigation (5, 7). There has not been a comprehensive investigation of the microbial water quality of drinking water sources in the transboundary waters of the Great Lakes since the IJC study 100 years ago (8).
In the century since the 1913 IJC study, advances in drinking water and sewage treatment practices have significantly reduced concerns about drinking water outbreaks. However, Hrudey and Hrudey (9) document many waterborne disease outbreaks that have continued to occur in developed countries despite advances in drinking water treatment systems. In fact, some of the world's most prominent drinking water outbreaks in recent years have occurred within the Great Lakes basin, in the communities of Walkerton, Ontario, Canada (10), and Milwaukee, Wisconsin (11). These outbreaks have highlighted the need to avoid complacency in assuming that modern drinking water utilities will always provide safe drinking water. Other drinking water outbreaks in communities on the Great Lakes have included a 1968 outbreak associated with a Lake Erie drinking water intake located 1,500 feet offshore of a small community south of Buffalo (12), a 1996 Cryptosporidium outbreak in Collingwood, Ontario, Canada (13), and a 2004 outbreak associated with groundwater along the shoreline of South Bass Island off the coast of Lake Erie (14).
The Milwaukee outbreak in 1993 showed that offshore drinking water intakes in the Great Lakes can be vulnerable to waterborne pathogen contamination. This outbreak was estimated to have caused more than 50 deaths, 4,000 hospitalizations, and 400,000 illnesses when Cryptosporidium contaminated the Milwaukee drinking water supply (11, 15, 16). The source water for Milwaukee at that time came from an intake located 1.4 miles (2.3 km) offshore in Lake Michigan at a depth of 42 feet (12.8 m) (15). The Milwaukee drinking water treatment plant provided conventional water treatment processes at the time (coagulation, sedimentation, filtration, and disinfection).
A wide variety of environmental changes are anticipated around the Great Lakes in the future associated with changing land uses, increasing urbanization, aging municipal water and wastewater infrastructure, fluctuating water levels, and diverse manifestations of climate change. An aging human population with more immunocompromised individuals is also anticipated. It will be important to have an understanding of waterborne pathogen occurrence in offshore source water to ensure water treatment plants continue to provide safe drinking water. It will also be important to have an understanding of indicators such as Escherichia coli for assessing water quality in offshore ecosystems in the Great Lakes. Much of what is known about E. coli as a water quality indicator has come from studies in riverine and shoreline lake ecosystems (e.g., beaches). In these settings, there is increasing evidence of the lack of association between the occurrence of E. coli and waterborne pathogens (17, 18), and of challenges associated with the resuspension (19, 20) or naturalization (21) of E. coli that can complicate prediction of pathogen occurrence and health risks. There has been little investigation of the ability of E. coli to predict pathogen occurrence in offshore ecosystems complicated by aspects such as thermally stratified water columns and complex horizontal and vertical water current regimes.
The objective of the present study was to investigate the occurrence of waterborne pathogens in the offshore source water of several drinking water treatment plants located in a rapidly urbanizing area in western Lake Ontario. We sought to establish a benchmark of waterborne pathogen occurrence that could be used to understand future trends in offshore source water quality. We also sought to assess the microbial water quality indicator E. coli for predicting pathogen occurrence in offshore ecosystems.
MATERIALS AND METHODS
Sampling sites.
The present study focused on three drinking water treatment plants (WTP1 to WTP3) in the vicinity of the mouth of the Credit River in western Lake Ontario (Fig. 1). Comparative reference sites were selected from nearby rivers (Credit River and Humber River), a sewage treatment plant (and its offshore outfall), and a Lake Ontario site located ∼10 km offshore. WTP1 treats up to 615,000 m3 of water per day using conventional drinking water treatment consisting of raw water disinfection, coagulation, flocculation, sedimentation, and filtration unit processes. The WTP1 intake is 1.6 km offshore at a depth of 11 m. There was some discrepancy between our offshore sampling location and the actual offshore intake for WTP1, so our surface/bottom sampling results are likely more conservative, representing water conditions a little farther offshore (about 400 m). WTP2 treats up to 923,000 m3 per day using conventional drinking water treatment consisting of raw water disinfection, coagulation, flocculation, sedimentation, and filtration unit processes, with additional advanced treatment including ozonation and membrane filtration. The WTP2 intake is 2 km offshore at a depth of 18 m. WTP3 treats up to 500,000 m3 per day using conventional drinking water treatment consisting of raw water disinfection, coagulation, flocculation, sedimentation, and filtration and disinfection unit processes. The WTP3 intake is 1.6 km offshore at a depth of 10 m. The sewage treatment plant (STP) provides secondary treatment for about 448,000 m3 of wastewater per day, and discharges dechlorinated final effluent through a 188 m diffuser located on the lake bottom about 1.25 km offshore. Wastewater samples were collected within the STP from the final effluent after the plant's dechlorination step. Reference sampling sites were also located in Lake Ontario several hundred meters offshore from the mouth of the Credit and Humber Rivers. The site near the mouth of the Humber River was located close to the outfall of another sewage treatment plant, so it does not represent a simple offshore dilution of the Humber River plume.
Fig 1.
Credit River study area on Lake Ontario depicting locations of WTPs and one STP. Water samples at offshore sites were collected by boat (surface and bottom). Water samples were collected by land-based sampling within water and sewage treatment plants and upstream of river mouths. An additional reference site was located ∼10 km offshore from the mouth of the Credit River.
Water sampling.
Water sampling was conducted every 2 weeks, subject to weather conditions, between April 2007 and December 2010. Additional sampling was conducted where possible to capture rain event days. A land-based sampling crew collected water samples at the WTPs from raw influent water lines within the WTPs. Wastewater samples were collected at the STP from final treated effluent as it left the plant (and after a dechlorination step). Sodium thiosulfate was added to wastewater sample containers to neutralize any chlorine residual remaining in the STP final effluent. Water samples from sampling lines in WTPs and the STP were collected in sterile, acid-washed 10-liter polypropylene carboys. Large water volumes from these sampling lines were also passed through different filtration apparatuses on site for protozoa (up to 100 liters) and viruses (up to 200 liters). A boat-based sampling crew collected offshore water samples from surface water (1 m below the surface) and bottom water (2 m above the lake bottom). Surface and bottom water samples were recovered using a 6-liter Van Dorn sampler and collected in sterile, acid-washed 10-liter carboys. Larger water volume sampling for protozoa and viral pathogens at offshore sites was done directly in the boat using a small pump to pass surface water through different filtration apparatus for protozoa (up to 100 liters) and viruses (up to 200 liters). The 10-liter carboys were returned to the Burlington laboratory within a few hours for same day processing. The protozoan filter cartridges and viral filtration units were couriered overnight on ice for next day processing. Raw water temperature and turbidity data were kindly shared by the three WTPs for our sampling dates.
Microbial indicator bacteria.
Water and wastewater samples were analyzed by membrane filtration for E. coli, total coliforms, enterococci, and clostridia. Standard water volumes of 100 ml were filtered for each indicator bacteria, and serial dilutions of water samples were performed where needed. Membrane filters (0.45-μm pore size; Fisher Scientific) were placed on selective media, and bacterial numbers were expressed as CFU/100 ml. Lab water samples were routinely filtered as negative controls and did not indicate any inadvertent sample contamination over the study period. E. coli was enumerated after 18 h of incubation at 44.5 ± 0.5°C on chromogenic differential coliform (DC) agar media supplemented with cefsulodin (Oxoid, USA). Duplicate water samples were collected for E. coli enumeration from 2008 to 2010 to assess sample variability, and E. coli concentrations were expressed as the average of the two samples. Total coliform bacteria were enumerated after 18 h of incubation at 37°C on DC agar media supplemented with cefsulodin (Oxoid). Enterococci were enumerated after 48 h of incubation at 37°C on chromogenic m-Enterococcus media (Accumedia). Clostridial bacteria were enumerated after 48 h of incubation in anaerobic bags at 44.5°C on m-CP agar (Accumedia) supplemented with d-cycloserine and polymyxin-3-sulfate, FeCl3·6H2O (4.5% solution), phenolphthalein diphosphate (0.5% solution), and indoxyl-β-d-glucoside (0.075% solution).
Water and wastewater samples were assessed for the presence of Bacteroidales that are associated with human fecal pollution (22). Water volumes of 300 ml (or occasionally less for very turbid samples) were filtered through 0.45-μm-pore-size membrane filters, and the filters were stored at −80°C. Each water or wastewater sample was analyzed for the presence of human-specific strains of Bacteroidales bacteria (human Bacteroidales DNA marker HF183), as well as for the presence of a broader range of Bacteroidales bacteria (universal Bacteroidales DNA marker BAC32). Membrane filters with total genomic DNA were removed from −80°C and then homogenized in a Mini-BeadBeater (BioSpec Products, Inc.) for 2 min. DNA was purified by using a Powersoil DNA isolation kit (MO BIO Laboratories, Inc.). A 1-μl extract was used as a template in a PCR assay using primer HF183F to amplify the human Bacteroidales DNA sequences and BAC32 to amplify universal Bacteroidales sequences if they were present in the sample. Primer BAC708R was the reverse primer for both reactions. For the PCR, the following agents and concentrations were used: 0.05 U of Hotmaster Taq/μl and 1× buffer (Intermedico), 0.8 mM deoxynucleoside triphosphate (dNTP) mixture, 0.06% bovine serum albumin (BSA), 1.56 pmol of each primer/μl, and water to 25 μl. The PCR cycling conditions were as follows: 2 min at 94°C; followed by 35 cycles of 20 s at 94°C, 10 s of annealing at 53°C for BAC32 or at 63°C for HF183 primers, and 50 s at 65°C; followed by a final single step at 65°C for 7 min. A human fecal DNA extract was run as a positive control for each set of reactions, along with sterile water as a negative control. A 5-μl portion of dye DNA mix was loaded into wells of a 1.25% agarose gel and run at 170 V for ∼1 h to resolve the bands, which were visualized by staining with ethidium bromide (0.5 μg/ml) and imaging under UV light using an Ingenius Syngene Bioimaging gel documentation system (Perkin-Elmer, MA). Determination of the presence or absence of a band corresponding to the expected human or universal Bacteroidales DNA marker was standardized by analyzing digitized gel images with computer software (BioNumerics) and using a band pattern recognition approach. Reference samples from fecal pollution sources were collected from around Toronto and southern Ontario to test the host specificity of the human Bacteroidales DNA marker assay.
Campylobacter.
