Abstract
The virulence of a KPC-producing Klebsiella pneumoniae sequence type 258 (ST258) strain representing those circulating in Greece was assessed in a mouse septicemia model. The strain was virtually avirulent (50% lethal dose, >108 and 5 × 107 CFU for immunocompetent and neutropenic animals, respectively). Also, it was highly susceptible to serum killing, rapidly phagocytosed in vitro, and classified as K41, which is not among the virulent capsular types. The findings indirectly support the notion that high ST258-associated mortality is largely due to inefficient antimicrobial treatment.
TEXT
KPC-producing Klebsiella pneumoniae (KPC-Kp) of sequence type 258 (ST258) is an important nosocomial pathogen. ST258 isolates have been widely spread, achieving epidemic status in many settings (1). These organisms can cause severe infections in hospitalized patients and are associated with unacceptably high mortality rates. Indeed, results from a continuing prospective survey of the large teaching hospitals in Athens—an epidemic setting—have so far indicated that the 28-day mortality rates of patients with bloodstream infections (BSIs) due to KPC-Kp range from 40 to 45%, while the mortality rate of patients with BSIs caused by carbapenemase-negative K. pneumoniae is approximately 20% (G. L. Daikos, unpublished data). It could be hypothesized that KPC-Kp ST258 is endowed with virulence traits that permit host invasion and affect infection outcomes. This notion, however, has been challenged by recent experimental findings (2), and some investigators clearly support the idea that the excessive mortality rate should be attributed primarily to the suboptimal efficacy of the available therapeutic regimens (1, 3). We sought to contribute to this discussion by evaluating the virulence of KPC-Kp ST258 in an animal model and assessing its interaction with innate immunity.
The isolates used here were from a collection of 110 well-characterized (STs, pulsed-field gel electrophoresis [PFGE] types, plasmid content, β-lactamase production, and resistance phenotypes) KPC-Kp ST258 isolates derived at random from bacteremia patients in Greek hospitals during 2009 to 2011. The data indicated marginal diversity in that 92 isolates exhibited similar type 1 PFGE patterns and the remaining 18 isolates were distributed between PFGE types 2 and 3. All of the isolates harbored similar KPC-2-encoding IncFIIk plasmids (∼110 kb) accompanied by three additional plasmids ∼200, ∼50, and ∼15 kb in size. They exhibited either multidrug or extensive drug resistance phenotypes. The imipenem and meropenem MICs ranged from 1 to >32 mg/liter. K. pneumoniae ATCC 43816, a K2 strain used in animal infection models, was included for comparison.
The pathogenic potential of the dominant type, 1, was assessed by using a representative isolate (L-78) in parallel with ATCC 43816 in a septicemia model utilizing immunocompetent and neutropenic female ICR mice (Harlan Sprague-Dawley). Neutropenia was induced by intraperitoneal (i.p.) administration of cyclophosphamide. Animals were infected by i.p. injection of bacterial suspensions (102 to 108 CFU) derived from log-phase broth cultures. Survival was recorded every 4 h for 4 days, and 50% lethal doses (LD50s) were determined. The procedures used were approved by the Hellenic Pasteur Institute's Committee of Animal Care and Use, the State Veterinary Section, and conformed to European Union guidelines.
