Abstract
The human intestinal tract is highly colonized by a vast number of microorganisms. Despite this permanent challenge, infections remain rare, due to a very effective barrier defense system. Essential effectors of this system are antimicrobial peptides and proteins (AMPs), which are secreted by intestinal epithelial and lymphoid cells, balance the gut microbial community, and prevent the translocation of microorganisms. Several antimicrobial proteins have already been identified in the gut. Nonetheless, we hypothesized that additional AMPs are yet to be discovered in this setting. Using biological screening based on antimicrobial function, here we identified competent antibacterial activity of high-mobility-group box 2 (HMGB2) against Escherichia coli. By recombinant expression, we confirmed this biologically new antimicrobial activity against different commensal and pathogenic bacteria. In addition, we demonstrated that the two DNA-binding domains (HMG boxes A and B) are crucial for the antibiotic function. We detected HMGB2 in several gastrointestinal tissues by mRNA analysis and immunohistochemical staining. In addition to the nuclei, we also observed HMGB2 in the cytoplasm of intestinal epithelial cells. Furthermore, HMGB2 was detectable in vitro in the supernatants of two different cell types, supporting an extracellular function. HMGB2 expression was not changed in inflammatory bowel disease but was detected in certain stool samples of patients, whereas it was absent from control individuals. Taken together, we characterized HMGB2 as an antimicrobial protein in intestinal tissue, complementing the diverse repertoire of gut mucosal defense molecules.
INTRODUCTION
Mammalian surfaces are in close contact with microbes. Especially the skin and the gut have to deal with high loads of bacteria, fungi, and viruses (1–3). The human gut is one of the most densely populated ecosystems and harbors approximately 100 trillion microbes (4). However, despite this enormous challenge, due to a very effective immune system, infections are rare events.
In the gut, the first line of defense against microorganisms is the intestinal epithelium, which is covered by a thick mucus layer containing highly glycosylated proteins (5, 6). In addition to physical protection, this barrier is strengthened by chemical defense molecules, including antimicrobial peptides and proteins (AMPs) (7). AMPs are effective weapons against a huge variety of microorganisms, including viruses, fungi, and bacteria, and are produced by all multicellular organisms (8–10). AMPs have a high level of diversity and can be subdivided into several classes based on their secondary structure (9). Two major characteristics of most AMPs are a positive net charge and an amphipathic structure (8, 9). These features seem to be crucial for the electrostatic attraction to negatively charged phospholipids of bacterial membranes and integration into the microbial cell membrane (11). Although the exact mode of action for many AMPs is not yet clear (8), there are various models illustrating microbial killing. Some AMPs act directly on the bacterial cell membrane by enzymatic or nonenzymatic mechanisms (12, 13). Others work via metal chelation, thereby limiting trace elements such as Mn2+ and Zn2+, which are essential for microbial survival (14, 15).
Well-investigated AMPs in humans are lysozyme, secretory phospholipase A2 (sPLA2), calprotectin, cathelicidin, and the defensins (2). Human defensins can be subdivided into α- and β-defensins based on the pattern of their disulfide bonds (8) and are expressed by granulocytes, epithelial cells, and small intestinal Paneth cells (7). A breakdown of the host antimicrobial barrier can have critical and clinically relevant consequences, as it seems to be central in the development of chronic inflammatory bowel diseases (IBDs). Crohn's disease (CD) of the small intestine, for example, is associated with different defects in small intestinal secretory Paneth cells, which permanently produce large amounts of different AMPs, including defensins, lysozyme, sPLA2, and others (16–18). In addition, attenuated induction of inducible β-defensins as well as decreased expression levels of human β-defensin 1 are associated with the colonic phenotype of CD (3, 17, 19). The disease-associated changes of AMP expression and function are mediated by a broad variety of different mechanisms, including host genetic and environmental alterations (20). Due to the resulting compromise of antimicrobial barrier function, commensal and pathogenic microbes are able to invade the mucosa and trigger and perpetuate a secondary inflammatory response. Single-nucleotide polymorphisms in defensin genes and alterations in their expression are also linked to several other diseases, including atopic dermatitis, periodontitis, cystic fibrosis, and asthma (21), underlining their important function in the human immune system.
While most AMPs are cytosolic proteins and partly stored in secretory granules, some nuclear proteins also exhibit antimicrobial activity. More than 70 years ago, Miller et al. demonstrated that histones, in addition to their important transcriptional regulatory roles, exhibit antibiotic function (22). Since then, antimicrobial activity has been found for several histones, histone fragments, or histone-like proteins from different species and tissues, including the human intestine (23–26). Furthermore, the nuclear protein high-mobility-group box 1 (HMGB1) was identified as an important player in inflammatory reactions (27). HMGB1 was originally described as a transcriptional regulator of several genes through various mechanisms, including direct interactions with nucleosomes and transcription factors (28). However, increasing evidence points to additional important functions of HMGB1 outside the nucleus. Recently, HMGB1 was isolated from human adenoid glands and testis tissue and was identified as a protein with antimicrobial activity (29, 30). In addition, Yanai and colleagues have shown that proteins of the HMGB family act as sentinels for immunogenic nucleic acids in the cytosol and are involved in the activation of Toll-like receptors by nucleic acids (31). HMGB1 can be passively released from necrotic cells, but it can also be actively secreted, which has been described for macrophages and monocytes (32–34).
In this study, we aimed to identify as-yet-unknown antimicrobial peptides using a biological functional approach. Using noninflamed colonic tissue, we isolated high-mobility-group box 2 (HMGB2) as a competent and broadly antimicrobial host defense molecule in gastrointestinal (GI) epithelial tissues.
MATERIALS AND METHODS
Bacterial and fungal strains.
The bacterial strains used were Escherichia coli K-12, Bifidobacterium adolescentis Ni3,29c (clinical isolate), Bifidobacterium breve PZ 1343 (from probiotic VSL#3), Lactobacillus acidophilus PZ 1130 (clinical isolate), Lactobacillus fermentum PZ 1162 (clinical isolate), Bacteroides vulgatus DSM 1447 (from the German Collection of Microorganisms and Cell Cultures [DSMZ]), Streptococcus salivarius subsp. thermophilus DSM 20617 (DSMZ), and Bifidobacterium longum DSM 20219T (DSMZ). In addition, Escherichia coli ATCC 25922, Pseudomonas aeruginosa ATCC 27853, Enterococcus faecalis ATCC 29212, and Candida albicans ATCC 10231 (pathogenic strains were provided by the Department for Laboratory Medicine, Robert Bosch Hospital Stuttgart, Stuttgart, Germany) were used.
Surgical and stool specimens.
Human surgical specimens were obtained during colectomy at Robert Bosch Hospital Stuttgart. Stool specimens were also taken at Robert Bosch Hospital Stuttgart. Patients gave written consent, and the study was approved by the local ethical committee at the University of Tübingen, Germany.
Protein extraction from intestinal tissue.
Human mucosal sigma tissue was disrupted by a ball mill, mixed with ice-cold extraction buffer (60% [vol/vol] acetonitrile and 1% [vol/vol] trifluoroacetic acid [TFA] in water), and incubated for 1.5 h on ice. Samples were centrifuged at 16,000 × g, the supernatant was dried by vacuum centrifugation, and the sediment was dissolved in 10% (vol/vol) ethanol. After a second centrifugation step, the pH of the supernatant was adjusted to 8.0 and then analyzed by high-performance liquid chromatography (HPLC) on an Agilent 1200 series system (Agilent, USA). To enrich positively charged proteins, samples were concentrated on a HiTrap Heparin HP 1-ml column (GE Healthcare, USA) in 0.01 M Tris–0.01 M citric acid in water (pH 8.0). Cationic proteins were eluted with 0.01 M Tris–0.01 M citric acid–2 M sodium chloride (NaCl) in water (pH 8.0). The eluate was dried by vacuum centrifugation, the sediment was dissolved in 10% (vol/vol) ethanol, and the pH was adjusted to 4.0. The cationic protein fraction was then further analyzed by HPLC using a Vydac 218TP-C18 column (250 by 4.6 mm, 5 μm; Grace, USA) with a flow rate of 0.5 ml/min. The gradient was increased from 95% buffer A (0.18% [vol/vol] TFA in water) and 5% buffer B (0.15% [vol/vol] TFA in acetonitrile) to 52% buffer B in 94 min. Fractions with a volume of 0.6 ml were collected, dried, and dissolved in 10% (vol/vol) ethanol.
Protein extraction from stool samples.
Stool samples were obtained from control individuals and Crohn's disease and ulcerative colitis (UC) patients. Five hundred milligrams of stool sample was mixed with 1,500 μl extraction buffer (60% [vol/vol] acetonitrile, 1% [vol/vol] TFA in water) and incubated for 2 h on ice. Samples were centrifuged at 16,000 × g for 20 min. One thousand microliters of supernatant was dried by vacuum centrifugation, and the sediment was dissolved in 100 μl water.
Radial diffusion assay.
