Abstract
Human parotid secretory protein (PSP; BPIF2A) is predicted to be structurally similar to bactericidal/permeability-increasing protein and lipopolysaccharide (LPS)-binding protein. Based on the locations of known antimicrobial peptides in the latter two proteins, potential active peptides in the PSP sequence were identified. One such peptide, GL13NH2 (PSP residues 141 to 153) was shown previously to interfere with LPS binding and agglutinate bacteria without bactericidal activity. By introducing three additional positively charged lysine residues, the peptide was converted to the novel bactericidal cationic peptide GL13K (MIC for Pseudomonas aeruginosa, 8 μg/ml [5.6 μM]). We investigated the antibiofilm activity of GL13K against static, monospecies biofilms of P. aeruginosa PAO1. Two-hour exposure of a 24-h biofilm to 64 μg/ml (44.8 μM) GL13K reduced biofilm bacteria by 102, and 100 μg/ml (70 μM) GL13K reduced bacteria by 103. Similar results could be achieved on 48-h-old biofilms. Lower concentrations of GL13K (32 μg/ml [22.4 μM]) were successful in reducing biofilm cell numbers in combination with tobramycin. This combination treatment also achieved total eradication of the biofilm in a majority (67.5%) of tested samples. An alanine scan of GL13K revealed the importance of the leucine residue in position six of the peptide sequence, where replacement led to a loss of antibiofilm activity, whereas the impact of replacing charged residues was less pronounced. Bacterial metalloproteases were found to partially inactivate GL13K but not a d amino acid version of the peptide.
INTRODUCTION
Bacteria organized in biofilm communities pose considerable clinical and industrial challenges. Microorganisms organized in biofilms become more recalcitrant to antibiotics due to the complex organization of the microbial community, differential gene expression among cells in the biofilm, and the presence of extracellular matrix material, including DNA and carbohydrate polymers (1). This challenge is compounded by an ever-increasing pool of antibiotic-resistant bacterial strains of clinical and veterinary importance (2). Thus, new approaches and compounds that can stand up to these challenges are needed.
Cationic antimicrobial peptides (CAMPs) have been considered possible alternatives to traditional antibiotics due to their interaction with bacterial membranes (3–6), which allows activity against metabolically dormant bacteria that are often found at the center of biofilms (7). Moreover, peptides that target the bacterial membrane are less likely to cause bacterial resistance since multifactorial resistance mechanisms are required at the cell membrane (5). To be clinically useful, a CAMP must show high selectivity for bacterial membranes with low toxicity to mammalian cell membranes.
We have recently designed the 13-amino-acid peptide GL13K, which was derived from human parotid secretory protein (PSP; BPIFA2) (8–10). PSP belongs to a family of bactericidal/permeability-increasing (BPI) fold proteins (11) that are expressed in the upper respiratory tract and oral cavity (12) and show predicted similarity to the BPI protein and lipopolysaccharide (LPS)-binding protein (LBP). Indeed, PSP causes bacterial agglutination (10) and binds LPS (9). These activities are mirrored by a 13-amino-acid peptide (GL13NH2) corresponding to amino acid residues 141 to 153 of PSP. This peptide aggregates both Gram-negative and Gram-positive bacteria and binds LPS but lacks bactericidal activity (8–10). In an effort to confer bactericidal activity, charged amino acids in positions 2, 5, and 11 of GL13NH2 were replaced by lysine residues, resulting in the peptide GL13K, with an overall positive charge of +5. GL13K exhibits bactericidal activity but not bacterium-agglutinating activity. The peptide retains the ability to block LPS action, with low toxicity against eukaryotic cells (8, 10). Recent studies on the mechanism of GL13K action indicate a carpet-like insertion in bacterial model membranes and release of bacterial membrane lipids in the form of micelles, leading to damage to the cell (13). The results also showed specificity for the bacterial membranes over eukaryotic membranes, fulfilling one of the requirements for clinically suitable CAMPs.
The goal of this study was to determine if GL13K is also active against bacteria in biofilm communities. We show that GL13K is effective against monospecies, static biofilms of the important opportunistic pathogen Pseudomonas aeruginosa. GL13K was able to significantly reduce the numbers of cells in biofilms grown under aerobic or anaerobic conditions. Combination of GL13K with the aminoglycoside tobramycin increased the likelihood that a biofilm was eradicated under continuous treatment. Our results demonstrate the potential of the PSP-derived peptide GL13K for the treatment of bacterial biofilms.
MATERIALS AND METHODS
Bacterial strains, growth media, and growth conditions.
The Pseudomonas aeruginosa strains PAO1 (14) and a bioluminescent derivate of PAO1, Xen41 (Xenogen, Alameda, CA), were used for all experiments. Luria broth (LB; Difco, Franklin Lakes, NJ) was used for overnight cultures and biofilm growth. Mueller-Hinton broth (MHB; Difco) was used for all experiments involving antibiotics and antimicrobial peptide testing. Anaerobic growth was achieved in an anaerobic chamber (80% N2, 10% CO2, 10% H2) at 37°C with addition of 1% KNO3 to LB and MHB (15).
Peptides.
The design of GL13K (GKIIKLKASLKLL-NH2) and the alanine-substituted GL13K peptides has previously been described (8, 10). A new peptide, d-GL13K, with d amino acids was synthesized. All peptides were obtained from outside vendors through the University of Minnesota Peptide Synthesis Facility. Peptides were synthesized on an ABI 433A peptide synthesizer and purified to >90% purity on a Beckman 126 high-pressure liquid chromatography (HPLC) system with an Agela (250- by 4.6-mm) C18 column as trifluoroacetate salt. All peptides were amidated at the C terminus. Peptides were stored as powder at −20°C and dissolved at 10 mg/ml in 0.01% acetic acid as working stock solutions, which were kept at 4°C.