Campylobacter species were detected and enumerated in a semiquantitative manner according to the method of Khan and Edge (23). One-liter water samples were centrifuged at 14,000 × g for 20 min, and the pellet was resuspended in 4 ml of saline (0.85%) solution to isolate Campylobacter spp. Then, 1 ml of the pellet suspension was inoculated into 9 ml of Bolton Broth (Oxoid) containing a selective antibiotic supplement (cefoperazone, cycloheximide, trimethoprim, and vancomycin). This inoculum was then serially diluted and incubated at 42°C for 48 h under microaerophilic conditions in a tri-gas (85% N2, 10% CO2, and 5% O2) incubator (Sanyo, Japan). The cultures from each tube with a turbid appearance were streaked onto modified Karmali agar plates (Oxoid) containing a selective antibiotic supplement (amphotericin B, cefoperazone, sodium pyruvate, and vancomycin), followed by incubation under the same microaerophilic conditions at 42°C for 24 to 48 h. A parallel approach was used to also investigate Campylobacter detection at an incubation temperature of 37°C. However, for the present study, we only considered thermophilic Campylobacter strains recovered from enrichment at 42°C as representing a pathogen, although the infectivity and virulence of these strains has not been assessed. Putative Campylobacter cultures on plates were confirmed by colony morphology and by a 16S ribosomal DNA (rDNA) genus-specific PCR assay (24). The PCR amplicons were electrophoresed on a 1% agarose gel matrix using 100-bp DNA size markers (PGC Scientifics). A species-specific multiplex-PCR assay based on the 16S-23S rDNA internal transcribed spacer (ITS) region was used to further determine the Campylobacter species as either C. jejuni, C. coli, C. lari, or other Campylobacter species (23). To account for small PCR fragments, PCR amplicons were electrophoresed on a 2% gel matrix. A theoretical detection limit for this method was considered to be ∼4 Campylobacter cells/liter based on processing one-fourth of the pellet centrifuged from 1-liter water samples in the initial enrichment step.
Cultivable enteric viruses.
Cultivable human enteric viruses were enumerated from a volume of up to 200 liters of water filtered through a NanoCeram cartridge (Argonide Corp., USA) at a filtration rate of ∼10 liters/min. The filtration apparatus was returned in a cooler (overnight carrier) to the INRS laboratory, and samples were processed within 24 h of receipt. Elution of viruses from the filters was done with beef extract, followed by organic flocculation and centrifugation (Beckman J6-H6) at 3,000 × g to concentrate the sample to 30 ml. The concentrates were kept frozen at −70°C until assayed. Viruses were detected by infection of MA-104 cells (2 passages: 11 and 7 days), followed by an immunoperoxidase method for detecting the presence of viruses (25, 26). The number of viruses in the original samples was calculated using the number of positive (cytopathic effect or immunoperoxidase positive) and negative wells after the second passage and estimating the most probable number of infectious unit per liter (MPN-IU/liter).
Cryptosporidium and Giardia.
The enumeration of Cryptosporidium oocysts and Giardia cysts from water was carried out according to U.S. Environmental Protection Agency method 1623 (27). Water collected for protozoan analyses was filtered on site using Filta-Max or Filta-Max Xpress filters (IDEXX, Westbrook, MN). The filters were shipped overnight in coolers with ice packs to the Alberta Provincial Laboratory for Public Health in Calgary, Alberta, Canada. The elution of parasites from the filters was carried out with phosphate-buffered saline (PBS) with 0.01% Tween 20 according to the manufacturer's protocols. Immunomagnetic separation (IMS) was carried out using a DynaBeads G-C combo kit (IDEXX), with a modification to the standard IMS protocol. Briefly, two washes of the bead-parasite complex in the Leighton tubes using PBS with 0.01% Tween 20 at pH 7.4 (PBST) were carried out after 1 h of incubation. The beads were captured by the magnet, the supernatant was poured off, and the beads were suspended in 12 ml of PBST, mixed by inverting the tube (three to five times). The tube was returned to the magnet for bead capture, with the process repeated. After discarding the PBST from the second wash, the standard protocol was resumed. Microscope slides were dried at 35°C on a slide warmer and fixed with methanol. Slides were stained with DAPI (4′,6′-diamidino-2-phenylindole; Sigma-Aldrich, Oakville, Ontario, Canada) and fluorescence-labeled monoclonal antibodies (EasyStain; Biotechnology Frontiers, North Ryde, New South Wales, Australia). Parasites were identified by fluorescence microscopy and confirmed to be Cryptosporidium or Giardia based on DAPI staining characteristics or by their size, shape, and morphology using differential interference contrast microscopy. An ongoing laboratory quality control program was maintained over the study period through sample spiking and use of method blanks that were inserted into the lab analysis process to ensure that carryover did not occur from one sample set to another. Sample spiking involved spiking various drinking water, raw water, and wastewater samples with commercially prepared spikes of oocysts or cysts that were flow cytometry sorted at specific concentrations. Although the recovery efficiency can be variable between water samples, this spiking program has found an average oocyst/cyst recovery efficiency of ca. 40 to 50%. Any slide that was positive by fluorescent antibody staining for Cryptosporidium oocysts or Giardia cysts was processed for molecular analysis. (Oo)cyst removal, (oo)cyst lysis, and DNA extraction were carried out as previously described (28). Replicate nested PCR-restriction fragment length polymorphism was carried out for Cryptosporidium according to the method of Ruecker et al. (29), with primers and conditions previously described (30). An additional replicate (sixth) was included for every sample and seeded with C. muris 18S rRNA plasmid DNA to determine possible sample inhibition.
Multiple seminested PCRs analyzing Giardia cysts in five replicates on each DNA extract were performed to target SSU rRNA gene loci. The seminested PCR for the SSU rRNA gene was constructed using an internal primer set (forward primer R11, 5′-CATCCGGTCGATCCTGCC-3′; reverse primer R4, 5′-GTCGAACCCTGATTCTCCGCCAGG-3′) and a developed external reverse primer (GiardiaM1R, 5′-TTGGATGTGCGGGCCGTCTCGCA-3′). The primary PCR product was amplified in a 50-μl volume with the following reaction components: 1× Taq buffer, 2 mM MgCl2, 0.8 mM dNTP, 1× Q-solution (Qiagen), 0.8 μM concentrations of primers, 0.04 U of native Taq (Invitrogen Canada, Inc., Burlington, Ontario, Canada)/μl, and 5 μl of DNA template. Reactions were performed under the following conditions: 96°C for 4 min; followed by 35 cycles of 96°C for 20 s, 59°C for 30 s, and 1 min at 72°C; followed in turn by a final extension of 7 min at 72°C. The secondary PCR was carried out in a 50-μl volume reaction using 5 μl of the primary PCR product as the template DNA. The reaction mixture and thermal cycling conditions were identical to the primary PCR except that 35 cycles were used. The PCR products were purified using a QIAquick PCR purification kit (Qiagen) and sequenced using an ABI dye terminator cycle sequencing kit (Applied Biosystems, Canada) according to the manufacturer's instructions. PCR products were sequenced in both forward and reverse directions using SSU rRNA primer sets. Sequencing was performed either in-house using BigDye (v1.1; Applied Biosystems) and a Prism 3100-Avant genetic analyzer (Applied Biosystems). Sequence data were analyzed with software, including Sequencing Analysis 5.1.1 (Applied Biosystems, Canada) and CLUSTAL-W (BioEdit, v7.0.0). Sequences showing single nucleotide polymorphisms were repeated by resequencing each primary PCR product conducted on separate days to ensure the accuracy of the reported sequence.
Statistical analyses.
Data for parameters were aggregated over the study period for each sampling location. Since untransformed and log10-transformed data were not normally distributed, most statistical analyses were performed using nonparametric tests. Statistical analyses were performed using SAS JMP software (31), and were considered significant at a P of <0.05. Chi-square contingency tests were used to test for significant differences in the frequency of occurrence of waterborne pathogens and fecal indicator bacteria between sampling locations. Wilcoxon signed-rank tests were used to test for significant differences in the numbers of waterborne pathogens and fecal indicator bacteria between sampling locations. Paired Wilcoxon signed-rank tests were conducted to compare results between sampling locations for water samples collected on the same sampling day. At each sampling location, chi-square tests were used to test for significant differences in the frequency of occurrence of pathogens and indicator bacteria between wet and dry weather sampling days, respectively. Wet weather days were defined as those where a precipitation event of >5 mm had occurred within 24 or 48 h preceding the day of water sample collection. Precipitation data were obtained from Environment Canada's on-line weather database for the Pearson International Airport weather station. At each sampling location, Wilcoxon signed-rank tests were used to test for significant differences in the numbers of pathogens and indicator bacteria between wet and dry weather sampling days.
Spearman rank correlation tests were used to explore the association between individual water quality parameters and the numbers of waterborne pathogens at WTPs, although pathogens and indicator bacteria were not detected in many of the offshore water samples. Logistic regression analyses in SAS JMP software were used to investigate whether parameters could be used to predict the occurrence of a pathogen at a WTP intake. Stepwise logistic regression analyses were used to select the optimal predictive parameters for each WTP intake. A classification and regression tree (CART) data mining analysis was also performed using SAS JMP software to identify models for predicting the occurrence of a pathogen at a WTP intake. Classification tree analysis is a nonparametric recursive partitioning methodology that splits a dependent variable (e.g., Cryptosporidium presence or absence) into homogeneous data groupings based on optimal independent variable (e.g., turbidity, E. coli concentration, etc.) splitting criteria (32, 33). The method tests the best discriminating split of the dependent variable using all possible independent variable splitting possibilities. The optimal classification tree results from a combination of optimizing predictive capacity and minimizing model complexity.
RESULTS
Between April 2007 and December 2010, 1,358 water and wastewater samples were collected for microbiological analyses in the Credit River study area. A subset of these water and wastewater samples were analyzed for semiquantitative enumeration of Campylobacter (1,314 samples), enumeration of Cryptosporidium and Giardia (397 samples), and enumeration of cultivable human enteric viruses (67 samples).
Campylobacter occurrence.
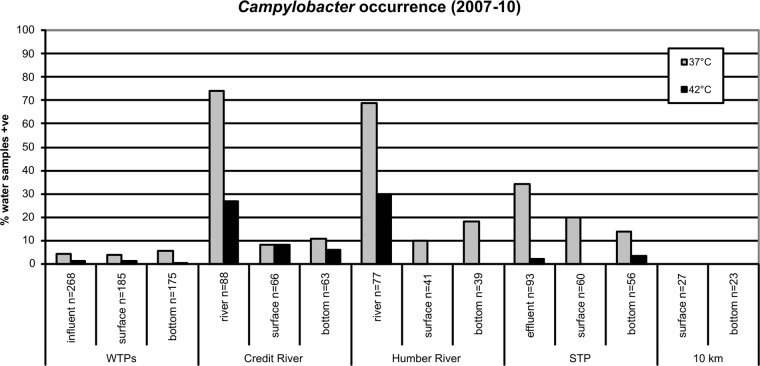
Campylobacter species were detected at all sampling sites investigated, except at the reference site located 10 km offshore in Lake Ontario (Fig. 2). Enrichment at 37°C was found to recover Campylobacter more frequently from water samples than at 42°C. However, enrichment at 42°C is more widely used for detection of thermophilic Campylobacter of human health concern. Only three water samples were positive for thermophilic Campylobacter over the study period across all WTP influent samples. Two water samples were positive in the influent at WTP1 (2%, n = 94), and one water sample was positive in the influent at WTP3 (1%, n = 94); thermophilic Campylobacter was not detected in the influent at WTP2 (n = 80). Offshore at intakes of the WTPs, thermophilic Campylobacter was only detected in the bottom water at WTP3 (2% of samples) and the surface water at WTP2 (3%). When thermophilic Campylobacter was detected in influent or offshore water at any WTP, it was only detected at our minimum detection limit of between about 4 and 40 cells/liter.