For measurement of antibacterial activity, a classical or modified radial diffusion assay (RDA) was used as previously described (35, 36). Briefly, E. coli K-12, E. coli ATCC 25922, P. aeruginosa ATCC 27853, E. faecalis ATCC 29212, and C. albicans ATCC 10231 were grown on Columbia blood agar plates for 18 to 24 h at 37°C, while anaerobic bacteria were grown for up to 48 h in an anaerobic jar (AnaeroGen; Oxoid, USA). Bacteria and fungi were then inoculated in liquid Trypticase soy broth (TSB) medium and incubated for 16 to 20 h at 37°C. Cells were washed and diluted to an attenuance value (optical density at 620 nm [OD620]) of 0.1, of which 300 μl of lactobacilli and 150 μl of all other anaerobic bacterial strains and pathogenic strains (P. aeruginosa, E. faecalis, and C. albicans) were used. For E. coli strains, about 4 × 105 CFU/ml were used. Antimicrobial testing was carried out in 10 mM sodium phosphate buffer containing 0.3 mg/ml of TSB powder and 1% (wt/vol) low-electroendoosmosis (EEO)-agarose (AppliChem, Germany) under aerobic (E. coli strains, P. aeruginosa, E. faecalis, and C. albicans) or anaerobic (other bacterial strains) conditions with lysozyme (Sigma-Aldrich, USA); recombinantly expressed HMGB2, A-box, B-box, acidic-tail, or synthetic carboxy-terminal peptide (EMC Microcollections, Germany); or HPLC-purified tissue extracts. The pH was adjusted to 5.7 or 7.4. To generate a reducing environment, 2 mM dithiothreitol (DTT) was added. After 3 h of incubation, an overlay gel containing 6% (wt/vol) TSB powder, 1% (wt/vol) low-EEO-agarose, and 10 mM sodium phosphate buffer was applied onto the first gel. After 18 to 20 h of incubation, the inhibition zone diameter was measured. All experiments were repeated at least 3 times.
Tricine-sodium dodecyl sulfate (SDS) gel electrophoresis and silver staining.
HPLC-purified tissue fractions showing antimicrobial activity were further analyzed. Twenty percent of each fraction was mixed with sample buffer (4% [wt/vol] SDS, 6 M urea, 12% [wt/vol] glycerol, 50 mM Tris, 0.01% [wt/vol] Serva Blue G [pH 6.8]), electrophoresis was done with Tricine-SDS gels (3.9% stacking gel, 9.7% spacer gel, and 21.4% separation gel) under nonreducing conditions according to methods described previously by Schägger et al. (37), and gels were visualized by silver staining.
Peptide mass fingerprinting.
For antimicrobially active fractions, protein bands were cut out of the gel and washed twice with peptide mass fingerprinting (PMF) buffer 1 (50 mM ammonium bicarbonate in water) and PMF buffer 2 (25 mM ammonium bicarbonate and 50% [vol/vol] acetonitrile in water). Gel slices were reduced with 10 mM DTT in PMF buffer 1, subsequently alkylated with 5 mM iodoacetamide in PMF buffer 1, washed, and dried by vacuum centrifugation. The in-gel digest was performed with 62.5 ng trypsin (V511; Promega, USA) in PMF buffer 1 at 37°C for 18 h. The peptides were eluted from the gel slices with 0.1% (vol/vol) TFA in water at room temperature for 45 min. Fragments were purified, concentrated, and cocrystallized with α-cyano-4-hydroxy cinnamic acid. Fragments were then analyzed by matrix-assisted laser desorption ionization–time of flight mass spectrometry (Ultraflex TOF/TOF; Bruker, Germany), and proteins were identified by PMF using Mascot (Matrix Science) and the NCBI database.
Determination of the MIC.
An assay to determine the MIC, slightly modified from a method described previously (38), was used. E. coli K-12 cells were grown on Columbia blood agar plates for 16 h at 37°C. Three to five colonies were used to inoculate TSB medium. After 2.5 to 3 h, cells were washed with 10 mM sodium phosphate containing 1% (wt/vol) TSB (pH 7.4) and diluted to an OD600 of 0.4. The bacterial suspension was diluted 1:100 with 10 mM sodium phosphate containing 1% (wt/vol) TSB (pH 7.4). Fifty microliters of water containing various concentrations of recombinant HMGB2 (0.6 μg/ml to 320 μg/ml) was mixed with 50 μl of the bacterial suspension to give approximately 5 × 105 CFU/ml. The sterility control contained 100 μl of 10 mM sodium phosphate containing 1% (wt/vol) TSB (pH 7.4), and the growth control contained 50 μl water mixed with 50 μl of the bacterial suspension. After 2 h of incubation at 37°C, 100 μl of 2× TSB was added. Dilutions of 1:100 and 1:1,000 were generated with water; 100 μl was plated onto Luria broth (LB) plates; and after 18 h of incubation at 37°C, colonies were counted. In the case of a 1:100 dilution, CFU/ml was calculated by using the equation CFU/ml = colonies × 10/10−2. In the case of a 1:1,000 dilution, CFU/ml was calculated by using the equation CFU/ml = colonies × 10/10−3. CFU/ml values were plotted against concentrations in μg/ml. The experiment was repeated three times.
Construction of expression vectors.
For recombinant expression, the gene sequence of human HMGB2 (NCBI database accession number NM_002129.3) was optimized for E. coli K-12 codon usage (Eurofins MWG Operon, Germany) (see Table S1 in the supplemental material) (designated rHMGB2 below), cloned into the pCR2.1 vector via TOPO-TA cloning, and amplified with E. coli TOP10 (Eurofins MWG Operon). This construct was used as the template for subcloning of HMGB2, A-box, B-box, and acidic-tail peptides by PCR with specific primers (see Table S2 in the supplemental material) and Taq polymerase. PCR products were ligated into the pSUMO3 vector (Invitrogen, USA) with T4 DNA ligase. The expression constructs were transformed into Mach1-T1 cells (Invitrogen), amplified, and confirmed by sequencing.
Recombinant expression of HMGB2, A-box, B-box, and acidic-tail proteins.
Recombinant expression of HMGB2, A-box, B-box, and acidic-tail proteins was performed by using the Champion pET SUMO protein expression system (Invitrogen). Based on the characteristics of the vector, the protein of interest was expressed as a fusion protein with the recognition site of the SUMO protease and an amino-terminal His tag. The constructed plasmids pSUMO3-rHMGB2, pSUMO3-A-Box, pSUMO3-B-Box, and pSUMO3-Acidic-tail were transformed into BL21(DE3) competent cells (Invitrogen) and plated onto LB plates containing kanamycin (50 μg/ml). The transformants were incubated at 37°C with shaking (250 rpm) in LB medium with kanamycin (50 μg/ml) for 16 to 20 h. The culture was then diluted 1:20 with LB medium (kanamycin [50 μg/ml]) and grown to an OD600 of 0.4 to 0.6, and protein expression was induced with 1 mM isopropyl-beta-d-thio-galactopyranoside. After 5 to 6 h, cells were harvested and resuspended in lysis buffer (50 mM potassium phosphate, 400 mM NaCl, 100 mM potassium chloride [KCl], 10% [vol/vol] glycerol, 0.5% [vol/vol] Triton X-100, 10 mM imidazole [pH 7.8]). The disruption of the cells was performed by 3 cycles of sonication on ice. After centrifugation at 16,000 × g at 4°C for 15 min, the supernatant was analyzed with a HisTrap HP column (GE Healthcare) with LEW buffer (50 mM monosodium phosphate [NaH2PO4], 300 mM NaCl, 25 mM imidazole [pH 7.8]). The polyhistidine-tagged fusion protein was then eluted with elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole [pH 8.0]) and dialyzed against cold phosphate-buffered saline (PBS). The fusion protein concentration was determined by using a NanoDrop 2000 spectrophotometer (Thermo Scientific, USA) at an A280 with the protein weight and extinction coefficient (see Table S3 in the supplemental material). The protein was then digested with SUMO protease 2 (LifeSensors, USA) at a ratio of 1 U protease to 125 to 250 μg fusion protein at 30°C for 16 to 18 h to cleave off the SUMO fusion protein and polyhistidine tail. Based on the characteristics of the vector, the protein of interest had no additional amino acids after the cleavage. To obtain pure recombinant protein, a second purification step was carried out with a HisTrap HP column, followed by a final purification step by HPLC (Vydac 218TP-C18 column, 250 by 4.6 mm, 5 μm; Grace) with a gradient of 5% buffer B to 52% buffer B in 50 min (buffer A, 0.18% [vol/vol] TFA in water; buffer B, 0.15% [vol/vol] TFA in acetonitrile). The fraction containing HMGB2, A-box, B-box, or acidic-tail protein was dried by vacuum centrifugation and dissolved in water. The protein concentration of HMGB2 was determined by using a NanoDrop 2000 spectrophotometer at an A280 with the calculated protein weight and extinction coefficient (see Table S3 in the supplemental material). The purity of HMGB2 was verified by SDS-PAGE with Coomassie staining and Western blotting. To monitor the size of the expressed protein, a Precision Plus Protein Kaleidoscope standard (Bio-Rad, USA) was used. Due to inappropriate amino acid composition of the acidic-tail protein, the concentrations of the A-box, B-box, and acidic-tail proteins were measured by using a bicinchoninic acid assay (39). The purity was also checked by SDS-PAGE with Coomassie staining. Amino acid sequences of all expressed proteins are shown in Table S4 in the supplemental material.
Western blotting.
One hundred micrograms of intestinal tissue extract was mixed with sample buffer (see the section on Tricine-SDS electrophoresis above), boiled for 5 min, and electrophoresed in a 12% SDS-polyacrylamide gel. The proteins were then transferred onto a Protran nitrocellulose membrane (Whatman, USA), washed with PBS and 0.1% (vol/vol) Tween 20 (PBST), blocked for 1 h in 3% (wt/vol) bovine serum albumin (BSA) in PBST, and incubated for 16 h at 4°C in 1% (wt/vol) BSA in PBST containing 0.042 μg/ml polyclonal rabbit HMGB2 antibody (ab11973; Abcam, United Kingdom). The membrane was washed and incubated for 1 h in 1% (wt/vol) nonfat powdered milk in PBST containing a 1:10,000 dilution of goat anti-rabbit IgG horseradish peroxidase (HRP)-conjugated secondary antibody (Jackson Immuno Research, USA). This was followed by 6 washing steps with PBST and 5 min of incubation with Super Signal West Dura extended-duration substrate (Thermo Scientific, USA). For visualization, a LAS-1000 cooled camera imaging system (Fujifilm, Japan) was used. Densitometric analyses were done with AIDA evaluation software (Raytest, Germany).