Biofilm formation.
Biofilms were grown in 200 μl LB on the pegs of a Calgary device (16) (Nunc Immuno-TSP; Thermo Fisher, Waltham, MA). The medium was inoculated 1:2,000 from an overnight culture, and biofilms were formed on the pegs of the device for 24 or 48 h at 35°C under constant shaking (Belly Dancer; Stovall Life Science, Greensboro, NC) in a humidified growth chamber. Biofilm plates incubated under anaerobic conditions were not shaken.
Peptide treatment of biofilm.
Biofilms formed on pegs were washed three times in 200 μl 0.9% NaCl before being transferred to a 96-well polypropylene plate (no. 4917; Thermo Fisher Scientific, Waltham, MA) and incubated in MHB containing GL13K. The peptide concentrations chosen were 100, 64, and 32 μg/ml, equal to concentrations of 70, 44.8, and 22.4 μM, respectively. After the treatment period, the pegs were again washed three times, aseptically removed, and placed in 200 μl of 0.9% NaCl. Cells were removed from the pegs by sonication in a water bath (Symphony; VWR Scientific, Radnor, PA) for 10 min.
For experiments in Fig. 2D, saliva was self-collected (8) and then clarified according to the protocol of Palmer et al. (17). Biofilms were then treated with the resulting 25% clarified saliva.
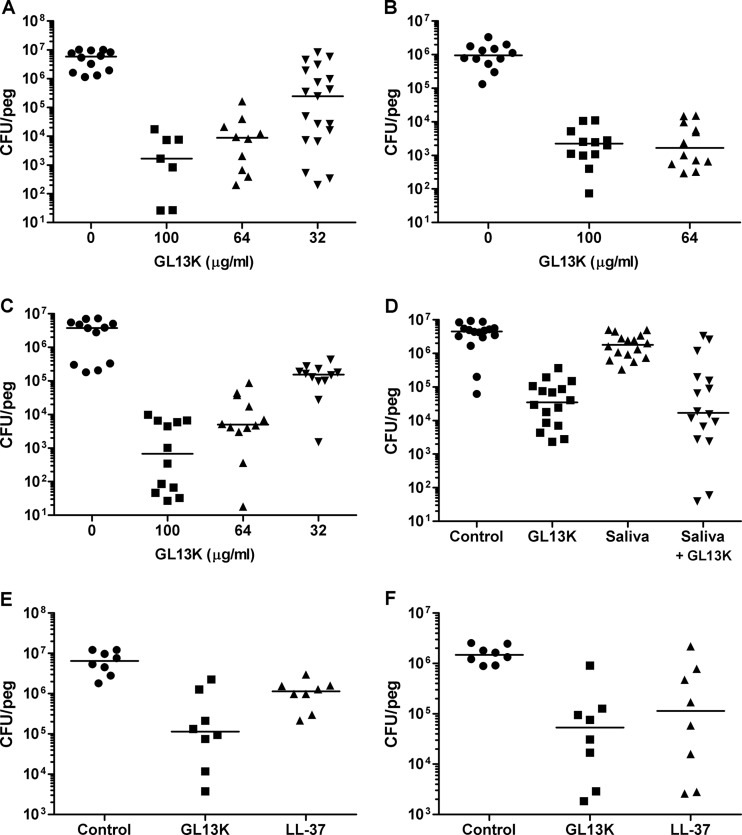
Fig 2.
Effect of GL13K on biofilms of P. aeruginosa PAO1. (A) Twenty-four-hour biofilms of P. aeruginosa incubated for 4 h with GL13K at the concentrations indicated. (B) Forty-eight hour biofilms of P. aeruginosa incubated for 4 h in the absence (control) or presence of GL13K. (C) Twenty-four-hour anaerobic biofilms of P. aeruginosa incubated in the absence (control) or presence of GL13K. (D) Twenty-four hour biofilms of P. aeruginosa incubated for 4 h in MHB (control) or 25% saliva (saliva) in the absence or presence of GL13K (100 μg/ml). (E) Comparison of GL13K and LL-37 in 10% fetal calf serum (24-h biofilms, 4-h treatment, peptides at 70 μM). (F) Comparison of GL13K and LL-37 in 25% clarified saliva (24-h biofilms, 4-h treatment, peptides at 70 μM). Each data point represents cell numbers from one individual peg determined in triplicate. Each line represents the median of 7 to 19 pegs. GL13K-treated samples are significantly different from control (P < 0.001). In panel F, GL13K is different from control (P < 0.001) and LL-37 is different from control (P < 0.01). One-way ANOVA with Bonferroni posttest was performed. One hundred, 64, and 32 μg/ml peptide corresponds to 70, 44.8, and 22.4 μM peptide, respectively.
Scanning electron microscopy.
Fixation and cryopreservation of biofilms were performed essentially as described by Erlandsen et al. (18). In brief, washed biofilms on pegs were fixed for 22 h in a mixture of 2% paraformaldehyde, 2% glutaraldehyde, and 4% sucrose in 0.15 M sodium cacodylate with 0.15% alcian blue 8GX. After 3 washes in phosphate-buffered saline (PBS), postfix was performed in 1% OsO4 with 1.5% potassium ferrocyanide in 0.15 M sodium cacodylate for 90 min. The pegs were washed three times in PBS before stepwise dehydration for 5 min per step in 50%, 70%, 85%, 95% (twice), and 100% (twice) ethanol. The pegs were removed and critical point dried in CO2. The pegs were then mounted on a metal platform and coated with platinum (Ion Tech argon ion beam coater). Electron microscopy was performed on a Hitachi S-4700 field emission scanning electron microscope operated at 2 keV. Images were collected as TIFF files and postprocessed with minor adjustments to sharpness and contrast in Aperture (Apple, Cupertino, CA).