Fig 2.
Percent occurrence of Campylobacter spp. in water samples collected from Credit River area study sites between April 2007 and December 2010. Water samples were analyzed in parallel using two different enrichment temperatures (37 and 42°C). WTPs, water treatment plants; STP, sewage treatment plant. 10 km, reference site located 10 km offshore from the mouth of the Credit River; influent, raw water entering WTPs; effluent, final treated wastewater leaving the STP; surface, water collected at the surface above offshore WTP intakes, offshore STP outfall, and offshore of river mouths or 10 km offshore; bottom, water collected at the bottom near offshore WTP intakes, offshore STP outfall, and offshore of river mouths or 10 km offshore.
Thermophilic Campylobacter was detected most frequently in water samples from reference sites on the Humber (30%) and Credit rivers (27%). However, the occurrence of thermophilic Campylobacter in river plumes dropped off quickly upon entering the Lake, since the occurrence was only 6 to 8% at the sampling location off the mouth of the Credit River, and they were not detected off the mouth of the Humber River. Thermophilic Campylobacter organisms were detected in 2% of the final treated effluent samples from the sewage treatment plant (STP). They were detected in 4% of bottom water samples collected at the offshore STP outfall but not in any surface water samples at this offshore outfall. The highest thermophilic Campylobacter concentration measured here (>4,000 cells/liter) was for two rain event water samples collected in surface and bottom waters offshore at the mouth of the Credit River on 8 January 2008. C. jejuni was detected in both of these water samples.
The only times of the year when thermophilic Campylobacter was detected in WTP offshore source water were in May (5% of samples), June (1%), and November (1%). Thermophilic Campylobacter was detected twice in the raw influent water at WTP1 on 14 May 2007 and 22 November 2007. Thermophilic Campylobacter was detected once in the raw influent water at WTP3 on 14 May 2007. At WTP offshore intakes, thermophilic Campylobacter was only detected twice in offshore surface water (C. coli on both 1 May 2007 and 16 June 2008 at WTP2) and once in offshore bottom water (C. jejuni on 1 May 2007 at WTP3). Four of the five thermophilic Campylobacter detections at WTPs were associated with rainfall events within 3 days preceding water sample collection.
Cultivable enteric virus occurrence.
Cultivable enteric viruses occurred in 50% of WTP3 influent samples (n = 18), followed by 28% of influent samples from WTP1 (n = 25), and 15% of influent samples from WTP2 (n = 13). Virus levels in WTP influents reached as high as 0.33 MPN-IU/liter in WTP1 influent on 15 March 2010. Samples collected from the STP effluent (n = 3), Credit River (n = 3), and the Humber River (n = 2) were all positive for viruses. The highest cultivable enteric virus levels measured were 36.7 MPN-IU/liter in STP final effluent and 10.3 MPN-IU/liter in the Humber River. Cultivable enteric viruses were only detected in influents at WTPs in colder months from October to March (44%, n = 41) compared to no detections from April to September (0%, n = 15). Detection of viruses in WTP influents did not show a significant precipitation event response, although sample sizes were small to draw conclusions. Only one of seven (14%) virus-positive water samples in the influent of WTP1 occurred after a >5 mm precipitation event in the 48 h preceding sample collection. Only three of nine (33%) WTP3 precipitation event water samples were positive for virus, whereas one of two (50%) WTP2 precipitation event water samples was positive for virus.
Cryptosporidium occurrence.
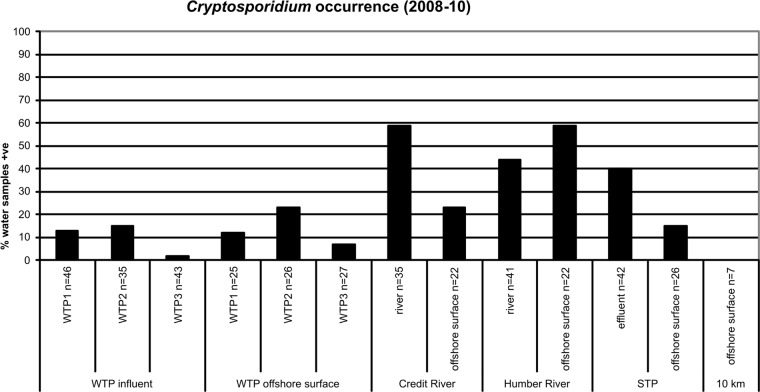
Cryptosporidium was detected at all sampling sites investigated, except at the reference site located 10 km offshore in Lake Ontario (Fig. 3). This pathogen was more commonly detected in influent and offshore surface waters for WTP2 than the other WTPs, although this difference was not significant (P > 0.05). Cryptosporidium was detected in 15% of WTP2 influent water samples compared to 13% for WTP1 and 2% for WTP3. It was detected in 23% of the surface water samples near the offshore WTP2 intake compared to 12% for WTP1 and 7% for WTP3. The highest numbers of Cryptosporidium oocysts enumerated in WTP influents were 0.40 oocysts/liter at WTP1, 0.03 oocysts/liter at WTP2, and 0.01 oocysts/liter at WTP3. The highest number of Cryptosporidium oocysts enumerated in surface water samples near an offshore intake was 0.10 oocysts/liter at WTP2.
Fig 3.
Percent occurrence of Cryptosporidium in water samples collected from Credit River area study sites between 2008 and 2010. WTPs, water treatment plants; STP, sewage treatment plant; 10 km, reference site located 10 km offshore from the mouth of the Credit River; influent, raw water entering WTPs; effluent, final treated wastewater leaving STP; surface, water collected at the surface above offshore WTP intakes, offshore STP outfall, and offshore of river mouths or 10 km offshore.
Cryptosporidium was most commonly detected in the Credit River (59%) and offshore at the mouth of the Humber River (59%). There was a drop in frequency of detection offshore at the mouth of the Credit River due to dilution of the river plume, although Cryptosporidium was still detected in 23% of water samples collected at this offshore site. Cryptosporidium was less commonly detected in the Humber River compared to the Credit River. The increase in Cryptosporidium occurrence offshore from the mouth of the Humber River is likely due to this sampling site representing a combination of impacts from both the Humber River and the offshore outfall of another STP (the Humber STP) near this location. The highest number of oocysts enumerated in the Humber River and Credit River were 1.20 oocysts/liter (30 November 2009) and 0.56 oocysts/liter (16 June 2008), respectively. Cryptosporidium could be regularly detected in final treated effluent from the STP (40%), and concentrations were as high as 0.43 oocysts/liter (25 March 2008) in this effluent.
Some temporal differences were noted with a relatively higher frequency of Cryptosporidium occurrence at WTP1 in April and at WTP2 in March. In addition, Cryptosporidium was significantly more common on wet weather days in influents at WTP1 and WTP2 (P < 0.05). This was not the case for WTP3, although there was only one Cryptosporidium-positive water sample in the influent of this plant. Almost all Cryptosporidium detections in offshore surface waters at WTP intakes and in WTP raw influents were associated with a precipitation event. These wet weather responses for Cryptosporidium were observed within 24 h after rain events at the WTP1 influent, the WTP2 influent, and surface water offshore at the WTP2 intake. The occurrence of Cryptosporidium was also more common at river sites on wet weather days, although this difference was only significant for the Credit River offshore surface water site (P < 0.05). There were no significant differences between wet and dry weather days for Cryptosporidium at the Humber River or STP sites (P > 0.05).
Most of the Cryptosporidium oocysts genotypes detected in the study area were not known to be associated with human disease (N. Neumann, unpublished data). However, C. hominis was detected in STP final effluent samples, surface water samples offshore of the mouth of the Humber River (near another STP outfall), and in one influent water sample for WTP1. This WTP1 influent water sample was collected on 16 September 2008 and indicated that human fecal waste can reach the offshore intake for this WTP. C. parvum was detected in the STP final effluent, the Credit River sampling site, and surface water offshore at the mouth of the Humber River.
Giardia occurrence.
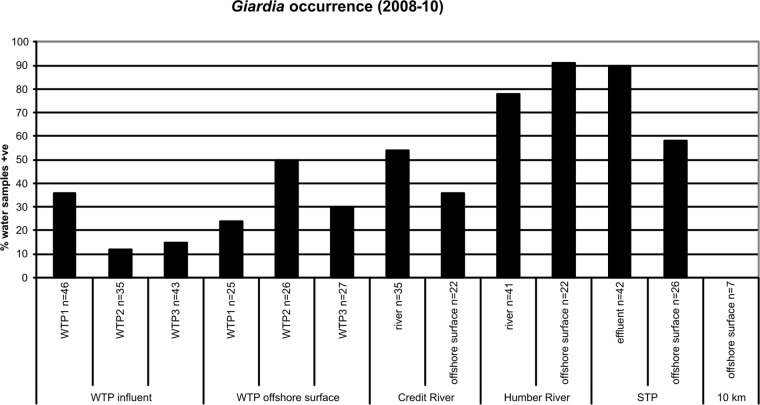
Giardia was detected at all sampling sites investigated, except at the reference site located 10 km offshore in Lake Ontario (Fig. 4). This pathogen was found significantly more often in the influent water for WTP1 (36%) than in the influent of WTP2 (12%) or WTP3 (15%) (P < 0.05). Giardia was significantly more common in the offshore surface waters above the WTP2 intake (50%) than in the WTP2 influent samples (P < 0.05). Giardia was detected in 24% of offshore surface water samples at the WTP1 intake compared to 30% for WTP3. The highest numbers of Giardia cysts enumerated in WTP influents were 0.70 cysts/liter at WTP1, 0.12 cysts/liter at WTP2, and 0.18 cysts/liter at WTP3. The highest number of Giardia cysts enumerated in offshore surface water samples was 0.13 cysts/liter at WTP1.
Fig 4.
Percent occurrence of Giardia in water samples collected from Credit River area study sites between 2008 and 2010. WTPs, water treatment plants; STP, sewage treatment plant; 10 km, reference site located 10 km offshore from the mouth of the Credit River; influent, raw water entering WTPs; effluent, final treated wastewater leaving STP; surface, water collected at the surface above offshore WTP intakes, offshore STP outfall, and offshore of river mouths or 10 km offshore.