Immunodot blotting.
One microliter of supernatant of cells from stimulation experiments (Caco2/TC7 cells or peripheral blood mononuclear cells [PBMCs]) or stool extracts was pipetted onto a Protran nitrocellulose membrane (Whatman). To determine the HMGB2 concentration in stool extracts, various amounts of recombinant HMGB2 were also applied onto the membrane and used as a standard. After 10 min of incubation, the membrane was washed for 10 min with PBST. The membrane was then treated like the Western blot membrane after the first washing step.
High-performance liquid chromatography analysis.
To analyze the purity of the recombinantly expressed A-box, B-box, and acidic-tail proteins, 15 μg of each peptide was analyzed by HPLC using a Vydac 218TP-C18 column (250 by 4.6 mm, 5 μm; Grace) and gradients from 5% buffer B to 52% buffer B in 50 min and from 52% buffer B to 95% buffer B in 5 min (buffer A, 0.18% [vol/vol] TFA in water; buffer B, 0.15% [vol/vol] TFA in acetonitrile).
Real-time PCR.
Intestinal biopsy specimens were taken at Robert Bosch Hospital (Stuttgart, Germany). After TRIzol extraction according to the manufacturer's protocol (Invitrogen), the total RNA was checked for quality and transcribed into cDNA by using oligo(dT) primers and the avian myeloblastosis virus (AMV) reverse transcriptase kit according to the manufacturer's protocol (Promega). cDNA corresponding to 10 ng of RNA served as the template for specific oligonucleotide primer pairs (see Table S2 in the supplemental material). mRNA copy numbers were measured by real-time PCR using a LightCycler 480 instrument (Roche Diagnostics, Germany). For quantification, an internal plasmid standard was used, as previously described (40).
Immunohistochemistry.
For HMGB2 immunostaining, we utilized a two-step immunoperoxidase procedure (EnVision; Dako, Denmark). Paraffin slides (3 μm) were deparaffinized and rehydrated. Antigen retrieval was performed for 30 min in a steamer with 0.01 M citrate buffer (pH 9.0). This was followed by a 10-min blocking step with Endogene peroxidase blocking solution (Dako) and immunohistochemical staining with polyclonal rabbit HMGB2 antibody (0.21 μg/ml) (ab11973; Abcam) in TBST (20 mM Tris base [pH 7.4], 0.14 M NaCl, 2.5 mM KCl, 0.1% Tween 20) for 1 h at room temperature. To uncover HMGB2, a second HRP-labeled antibody (Dako) was used and detected with 3′-diaminobenzidine tetrahydrochloride (Dako). Hematoxylin was used for counterstaining.
Cell culture experiments.
The colon cell line Caco2/TC7 (kindly provided by Oliver Burk) was cultivated in Dulbecco's modified Eagle medium (DMEM; Gibco Life Technologies, Germany) in a humidified atmosphere at 37°C in 5% CO2. Fetal calf serum (FCS) (10% [vol/vol]; PAA Laboratories, Austria), 1% (vol/vol) penicillin-streptomycin (Gibco Life Technologies), 1% (vol/vol) nonessential amino acids (NEAA) (Gibco Life Technologies), and 1% (vol/vol) sodium pyruvate (NaP) (Gibco Life Technologies) were added. For stimulation experiments, 45,000 cells were seeded per well into 24-well culture plates (Thermo Scientific, USA). After 24 h, cells were washed with PBS (Gibco Life Technologies) and incubated in DMEM with 1% (vol/vol) NEAA (Gibco Life Technologies) and 1% (vol/vol) NaP (Gibco Life Technologies) for 20 h. After 20 h, cells were washed with PBS (Gibco Life Technologies) and 500 μl DMEM with 1% (vol/vol) NEAA (Gibco Life Technologies), and 1% (vol/vol) NaP (Gibco Life Technologies) containing either 50 ng/ml interleukin-1β (IL-1β) (Sigma, USA), interleukin-4 (ImmunoTools, Germany), interleukin-13 (Sigma, USA), interleukin-22 (ImmunoTools), tumor necrosis factor alpha (TNF-α) (Sigma), or gamma interferon (IFN-γ) (Abcam) was added. Control cells were treated with DMEM with identical supplements, excluding cytokines. After 3 h, 24 h, and 48 h, supernatants of two wells under the same conditions were combined and dried by vacuum centrifugation. The sediment was dissolved in 100 μl water and used for immunodot blotting.
Isolation and stimulation of peripheral blood mononuclear cells.
Fifty milliliters of blood in S-Monovette tubes (S-Monovette, Sarstedt, Germany) was combined and mixed 1:1 with PBS (Gibco Life Technologies). Biocoll separating solution (Biochrom, Germany) was filled in a tube, carefully overlaid with a blood-PBS mixture, and subsequently centrifuged at 300 × g for 25 min without break. PBMCs were collected from the interphase of PBS, combined in a new tube, and centrifuged at 300 × g for 5 min. The pellet was dissolved in 10 ml PBS (Gibco Life Technologies), followed by centrifugation at 300 × g for 5 min. To culture the PBMCs, 500 ml of RPMI 1640 medium (Gibco Life Technologies) was supplemented with 10% (vol/vol) FCS (PAA Laboratories), 0.1 g/liter penicillin-streptomycin (Gibco Life Technologies), 10 mM HEPES (pH 7.4) (Merck, Germany), 2 mM l-glutamine (Biochrom), 0.13 mM l-asparagine (Serva Electrophoresis, Germany), 0.05 mM 2-mercaptoethanol (Merck), 1 mM NaP (Gibco Life Technologies), and 3 ml 100× nonessential amino acids (Biochrom), and the pellet was dissolved in 60 ml of this medium. Finally, 2.5 μg phytohemagglutinin L (Roche, Germany) per ml was added, and PBMCs were cultivated in a humidified atmosphere at 37°C in 5% CO2. After 48 h or 72 h, cells were centrifuged at 2,500 × g, and the supernatant was used for immunodot blotting.
RESULTS
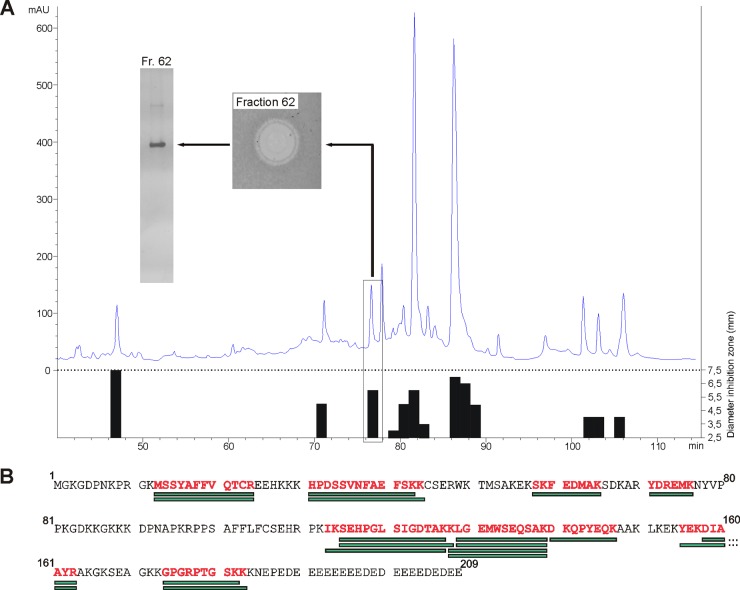
The human intestine is permanently challenged by large amounts of microorganisms. Hence, an effective antimicrobial barrier is crucial in maintaining a beneficial homeostasis toward a diverse microbial community to prevent translocation of microorganisms into the intestinal mucosa. Although various and diverse antimicrobial peptides that protect the gut epithelium have already been identified, we hypothesized that additional as-yet-unidentified proteins are also involved in this defense. Therefore, our aim was to identify new AMPs from intestinal tissue. We extracted proteins from human sigmoid mucosa and separated cationic from noncationic proteins by cation-exchange chromatography. Positively charged proteins and peptides were further purified by reversed-phase HPLC based on their hydrophobicity. As a first selection based on antimicrobial function, the fractions were screened for activity against the E. coli K-12 laboratory strain by a radial diffusion assay (35). Of 100 fractions, 13 showed considerable inhibition of bacterial growth of E. coli K-12 (Fig. 1A). To investigate if the observed activity is caused by single proteins or a protein mixture, we checked the purity of the active fractions by Tricine-SDS gels and silver staining. If fractions contained one single protein, it was likely that this protein was responsible for the observed antimicrobial activity. In such cases or when only a few bands were detected by staining, they were cut out of the gel, digested with the protease trypsin, and analyzed by matrix-assisted laser desorption ionization mass spectrometry using peptide mass fingerprinting. This workflow led to the identification of histones (H1b, H1d, H2a, and H2b), RNase 3, ribosomal protein S19, and high-mobility-group box 1 (data not shown), for which antimicrobial activity has already been described (24, 26, 29, 30, 41, 42). Of note, we did not identify defensins in antimicrobially active fractions. A possible explanation for this is that some bands that could not be identified by PMF contained defensins or that defensins were present primarily in the fractions with multiple peptide bands and subsequently were not analyzed. In one antimicrobially active fraction that eluted after 76 min at about 32.9% acetonitrile (Fig. 1A), we detected two protein bands that could both be identified via PMF as HMGB2 (Fig. 1B). This finding established the hypothesis that HMGB2 has antimicrobial activity, which is a new biological function of this protein.
Fig 1.