Bacterial cell viability.
Biofilm cells were recovered from pegs as described above. Cell viability was determined by enumerating bacterial CFU on culture plates containing LB with 1.5% agar. The cells were enumerated by dilution plating in triplicate from a group of three or four pegs per treatment.
Bacterial viability was also determined by quantifying cellular ATP using the BacTiter-Glo assay (Promega, Madison, WI) according to the manufacturer's instructions; biofilms were incubated with BacTiter-Glo reagent for 10 min to achieve full cell lysis.
Viability of P. aeruginosa Xen41 was directly monitored by recording luminescence in a Synergy HT microplate reader (BioTek, Winooski, VT). Bioluminescence of P. aeruginosa Xen41 biofilms was recorded as relative light units by placing the pegs in 0.9% NaCl in black-walled 96-well plates (Costar 3631; Corning International, Corning, NY) that were read in a BioTek microplate reader. Planktonic P. aeruginosa Xen41 was similarly analyzed in microplates.
Peptide stability and degradation assay.
To determine if bacteria degrade GL13K, a 24-h biofilm was incubated with 100 μg/ml (70 μM) GL13K in MHB for 4 h. The spent medium, containing biofilm-exposed GL13K, was then centrifuged, and 10 μl of the supernatant was mixed with 10 μl of Xen41 cells (106 cells in 10 mM Na-phosphate buffer) and 80 μl of 10 mM Na-phosphate buffer, pH 7.4, and incubated for 10 min at 35°C. Fresh medium containing GL13K that had not been preexposed to biofilm served as a positive control. A decrease in P. aeruginosa Xen41 bioluminescence was used as a measure of GL13K activity in each medium.
In the experiments to test the effect of metalloproteases on GL13K stability, 1 mM EDTA was added to the biofilm medium during peptide incubation. In addition, the stability of GL13K was assessed in supernatants of P. aeruginosa overnight cultures that were either (i) sterile filtered through a 0.22-μm filter or (ii) autoclaved for 20 min. Peptides were then added to the treated P. aeruginosa supernatants to test their effect on 24-h biofilms.
Biofilm eradication assay.
Biofilms were grown as described above. Twenty-four-hour biofilms were washed three times and then exposed to the peptide (32 μg/ml; 22.4 μM) or peptide-tobramycin combination (32 μg/ml–1 μg/ml) for 24 h. Following incubation, the pegs were washed and then transferred to a new 96-well plate containing fresh LB with no antimicrobials and incubated for 24 h, after which eradication was scored as “growth” or “no growth” in the wells. The lowest peptide concentration that led to “no growth” was defined as the minimal biofilm eradication concentration (MBEC) (16). In the experiment with repeated peptide treatments, the pegs were transferred to fresh MHB containing peptide and/or tobramycin every 2 h for a period of 8 h, followed by 16 h of incubation with fresh peptide. At the end of this incubation, the pegs were washed and then transferred to a new 96-well plate containing fresh LB with no antimicrobials and incubated for 24 h, after which eradication was scored as “growth” or “no growth” in the wells.
Statistical analysis.
Statistical analysis was performed by one-way analysis of variance (ANOVA) with Dunnett's multiple comparison posttest using Prism (version 5.00 for Windows; GraphPad Software, San Diego, CA). A P value <0.05 was considered statistically significant. Fisher's exact test was used for the analysis of the biofilm eradication assay in GraphPad.
RESULTS
Biofilm formation and microscopic characterization.
A Calgary device was used to form biofilms of P. aeruginosa on plastic pegs suspended from the lid of a 96-well microtiter plate (16). Cell densities reached 1.1 × 107 ± 6.8 × 106 CFU/peg after 24 h (mean ± standard error of the mean [SEM], n = 35) and 7 × 106 ± 5.4·106 CFU/peg after 48 h (mean ± SEM, n = 11), with a change of medium after 24 h (2.4 × 106 CFU/peg with no change of medium) in accordance with previously reported cell densities for this experimental system (16). To determine the structure of the biofilms, they were analyzed by scanning electron microscopy. Analysis of untreated biofilms showed well-developed, complex biofilms with channel structures at the air-liquid interphase that were found to be up to 10 to 20 cell layers deep (Fig. 1A and B).
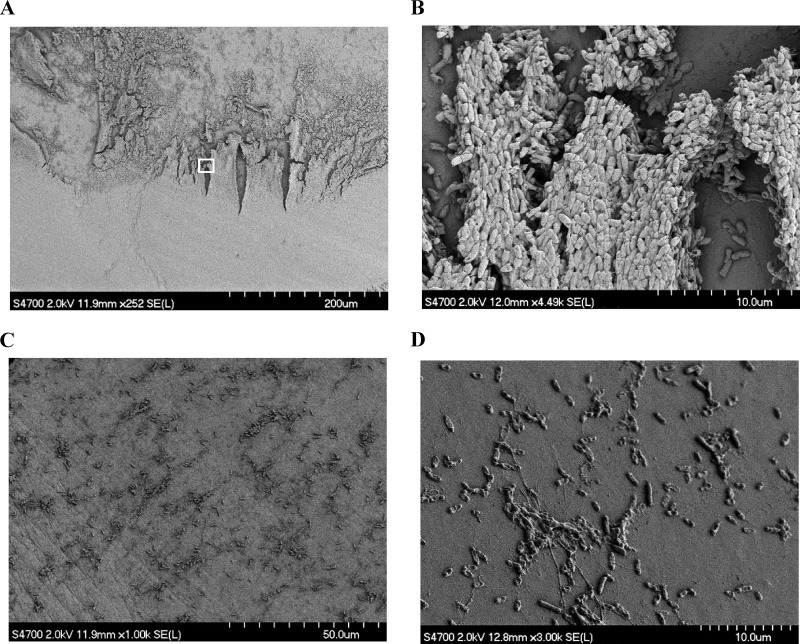
Fig 1.