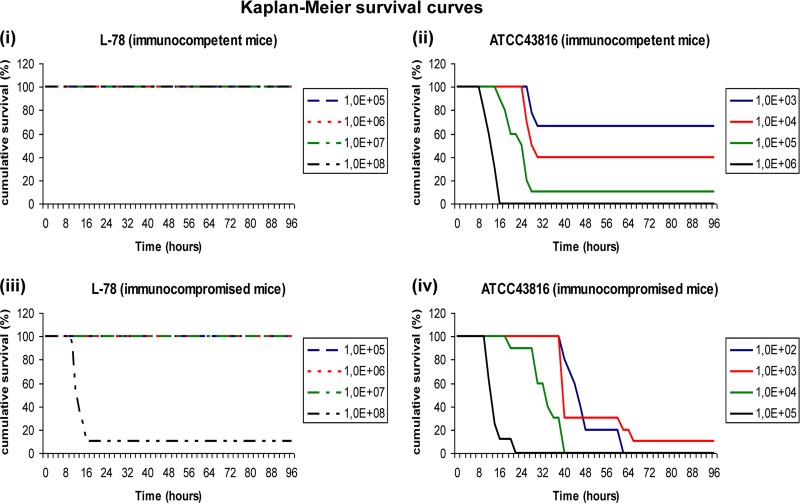
The low virulence of the ST258 isolate was reflected in the LD50s (Fig. 1). L-78 was unable to kill immunocompetent animals even when they were infected with the larger inoculum (LD50, >108 CFU). The smaller inoculum of L-78 that killed immunocompromised animals within 4 days was 108 CFU (LD50, 5 × 107 CFU). The respective LD50s of ATCC 43816 were 5 × 103 and <102 CFU (Fig. 1).
Fig 1.
Kaplan-Meier survival curves of immunocompetent (i and ii) and neutropenic (iii and iv) mice infected with K. pneumoniae ST258 strain L-78 (i and iii) or comparator strain ATCC 43816 (ii and iv). Note that the range of inocula of the latter strain was smaller.
To gain further information regarding the low virulence of the ST258 isolate, we evaluated (i) its susceptibility to human serum by the method of Hughes et al. (4) and (ii) the rates of phagocytosis by the BALB/c mouse macrophage cell line J774A.1 (LGC Promochem) as described previously (5), by using bacterial cells labeled with carboxyfluorescein diacetate succinimidyl ester (CFDA-SE; Invitrogen). Internalization of bacterial cells was monitored for 2 h by flow cytometry with a BD FACSCalibur (Becton Dickinson). ATCC 43816 was tested in parallel in both experiments.
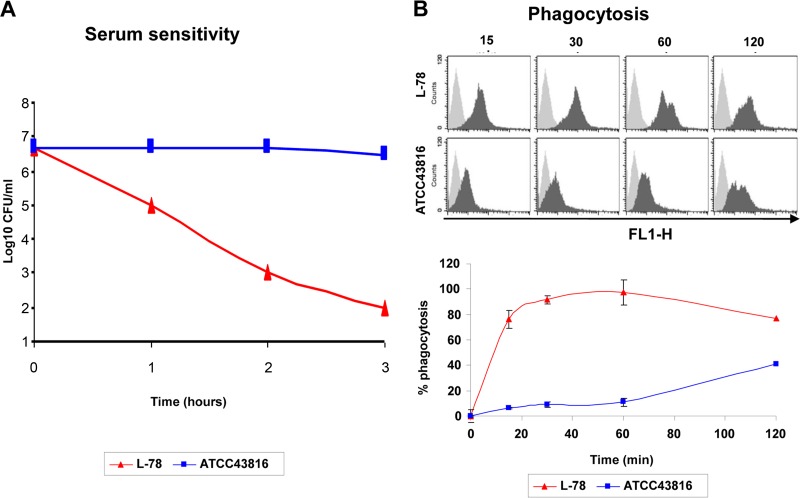
L-78 was rapidly killed by human serum (4.5 Δlog10 CFU reduction within 3 h). Serum-resistant variants were not observed after 24 h of incubation. Bactericidal activity was abolished when heat-inactivated serum (56°C for 30 min) was used, suggesting that bacterial killing was complement mediated. Similar results were obtained with 10 additional ST258 isolates representing all three PFGE types. The viable counts of ATCC 43816 bacteria remained essentially unaffected after incubation for 3 h (Fig. 2).
Fig 2.
(A) Bactericidal activity of human serum against ST258 and ATCC 43816. (B) Representative fluorescence-activated cell sorting histogram plots from three independent experiments (top) and rates of phagocytosis of ST258 and ATCC 43816 by J774A.1 murine macrophages (bottom). Macrophages were exposed to CFDA-SE-labeled bacterial cells (multiplicity of infection = 25) for 15, 30, 60, or 120 min at 37°C and analyzed by flow cytometry. For each sample, 10,000 cells were analyzed. The gray histograms at the top represent cells in the absence of bacteria (negative control), and the black histograms represent cells in the presence of bacteria. The results at the bottom are expressed as the percentages of macrophages that internalized ST258 or ATCC 43816 (percent phagocytosis) versus time. The results shown are the means of three independent experiments ± the standard deviations.