(A) Purification of HMGB2 from colonic mucosal extracts. Mucosa extract was separated by reverse-phase HPLC, and the antimicrobial activity of collected protein fractions against Escherichia coli K-12 was tested. Antimicrobial activity determined by RDA is illustrated as an inhibition zone diameter (black bars). HMGB2 containing fraction 62 is marked with a frame; the antimicrobial inhibition zone and gel electrophoresis run of this fraction are shown. mAU, milli-absorbance unit. (B) Amino acid sequence of HMGB2 and results of PMF analysis of fraction 62. Green boxes represent peptide masses which were calculated from detected masses and could be assigned to theoretical peptide fragments of HMGB2. Red letters indicate sequences of HMGB2 that could be covered by experimentally obtained fragments.
Characterization of the antimicrobial activity spectrum of HMGB2.
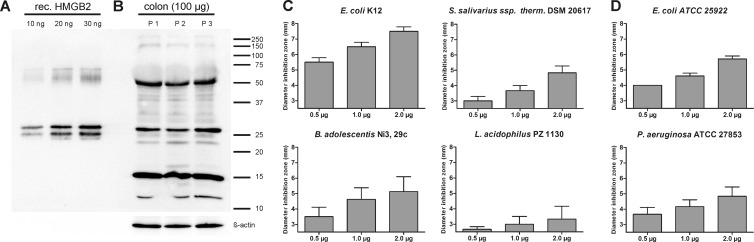
To verify the antimicrobial activity of HMGB2 and to investigate its antimicrobial activity in more detail, we expressed the protein recombinantly in E. coli with a pET-SUMO expression system. The purity of HMGB2 was checked by polyacrylamide gels and Coomassie staining. Additionally, we performed Western blotting to verify the presence of HMGB2 with a specific antibody (Fig. 2A). Recombinantly expressed HMGB2 showed a double band at around 25 kDa (expected size, 24.03 kDa) (UniProtKB accession number P26583), probably due to different HMGB2 variants or posttranslational modifications. We also detected a double band at around 50 kDa, probably caused by a dimeric form, which has already been described for HMGB1 (43). HMGB2, similar to HMGB1, has three cysteine residues and can thus most likely dimerize via intermolecular disulfide bridges.
Fig 2.
(A) Western blot analysis of recombinantly expressed HMGB2 separated by a 12% SDS-polyacrylamide gel and treated with HMGB2-specific antibody. Ten, 20, and 30 ng of recombinant peptide were analyzed. (B) Western blot analysis of colonic mucosal protein extracts from 3 patients separated by a 12% SDS-polyacrylamide gel and treated with HMGB2-specific antibody. Equal loading was verified with β-actin antibody after stripping. (C) Antimicrobial activity of 0.5, 1, and 2 μg of recombinantly expressed HMGB2 measured by RDA against Escherichia coli K-12, Streptococcus salivarius subsp. thermophilus, Bifidobacterium adolescentis, and Lactobacillus acidophilus. The diameter of inhibition zones is shown; experiments were repeated at least three times. Data are presented as means, and error bars indicate standard errors of the means. The baseline at 2.5 mm represents the diameter of a sample well, corresponding to no antimicrobial activity. (D) Antimicrobial activity of 0.5, 1, and 2 μg of recombinantly expressed HMGB2 measured by RDA against Escherichia coli ATCC 25922 and Pseudomonas aeruginosa. The diameter of inhibition zones is shown; experiments were repeated at least three times. Data are presented as means, and error bars indicate standard errors of the means. The baseline at 2.5 mm represents the diameter of a sample well, corresponding to no antimicrobial activity.
To confirm the presence of HMGB2 protein in colonic tissue, we analyzed intestinal sigma extract by Western blotting and detected HMGB2 at around 25 kDa and the dimerization product at 50 kDa (Fig. 2B), which is in accordance with the recombinantly expressed protein. In addition, two bands, potentially degradation products, were detected at around 12 kDa and 15 kDa. To characterize the antimicrobial activity of HMGB2, we tested the recombinantly produced protein against several bacterial strains that are part of the normal human microbiota. We found activity against E. coli K-12, B. adolescentis, and S. salivarius subsp. thermophilus, while activity against L. acidophilus was rather weak but still detectable (Fig. 2C). All growth-inhibitory effects were concentration dependent. For bacterial strains of L. fermentum, B. vulgatus, B. longum, and B. breve, we did not detect any growth-inhibitory effect at the tested concentrations of HMGB2 (data not shown). To analyze if HMGB2 also exhibits antimicrobial activity against pathogenic bacteria and fungi, several strains were treated with the recombinantly expressed protein. We detected inhibition zones for E. coli ATCC 25922 and P. aeruginosa (Fig. 2D), whereas no activity against E. faecalis was detected (data not shown). C. albicans could be inhibited only if large amounts (10 μg) of HMGB2 were used (data not shown).
Further characterization of the antimicrobial activity of HMGB2.
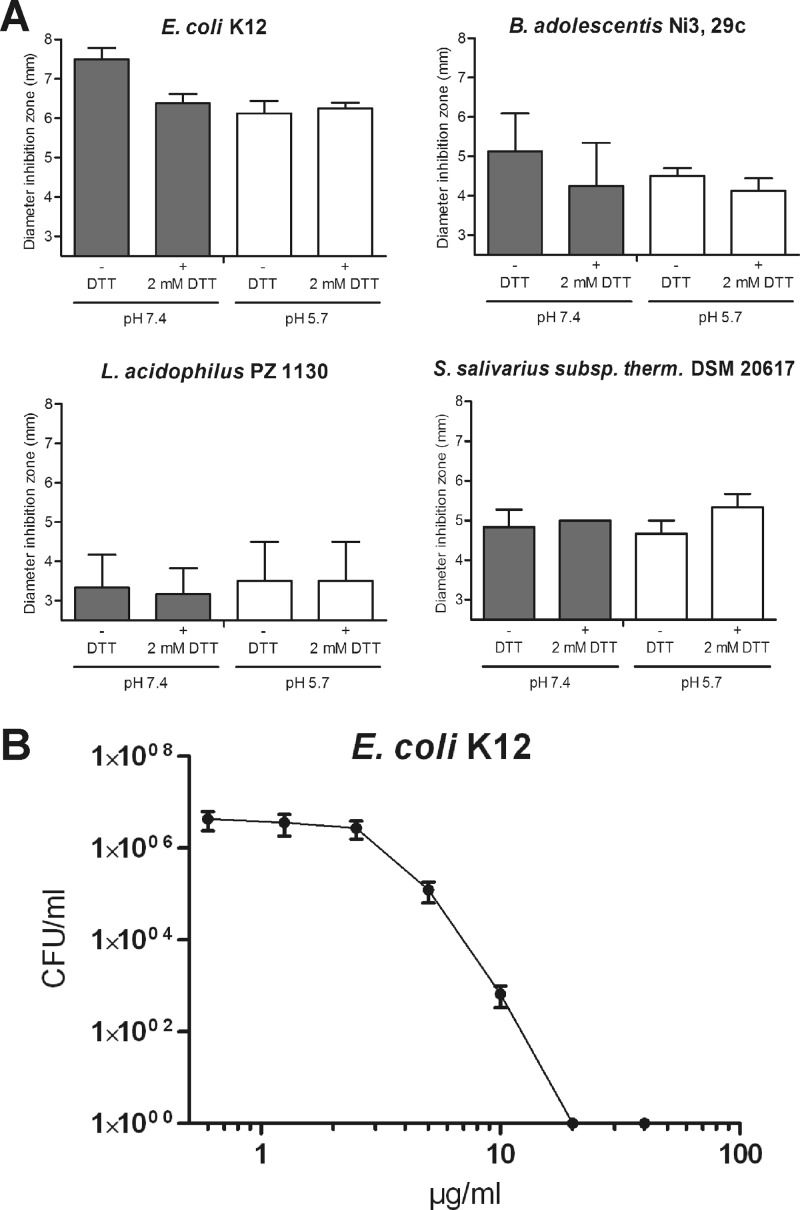
The human intestine is characterized by different milieu conditions with various pHs and redox potentials (44). Our group recently demonstrated how a reducing environment can strongly influence the antimicrobial activity of a specific defensin (36, 45). In detail, we found that human β-defensin 1 becomes active against bifidobacteria, lactobacilli, and the opportunistic fungus Candida albicans under reducing conditions. Hence, we studied whether changes in pH and the generation of a reducing environment affect the antibacterial activity of HMGB2. In modified radial diffusion assays, we tested 2 μg of protein under standard conditions (pH 7.4), reducing conditions (pH 7.4 and 2 mM DTT), acidic conditions (pH 5.7), and acidic, reducing conditions (pH 5.7 and 2 mM DTT) against several bacterial strains (Fig. 3A). However, altering these environmental conditions did not lead to evident changes in the activity of HMGB2. Only for E. coli did we find slightly decreased activity under conditions different from those of the standard environment.
Fig 3.
(A) Antimicrobial activity of 2 μg of recombinantly expressed HMGB2 measured by RDA against Escherichia coli K-12, Bifidobacterium adolescentis, Lactobacillus fermentum, and Streptococcus salivarius subsp. thermophilus at pH 7.4 and pH 5.7 under reducing (+ 2 mM DTT) and nonreducing (− DTT) conditions in growth medium. The diameter of inhibition zones is shown; experiments were repeated at least three times. Data are presented as means, and error bars indicate standard errors of the means. The baseline at 2.5 mm represents the diameter of a sample well, corresponding to no antimicrobial activity. (B) Determination of the MIC of recombinantly expressed HMGB2 against Escherichia coli K-12. Bacteria and protein were incubated and plated onto agar plates. After 18 h of incubation, CFU/ml were determined and plotted against the HMGB2 concentration. Data are presented as means of three experiments, and error bars indicate standard errors of the means.