Scanning electron microscopy of 24-h P. aeruginosa biofilm formed on the pegs of a Calgary device. (A) Biofilm on a representative control peg not exposed to antimicrobials. (B) Magnification (×20) of the area indicated by a white box in panel A. (C and D) Images of pegs treated with 100 μg/ml (70 μM) GL13K for 4 h at low (C) and high (D) magnifications.
Effect of GL13K on biofilms.
The MIC of GL13K against planktonic P. aeruginosa is 8 μg/ml (5.6 μM) (8). Biofilms cultured for 24 h and then exposed to 100, 64, or 32 μg/ml (70, 44.8, or 22.4 μM, respectively) of GL13K for 4 h showed cell numbers that were reduced by 99.9%, 99%, and 10%, respectively (Fig. 2A). A similar reduction in cell number was seen with a 2-h treatment (data not shown). The peptide was similarly effective against 48-h-old biofilms (Fig. 2B). Consistent with the reduction in cell numbers, no biofilm structure was visible by scanning electron microscopy of the pegs after treatment with 100 μg/ml (70 μM) of GL13K for 4 h (Fig. 1C and D).
A challenge for biofilm eradication is that cells inside the biofilm are marginally metabolically active and the inside of the biofilm is often anaerobic despite the presence of well-developed channel structures that supply cells with nutrients (19, 20) (Fig. 1A and B). Recently it became apparent that the normally aerobic P. aeruginosa is found in lungs of cystic fibrosis patients in anaerobic, biofilm-like communities that utilize alternative electron acceptors such as nitrate (15). We therefore tested the effect of GL13K on a biofilm that was established and treated under anaerobic conditions (Fig. 2C). The reduction in cell numbers was similar to that observed under aerobic conditions (Fig. 2A), indicating that GL13K was active against slow-growing cells and cells cultured under anaerobic conditions.
Activity of the peptide in the presence of mucosal secretions is also a practical consideration for application of an antimicrobial peptide against biofilms in the oral cavity, airways, or other mucosal surfaces. We therefore tested GL13K activity against a biofilm in the presence of 25% clarified saliva. Saliva alone did not reduce cell numbers in the biofilm, but importantly GL13K retained its activity in the presence of saliva (Fig. 2D). We compared the efficacy of GL13K with the paradigm human antimicrobial peptide LL-37 in 25% saliva or with the addition of 10% serum. The peptides are comparable at equimolar concentrations, with GL13K having a slightly better performance than LL-37 in 10% serum (Fig. 2E). The difference between the GL13K and LL-37 treatments was not significant. Results in saliva were comparable—the difference between GL13K and LL-37 again was not significant—yet both were significantly different from the control (Fig. 2F).
Stability of GL13K during biofilm treatment.
Reduction in cell numbers by 3 orders of magnitude at the 4-h time point suggested that GL13K has the potential for biofilm eradication, an important goal in antimicrobial treatment of biofilm. To test this, we determined the minimal biofilm eradication concentration (MBEC) after a single treatment with GL13K for 24 h. In this assay, GL13K failed to eradicate the biofilm in a single treatment with concentrations up to 512 μg/ml (358.4 μM) (not shown). The inability to eradicate a biofilm at peptide concentrations 2- to 5-fold higher than the concentration that reduced biofilm bacteria by 99.9% suggested that some bacteria in the biofilm remain uniquely resistant to peptide treatment. To investigate biofilm dynamics during treatment with GL13K, we monitored biofilm bacteria in real time using the bioluminescent strain P. aeruginosa Xen41. A preliminary experiment verified that GL13K kills P. aeruginosa Xen41 in a dose-dependent fashion, as expressed by reduced bacterial luminescence (Fig. 3A). In time course experiments, biofilm bacteria were suppressed for up to 6 h at 100 μg/ml (70 μM), but the biofilm had recovered to control levels after 24 h of incubation with GL13K (Fig. 3B). At lower GL13K concentrations, biofilm bacteria monitored via luminescence started to increase earlier. Parallel monitoring of cell numbers in the planktonic phase confirmed this observation, with cell numbers steadily increasing in the wells during the incubation period (data not shown).
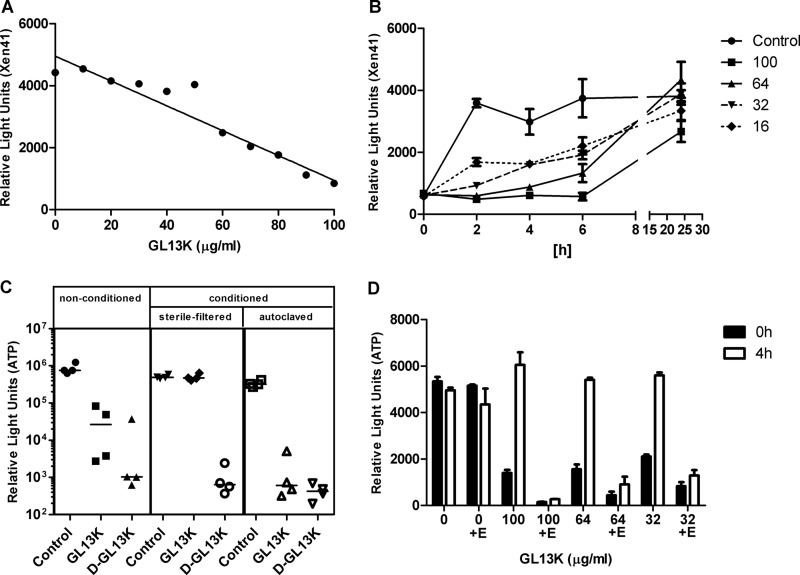
Fig 3.