Giardia was most commonly detected in surface waters offshore at the mouth of the Humber River (91%) and in the final effluent of the STP (90%). Giardia was more common in the Humber River (78%) than in the Credit River (54%). It was still detected relatively frequently in surface water at offshore sites off the mouth of the Credit River (36%) and at the offshore STP outfall (58%). The increase in Giardia occurrence offshore from the mouth of the Humber River is likely due to this sampling site representing a combination of impacts from both the Humber River and the offshore outfall of the Humber STP near this location. The highest number of Giardia cysts enumerated in the STP effluent was 28.3 cysts/liter, followed by the Humber River (5.40 cysts/liter) and the Credit River (0.90 cysts/liter).
Giardia was detected in WTP influents and offshore surface waters above intakes in all seasons, although it was generally more common in the spring months. Giardia occurred in 100% of April influent samples at WTP1 (n = 7) over 3 years and in 50% of March influent samples at WTP2 (n = 4). Giardia was also more common in the surface waters above offshore WTP intakes in the spring months of April and May. Giardia was often more common at WTP and other study sites on wet weather days than on dry weather days, although wet weather responses for this pathogen were not as notable as for Cryptosporidium. Giardia was very commonly associated with the Humber River and the STP final effluent irrespective of weather conditions.
A variety of Giardia assemblages were detected in the present study (N. Neumann, unpublished data). In particular, Giardia intestinalis assemblages A and B were observed across most of the sampling sites in the present study, suggesting that human-infectious forms of G. intestinalis are widespread in Lake Ontario. G. intestinalis assemblage A was the most prominent assemblage detected, and it was found in the STP final effluent, surface waters offshore at the STP outfall, and surface waters offshore at all WTP intakes, as well as in the influent of WTP1 and WTP3. G. intestinalis assemblage B was also widespread in Lake Ontario, being found in the surface waters offshore at all WTP intakes, as well as in the influent of WTP1 and WTP3.
Fecal indicator bacterium occurrence.
A summary of fecal indicator bacterium results for study sites is presented in Table 1. The concentration of E. coli in WTP influent samples was usually below detection, although spikes were seen occasionally in WTP influent water samples, with maximum concentrations as high as 112 CFU/100 ml (WTP1), 40 CFU/100 ml (WTP2), and 20 CFU/100 ml (WTP3). It was notable that WTP2 influent samples usually had lower levels of E. coli and other indicator bacteria such as total coliforms than did the other WTPs. Only 7% of the water samples at WTP2 were positive for E. coli compared to 36 and 49% positive for E. coli at WTP1 and WTP3, respectively. The detection of E. coli was somewhat variable between duplicate water samples, ranging from 82% positive concordance at the WTP3 intake offshore to 98% positive concordance at the WTP2 intake.
Table 1.
Enumeration of fecal indicator bacteria at study sites between 2007 and 2010
| Site | na | %Hub | 90th percentile (CFU/100 ml) |
Maximum (CFU/100 ml) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total coliforms | E. coli | Enterococci | Clostridia | Total coliforms | E. coli | Enterococci | Clostridia | |||
| WTP (influent) | ||||||||||
| WTP1 | 103 | 19 | 119 | 5 | 6 | 15 | 320 | 112c | 143 | 89 |
| WTP2 | 91 | 3 | 35 | 0 | 2 | 4 | 10,000 | 40 | 32 | 200 |
| WTP3 | 104 | 2 | 180 | 7 | 8 | 10 | 4,170 | 20 | 41 | 130 |
| WTP (bottom) | ||||||||||
| WTP1 | 45 | 7 | 332 | 9 | 13 | 4 | 850 | 26 | 40 | 241 |
| WTP2 | 67 | 6 | 525 | 7 | 17 | 4 | 1,900 | 27 | 65 | 10 |
| WTP3 | 68 | 6 | 556 | 18 | 26 | 4 | 5,000 | 58 | 210 | 15 |
| WTP (surface) | ||||||||||
| WTP1 | 49 | 8 | 334 | 13 | 34 | 6 | 679 | 70 | 118 | 23 |
| WTP2 | 71 | 6 | 812 | 19 | 17 | 9 | 1,620 | 222 | 400 | 71 |
| WTP3 | 72 | 10 | 480 | 14 | 11 | 4 | 1,690 | 86 | 52 | 11 |
| Credit River | ||||||||||
| River | 90 | 31 | 100,000 | 1,883 | 1,122 | 218 | 430,000 | 8,400 | 4,160 | 540 |
| Surface | 67 | 5 | 2,832 | 177 | 190 | 30 | 78,000 | 1,080 | 624 | 91 |
| Bottom | 64 | 8 | 2,750 | 83 | 53 | 13 | 24,000 | 179 | 198 | 42 |
| Humber River | ||||||||||
| River | 85 | 47 | 210,000 | 2,942 | 1,754 | 314 | 1,190,000 | 7,270 | 4,200 | 820 |
| Surface | 43 | 69 | 3,840 | 378 | 98 | 256 | 22,800 | 648 | 200 | 376 |
| Bottom | 41 | 63 | 7,200 | 154 | 66 | 173 | 12,400 | 1,600 | 516 | 248 |
| STP | ||||||||||
| Effluent | 102 | 74 | 39,020 | 251 | 246 | 1,183 | 640,000 | 2,960 | 1,220 | 16,000 |
| Surface | 62 | 19 | 1,336 | 68 | 15 | 60 | 8,900 | 288 | 300 | 132 |
| Bottom | 58 | 16 | 1,804 | 29 | 80 | 36 | 8,400 | 74 | 210 | 110 |
| 10 km | ||||||||||
| Surface | 29 | 7 | 390 | 2 | 11 | 1 | 690 | 6 | 29 | 1 |
| Bottom | 24 | 0 | 4,692 | 4 | 79 | 18 | 7,360 | 24 | 160 | 26 |
n, number of water samples analyzed for E. coli (slightly fewer samples were analyzed for the other indicator bacteria).
%Hu, percent positive for the HF183 human Bacteroidales DNA marker.
A concentration of 112 E. coli CFU/100 ml was measured on 14 March 2011. Otherwise, the maximum E. coli concentration measured during the 2007 to 2010 study period was 66 E. coli CFU/100 ml.
Although E. coli spikes in WTP influent waters often occurred during colder months, spikes in surface waters at the offshore WTP intakes often occurred in the summer months associated with rain events. The highest offshore surface water spikes at the WTP2 intake 2 km offshore were on 16 May 2007 (188 CFU/100 ml), 16 June 2008 (222 CFU/100 ml), and 11 August 2009 (140 CFU/100 ml). E. coli concentrations at other WTP offshore surface settings were as high as 70 CFU/100 ml (WTP1) and 86 CFU/100 ml (WTP3). E. coli concentrations were typically higher in the surface waters than bottom waters at offshore WTP intakes, which were separated by a thermocline typically residing above a depth of ∼10 m over the summer months. Among reference sites, the Humber River generally had the highest levels of E. coli. The numbers of total coliforms, E. coli, and enterococci were higher at the river reference sites than in the STP final effluent. In contrast, the numbers of clostridia were higher in the STP final effluent than the river reference and WTP sites.
The HF183 human Bacteroidales DNA marker was detected in 74% of wastewater samples from the STP final effluent (Table 1), but it was not detected in the following fresh fecal samples: dog (n = 16), cat (n = 17), gull (n = 85), Canada geese (n = 58), mallard duck (n = 10), cow (n = 31), pig (n = 8), and chicken (n = 14). However, the human marker was detected in fresh fecal samples from a Toronto dog (1 of 17 samples) and a Niagara chicken (1 of 15 samples). The HF183 human Bacteroidales DNA marker was most frequently detected in the final effluent of the Lakeview area STP (74%), followed by the offshore site at the mouth of the Humber River (surface = 69%, bottom = 63%) located near the outfall of the Humber STP. The HF183 DNA marker was detected more often in the Humber River than the Credit River. The human DNA Bacteroidales marker was detected less frequently at offshore sampling locations, although it was detected in the influent for each WTP. The human marker was detected more often in the influent for WTP1 (19%) compared to WTP2 (3%) and WTP3 (2%).
Predicting waterborne pathogen occurrence at WTPs.
E. coli was not predictive of thermophilic Campylobacter occurrence in WTP influent waters. Only three influent water samples were positive for thermophilic Campylobacter over the study period across all WTPs. E. coli concentration was below detection on two of these occasions, and only 1 CFU/100 ml on the other. A spike in concentration of clostridia (14 CFU/100 ml) proved more useful for predicting the 22 November 2007 Campylobacter event at WTP1. However, this indicator was not measured for the other two sampling occasions in May 2007. Turbidity was relatively elevated at 0.57 and 0.88 nephelometric turbidity unit(s) (NTU) for WTP1 Campylobacter detections, whereas the turbidity was only 0.38 NTU for the WTP3 Campylobacter detection. The May 2007 Campylobacter contamination events were not associated with rainfall in the 3 days preceding water sampling. However, Campylobacter detection at WTP1 in November 2007 was preceded by 29.2 mm of rain the day before water sampling.
E. coli was not predictive of cultivable enteric virus occurrence in WTP influent waters, although other parameters had some predictive value for viruses in WTP1 and WTP3 influents. Clostridia (R2 = 0.57) and enterococci (R2 = 0.41) were significantly correlated with virus concentrations in the WTP1 influent (P < 0.05). Rainfall 3 days before sampling (R2 = 0.43), cumulative rainfall 48 h before sampling (R2 = 0.38), and cumulative rainfall 72 h before sampling (R2 = 0.37) were also significantly correlated with virus concentrations at WTP1. A stepwise logistic regression analysis found clostridium concentration to be significant for predicting occurrence of cultivable enteric virus in the influent of WTP1 (P < 0.05) as characterized by Y = 0.3842 (clostridium CFU/100 ml) − 2.6302 (R2 = 0.31). For the WTP3 influent, enterococci (R2 = 0.58) and rainfall 24 h before sampling (R2 = 0.49) were the only parameters significantly correlated with virus concentrations (P < 0.05). A stepwise logistic regression analysis did not yield a significant model for predicting cultivable enteric virus occurrence in the influent of WTP3 (P > 0.05). The correlation coefficients for commonly used indicators such as E. coli, total coliforms, and turbidity were not significant for viruses in the influent of any WTP (P > 0.05). CART classification trees were not found for predicting viruses in the influents of WTPs.