To confirm the antimicrobial activity of HMGB2 obtained by RDA, we used an assay to determine the MIC. E. coli K-12 bacteria were incubated with HMGB2 and plated onto agar plates. After an overnight incubation, the colonies were counted, and the CFU/ml was calculated. A concentration-dependent activity of the protein was observed, validating that HMGB2 can indeed inhibit bacterial growth of E. coli K-12. A concentration of 832 nM (20 μg/ml) HMGB2 was sufficient for total repression of bacterial growth.
A-box and B-box proteins are crucial for the antimicrobial activity of HMGB2.
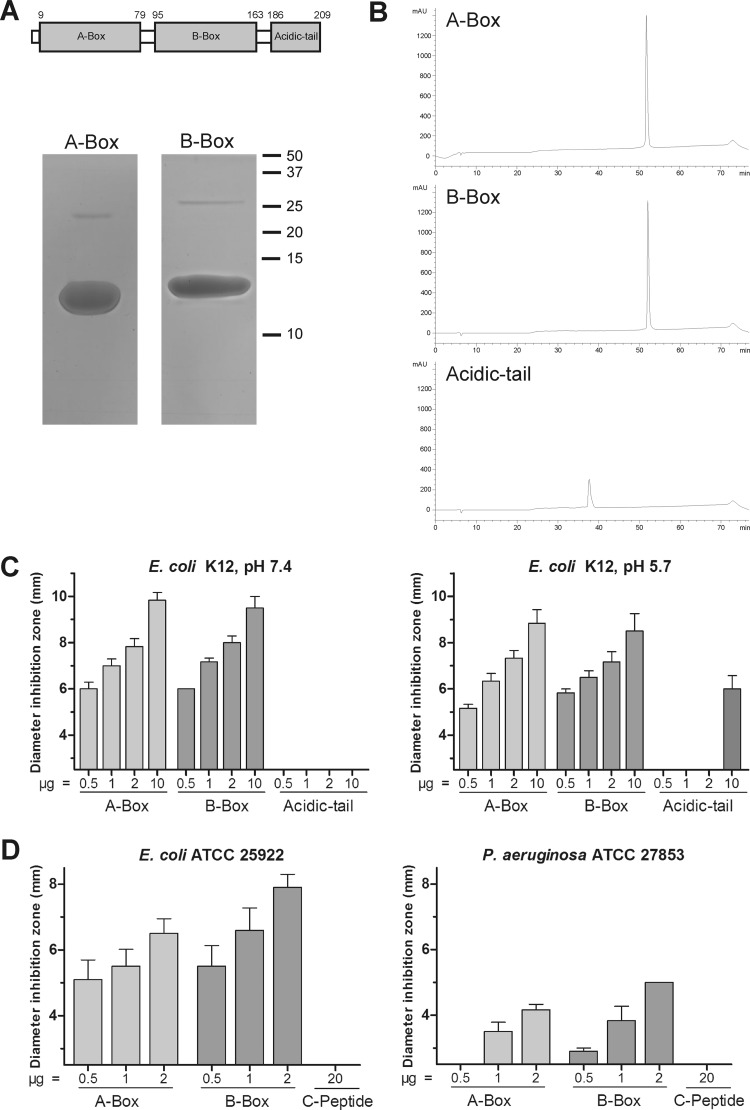
As we identified antimicrobial activity as a novel function of HMGB2, we were interested to determine which part of the protein is important for this function. To address this question, we expressed the two DNA-binding domains HMG box A (A box) and HMG box B (B box) and the acidic carboxy-terminal tail (acidic tail) recombinantly (Fig. 4A). After purification by HPLC, the peptides were analyzed by SDS-PAGE and Coomassie staining. A-box and B-box proteins could be detected at approximately 12 kDa and 13 kDa (expected, 9.8 and 10.6 kDa; ExPASy ProtParam) (Fig. 4A). The acidic-tail peptide could not be visualized by Coomassie staining, probably due to the extraordinary amino acid composition (see Table S4 in the supplemental material). The peptide contains mainly acidic amino acids (aspartic acid and glutamic acid), which could explain the detection problems with this staining method. Nonetheless, we analyzed all three peptides by HPLC and confirmed their purity (Fig. 4B).
Fig 4.
(A) Schematic model of HMGB2. (Top) Numbers indicate amino acid positions at the beginning and end of the domains. (Bottom) Five micrograms of recombinantly expressed A-box and B-box peptides was separated by 18% SDS-polyacrylamide gels and stained with Coomassie. (B) Fifteen micrograms of recombinantly expressed A-box, B-box, and acidic-tail peptides was separated by reverse-phase HPLC. Absorbance was monitored at 214 nm. (C) Antimicrobial activity of various concentrations of recombinantly expressed A-box, B-box, and acidic-tail peptides measured by RDA at pH 7.4 and pH 5.7 against Escherichia coli K-12. Experiments were repeated at least three times. Data are presented as means; error bars indicate standard errors of the means. The baseline at 2.5 mm represents the diameter of sample wells, corresponding to no antimicrobial activity. (D) Antimicrobial activity of various concentrations of recombinantly expressed A-box and B-box peptides and synthetic C-peptide measured by RDA at pH 7.4 against Escherichia coli ATCC 25922 and Pseudomonas aeruginosa. Experiments were repeated three times. The baseline at 2.5 mm represents the diameter of sample wells.
The recombinantly expressed A-box, B-box, and acidic-tail peptides were used for antimicrobial activity assays against E. coli K-12 at pH 7.4 and 5.7 (Fig. 4B). A-box and B-box peptides showed concentration-dependent antimicrobial activity and effectively inhibited growth of E. coli K-12 at pH 7.4 and 5.7. In contrast, the acidic-tail peptide did not show any activity at pH 7.4 and was active only at amounts of 10 μg at pH 5.7. To exclude potential errors during detection and determination of the concentration of the acidic-tail peptide, we also tested a chemically synthesized peptide, which contained the last 22 amino acids of the carboxy terminus (C-peptide) of HMGB2. The C-peptide did not show any antimicrobial activity against E. coli K-12, E. coli ATCC 25922, and P. aeruginosa, whereas the A-box and B-box peptides exhibited growth-inhibitory effects (Fig. 4C and D and data not shown). Consequently, both the A box and B box seem to be the crucial domains for the antimicrobial activity of HMGB2.
HMGB2 is expressed in gastric and intestinal tissue.
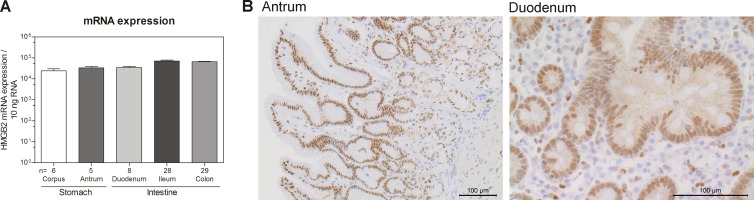
Proteins of the HMGB family were originally identified as chromatin-associated proteins from calf thymus cells (46). As we identified HMGB2 from colonic tissue extracts, we wanted to compare the expression levels of HMGB2 mRNA in different parts of the gastrointestinal (GI) tract. In the stomach, the number of HMGB2 mRNA copies per 10 ng total RNA was around 24,000 in the corpus and around 33,000 in the antrum (Fig. 5A). In the intestine, expression levels were comparable to those in gastric tissue, with around 34,000 copies in the duodenum, around 71,000 copies in the ileum, and 65,000 copies in the colon. To confirm this expression at the protein level and to investigate its localization, we performed immunohistochemical staining of gastrointestinal tissue with a specific antibody directed against HMGB2 (Fig. 5B). As expected, HMGB2 was detected in stomach and intestinal tissue and was located mainly in the nucleus. However, localization revealed moderate staining in inflammatory cells and the strongest staining in epithelial cells, which might support its function in epithelial protection. In addition to its primarily nuclear localization, in the duodenum, ileum, and colon, we observed weak staining for HMGB2 in the cytoplasm, again supporting the hypothesis of a second function of the protein aside from its role in the nucleus (Fig. 5B and data not shown).
Fig 5.
(A) mRNA transcript levels of HMGB2 per 10 ng total RNA in corpus, antrum, duodenum, ileum, and colon biopsy specimens from control individuals. Data are presented as means; error bars indicate standard errors of the means. (B) Immunohistochemical analysis of antrum and duodenum biopsy specimens from control individuals with HMGB2-specific antibody. Representative pictures are shown and were taken at ×20 and ×40 magnifications. HMGB2, high-mobility-group box 2.
HMGB2 expression is unchanged in IBD.
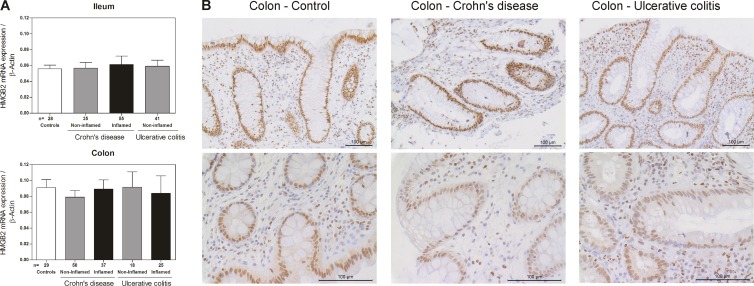
Altered expression of AMPs is associated with infectious diseases, and reduced expression of defensins is associated with inflammatory bowel diseases, especially Crohn's disease (17). Here we identified antimicrobial activity as a new function of HMGB2. To analyze whether HMGB2 expression might also be altered in patients who suffer from IBD, its mRNA level was quantified in IBD patients and controls. However, no significant mRNA differences could be detected between CD, UC, and control patients after normalization to the housekeeping gene β-actin (Fig. 6A). Similarly, no protein differences in immunohistochemical staining for HMGB2 of colonic tissues from IBD patients and controls were observed (Fig. 6B).