Stability of GL13K during biofilm treatment. (A) Luminescence of planktonic Xen41 cells (106 CFU in Na-phosphate buffer) after incubation for 10 min with GL13K. (B) Biofilms of P. aeruginosa Xen41 were incubated for 24 h without peptide (control) or with 100, 64, 32, or 16 μg/ml GL13K as indicated (70, 44.8, 22.4, or 11.2 μM, respectively). Surviving bacteria were quantitated by luminescence. Data are shown as means ± standard deviations (SD) (n = 3). (C) GL13K activity in nonconditioned (fresh) medium (left) and conditioned (spent) medium that was either sterile filtered (middle) or autoclaved (right). A viability assay (ATP) of 24-h P. aeruginosa PAO1 biofilms after 4 h of treatment with 100 μg/ml (70 μM) peptide added to each modified MHB medium was performed. The data are shown as the median of four samples for each condition. All peptide treatments with the exception of GL13K in sterile-filtered conditioned medium are significantly different from control (P < 0.001). (D) Biofilms were incubated with GL13K for 0 h or 4 h in the presence and absence of EDTA (E; 1 mM). Residual GL13K activity was quantitated by a Xen41 kill assay (see panel A). The bars (means ± SD, n = 3) indicate relative survival of the Xen41 bacteria.
Stability of GL13K.
Growth of bacteria in the liquid phase despite an initial GL13K concentration that was a multiple of the MIC suggested a possible loss of peptide from the medium. To determine if GL13K was potentially inactivated by soluble factors, the spent medium from stationary-phase P. aeruginosa was centrifuged to remove bacteria. The spent medium supernatant was then sterile filtered or heat inactivated and used to test GL13K activity. Figure 3C shows that GL13K lost activity in sterile-filtered medium but remained active in heat-inactivated medium. This result suggested that a heat-sensitive factor is responsible for the inactivation of GL13K during incubation with P. aeruginosa. To test if proteolytic degradation could play a role in degradation, we generated an all-d-amino-acid version of GL13K. This peptide (d-GL13K) was not susceptible to degradation and remained active in both sterile filtered and heat-inactivated P. aeruginosa culture supernatant (Fig. 3C).
P. aeruginosa produces metalloproteases (21) that are capable of degrading antimicrobial peptides, potentially including GL13K. To confirm the involvement of metalloproteases in the observed GL13K degradation, biofilms were exposed to different concentrations of GL13K with or without 1 mM EDTA for 4 h. Control medium was not exposed to biofilms (control = 0 h). The GL13K-containing biofilm-exposed medium and GL13K-containing control medium were then incubated with fresh planktonic P. aeruginosa Xen41. Biofilm-exposed medium lost its ability to kill P. aeruginosa, confirming our previous observations. This loss of GL13K activity was prevented by the addition of EDTA (Fig. 3D), confirming that the metalloproteases of P. aeruginosa could be involved in GL13K degradation. EDTA alone had no effect on P. aeruginosa viability, whereas control medium with the peptide and EDTA was consistently more effective than control medium with the peptide alone, suggesting that the peptide is also degraded during incubation with P. aeruginosa Xen41. We hypothesized that the increased stability of d-GL13K would lead to an improvement in the effect on biofilms, which we could confirm in the kinetics experiment with Xen41, where luminescence activity remained suppressed over 24 h (not shown). Replacing only the Lys residues with d amino acids resulted in a peptide with reduced solubility, possibly due to incorrect folding (not shown).
Combination treatment for biofilm eradication.
A recent report described the successful combination of colistin and the aminoglycoside tobramycin for treatment of cystic fibrosis (22). Thus, as an alternative approach for biofilm eradication, we tested a combination of GL13K and tobramycin. Initial checkerboard MBEC assays were used to determine the optimal concentrations of the two compounds (data not shown). A single 24-h treatment with GL13K or tobramycin failed to eradicate biofilms on individual pegs (not shown). As an alternative, we serially treated 24-h biofilms four times for 2 h each with fresh antimicrobials before a final incubation with fresh antimicrobials for 16 h (total of five treatments over 24 h). GL13K and d-GL13K were used alone or in combination with tobramycin (Table 1). As expected from our experiments monitoring bioluminescence in biofilms, GL13K (32 μg/ml, 22.4 μM) did not eradicate biofilms, whereas the d-GL13K peptide was able to eradicate biofilms on 15.6% of treated pegs. The combination treatment with GL13K (32 μg/ml, 22.4 μM) and tobramycin (1 μg/ml) led to the eradication of 67.5% of the biofilms. The combination of d-GL13K and tobramycin was equally successful. Indeed either GL13K or d-GL13K in combination with tobramycin was significantly more effective than treatment with GL13K, d-GL13K, or tobramycin alone.
Table 1.