E. coli was not predictive of Cryptosporidium and Giardia occurrence in WTP influents. There was no significant correlation between E. coli and Cryptosporidium or Giardia concentrations in WTP influents (P > 0.05). E. coli concentrations were often below detection when these pathogens were detected, and spikes in E. coli concentrations often did not coincide with pathogen occurrence (Fig. 5 to 7). For example, E. coli was not detected when the highest concentrations of Giardia and Cryptosporidium occurred at WTP2 on 21 April 2009. A concentration of only 1 E. coli CFU/100 ml was measured when the highest concentrations of Giardia and Cryptosporidium occurred at WTP1 on 4 May 2009. Across all three WTPs, E. coli was below detection for 58% of the Cryptosporidium-positive water samples (n = 12) and 54% of the Giardia-positive water samples (n = 26). In addition, there were numerous cases where detection of E. coli did not coincide with the occurrence of Cryptosporidium or Giardia. Across all three WTPs, Cryptosporidium and Giardia were not detected for 88 and 72%, respectively, of E. coli-positive water samples (n = 43).
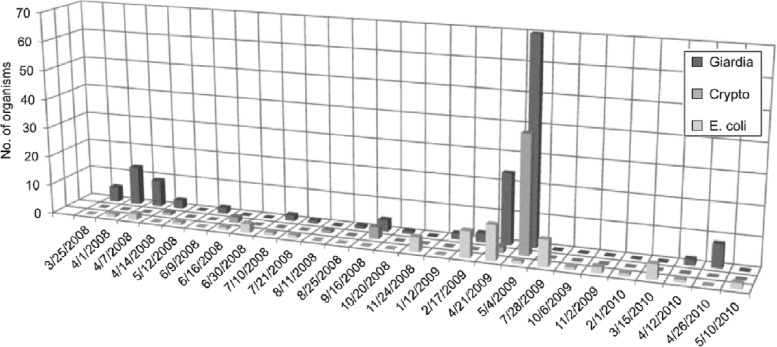
Fig 5.
Numbers of E. coli (CFU/100 ml) and Cryptosporidium and Giardia [(oo)cysts/100 liter] in water samples collected from the influent of WTP1 between 2008 and 2010. The sampling dates were removed when no pathogens or E. coli were detected in water samples.
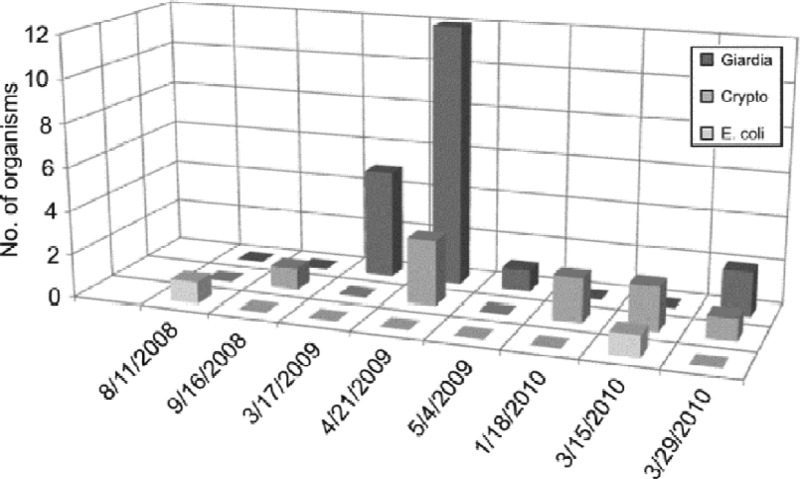
Fig 7.
Numbers of E. coli (CFU/100 ml) and Cryptosporidium and Giardia [(oo)cysts/100 liter] in water samples collected from the influent of WTP3 between 2008 and 2010. The sampling dates were removed when no pathogens or E. coli were detected in water samples.
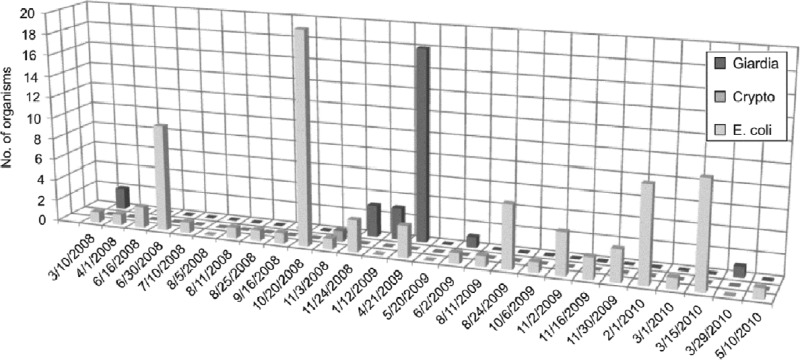
Fig 6.
Numbers of E. coli (CFU/100 ml) and Cryptosporidium and Giardia [(oo)cysts/100 liter] in water samples collected from the influent of WTP2 between 2008 and 2010. The sampling dates were removed when no pathogens or E. coli were detected in water samples.
Although parameters other than E. coli had some predictive value for Cryptosporidium and Giardia in WTP influents, these associations were typically specific for a particular WTP and pathogen. None of the indicator bacteria, turbidity, or rainfall parameters were significantly correlated with Cryptosporidium concentrations for WTP1 (P > 0.05). A stepwise logistic regression analysis did not find a significant model for predicting Cryptosporidium occurrence in the influent of WTP1 (P > 0.05). However, a CART classification tree found that five of six Cryptosporidium detections in the influent of WTP1 occurred when cumulative 72 h rainfall was ≥25.8 mm (R2 = 0.37). It was also notable that all six Cryptosporidium detections at WTP1 occurred when the turbidity was ≥0.34 NTU and that five of these six detections were above the median turbidity of 0.35 NTU for WTP1. Clostridia (R2 = 0.47) and temperature (R2 = −0.38) were the only parameters significantly correlated with Cryptosporidium concentrations for WTP2 (P < 0.05). A stepwise logistic regression analysis found clostridia concentration to be significant for predicting Cryptosporidium occurrence in the influent of WTP2 (P < 0.05) as characterized by Y = 0.7315 (clostridium CFU/100 ml) − 3.2326 (R2 = 0.36). A CART classification tree found four of five Cryptosporidium detections in the influent of WTP2 occurred when clostridia was ≥2 CFU/100 ml, and three of these four detections occurred when the cumulative 48-h rainfall before sampling was ≥4.6 mm (R2 = 0.34). It was also notable that four of five Cryptosporidium detections occurred when the turbidity was ≥0.36 NTU, although only three of the five detections were above the median turbidity of 0.39 NTU for WTP2. There were too few detections (n = 1) of Cryptosporidium in the influent of WTP3 to permit statistical analyses. A single oocyst was detected in the WTP3 influent on 3 November 2008. The E. coli concentration for this sample was only 1 CFU/100 ml, although the enterococcus and clostridium concentrations had spiked a little higher at 19 and 8 CFU/100 ml, respectively, on this day, and the turbidity was relatively high at 3.1 NTU. No rainfall had occurred in the 3 days prior to collecting this water sample at WTP3.
Turbidity (R2 = 0.41) was the only individual parameter significantly correlated with Giardia concentrations for WTP1 (P < 0.05). A stepwise logistic regression analysis did not find a significant model for predicting Giardia occurrence in the influent of WTP1 (P > 0.05). A CART classification tree found that all 16 Giardia detections in the influent of WTP1 occurred when the universal Bacteroidales DNA marker was also detected in water samples (R2 = 0.61). Furthermore, 15 of 16 Giardia detections occurred when the turbidity was ≥0.35 NTU, which was the median turbidity for WTP1. Temperature (R2 = −0.53) was the only individual parameter significantly correlated with Giardia concentrations for WTP2 (P < 0.05). Of note was that Cryptosporidium was also significantly correlated (R2 = 0.39) with Giardia in the influent of WTP2 (P < 0.05). A stepwise logistic regression analysis did not find any significant model for predicting Giardia occurrence in the influent of WTP2 (P > 0.05). A CART classification tree found that all four Giardia detections in the influent at WTP2 occurred when the water temperature was <9°C. All four Giardia detections also occurred when the turbidity was ≥0.36 NTU, with three of four detections above the median turbidity of 0.39 NTU for WTP2. None of the indicator bacteria, turbidity, or rainfall parameters were significantly correlated with Giardia concentrations for WTP3 (P > 0.05). A stepwise logistic regression analysis did not find a significant relationship for predicting Giardia occurrence in the influent of WTP3 (P > 0.05). A CART classification tree found that all six Giardia detections in the influent of WTP3 occurred when rainfall 2 days before sample collection was <0.4 mm and turbidity was ≥0.31 NTU. However, only three of six Giardia detections occurred above the median turbidity of 0.47 NTU for WTP3.
DISCUSSION
We collected 1,358 water and wastewater samples to investigate waterborne pathogen occurrence at three offshore drinking water intakes in western basin of Lake Ontario. Campylobacter, cultivable enteric viruses, Cryptosporidium, and Giardia were detected in the offshore source water of each drinking water treatment plant (WTP) in our study area even though some intakes were located 2 km offshore. However, pathogens generally occurred infrequently and in relatively low concentrations in the influents at these WTPs. A preliminary quantitative microbial risk assessment using data from WTP2 and WTP3 indicated that the concentrations of pathogens detected here would not pose a challenge for normal water treatment operations at these WTPs (34).
Thermophilic Campylobacter was never detected more commonly than 2 to 3% in the influent water samples at WTPs. The concentration of Campylobacter in Lake Ontario offshore water samples was always at our lowest limit of detection (about 4 to 40 cells/liter) and likely below the levels associated with an infectious dose of Campylobacter even before water treatment (35, 36, 37, 38). Thermophilic Campylobacter was detected most frequently in the Credit and Humber Rivers, although its occurrence dropped off sharply at sampling sites offshore of the mouths of these rivers. It is likely that there is a substantial dilution and die-off bacterial pathogens upon entering Lake Ontario. However, Campylobacter can survive for several weeks or more in colder waters (39), enter into viable but nonculturable states (40), and find protection within protozoa (41). Such transformations could make them less detectable by culture-based detection methods and enhance their persistence in offshore ecosystems. These considerations would only seem relevant if bacterial pathogens were delivered offshore in river plumes in a matter of hours at a time when normal water treatment operations were down.
Cultivable enteric viruses were detected in 15 to 50% of influent water samples at WTPs, with a maximum concentration measured at 0.33 MPN-IU/liter at WTP1. Enteric viruses were detected less frequently in the offshore source water of two Milwaukee WTPs on Lake Michigan (6.8 and 10.9% of water samples), although maximum cultivable virus concentrations were similar to those in our study (42). Payment et al. (43) detected cultivable viruses in 39% of source water samples from 45 WTPs along the St. Lawrence River near Montreal, and maximum virus concentrations in these riverine water samples were mostly higher than our Lake Ontario study (up to 62 MPN-IU/liter). It was notable that viruses were only detected at Lake Ontario WTPs in colder months between October and March. Both Payment et al. (43) and Sedmak et al. (42) also reported that viruses were detected more frequently in winter months. Most virus detections at the Milwaukee intakes were from December to March, with significant spikes in virus titers at the Howard WTP typically observed in the months of March and April (42). The Howard plant was associated with the spring 1993 Cryptosporidium outbreak. Greer et al. (44) alluded to the possibility of offshore drinking water intakes being the source of winter norovirus outbreaks in the Toronto Area. However, given the relatively low concentrations of cultivable viruses detected in influent waters in our study, and the effectiveness of chlorination alone at WTPs, additional research would be needed to attribute any significance to the winter occurrence of viruses at the offshore intakes in our Lake Ontario study area.