Fig 6.
(A) mRNA expression of HMGB2 in ileal and colonic biopsy specimens from patients with Crohn's disease and ulcerative colitis and controls. In patients, specimens were subdivided into macroscopically inflamed and noninflamed mucosa. mRNA transcript levels per 10 ng total RNA are normalized to the expression level of β-actin in the corresponding biopsy specimens. Data are presented as means; error bars indicate standard errors of the means. (B) Immunohistochemical analysis of colonic biopsy specimens from patients with Crohn's disease and ulcerative colitis and controls with HMGB2-specific antibody. Samples from Crohn's disease and ulcerative colitis patients were taken from inflamed mucosa. Representative pictures are shown and were taken at ×20 and ×40 magnifications.
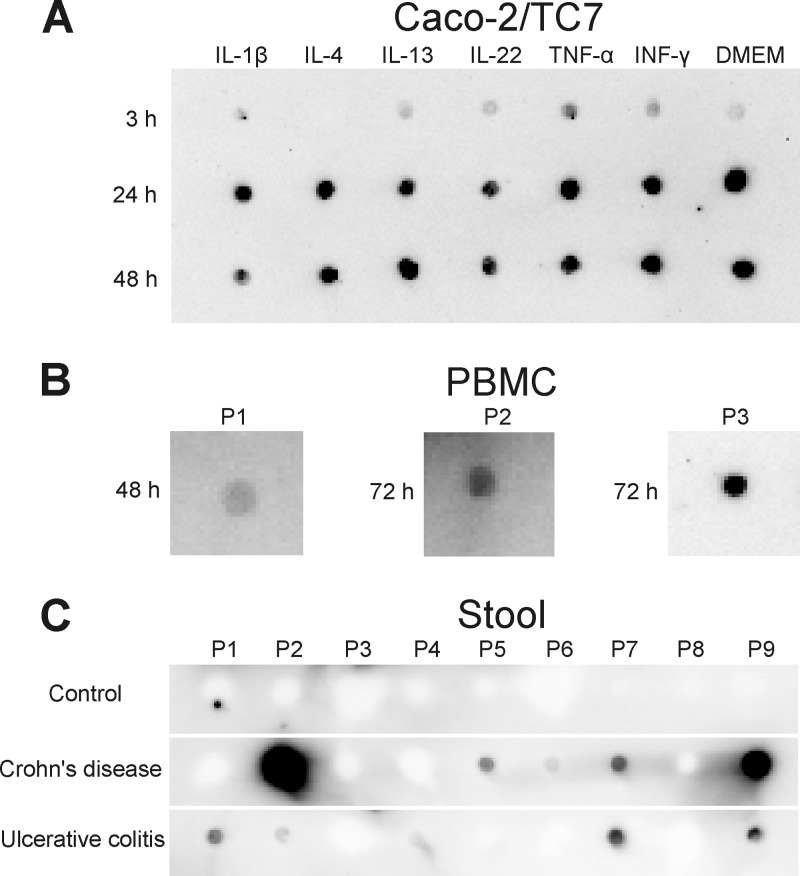
HMGB2 is known primarily as an intracellular protein, although antimicrobial defense is a process occurring extracellularly. To investigate if HMGB2 can be localized extracellularly, we investigated supernatants of the intestinal epithelial cell line Caco2/TC7 and of primary peripheral blood mononuclear cells for the presence of HMGB2. As shown in Fig. 7A and B, for both cell types, HMGB2 was detected in the supernatants. In Caco2/TC7 cell supernatants, HMGB2 was detectable independent of stimulation by cytokines, with a weak signal after 3 h and with a more intense signal after 24 h and 48 h. In addition, we detected HMGB2 in stool extracts from 5 out of 9 CD patients and 5 out of 9 UC patients but none out of the 9 analyzed control individuals (Fig. 7C). The HMGB2 amounts did not correlate with the disease severity of the patients and varied strongly. The concentration of detected HMGB2 ranged from 0.1 μg/ml protein extract in UC patients to 6.4 μg/ml protein extract in CD patients. As we have measured significant antimicrobial activity of HMGB2 at concentrations of 5 μg/ml (Fig. 3B), these experiments can be interpreted as a proof of concept that extracellular HMGB2 might indeed have a possible physiological function in antimicrobial host defense.
Fig 7.
(A) Stimulation experiments with Caco-2/TC7 cells with IL-1β, IL-4, IL-13, IL-22, TNF-α, and IFN-γ. Cells were treated with cytokines, and the control was treated only with culture medium (DMEM). After 3 h, 24 h, and 48 h, supernatants were taken, and 1 μl was analyzed by immunodot blotting with HMGB2-specific antibody. (B) Supernatants of PBMCs that were isolated from blood of 3 healthy donors and treated with phytohemagglutinin. Supernatants were taken after 48 h or 72 h, and 1 μl was analyzed by immunodot blotting with HMGB2-specific antibody. (C) One microliter of stool extracts from controls and Crohn's disease and ulcerative colitis patients analyzed by immunodot blotting with HMGB2-specific antibody. P1, patient 1.
DISCUSSION
To maintain peace in the heavily colonized human intestine, it is crucial to limit microbial growth at the mucosal surface. Besides a tight physical barrier provided by epithelial cells, antimicrobial peptides are major players in the border patrol against microorganisms. Although several AMPs have already been isolated from the intestinal mucosa, we hypothesized that there are still some that have not yet been identified. Therefore, we performed a functional screening and, as expected, identified some known AMPs (histones, RNase 3, ribosomal protein S19, and HMGB1). In addition to the previously described AMP HMGB1, we extracted and identified HMGB2 as a novel high-mobility-group box protein with potent antimicrobial activity. HMGB1 and -2 (formerly called HMG1 and HMG2, respectively) are highly conserved proteins with significant sequence similarity (>80% amino acid identity) (47). Both proteins contain two DNA-binding domains (high-mobility-group boxes A and B) and an acidic carboxy terminus (48). HMGB proteins can regulate the transcription of several genes by various mechanisms. They can interact with nucleosomes (49, 50), general transcription factors (51), and specific transcription factors like steroid hormone receptors and p53, p73, NF-κB, POU, and HOX proteins (52). Many functions of HMGB2 have been studied in mouse models. While a HMGB2 knockout leads to viable mice, with reduced fertility in male mice, a HMGB1 knockout is lethal, and mice die within 24 h after birth due to hypoglycemia (53, 54). To date, only a few studies have investigated the expression of HMGB2 in human tissues, with differing results. According to the data summarized at the GeneCards website (http://www.genecards.org/), HMGB2 expression was found in colonic tissue by using a microarray (http://www.biogps.org/). A SAGE (Serial Analysis of Gene Expression) study (http://www.cgap.nci.nih.gov/sage), on the other hand, did not detect any expression. In contrast, here, in a more targeted analysis, we detected HMGB2 expression in all studied gastrointestinal tissues.
There are only limited data available on an extranuclear role of HMGB2. In contrast, in the last years, many studies emerged which support important extranuclear functions of HMGB1. HMGB1 can be actively secreted by several cell types, including activated murine macrophages and human monocytes, via a Golgi/endoplasmic reticulum (ER)-independent pathway. A first stimulus (e.g., TNF-α) thereby leads to the acetylation of specific lysine residues and the accumulation of HMGB1 in the cytoplasm. Cytoplasmic HMGB1 can then be taken up by secretory endolysosomes (in hematopoietic cells). A second signal leads to the fusion of the endolysosomes with the cell membrane and the secretion of HMGB1 (33, 55). In addition to active secretion, the protein can be passively released by necrotic cells attracting immune cells and transmitting a damage signal to neighboring cells (32). Furthermore, HMGB1 seems to be an important player in several inflammatory conditions, including sepsis. HMGB1 levels are increased in the serum of sepsis patients as well as in a murine sepsis model (34, 56). The administration of anti-HMGB1 antibodies at a late state (24 h) could rescue mice that have already developed signs of sepsis. In addition to such proinflammatory and potentially fatal functions, HMGB1 has antimicrobial activity (29, 30). Gong and colleagues showed that amino acids 201 to 205 in the acidic carboxy-terminal region seem to be crucial for this function (57). As we identified antimicrobial activity for HMGB2, we were interested in whether the carboxy terminus of HMGB2 is also the crucial domain for this function and consequently expressed the A box, B box, and acidic tail recombinantly. In contrast to the findings of Gong et al., the acidic-tail protein did not show significant antimicrobial activity and was active only at a concentration of 10 μg at pH 5.7. A chemically synthesized peptide which consisted of the last 22 amino acids of the C terminus of HMGB2 did not show any antibacterial effects at all. Conversely, the A box and B box could effectively inhibit the growth of two E. coli strains and P. aeruginosa. Because of the high level of similarity of HMGB1 and -2, this contradictory result was surprising. However, the A-box and B-box peptides are both highly cationic (net charges of +8 for the A box and +12 for the B box), which is a characteristic feature of many known AMPs (9) and would therefore support their importance in this context.
Due to the limited available expression data for HMGB2 in human tissues, we analyzed its expression in the GI tract by real-time PCR and immunohistochemistry. Overall, we found significant amounts of both transcript and protein expression throughout the GI tract. We detected 24,000 to 33,000 copies (per 10 ng total RNA) in gastric tissue and 34,000 to 71,000 copies (per 10 ng total RNA) in intestinal tissue. Furthermore, the protein was detectable in all analyzed tissues in epithelial and inflammatory cells and was located mainly in the nucleus, which fits its important nuclear functions. In addition to its nuclear localization, HMGB2 was also present in the cytoplasm of duodenal, ileal, and colonic epithelial cells in minor amounts.