Eradication assay of established biofilmsa
| Treatment | No. of biofilms: |
P vs: |
||
|---|---|---|---|---|
| Showing growth | Eradicated | Control | Tobramycin | |
| Control | 64 | 0 | NAb | NA |
| GL13K, 32 μg/ml | 64 | 0 | NA | NA |
| Tobramycin, 1 μg/ml | 56 | 8 | 0.0062 | NA |
| GL13K, 32 μg/ml, + tobramycin, 1 μg/ml | 26 | 54 | <0.0001 | <0.0001 |
| d-GL13K, 32 μg/ml | 27 | 5 | 0.0033 | 0.7546 |
| d-GL13K, 32 μg/ml, + tobramycin, 1 μg/ml | 11 | 21 | <0.0001 | <0.0001 |
Twenty-four-hour biofilms were exposed 4 times for 2 h to GL13K, d-GL13K, tobramycin, or a combination of a peptide and tobramycin, followed by a final 16 h of incubation with fresh antimicrobials. Pegs were scored for bacterial growth after 24 h, as described in Materials and Methods. Biofilms on pegs that did not exhibit growth of P. aeruginosa were considered eradicated. Statistical analysis was performed with Fisher's exact test. Thirty-two micrograms/milliliter of peptide equals 22.4 μM.
NA, not applicable.
Structural analysis of GL13K.
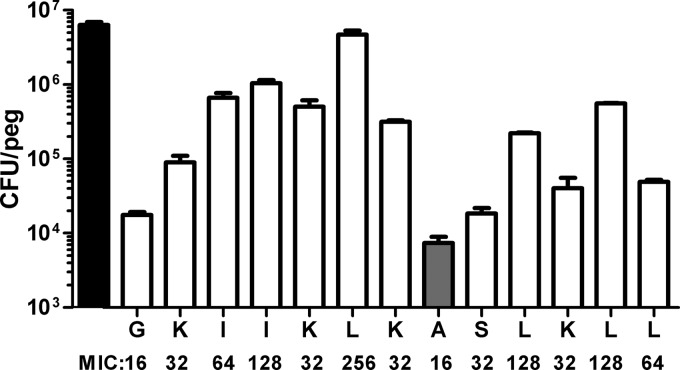
GL13K was developed from GL13NH2 by replacing targeted charged amino acid residues with lysine residues (8). The contribution of individual amino acids to GL13K activity was characterized by replacing individual amino acids with alanine in 12 new peptide variants (the original alanine in position 8 remained unchanged) (8). The 12 new alanine-substituted peptides and the original GL13K peptide were tested for activity against biofilms (Fig. 4). Although, GL13K was created by introducing additional positive charges in GL13NH2, it was the replacement of hydrophobic amino acid residues, not the charged amino acid residues, that had the most pronounced effect on antibiofilm activity. In particular, the replacement of leucine in position 6 by alanine almost completely eliminated antibiofilm activity. The antibiofilm effect of these peptides was roughly correlated with their MICs against planktonic bacteria (Fig. 4).
Fig 4.
Leucine in position 6 is essential for GL13K effect on biofilm. Amino acids in GL13K with the exception of alanine in position 8 were changed to alanine. Numbers of cells in 24-h biofilms after 4 h of treatment with peptide variants and GL13K (alanine in position 8; gray bar) is shown. The original GL13K sequence is represented on the x axis. Black bar, control, no peptide added. Data are shown as means ± SD (n = 2 to 9). Results for all amino acid replacements are significantly different from control (P < 0.001). The result for L-6 is significantly different from those for all the other amino acid replacements (P < 0.001).
DISCUSSION
The importance of biofilms is exemplified by estimates that 80% of infections in the United States have a biofilm etiology (23, 24). Traditional antibiotics often require 10- to 1,000-fold-higher concentrations to be effective against biofilm than are required for effectiveness against planktonic cells due to the presence of the extracellular matrix and low growth rates exhibited by cells within the biofilm (25). The increase in antibiotic resistance among medically important microorganisms is well documented and adds to this challenge (26, 27).
CAMPs offer an alternative approach to combat increasing antibiotic resistance and the biofilm lifestyle. Their activity against bacterial membranes is less specific than that of other antibiotic agents, thus lowering the probability of bacterial resistance (5). CAMPs also provide activity against metabolically less-active cells, circumventing the restriction of many traditional antibiotics that require bacterial growth (28). It can be predicted that no single CAMP will be suitable for all applications or as the sole antibiotic. Thus, we have embarked on the development of a new CAMP that is based on the sequence of the human salivary protein parotid secretory protein (8–10). The 13-amino-acid peptide GL13K exhibits bactericidal activity against planktonic bacteria and low toxicity against mammalian cells (8). We show in this study that GL13K is also highly effective against P. aeruginosa biofilms formed on the pegs of a Calgary device, a static biofilm system (16). Overall, we could achieve a 99.9% reduction in cell numbers in a 4-h treatment with 100 μg/ml (70 μM) GL13K. These results compare favorably with those for other antimicrobial peptides: Although colistin can achieve a greater rate of reduction in biofilm cell numbers, the toxicity of the peptide remains a concern (29). Recently described peptoids were able to reduce established P. aeruginosa biofilms by 1 to 2 logs at 100 μM (30), whereas Trp- and Arg-containing peptides were able to reduce 24-h Escherichia coli biofilms by 90% at that concentration (31). That biofilm cells are more resilient to the peptide is shown by the fact that a comparably sized population of P. aeruginosa stationary-phase cells can be easily killed within 5 min by the peptide at the concentration of 70 μM (8).
Evaluation of the formed biofilm by scanning electron microscopy revealed far greater structural diversity than previously reported (16). The air-liquid interphase in particular revealed intricate structures of ridges and channels, with cells packed in a depth of up to 10 to 20 cells in some places. Based on the anatomy of the formed biofilm, we see the experimental system as a valid starting point for the characterization of GL13K antibiofilm activity. Indeed, the complexity of the biofilm may contribute to the observed variability of GL13K efficacy on individual pegs. Although the steps involved in preparing the samples for microscopy may result in loss of biofilm matrix, scanning electron microscopy is accepted for characterization of biofilms (32), and imaging of the biofilms directly on the pegs by fluorescent confocal microscopy indicated a strong reduction of the fluorescent signal in the peptide-treated samples, validating the results obtained with electron microscopy. In addition, we also looked at biofilms formed on a glass slide, where again the biofilm mass was significantly reduced in the peptide-treated samples compared to control (data not shown). The data support a significant weakening of the biofilm by the peptide treatment, which leads to a reduction in viable cell numbers and, we hypothesize, to greater structural weakness of the biofilm, resulting in easier removal.