Cryptosporidium was detected in the influents of each WTP in our study, occurring in up to 15% of WTP2 water samples. However, Cryptosporidium concentrations rarely exceeded 0.1 oocysts/liter, and the maximum WTP influent concentration in our study was 0.4 oocysts/liter. Although the methods used by Wallis et al. (45) only detected Cryptosporidium in 4.5% of raw water samples (n = 1,173) from 72 WTP facilities across Canada, other studies have reported higher values. Payment et al. (43) detected Cryptosporidium in 37% of raw water samples (n = 415) from 45 WTP facilities along the St. Lawrence River near Montreal. Maximum Cryptosporidium concentrations in the St. Lawrence riverine water samples were often higher (up to 15.6 oocysts/liter) than at Lake Ontario WTPs. Rose (46) summarized studies of surface waters across North America and reported frequencies of occurrence and concentrations of Cryptosporidium that were often much higher than the influents of Lake Ontario WTPs.
The arithmetic mean concentrations of Cryptosporidium at all offshore Lake Ontario sampling sites placed them within or below the Bin 2 category of the U.S. Environmental Protection Agency (EPA)'s “Long Term 2 Enhanced Surface Water Treatment Rule” (LT2ESWTR). Risk to human health is considered to be relatively low under a Bin 2 classification (arithmetic mean, >0.075 oocysts/liter but <1.0 oocysts/liter). It is notable that WTPs on the United States side of the Great Lakes must follow U.S. EPA LT2ESWTR to characterize Cryptosporidium occurrence in source water in order to understand potential implications for water treatment. However, there is no requirement for Cryptosporidium surveillance at WTPs on the Canadian side of the Great Lakes.
It is increasingly recognized that not only the numbers of Cryptosporidium oocysts in source water surveillance but also their genotypes are important for understanding potential health risks (47). Most Cryptosporidium genotypes detected here were from animal fecal pollution sources (N. Neumann, unpublished data). However, genotypes of Cryptosporidium of human health concern were detected at several sampling sites, most notably on one occasion in the influent water of WTP1. The occurrence of C. hominis at WTP1 indicated that there are transport mechanisms to deliver human sewage to this WTP intake. This was consistent with the relatively higher frequency of detection of the human Bacteroidales DNA marker in the influent of WTP1 as well. Bower et al. (48) also detected the human Bacteroidales DNA marker as far as 2 km from shore in Lake Michigan near Milwaukee during sewage overflow events. Newton et al. (49) applied multiple molecular methods to detect evidence of human fecal pollution out to at least 8 km offshore from Milwaukee.
Giardia was the most commonly detected pathogen in the offshore waters of our study area. The frequency of Giardia in WTP influent water samples was as high as 36% (WTP1), and Giardia was commonly detected in the offshore surface waters above the WTP2 intake (50%) and the STP outfall (58%). However, Giardia concentrations rarely exceeded 0.1 cysts/liter, and the maximum WTP influent concentration in our study was 0.7 cysts/liter. Although the methods of Wallis et al. (45) detected Giardia in 21% of raw water samples (n = 1,173) from 72 WTP facilities across Canada, other studies have reported higher values. Payment et al. (43) detected Giardia in 60% of raw water samples (n = 415) from 45 WTP facilities along the St. Lawrence River near Montreal. Maximum Giardia concentrations in the St. Lawrence riverine water samples were often higher (up to 38.3 cysts/liter) than at Lake Ontario WTPs. LeChevallier and Norton (50) summarized studies of raw water samples from North America WTPs and reported that Giardia was detected in 53.9% of samples (n = 347). The frequency of occurrence and concentrations of Giardia in these sources of drinking water was often higher than in the influents of Lake Ontario WTPs.
Although the host-specific nature of Giardia is not as robust as for Cryptosporidium, determining the genotypes of Giardia can be important for understanding the potential for human health risks. Giardia assemblages A and B were the most commonly found assemblages in WTP influents and at offshore sampling locations in our study. Since almost all cases of human giardiasis are associated with these two assemblages (51), this would suggest that human-infectious forms of Giardia were widespread in Lake Ontario. The occurrence of Giardia was commonly associated with occurrence of human sewage in our study. Both Giardia and the human Bacteroidales DNA marker were most commonly detected in the STP final effluent and at the mouth of the Humber River. Giardia was detected in 90% of STP final effluent samples, with concentrations as high as 28.3 cysts/liter. Feng and Xiao (51) also reported that Giardia is commonly found in sewage treatment plant effluents. It is possible that the regular occurrence of low numbers of Giardia in offshore settings in our study area could have come from offshore STP outfalls or from sewage sources in the Humber and Credit Rivers.
We have provided here a benchmark of waterborne-pathogen occurrence that can be used to understand future trends in source water quality for three WTPs on Lake Ontario. The impacts from human sewage, as well as the occurrence of waterborne pathogens, were found to differ between three closely located WTPs. This was likely the result of various influences from river runoff or STP effluent outfalls that would be unique to each WTP intake. This variability raises concern about simply extrapolating these results to other WTPs and source water conditions around the Great Lakes. It also raises questions about the ability to apply generic mathematical/statistical models for predicting pathogen occurrence at specific drinking water intakes around the Great Lakes. It will be important to continue to collect data on waterborne-pathogen occurrence at specific WTP intakes to validate the use of predictive modeling tools.
In our study, E. coli measured by standard method was not predictive of pathogen occurrence in the influents of WTPs. Concentrations of E. coli were often below detection when pathogens were detected, and spikes in E. coli concentrations often did not coincide with pathogen occurrence. It is possible that E. coli may be useful for predicting pathogen occurrence in offshore river plumes for a few hours following precipitation events when there would be less time for differential settling or die-off with pathogens such as viruses and protozoa. However, over longer periods, one might expect such predictive relationships to break down given the longer persistence of pathogens such as Giardia cysts and Cryptosporidium oocysts (46). E. coli has also been reported as a poor surrogate for Cryptosporidium in surface waters by Nieminski et al. (52), who suggested that direct monitoring of source vulnerability for Cryptosporidium would have more significant value. McLellan et al. (53) found that E. coli numbers declined more quickly than the numbers of fecal coliform bacteria in offshore waters outside the Milwaukee Harbor on Lake Michigan. Fecal coliforms were suggested as a more conservative tracer of sewage and potentially a better indicator than E. coli for protozoan pathogens. Similarly, Bower et al. (48) detected Bacteroides spp. by PCR methods when culturable E. coli was absent from Lake Michigan water samples, suggesting these DNA markers might be a more sensitive indicator of offshore fecal pollution. Newton et al. (49) applied a more sensitive microbial community sequencing approach to detect evidence of fecal pollution eight kilometers offshore in Lake Michigan, where E. coli and enterococci were below detection by traditional culture methods.
It was notable that E. coli was not predictive of pathogen occurrence at a WTP that had a problem with its prechlorination process used to limit mussel infestations on the surfaces of its offshore intake crib. WTP2 frequently had inadvertent chlorination in its “raw” offshore water sampling line. This chlorination compromised the use of E. coli for “raw” water quality assessment, particularly for chlorine-resistant pathogens such as Cryptosporidium. Although the influent of WTP2 had the lowest frequency of indicator bacteria (E. coli was usually below detection), it had the highest frequency of Cryptosporidium among the three WTPs. Caution is needed to ensure prechlorination at offshore intake cribs does not inadvertently kill or render E. coli (or other indicator bacteria) unculturable in “raw” water sampling lines, so as to mask the presence of more chlorine-resistant pathogens. Winter et al. (54) also reported that inadvertent chlorination may have influenced their assessment of algae in raw water samples at a Lake Ontario WTP.
It is important to note that most studies measure indicator bacteria such as E. coli and waterborne pathogens such as protozoa from different water sample volumes. We followed standard methods for E. coli (and other fecal indicator bacteria) that are based on filtering 100 ml. Our methods for detecting protozoa and viruses were based on typically larger water volumes (e.g., 100 liters). Such different sample volumes may be more problematic when applied in offshore ecosystems where fecal indicator bacteria and waterborne pathogens are more likely to occur infrequently and close to the detection limits of filtration methods. Loge et al. (55) and Emelko et al. (56) discuss concerns about differing detection sensitivity and uncertainty associated with using different water sample volumes in pathogen detection studies. At present, it should be recognized that typically used detection methods for fecal indicator bacteria and protozoa and viral pathogens present biases from different water sample volumes and recovery efficiencies that may be more significant for studies in offshore ecosystems.
In our study, no single parameter was found to be predictive of pathogen occurrence at all three WTPs on Lake Ontario. The logistic and classification tree models that could be developed were different for each pathogen and WTP intake. These predictive models were generally developed based on few pathogen detections, and so they should be viewed cautiously, and more as a guide for future research. Clostridia (CFU/100 ml), turbidity, and rainfall parameters showed more potential for predicting pathogen occurrence at offshore intakes than E. coli. Among the indicator bacteria studied, clostridia was found to provide some ability to predict occurrence of enteric viruses at WTP1 and Cryptosporidium at WTP2. It is possible that the greater persistence of Clostridium spp. and their reduced susceptibility to chlorine relative to E. coli might make this a better water quality indicator for pathogens such as Cryptosporidium in offshore ecosystems. In particular, Clostridium perfringens might be a good candidate (57, 58). Continued research is required to identify practical alternative indicators of water quality in offshore ecosystems in the Great Lakes.
Until more reliable water quality indicators are developed for offshore ecosystems, it will be important to understand the hydrodynamic and ecological factors influencing the occurrence, transport, and fate of pathogens entering the Great Lakes. Brookes et al. (59) developed a conceptual model for lakes and reservoirs that identified the importance of processes such as river inflows, settling, and resuspension, as well as pathogen inactivation by grazing, UV, and temperature, as guidance for pathogen monitoring and risk assessment. In Lake Ontario, we found that river flows could deliver high concentrations of indicator bacteria to surface waters above an offshore WTP intake in response to a rain event (e.g., E. coli > 200 CFU/100 ml). However, the summer thermocline served as a barrier to the transport of indicator bacteria down to WTP intakes on the lake bottom. In general, the fate of river inflows will be determined by their density relative to the lake, such that warm inflows will flow out over the surface of the lake, whereas colder dense inflows will sink and flow out along the lake bottom (59). Colder dense inflows of rivers in our study area may have contributed to pathogen occurrence at offshore intakes in some months. These hydrodynamic events will also be influenced by upwelling and downwelling events. Previous research in our study area has shown that taste and odor events in drinking water can occur from a sequential series of upwelling and downwelling events, typically in the late summer period (60–62). Any pathogens occurring in offshore surface waters at the time of a downwelling event could presumably also be delivered to WTP intakes on the lake bottom. In addition, since waves and currents can resuspend offshore bottom sediments (62), it is possible that indicator bacteria or pathogens in offshore sediments could be drawn into intakes.