Given that among gram-negative bacteria a common mechanism used to resist complement-mediated killing is the activation of complement components by their lipopolysaccharides (LPS) away from the cell surface, the LPS pattern of L-78 was characterized (6). Upon SDS-PAGE of proteinase K-treated bacterial cell preparations, followed by silver staining, L-78 appeared to possess high-molecular-weight LPS similar in size to those of ATCC 43816 (not shown). Further studies of the interaction between LPS and complement components were not conducted. Yet, it was evident that the LPS of ST258 was unable to block killing by human serum.
Phagocytosis experiments showed that L-78 was internalized at high rates (∼76% within 15 min), while ATCC 43816 exhibited only marginal phagocytosis within the first 15 min, reaching a maximal rate (∼40%) after 2 h (Fig. 2). The phagocytosis profile of three additional ST258 isolates (PFGE types 1, 2, and 3) was similar to that of L-78 (70 to 95% within 15 min). The antiphagocytic property of capsule is a major virulence factor. Capsular typing of the three ST258 isolates performed at the Statens Serum Institut, Copenhagen, Denmark, classified the microorganisms as K41 (7), which is not among the known virulent capsular types.
Twelve isolates, including L-78, were examined for the presence of pathogenicity-associated genes by specific PCR assays (8, 9). ST258 isolates possessed ureA (urease production), uge, and wabG genes (involved in LPS core synthesis) but lacked the K1-associated magA, allS, and kfuBC genes. All were positive for the adhesin-encoding fimH and mrkD genes, which facilitate epithelial colonization, while they lacked cf29a, which encodes a nonfimbrial adhesin. Also, the plasmid-borne rmpA gene (which encodes an activator of capsule biosynthesis) was not detected. ST258 isolates were PCR positive for entB/entE (enterobactin) and ybtS (yersiniabactin) but negative for iroN (salmocelin), which is implicated in iron acquisition (9, 10). This pattern does not contradict the other findings presented here. It should be noted that the set of genes examined is most likely incomplete; recently available genomic data have indicated the presence of unique ST258 sequences whose putative polypeptides might contribute not only to host-pathogen interactions but also to the exceptional pace and extent of dissemination of these microorganisms (11).
In conclusion, the experimental data presented here suggest that KPC-Kp ST258 exhibits very low virulence, in line with several previous clinical observations, thus suggesting that it should be considered merely an opportunistic pathogen. Indeed, these microorganisms affect mainly hospitalized patients with severe underlying diseases and comorbidities (1, 12). Furthermore, only a minority (10%) of KPC-Kp ST258-colonized patients develops infections (13, 14). In addition, and perhaps not insignificantly, cases of community-acquired infections (such as had eventually happened with nosocomial extended-spectrum β-lactamase-producing Escherichia coli) due to KPC-Kp ST258 have not been documented, despite its increasingly pronounced presence in health care settings for at least 10 years.
Low virulence stands in contrast to the high mortality rate attributed to KPC-Kp ST258. This “paradox” may be explained by the low efficacy of the antimicrobials used against KPC-Kp infections and the severity of the underlying conditions of the patients, some of whom are profoundly immunocompromised. Both factors may result in uncontrollable proliferation and excessive microbial loads. In such an adverse scenario, we may hypothesize that KPC-Kp ST258 overwhelms the innate defenses in a quantitative fashion rather than by exploiting specific virulence factors.
ACKNOWLEDGMENTS
We thank T. L. Pitt and P. T. Tassios for helpful suggestions.