As the barrier function mediated by AMPs seems to be defective in inflammatory bowel diseases, we also analyzed whether HMGB2 expression is altered in patients who suffer from such disorders. However, no significant differences in mRNA expression levels could be detected between Crohn's disease, ulcerative colitis, and control patients. This stability of HMGB2 expression is remarkable compared to other peptides like HBD2 and suggests no major transcriptional regulation by inflammatory pathways. In addition to HMGB2, there are other proteins, such as glyceraldehyde-3-phosphate dehydrogenase, that are constitutively expressed in all kinds of tissues and also exhibit antimicrobial activity (58).
A remaining question in our study is how the antimicrobial function of HMGB2 might be linked to the defense against microorganisms in the GI tract, as the protein is located mainly in the nucleus. One point to consider is that the epithelial cells in the stomach and intestine have a high rate of turnover (59). Thus, passive release of intracellular peptides by shed cells of the constantly renewing epithelium could likely be of biological relevance in this setting. The content of dying epithelial cells, including nuclear proteins such as histones, HMGB1/2, and others, is thereby released into the lumen. Consequently, intracellular components such as HMGB2 might thus be recycled and support the actively secreted AMPs in their war against pathogenic but also commensal microorganisms. In addition to this passive release caused by permanent cell turnover, Vitali and colleagues recently demonstrated that HMGB1 can be actively secreted by cells in inflamed intestinal tissues (60). The protein was detectable in stool samples of children suffering from inflammatory bowel disease and was suggested as a new marker for IBD. In healthy cells, HMGB1 was located only in the nucleus. In inflamed tissues, however, the protein was additionally present in the cytoplasm, which is probably important for the secretion of HMGB1. Furthermore, Pusterla and coworkers showed that HMGB2 can also be secreted by myeloid cells and, like HMGB1, has chemoattractant activity (61). In line with these observations, we were interested in whether HMGB2 is also detectable in intestinal luminal contents, and we analyzed a small cohort of stool samples from IBD and control patients. While HMGB2 could not be detected in stool samples from control patients, the protein was traceable in some IBD samples, with amounts of up to 6.4 μg per ml protein extract. The antimicrobial activity assays that were executed during this study showed that at these concentrations, HMGB2 can indeed impair the growth of bacterial cells and therefore can influence the microbial composition of the gut. In addition, HMGB2 protein was evident in the supernatants of two different in vitro-cultured cell types, indicating that HMGB2 can end up being present extracellularly. The intra- and extracellular presence of this new antimicrobial protein therefore further supports its relevance in the defense against intestinal microorganisms.
As in this study, every year new findings in the field of AMPs demonstrate how the strategic search for new effector molecules still offers promising research avenues. The complex antibiotic innate defense in our gut relies on a multilayered system in which HMGB2 now represents a new player. Epithelial shedding and therefore the release and recycling of some nuclear peptides such as HMGB2 as antimicrobials could likely be functionally and potentially clinically relevant in mucosal host defense.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the Robert Bosch Foundation (Stuttgart, Germany) and the Deutsche Forschungsgemeinschaft (WE4336/5-1 and WE4336/2-1). We declare that we have no conflicts of interest.
We thank Marion Schiffmann, Michelle Katajew, Jutta Bader, and Kathleen Siegel for excellent technical help; Maureen Ostaff for inspiring discussions; and Christoph Pöhlmann for providing pathogenic bacterial and fungal strains.
Footnotes
Published ahead of print 22 July 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00805-13.
REFERENCES
- 1.Harder J, Schröder J-M, Gläser R. 2013. The skin surface as antimicrobial barrier: present concepts and future outlooks. Exp. Dermatol. 22:1–5 [DOI] [PubMed] [Google Scholar]
- 2.Gallo RL, Hooper LV. 2012. Epithelial antimicrobial defence of the skin and intestine. Nat. Rev. Immunol. 12:503–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wehkamp J, Schmid M, Stange EF. 2007. Defensins and other antimicrobial peptides in inflammatory bowel disease. Curr. Opin. Gastroenterol. 23:370–378 [DOI] [PubMed] [Google Scholar]
- 4.Ley RE, Peterson DA, Gordon JI. 2006. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124:837–848 [DOI] [PubMed] [Google Scholar]
- 5.Hecht G. 1999. Innate mechanisms of epithelial host defense: spotlight on intestine. Am. J. Physiol. 277:C351–C358 [DOI] [PubMed] [Google Scholar]
- 6.Meyer-Hoffert U, Hornef MW, Henriques-Normark B, Axelsson L-G, Midtvedt T, Pütsep K, Andersson M. 2008. Secreted enteric antimicrobial activity localises to the mucus surface layer. Gut 57:764–771 [DOI] [PubMed] [Google Scholar]
- 7.Bevins CL, Salzman NH. 2011. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat. Rev. Microbiol. 9:356–368 [DOI] [PubMed] [Google Scholar]
- 8.Ganz T. 2003. The role of antimicrobial peptides in innate immunity. Integr. Comp. Biol. 43:300–304 [DOI] [PubMed] [Google Scholar]
- 9.Zasloff M. 2002. Antimicrobial peptides of multicellular organisms. Nature 415:389–395 [DOI] [PubMed] [Google Scholar]
- 10.Gallo RL, Nizet V. 2003. Endogenous production of antimicrobial peptides in innate immunity and human disease. Curr. Allergy Asthma Rep. 3:402–409 [DOI] [PubMed] [Google Scholar]
- 11.Cederlund A, Gudmundsson GH, Agerberth B. 2011. Antimicrobial peptides important in innate immunity. FEBS J. 278:3942–3951 [DOI] [PubMed] [Google Scholar]
- 12.Harwig SS, Tan L, Qu XD, Cho Y, Eisenhauer PB, Lehrer RI. 1995. Bactericidal properties of murine intestinal phospholipase A2. J. Clin. Invest. 95:603–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kagan BL, Selsted ME, Ganz T, Lehrer RI. 1990. Antimicrobial defensin peptides form voltage-dependent ion-permeable channels in planar lipid bilayer membranes. Proc. Natl. Acad. Sci. U. S. A. 87:210–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kehl-Fie TE, Chitayat S, Hood MI, Damo S, Restrepo N, Garcia C, Munro KA, Chazin WJ, Skaar EP. 2011. Nutrient metal sequestration by calprotectin inhibits bacterial superoxide defense, enhancing neutrophil killing of Staphylococcus aureus. Cell Host Microbe 10:158–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corbin BD, Seeley EH, Raab A, Feldmann J, Miller MR, Torres VJ, Anderson KL, Dattilo BM, Dunman PM, Gerads R, Caprioli RM, Nacken W, Chazin WJ, Skaar EP. 2008. Metal chelation and inhibition of bacterial growth in tissue abscesses. Science 319:962–965 [DOI] [PubMed] [Google Scholar]
- 16.Wehkamp J, Stange EF. 2010. Paneth's disease. J. Crohns Colitis 4:523–531 [DOI] [PubMed] [Google Scholar]
- 17.Wehkamp J, Salzman NH, Porter E, Nuding S, Weichenthal M, Petras RE, Shen B, Schaeffeler E, Schwab M, Linzmeier R, Feathers RW, Chu H, Lima H, Fellermann K, Ganz T, Stange EF, Bevins CL. 2005. Reduced Paneth cell α-defensins in ileal Crohn's disease. Proc. Natl. Acad. Sci. U. S. A. 102:18129–18134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koslowski MJ, Teltschik Z, Beisner J, Schaeffeler E, Wang G, Kübler I, Gersemann M, Cooney R, Jewell D, Reinisch W, Vermeire S, Rutgeerts P, Schwab M, Stange EF, Wehkamp J. 2012. Association of a functional variant in the Wnt co-receptor LRP6 with early onset ileal Crohn's disease. PLoS Genet. 8:e1002523. 10.1371/journal.pgen.1002523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peyrin-Biroulet L, Beisner J, Wang G, Nuding S, Oommen ST, Kelly D, Parmentier-Decrucq E, Dessein R, Merour E, Chavatte P, Grandjean T, Bressenot A, Desreumaux P, Colombel J-F, Desvergne B, Stange EF, Wehkamp J, Chamaillard M. 2010. Peroxisome proliferator-activated receptor gamma activation is required for maintenance of innate antimicrobial immunity in the colon. Proc. Natl. Acad. Sci. U. S. A. 107:8772–8777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jäger S, Stange EF, Wehkamp J. 2010. Antimicrobial peptides in gastrointestinal inflammation. Int. J. Inflam. 2010:910283. 10.4061/2010/910283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prado-Montes de Oca E. 2010. Human beta-defensin 1: a restless warrior against allergies, infections and cancer. Int. J. Biochem. Cell Biol. 42:800–804 [DOI] [PubMed] [Google Scholar]
- 22.Miller BF, Abrams R, Dorfman A, Klein M. 1942. Antibacterial properties of protamine and histone. Science 96:428–430 [DOI] [PubMed] [Google Scholar]