The transport and fate of pathogens entering the Great Lakes will also be influenced by seasonal phenomena, such as increased river flows in spring runoff periods. At these times, colder temperatures would enhance pathogen survival, the water column would be more isothermal to facilitate mixing with bottom waters, and the influence of a thermal bar could confine river plumes to nearshore waters near WTP intakes. The thermal bar occurs each spring (and fall) as nearshore waters warm and a downwelling plume of maximum density water (4°C) is formed that proceeds out into the lake until stable stratification prevails (63, 64). The downwelling plume occurs where nearshore and offshore waters on either side of the 4°C isotherm meet, and this thermal bar creates a barrier inhibiting exchange of water between the nearshore and offshore regions. Menon et al. (65) investigated the thermal bar in Lake Ontario east of Oshawa during a research cruise on board the Limnos and suggested that the spring thermal bar had the potential to confine bacteria to nearshore areas and to present potential for increased health risks. Makarewicz et al. (66) observed a similar thermal bar entrapment of nutrients and turbidity in Lake Ontario. It is notable that the 1993 Cryptosporidium outbreak in Milwaukee, Wisconsin, the offshore spikes in culturable viruses detected by Sedmak et al. (42) at the Howard WTP in Milwaukee, and the 1996 Cryptosporidium outbreak in Collingwood, Ontario occurred around the March-April spring period. Similarly, we detected pathogens such as Giardia more frequently at WTP1 and WTP2 in this March-April period in our study. Winter et al. (54) found chloride levels at WTP1 and WTP2 to peak in March, likely reflecting road salt use and urban runoff. This spring period likely represents a potentially vulnerable time for some WTPs around the Great Lakes.
It is uncertain whether past experience with nearshore water dynamics, water treatment practices and use of water quality indicators such as E. coli will be a reliable guide for the safe operation of drinking water treatment plants on the Great Lakes into the future. Our study area in western Lake Ontario is rapidly urbanizing, which will present a variety of stormwater and municipal wastewater challenges. In addition, climate change projections for more extreme precipitation events and milder winters could significantly change river runoff patterns and offshore water column mixing dynamics. Curreiro et al. (67) found that 51% of waterborne disease outbreaks in the United States were associated with precipitation events above the 90th percentile, and both the Milwaukee outbreak in 1993 and the Walkerton outbreak in 2000 were associated with extreme precipitation events. Patz et al. (68) projected that climate change and associated precipitation changes can be anticipated to lead to increased flooding, more combined sewer overflow events, and higher risks for waterborne disease in the Great Lakes in the future. More frequent turbidity events associated with algal blooms or the erosion of sediments from extreme weather events could present challenges for pathogen removal at drinking water treatment plants through clogging of filtration systems, as well as changing coagulant and chlorine demands. These environmental changes in source water quality are likely to coincide with demographic changes from an aging and more frequently immunocompromised population around the Great Lakes. The elderly, very young, pregnant women, and those immunocompromised from AIDS, cancer chemotherapy, or organ transplants are more sensitive and at significantly greater risk from waterborne pathogens (69, 70).
In the future, WTPs around the Great Lakes will need data on the occurrence of key waterborne pathogens such as Cryptosporidium in their source water considering the limitations of water quality monitoring tools such as determining E. coli levels. WTPs should know what pathogens could be knocking at their treatment door when they have lapses in water treatment operations at times. This is particularly the case for WTPs with intakes located close to shorelines or river mouths. It will be especially important to maintain optimal drinking water treatment operations during potentially vulnerable periods such as spring runoff around the Great Lakes. It will also be important to consider source water quality in assessments of the state of drinking water quality around the Great Lakes. A number of Great Lakes Programs (e.g., State of the Great Lakes reporting, Areas of Concern beneficial use impairments, etc.) monitor the status and trends of drinking water quality around the Great Lakes by focusing on treated water quality and compliance with finished drinking water standards. By focusing on treated water quality, this approach does not enable assessment of changing environmental conditions and trends in source water quality around the Great Lakes.
ACKNOWLEDGMENTS
We thank Mark Schiller and Jane Bonsteel, Region of Peel, for their support of this study. We also thank the Ontario Ministry of Environment and Michael D'Andrea and Bill Snodgrass, City of Toronto, for support, as well as Alyssa Loughborough, Flora Suen, Franco Solimano, Erin Hennessey, Jessica Rando, and many students that helped with water sample collection and laboratory analyses for this study. We thank Environment Canada's Technical Operations for providing much assistance in collecting water samples and Lucas Neilson for help with the figures. Finally, we thank the staff at the WTPs and the STP, who kindly shared data and facilitated water and wastewater sampling at their plants.
Footnotes
Published ahead of print 8 July 2013
REFERENCES
- 1.Keating M. 2004. Our Great Lakes. Catalogue no. En164-2/2004e. Great Lakes and Corporate Affairs Branch, Environment Canada, and Great Lakes National Program Office, U.S. Environmental Protection Agency, Washington, DC [Google Scholar]
- 2.Canadian Public Health Association 1986. Comprehensive survey of the status of Great Lakes drinking water. Canadian Public Health Association, Ottawa, Ontario, Canada [Google Scholar]
- 3.Patel M. 2008. The long haul: integrating water, sewage, public health, and city-building, p 92–101 In Reeves W, Palassio C. (ed), HTO: Toronto's water from Lake Iroquois to Lost Rivers to low-flow toilets. Coach House Books, Toronto, Ontario, Canada [Google Scholar]
- 4.Beatty WK. 1982. When cholera scourged Chicago. Chicago Hist. 11:2–13 [PubMed] [Google Scholar]
- 5.International Joint Commission 1914. Progress report of the International Joint Commission on the Pollution of Boundary Waters, including a report of the sanitary experts. U.S. Government Printing Office, Washington, DC [Google Scholar]
- 6.International Joint Commission 1918. Final report of the International Joint Commission on the Pollution of Boundary Waters Reference. U.S. Government Printing Office, Washington, DC [Google Scholar]
- 7.Dreelin EA. 2008. Bacteriological monitoring in the Great Lakes: a historical perspective to inform the present, p 39–57 In Rose JB, Dreelin EA. (ed), Effective cross-border monitoring systems for waterborne microbial pathogens. IWA Publishing, London, United Kingdom [Google Scholar]
- 8.Rose JB, Dreelin EA. (ed). 2008. Effective cross-border monitoring systems for waterborne microbial pathogens. IWA Publishing, London, United Kingdom [Google Scholar]
- 9.Hrudey SE, Hrudey EJ. 2004. Safe drinking water. IWA Publishing, London, United Kingdom [Google Scholar]
- 10.O'Connor DR. 2002. Report of the Walkerton Enquiry. Queen's Printer for Ontario, Toronto, Ontario, Canada [Google Scholar]
- 11.MacKenzie WR, Hoxie NJ, Proctor ME. 1994. Massive waterborne outbreak of Cryptosporidium infection associated with a filtered public water supply. N. Engl. J. Med. 331:161–167 [DOI] [PubMed] [Google Scholar]
- 12.Borden HH, Harris RW, Mosher WE. 1970. A waterborne outbreak of gastroenteritis in western New York State. Am. J. Public Health 60:283–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frost FJ, Muller T, Craun GF, Fraser D, Thompson D, Notenboom R, Calderon RL. 2000. Serological analysis of a cryptosporidiosis epidemic. Int. J. Epidemiol. 29:376–379 [DOI] [PubMed] [Google Scholar]
- 14.Fong T, Mansfield LS, Wilson DL, Schwab DJ, Molloy SL, Rose JB. 2007. Massive microbiological groundwater contamination associated with a waterborne outbreak in Lake Erie South Bass Island, Ohio. Environ. Health Perspect. 115:856–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fox KR, Lytle DA. 1996. Milwaukee's crypto outbreak: investigation and recommendations. J. AWWA 1996:87–94 [Google Scholar]
- 16.Hoxie NJ, Davis JP, Vergeront JM, Nashold RD, Blair KA. 1997. Cryptosporidiosis-associated mortality following a massive waterborne outbreak in Milwaukee, Wisconsin. Am. J. Public Health 87:2032–2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Payment P, Locas A. 2011. Pathogens in water: value and limits of correlation with microbial indicators. Groundwater 49:4–11 [DOI] [PubMed] [Google Scholar]
- 18.Wu J, Long SC, Das D, Dorner SM. 2011. Are microbial indicators and pathogens correlated? A statistical analysis of 40 years of research. J. Water Health 9:265–278 [DOI] [PubMed] [Google Scholar]
- 19.Whitman RL, Nevers MB. 2003. Foreshore sand as a source of Escherichia coli in nearshore water of a Lake Michigan beach. Appl. Environ. Microbiol. 69:5555–5562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edge TA, Hill S. 2007. Multiple lines of evidence to identify the sources of fecal pollution at a freshwater beach in Hamilton Harbour, Lake Ontario. Water Res. 41:3585–3594 [DOI] [PubMed] [Google Scholar]
- 21.Ishii S, Ksoll WB, Hicks RE, Sadowsky MJ. 2006. Presence and growth of naturalized Escherichia coli in temperate soils from Lake Superior watersheds. Appl. Environ. Microbiol. 72:612–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kildare BJ, Leutenegger CM, McSwain BS, Bambic DG, Rajal VB, Wuertz S. 2007. 16S rRNA-based assays for quantitative detection of universal, human-, cow-, and dog-specific fecal Bacteroidales: a Bayesian approach. Water Res. 41:3701–3715 [DOI] [PubMed] [Google Scholar]
- 23.Khan IUH, Edge TA. 2007. Development of a novel triplex PCR assay for the detection and differentiation of thermophilic species of Campylobacter using 16S-23S rDNA internal transcribed spacer (ITS) region. J. Appl. Microbiol. 103:2561–2569 [DOI] [PubMed] [Google Scholar]