Our repeat treatment eradication effort increased the success rate of the treatment to 67.5%. As observed, repeat treatment with a low concentration of tobramycin that is insufficient to eradicate biofilms in a single treatment is somewhat effective, with an eradication rate of 12.5%. The d version of GL13K was able to reach the same efficiency as tobramycin at a concentration of 32 μg/ml (22.4 μM), clearly superior to GL13K, yet there was no difference between GL13K and d-GL13K in combination with tobramycin, with both of them significantly (P < 0.0001) improving the eradication rate over tobramycin alone to 67.5% and 65.6%, respectively. It is not clear at this point why d-GL13K did not show a better performance in this assay over GL13K, since it is clearly superior standing alone. It is apparent that eradication is possible under these conditions, yet eradication of all pegs in the population remains a challenge. This is consistent with the view that variability of the biofilms on individual pegs contributes to this effect, which is supported by observations of other groups (33).
Variations within individual biofilms can contribute to the difficulty in eradicating them. Thus, Pamp et al. showed that colistin preferentially killed the metabolically inactive cells in the center of a P. aeruginosa biofilm, whereas metabolically active cells on the biofilm surface were able to withstand colistin treatment by modification of LPS, which easily contributes to the resilience of the biofilm (7). The combination treatment with tobramycin offers an avenue to killing the metabolically active cells in the outer layer of the biofilm. Indeed, it is likely that complete biofilm eradication by antibiotics is not necessary in vivo. Instead, a significant reduction of biofilm bacteria may be sufficient to allow the immune system to gain the advantage and complete the eradication.
Unlike other antibiotics, antimicrobial peptides are typically susceptible to degradation by proteases (28). We found evidence for this in our model system, since GL13K activity diminished rapidly when the peptide was exposed to biofilm. EDTA inhibited this loss of activity, implicating metalloproteinases in peptide degradation. A potential solution for this problem is the use of the all-d-amino-acid version d-GL13K, which retained activity under conditions that inactivated the l-amino-acid peptide. The greater stability of d-GL13K may also be responsible for the increased effect of d-GL13K over GL13K for biofilm eradication in the repeat treatment experiments.
The mechanism of action of GL13K remains to be elucidated. However, characterization of the amino acids responsible for antibiofilm activity identified a leucine residue in position 6 that was necessary for peptide activity against biofilm. The amino acids in that region around position 6 of the peptide appear to contribute to peptide activity, in particular amino acids 3 to 7. This region contains three hydrophobic and two charged amino acids, indicating that the correct balance of charge and hydrophobic features in that part of the peptide is necessary for full activity. The bactericidal activity of GL13K was achieved by replacing four amino acids with Lys residues (8). Thus, it is somewhat surprising that the loss of a hydrophobic residue and not a basic residue appears to have the greater effect on peptide activity. This indicates that the loss of one positive charge does not impact the peptide significantly but that the remaining positive charges are sufficient to attach to the membrane. At this point, hydrophobicity appears to become more important, and one can hypothesize that the peptide in the alanine-6 variant is not able to sufficiently infiltrate the membrane. However, overall hydrophobicity is not the only explanation, since other leucine-to-alanine replacements do not impact activity to the same extent. Since the leucine in position 6 is centrally located, one can hypothesize that leucine-6 is important for the arrangement of the peptide, perhaps as a multimer, in the membrane. All peptide variants are active against planktonic cells in a kill assay, although the MIC of the leucine-6 variant is increased to 256 μg/ml (179.2 μM).
A common limitation of CAMPs is their sensitivity to physiological salt concentrations, which may limit their use in biological fluids (34, 35). We previously found that GL13K remained active against planktonic E. coli in the presence of 150 mM NaCl or 45% saliva. In addition, GL13K retains antilipopolysaccharide activity after intraperitoneal (i.p.) injection in vivo (8). The finding that the antibiofilm activity was not affected by 25% saliva suggests that applications on human mucosal surfaces are possible. Thus, with its high antibiofilm activity, activity in human secretions, and low toxicity, GL13K is a promising candidate for antimicrobial therapy of bacterial biofilms. This is of particular interest since saliva alone had no effect on the biofilm, despite the large number of antimicrobial peptides found in saliva (24). This finding is consistent with the observed accumulation of oral biofilm in vivo in the presence of continuous salivary flow. Thus, mechanical removal is necessary to control daily biofilm growth. GL13K and similar peptides may have a role in controlling biofilm growth in vivo in some patient populations.
ACKNOWLEDGMENTS
This work was supported by PHS grant R01DE017989 from the National Institute of Dental and Craniofacial Research.
We thank Chris Fretham of the University of Minnesota Characterization Facility for assistance with electron microscopy.