Footnotes
Published ahead of print 15 July 2013
REFERENCES
- 1.Tzouvelekis LS, Markogiannakis A, Psichogiou M, Tassios PT, Daikos GL. 2012. Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin. Microbiol. Rev. 25:682–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siu LK, Lin JC, Gomez E, Eng Chiang RT. 2012. Virulence and plasmid transferability of KPC Klebsiella pneumoniae at the Veterans Affairs Healthcare System of New Jersey. Microb. Drug Resist. 18:380–384 [DOI] [PubMed] [Google Scholar]
- 3.Satlin MJ, Calfee DP, Chen Fauntleroy L KA, Wilson SJ, Jenkins SG, Feldman EJ, Roboz GJ, Shore TB, Helfgott DC, Soave R, Kreiswirth BN, Walsh TJ. 2013. Emergence of carbapenem-resistant Enterobacteriaceae as causes of bloodstream infections in patients with hematologic malignancies. Leuk. Lymphoma 54:799–806 [DOI] [PubMed] [Google Scholar]
- 4.Hughes C, Phillips R, Roberts AP. 1982. Serum resistance among Escherichia coli strains causing urinary tract infection in relation to O type and the carriage of hemolysin, colicin, and antibiotic resistance determinants. Infect. Immun. 35:270–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sokolovska A, Becker CE, Stuart LM. 2012. Measurement of phagocytosis, phagosome acidification, and intracellular killing of Staphylococcus aureus. Curr. Protoc. Immunol. Chapter 14:Unit14.30. 10.1002/0471142735.im1430s99 [DOI] [PubMed] [Google Scholar]
- 6.Livermore DM, Pitt TL. 1986. Dissociation of surface properties and “intrinsic” resistance to β-lactams in Pseudomonas aeruginosa. J. Med. Microbiol. 22:217–224 [DOI] [PubMed] [Google Scholar]
- 7.Joseleau J-P, Lapyere M, Vignon M, Dutton GGS. 1978. Chemical and n.m.r.-spectroscopic investigation of the capsular polysaccharide of Klebsiella serotype K41. Carbohydr. Res. 67:197–212 [Google Scholar]
- 8.Brisse S, Fevre C, Passet V, Issenhuth-Jeanjean S, Tournebize R, Diancourt L, Grimont P. 2009. Virulent clones of Klebsiella pneumoniae: identification and evolutionary scenario based on genomic and phenotypic characterization. PLoS One 4:e4982. 10.1371/journal.pone.0004982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bachman MA, Oyler JE, Burns SH, Caza M, Lépine F, Dozois CM, Weiser JN. 2011. Klebsiella pneumoniae yersiniabactin promotes respiratory tract infection through evasion of lipocalin 2. Infect. Immun. 79:3309–3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Podschun R, Ullmann U. 1998. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 11:589–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chmelnitsky I, Shklyar M, Hermesh O, Navon-Venezia S, Edgar R, Carmeli Y. 2013. Unique genes identified in the epidemic extremely drug-resistant KPC-producing Klebsiella pneumoniae sequence type 258. J. Antimicrob. Chemother. 68:74–83 [DOI] [PubMed] [Google Scholar]
- 12.Gasink LB, Edelstein PH, Lautenbach E, Synnestvedt M, Fishman NO. 2009. Risk factors and clinical impact of Klebsiella pneumoniae carbapenemase-producing K. pneumoniae. Infect. Control Hosp. Epidemiol. 30:1180–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borer A, Saidel-Odes L, Eskira S, Nativ R, Riesenberg K, Livshiz-Riven I, Schlaeffer F, Sherf M, Peled N. 2012. Risk factors for developing clinical infection with carbapenem-resistant Klebsiella pneumoniae in hospital patients initially only colonized with carbapenem-resistant K. pneumoniae. Am. J. Infect. Control 40:421–425 [DOI] [PubMed] [Google Scholar]
- 14.Schechner V, Kotlovsky T, Kazma M, Mishali H, Schwartz D, Navon-Venezia S, Schwaber MJ, Carmeli Y. 2013. Asymptomatic rectal carriage of blaKPC producing carbapenem-resistant Enterobacteriaceae: who is prone to become clinically infected? Clin. Microbiol. Infect. 19:451–456 [DOI] [PubMed] [Google Scholar]