- 24.Linton D, Owen RJ, Stanley J. 1996. Rapid identification by PCR of the genus Campylobacter and of five Campylobacter species enteropathogenic for man and animals. Res. Microbiol. 147:707–718 [DOI] [PubMed] [Google Scholar]
- 25.Payment P, Trudel M. 1985. Immunoperoxidase method with human immune serum globulin for broad-spectrum detection of cultivable human enteric viruses: application to enumeration of cultivable viruses in environmental samples. Appl. Environ. Microbiol. 50:1308–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Payment P, Trudel M. 1987. Detection and quantitation of human enteric viruses in waste waters: increased sensitivity using a human immune serum globulin-immunoperoxidase assay on MA-104 cells. Can. J. Microbiol. 33:568–570 [DOI] [PubMed] [Google Scholar]
- 27.U.S. Environmental Protection Agency 2005. Method 1623: Cryptosporidium and Giardia in water by filtration/IMS/FA. EPA document 815-R-05-002 U.S. Environmental Protection Agency, Office of Water, Washington, DC [Google Scholar]
- 28.Ruecker NJ, Bounsombath N, Wallis P, Ong CSL, Isaac-Renton JL, Neumann NF. 2005. Molecular forensic profiling of Cryptosporidium species and genotypes in raw water. Appl. Environ. Microbiol. 71:8991–8994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruecker NJ, Hoffman RM, Chalmers RM, Neumann NF. 2011. Detection and resolution of Cryptosporidium species and species mixtures by genus-specific nested PCR-RFLP, direct sequencing, and cloning. Appl. Environ. Microbiol. 77:3998–4007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao L, Singh A, Limor J, Graczyk TK, Gradus S, Lal A. 2001. Molecular characterization of Cryptosporidium oocysts in samples of raw surface water and wastewater. Appl. Environ. Microbiol. 67:1097–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.SAS Institute 2010. JMP statistics and graphics guide. SAS Institute, Inc, Cary, NC [Google Scholar]
- 32.Breiman L, Freidman J, Olshen R, Stone C. 1984. Classification and regression trees. Wadsworth International, Pacific Grove, CA [Google Scholar]
- 33.Wilkes G, Edge T, Gannon V, Jokinen C, Lyautey E, Medeiros D, Neumann N, Ruecker N, Topp E, Lapen DR. 2009. Seasonal relationships among indicator bacteria, pathogenic bacteria, Cryptosporidium oocysts, Giardia cysts, and hydrological indices for surface waters within an agricultural landscape. Water Res. 43:2209–2223 [DOI] [PubMed] [Google Scholar]
- 34.Locas A, Payment P. 2010. Quantitative microbial risk assessment (QMRA) for assessing source water risks in the Region of Peel: report prepared for the Region of Peel. Centre INRS/Institut Armand-Frappier, Laval, Quebec, Canada [Google Scholar]
- 35.Black RE, Levine MM, Clements ML, Hughes TP, Blaser MJ. 1988. Experimental Campylobacter jejuni infection in humans. J. Infect. Dis. 157:472–479 [DOI] [PubMed] [Google Scholar]
- 36.Teunis P, Havelaar A, Vliegenthart J, Roessink G. 1997. Risk assessment of Campylobacter species in shellfish: identifying the unknown. Water Sci. Technol. 35:29–34 [Google Scholar]
- 37.Teunis P, Van den Brandhof W, Nauta M, Wagenaar J, Van den Kerkhof H, Van Pelt W. 2005. A reconsideration of the Campylobacter dose-response relation. Epidemiol. Infect. 133:583–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Health Canada 2012. Guidelines for Canadian Recreational Water Quality, 3rd ed. Document H129-15/2012E Water, Air, and Climate Change Bureau, Healthy Environments and Consumer Safety Branch, Health Canada, Ottawa, Ontario, Canada [Google Scholar]
- 39.Blaser MJ, Hardesty HL, Powers B, Wang WL. 1980. Survival of Campylobacter fetus subsp. jejuni in biological milieus. J. Clin. Microbiol. 11:309–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rollins DM, Colwell RR. 1986. Viable but nonculturable stage of Campylobacter jejuni and its role in survival in the natural aquatic environment. Appl. Environ. Microbiol. 52:531–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Axelsson-Olsson D, Waldenstrom J, Broman T, Olsen B, Holmberg M. 2005. Protozoan Acanthamoeba polyphaga as a potential reservoir for Campylobacter jejuni. Appl. Environ. Microbiol. 71:987–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sedmak G, Bina D, MacDonald J, Couillard L. 2005. Nine-year study of the occurrence of culturable viruses in source water for two drinking water treatment plants and the influent and effluent of a wastewater treatment plant in Milwaukee, Wisconsin (August 1994 through July 2003). Appl. Environ. Microbiol. 71:1042–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Payment P, Berte A, Prevost M, Menard B, Barbeau B. 2000. Occurrence of pathogenic microorganisms in the Saint Lawrence River (Canada) and comparison of health risks for populations using it as their source of drinking water. Can. J. Microbiol. 46:565–576 [PubMed] [Google Scholar]
- 44.Greer AL, Drews SJ, Fisman DN. 2009. Why “winter” vomiting disease? Seasonality, hydrology, and norovirus epidemiology in Toronto, Canada. EcoHealth 6:192–199 [DOI] [PubMed] [Google Scholar]
- 45.Wallis PM, Erlandsen SL, Isaac-Renton JL, Olson ME, Robertson WJ, van Keulen H. 1996. Prevalence of Giardia cysts and Cryptosporidium oocysts and characterization of Giardia spp. isolated from drinking water in Canada. Appl. Environ. Microbiol. 62:2789–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rose JB. 1997. Environmental ecology of Cryptosporidium and public health implications. Annu. Rev. Public Health 18:135–161 [DOI] [PubMed] [Google Scholar]
- 47.Ruecker NJ, Matsune JC, Wilkes G, Lapen DR, Topp E, Edge TA, Sensen CW, Xiao L, Neumann NF. 2012. Molecular and phylogenetic approaches for assessing sources of Cryptosporidium contamination in water. Water Res. 46:5135–5150 [DOI] [PubMed] [Google Scholar]
- 48.Bower PA, Scopel CO, Jensen ET, Depas MM, McLellan SL. 2005. Detection of genetic markers of fecal indicator bacteria in Lake Michigan and determination of their relationship to Escherichia coli densities using standard microbiological methods. Appl. Environ. Microbiol. 71:8305–8313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Newton RJ, Bootsma MJ, Morrison HG, Sogin ML, McLellan SL. 2013. A microbial signature approach to identify fecal pollution in the waters off an urbanized coast of Lake Michigan. Microb. Ecol. 65:1011–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.LeChevallier MW, Norton WD. 1995. Giardia and Cryptosporidium in raw and finished water. J. Am. Water Works Assoc. 87:54–68 [Google Scholar]
- 51.Feng Y, Xiao L. 2011. Zoonotic potential and molecular epidemiology of Giardia species and giardiasis. Clin. Microbiol. Rev. 24:110–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nieminski E, Durrant GC, Hoyt MB, Owens ME, Peterson L, Peterson S, Tanner WD, Rose J, Clancy JL. 2010. Is Escherichia coli an appropriate surrogate for Cryptosporidium occurrence in water? J. Am. Water Works Assoc. 102:65–78 [Google Scholar]
- 53.McLellan SL, Hollis EJ, Depas MM, Van Dyke M, Harris J, Scopel CO. 2007. Distribution and fate of Escherichia coli in Lake Michigan following contamination with urban stormwater and combined sewer overflows. J. Great Lakes Res. 33:566–580 [Google Scholar]
- 54.Winter JG, Howell ET, Nakamoto LK. 2012. Trends in nutrients, phytoplankton, and chloride in nearshore waters of Lake Ontario: synchrony and relationships with physical conditions. J. Great Lakes Res. 38:124–132 [Google Scholar]
- 55.Loge FJ, Thompson DE, Call DR. 2002. PCR detection of specific pathogens in water: a risk-based analysis. Environ. Sci. Technol. 36:2754–2759 [DOI] [PubMed] [Google Scholar]
- 56.Emelko MB, Schmidt PJ, Robertson JA. 2008. Quantification of uncertainty in microbial data: reporting and regulatory implications. J. Am. Water Works Assoc. 100:94–104 [Google Scholar]
- 57.Payment P, Franco E. 1993. Clostridium perfringens and somatic coliphages as indicators of the efficiency of drinking water treatment for viruses and protozoan cysts. Appl. Environ. Microbiol. 59:2418–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brookes JD, Hipsey M, Burch MD, Regel RH, Linden LG, Ferguson CM, Antenucci JP. 2005. Relative value of surrogate indicators for detecting pathogens in lakes and reservoirs. Environ. Sci. Technol. 39:8614–8621 [DOI] [PubMed] [Google Scholar]
- 59.Brookes JD, Antenucci J, Hipsey M, Burch MD, Ashbolt NJ, Ferguson C. 2004. Fate and transport of pathogens in lakes and reservoirs. Environ. Int. 30:741–759 [DOI] [PubMed] [Google Scholar]
- 60.Rao YR, Skafel MG, Howell T, Murthy RC. 2003. Physical processes controlling taste and odor episodes in Lake Ontario drinking water. J. Great Lakes Res. 29:70–78 [Google Scholar]
- 61.Watson SB, Charlton M, Yerubandi R, Howell T, Ridal J, Brownlee B, Marvin C, Millard S. 2007. Off flavour in large waterbodies: physics, chemistry and biology in synchrony. Water Sci. Technol. 55:1–8 [DOI] [PubMed] [Google Scholar]
- 62.Rao YR, Milne JE, Marvin CH. 2012. Hydrodynamics and water quality in western Lake Ontario. J. Great Lakes Res. 38:91–98 [Google Scholar]
- 63.Holland PR, Kay A. 2003. A review of the physics and ecological implications of the thermal bar circulation. Limnologica 33:153–162 [Google Scholar]
- 64.Rao YR, Schwab DJ. 2007. Transport and mixing between the coastal and offshore waters in the Great Lakes: a review. J. Great Lakes Res. 33:202–218 [Google Scholar]
- 65.Menon AS, Dutka BJ, Jurkovic AA. 1970. Preliminary studies on the effect of the Lake Ontario thermal bar in confining bacteria to the near-shore region. Document KR.70-2 Division of Public Health Engineering, Department of National Health and Welfare, Kingston, Ontario, Canada [Google Scholar]
- 66.Makarewicz JC, Lewis TW, Boyer GL. 2012. Nutrient enrichment and depletion on the shoreside of the spring thermal front. J. Great Lakes Res. 38:72–77 [Google Scholar]
- 67.Curreiro FC, Patz JA, Rose JB, Lele S. 2001. The association between extreme precipitation and waterborne disease outbreaks in the United States 1948–1994. Am. J. Public Health 91:1194–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Patz JA, Vavrus SJ, Uejio CK, McLellan SL. 2008. Climate change and waterborne disease risk in the Great Lakes region of the U.S. Am. J. Prev. Med. 35:451–458 [DOI] [PubMed] [Google Scholar]
- 69.Gerba CP, Rose JB, Haas CN. 1996. Sensitive populations: who is at the greatest risk? Int. J. Food Microbiol. 30:113–123 [DOI] [PubMed] [Google Scholar]
- 70.Mor SM, DeMaria A, Griffiths JK, Naumova EN. 2009. Cryptosporidiosis in the elderly population of the United States. Clin. Infect. Dis. 48:698–705 [DOI] [PMC free article] [PubMed] [Google Scholar]