- 23.Park CB, Kim MS, Kim SC. 1996. A novel antimicrobial peptide from Bufo bufo gargarizans. Biochem. Biophys. Res. Commun. 218:408–413 [DOI] [PubMed] [Google Scholar]
- 24.Howell SJ, Wilk D, Yadav SP, Bevins CL. 2003. Antimicrobial polypeptides of the human colonic epithelium. Peptides 24:1763–1770 [DOI] [PubMed] [Google Scholar]
- 25.Park IY, Park CB, Kim MS, Kim SC. 1998. Parasin I, an antimicrobial peptide derived from histone H2A in the catfish, Parasilurus asotus. FEBS Lett. 437:258–262 [DOI] [PubMed] [Google Scholar]
- 26.Rose FR, Bailey K, Keyte JW, Chan WC, Greenwood D, Mahida YR. 1998. Potential role of epithelial cell-derived histone H1 proteins in innate antimicrobial defense in the human gastrointestinal tract. Infect. Immun. 66:3255–3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang H, Yang H, Tracey KJ. 2004. Extracellular role of HMGB1 in inflammation and sepsis. J. Intern. Med. 255:320–331 [DOI] [PubMed] [Google Scholar]
- 28.Bianchi ME, Agresti A. 2005. HMG proteins: dynamic players in gene regulation and differentiation. Curr. Opin. Genet. Dev. 15:496–506 [DOI] [PubMed] [Google Scholar]
- 29.Zetterström CK, Bergman T, Rynnel-Dagöö B, Erlandsson Harris H, Soder O, Andersson U, Boman HG. 2002. High mobility group box chromosomal protein 1 (HMGB1) is an antibacterial factor produced by the human adenoid. Pediatr. Res. 52:148–154 [DOI] [PubMed] [Google Scholar]
- 30.Zetterström CK, Strand M-L, Söder O. 2006. The high mobility group box chromosomal protein 1 is expressed in the human and rat testis where it may function as an antibacterial factor. Hum. Reprod. 21:2801–2809 [DOI] [PubMed] [Google Scholar]
- 31.Yanai H, Ban T, Wang Z, Choi MK, Kawamura T, Negishi H, Nakasato M, Lu Y, Hangai S, Koshiba R, Savitsky D, Ronfani L, Akira S, Bianchi ME, Honda K, Tamura T, Kodama T, Taniguchi T. 2009. HMGB proteins function as universal sentinels for nucleic-acid-mediated innate immune responses. Nature 462:99–103 [DOI] [PubMed] [Google Scholar]
- 32.Scaffidi P, Misteli T, Bianchi ME. 2002. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature 418:191–195 [DOI] [PubMed] [Google Scholar]
- 33.Gardella S, Andrei C, Ferrera D, Lotti LV, Torrisi MR, Bianchi ME, Rubartelli A. 2002. The nuclear protein HMGB1 is secreted by monocytes via a non-classical, vesicle-mediated secretory pathway. EMBO Rep. 3:995–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang H, Bloom O, Zhang M, Vishnubhakat JM, Ombrellino M, Che J, Frazier A, Yang H, Ivanova S, Borovikova L, Manogue KR, Faist E, Abraham E, Andersson J, Andersson U, Molina PE, Abumrad NN, Sama A, Tracey KJ. 1999. HMG-1 as a late mediator of endotoxin lethality in mice. Science 285:248–251 [DOI] [PubMed] [Google Scholar]
- 35.Lehrer RI, Rosenman M, Harwig SS, Jackson R, Eisenhauer P. 1991. Ultrasensitive assays for endogenous antimicrobial polypeptides. J. Immunol. Methods 137:167–173 [DOI] [PubMed] [Google Scholar]
- 36.Schroeder BO, Wu Z, Nuding S, Groscurth S, Marcinowski M, Beisner J, Buchner J, Schaller M, Stange EF, Wehkamp J. 2011. Reduction of disulphide bonds unmasks potent antimicrobial activity of human β-defensin 1. Nature 469:419–423 [DOI] [PubMed] [Google Scholar]
- 37.Schägger H, von Jagow G. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368–379 [DOI] [PubMed] [Google Scholar]
- 38.Wiegand I, Hilpert K, Hancock REW. 2008. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 3:163–175 [DOI] [PubMed] [Google Scholar]
- 39.Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150:76–85 [DOI] [PubMed] [Google Scholar]
- 40.Wehkamp J, Harder J, Weichenthal M, Schwab M, Schäffeler E, Schlee M, Herrlinger KR, Stallmach A, Noack F, Fritz P, Schröder JM, Bevins CL, Fellermann K, Stange EF. 2004. NOD2 (CARD15) mutations in Crohn's disease are associated with diminished mucosal alpha-defensin expression. Gut 53:1658–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim HS, Cho JH, Park HW, Yoon H, Kim MS, Kim SC. 2002. Endotoxin-neutralizing antimicrobial proteins of the human placenta. J. Immunol. 168:2356–2364 [DOI] [PubMed] [Google Scholar]
- 42.Rosenberg HF, Domachowske JB. 1999. Eosinophils, ribonucleases and host defense: solving the puzzle. Immunol. Res. 20:261–274 [DOI] [PubMed] [Google Scholar]
- 43.Yang H, Lundbäck P, Ottosson L, Erlandsson-Harris H, Venereau E, Bianchi ME, Al-Abed Y, Andersson U, Tracey KJ, Antoine DJ. 2012. Redox modification of cysteine residues regulates the cytokine activity of high mobility group box-1 (HMGB1). Mol. Med. 18:250–259 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44.Wilson M. 2004. Microbial inhabitants of humans: their ecology and role in health and disease, p 263–267 Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 45.Jaeger SU, Schroeder BO, Meyer-Hoffert U, Courth L, Fehr SN, Gersemann M, Stange EF, Wehkamp J. 10 April 2013. Cell-mediated reduction of human β-defensin 1: a major role for mucosal thioredoxin. Mucosal Immunol. [Epub ahead of print.] 10.1038/mi.2013.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goodwin GH, Sanders C, Johns EW. 1973. A new group of chromatin-associated proteins with a high content of acidic and basic amino acids. Eur. J. Biochem. 38:14–19 [DOI] [PubMed] [Google Scholar]
- 47.Müller S, Ronfani L, Bianchi ME. 2004. Regulated expression and subcellular localization of HMGB1, a chromatin protein with a cytokine function. J. Intern. Med. 255:332–343 [DOI] [PubMed] [Google Scholar]
- 48.Knapp S, Müller S, Digilio G, Bonaldi T, Bianchi ME, Musco G. 2004. The long acidic tail of high mobility group box 1 (HMGB1) protein forms an extended and flexible structure that interacts with specific residues within and between the HMG boxes. Biochemistry 43:11992–11997 [DOI] [PubMed] [Google Scholar]
- 49.Bonaldi T, Längst G, Strohner R, Becker PB, Bianchi ME. 2002. The DNA chaperone HMGB1 facilitates ACF/CHRAC-dependent nucleosome sliding. EMBO J. 21:6865–6873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Travers AA. 2003. Priming the nucleosome: a role for HMGB proteins? EMBO Rep. 4:131–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Das D, Scovell WM. 2001. The binding interaction of HMG-1 with the TATA-binding protein/TATA complex. J. Biol. Chem. 276:32597–32605 [DOI] [PubMed] [Google Scholar]
- 52.Agresti A, Bianchi ME. 2003. HMGB proteins and gene expression. Curr. Opin. Genet. Dev. 13:170–178 [DOI] [PubMed] [Google Scholar]
- 53.Ronfani L, Ferraguti M, Croci L, Ovitt CE, Schöler HR, Consalez GG, Bianchi ME. 2001. Reduced fertility and spermatogenesis defects in mice lacking chromosomal protein Hmgb2. Development 128:1265–1273 [DOI] [PubMed] [Google Scholar]
- 54.Calogero S, Grassi F, Aguzzi A, Voigtländer T, Ferrier P, Ferrari S, Bianchi ME. 1999. The lack of chromosomal protein Hmg1 does not disrupt cell growth but causes lethal hypoglycaemia in newborn mice. Nat. Genet. 22:276–280 [DOI] [PubMed] [Google Scholar]
- 55.Bonaldi T, Talamo F, Scaffidi P, Ferrera D, Porto A, Bachi A, Rubartelli A, Agresti A, Bianchi ME. 2003. Monocytic cells hyperacetylate chromatin protein HMGB1 to redirect it towards secretion. EMBO J. 22:5551–5560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang H, Ochani M, Li J, Qiang X, Tanovic M, Harris HE, Susarla SM, Ulloa L, Wang H, DiRaimo R, Czura CJ, Wang H, Roth J, Warren HS, Fink MP, Fenton MJ, Andersson U, Tracey KJ. 2004. Reversing established sepsis with antagonists of endogenous high-mobility group box 1. Proc. Natl. Acad. Sci. U. S. A. 101:296–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gong W, Li Y, Chao F, Huang G, He F. 2009. Amino acid residues 201-205 in C-terminal acidic tail region plays a crucial role in antibacterial activity of HMGB1. J. Biomed. Sci. 16:83. 10.1186/1423-0127-16-83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wagener J, Schneider JJ, Baxmann S, Kalbacher H, Borelli C, Nuding S, Küchler R, Wehkamp J, Kaeser MD, Mailänder-Sanchez D, Braunsdorf C, Hube B, Schild L, Forssmann W-G, Korting H-C, Liepke C, Schaller M. 2013. A peptide derived from the highly conserved protein GAPDH is involved in tissue protection by different antifungal strategies and epithelial immunomodulation. J. Investig. Dermatol. 133:144–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Creamer B, Shorter RG, Bamforth J. 1961. The turnover and shedding of epithelial cells. I. The turnover in the gastro-intestinal tract. Gut 2:110–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vitali R, Stronati L, Negroni A, Di Nardo G, Pierdomenico M, del Giudice E, Rossi P, Cucchiara S. 2011. Fecal HMGB1 is a novel marker of intestinal mucosal inflammation in pediatric inflammatory bowel disease. Am. J. Gastroenterol. 106:2029–2040 [DOI] [PubMed] [Google Scholar]
- 61.Pusterla T, de Marchis F, Palumbo R, Bianchi ME. 2009. High mobility group B2 is secreted by myeloid cells and has mitogenic and chemoattractant activities similar to high mobility group B1. Autoimmunity 42:308–310 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.