Footnotes
Published ahead of print 5 August 2013
REFERENCES
- 1.Parsek MR, Singh PK. 2003. Bacterial biofilms: an emerging link to disease pathogenesis. Annu. Rev. Microbiol. 57:677–701 [DOI] [PubMed] [Google Scholar]
- 2.Chen LF, Chopra T, Kaye KS. 2009. Pathogens resistant to antibacterial agents. Infect. Dis. Clin. North Am. 23:817–845, vii [DOI] [PubMed] [Google Scholar]
- 3.Yount NY, Yeaman MR. 2012. Emerging themes and therapeutic prospects for anti-infective peptides. Annu. Rev. Pharmacol. Toxicol. 52:337–360 [DOI] [PubMed] [Google Scholar]
- 4.Jacob L., Zasloff M. 1994. Potential therapeutic applications of magainins and other antimicrobial agents of animal origin. Ciba Found. Symp. 186:197–223 [DOI] [PubMed] [Google Scholar]
- 5.Yeung AT, Gellatly SL, Hancock RE. 2011. Multifunctional cationic host defence peptides and their clinical applications. Cell Mol. Life Sci. 68:2161–2176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bechinger B. 1999. The structure, dynamics and orientation of antimicrobial peptides in membranes by multidimensional solid-state NMR spectroscopy. Biochim. Biophys. Acta 1462:157–183 [DOI] [PubMed] [Google Scholar]
- 7.Pamp SJ, Gjermansen M, Johansen HK, Tolker-Nielsen T. 2008. Tolerance to the antimicrobial peptide colistin in Pseudomonas aeruginosa biofilms is linked to metabolically active cells, and depends on the pmr and mexAB-oprM genes. Mol. Microbiol. 68:223–240 [DOI] [PubMed] [Google Scholar]
- 8.Abdolhosseini M, Nandula SR, Song J, Hirt H, Gorr SU. 2012. Lysine substitutions convert a bacterial-agglutinating peptide into a bactericidal peptide that retains anti-lipopolysaccharide activity and low hemolytic activity. Peptides 35:231–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abdolhosseini M, Sotsky JB, Shelar AP, Joyce PB, Gorr SU. 2012. Human parotid secretory protein is a lipopolysaccharide-binding protein: identification of an anti-inflammatory peptide domain. Mol. Cell. Biochem. 359:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gorr SU, Abdolhosseini M, Shelar A, Sotsky J. 2011. Dual host-defence functions of SPLUNC2/PSP and synthetic peptides derived from the protein. Biochem. Soc. Trans. 39:1028–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bingle CD, Seal RL, Craven CJ. 2011. Systematic nomenclature for the PLUNC/PSP/BSP30/SMGB proteins as a subfamily of the BPI fold-containing superfamily. Biochem. Soc. Trans. 39:977–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bingle CD, Gorr SU. 2004. Host defense in oral and airway epithelia: chromosome 20 contributes a new protein family. Int. J. Biochem. Cell Biol. 36:2144–2152 [DOI] [PubMed] [Google Scholar]
- 13.Balhara V, Schmidt R, Gorr SU, Dewolf C. 2013. Membrane selectivity and biophysical studies of the antimicrobial peptide GL13K. Biochim. Biophys. Acta 1828:2193–2203 [DOI] [PubMed] [Google Scholar]
- 14.Holloway BW. 1969. Genetics of Pseudomonas. Bacteriol. Rev. 33:419–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoon SS, Hennigan RF, Hilliard GM, Ochsner UA, Parvatiyar K, Kamani MC, Allen HL, DeKievit TR, Gardner PR, Schwab U, Rowe JJ, Iglewski BH, McDermott TR, Mason RP, Wozniak DJ, Hancock RE, Parsek MR, Noah TL, Boucher RC, Hassett DJ. 2002. Pseudomonas aeruginosa anaerobic respiration in biofilms: relationships to cystic fibrosis pathogenesis. Dev. Cell 3:593–603 [DOI] [PubMed] [Google Scholar]
- 16.Ceri H, Olson ME, Stremick C, Read RR, Morck D, Buret A. 1999. The Calgary biofilm device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J. Clin. Microbiol. 37:1771–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palmer RJ, Jr, Kazmerzak K, Hansen MC, Kolenbrander PE. 2001. Mutualism versus independence: strategies of mixed-species oral biofilms in vitro using saliva as the sole nutrient source. Infect. Immun. 69:5794–5804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Erlandsen SL, Kristich CJ, Dunny GM, Wells CL. 2004. High-resolution visualization of the microbial glycocalyx with low-voltage scanning electron microscopy: dependence on cationic dyes. J. Histochem. Cytochem. 52:1427–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Werner E, Roe F, Bugnicourt A, Franklin MJ, Heydorn A, Molin S, Pitts B, Stewart PS. 2004. Stratified growth in Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 70:6188–6196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Borriello G, Werner E, Roe F, Kim AM, Ehrlich GD, Stewart PS. 2004. Oxygen limitation contributes to antibiotic tolerance of Pseudomonas aeruginosa in biofilms. Antimicrob. Agents Chemother. 48:2659–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmidtchen A, Frick IM, Andersson E, Tapper H, Bjorck L. 2002. Proteinases of common pathogenic bacteria degrade and inactivate the antibacterial peptide LL-37. Mol. Microbiol. 46:157–168 [DOI] [PubMed] [Google Scholar]
- 22.Herrmann G, Yang L, Wu H, Song Z, Wang H, Hoiby N, Ulrich M, Molin S, Riethmuller J, Doring G. 2010. Colistin-tobramycin combinations are superior to monotherapy concerning the killing of biofilm Pseudomonas aeruginosa. J. Infect. Dis. 202:1585–1592 [DOI] [PubMed] [Google Scholar]
- 23.Davies D. 2003. Understanding biofilm resistance to antibacterial agents. Nat. Rev. Drug Discov. 2:114–122 [DOI] [PubMed] [Google Scholar]
- 24.Fux CA, Stoodley P, Hall-Stoodley L, Costerton JW. 2003. Bacterial biofilms: a diagnostic and therapeutic challenge. Expert Rev. Anti Infect. Ther. 1:667–683 [DOI] [PubMed] [Google Scholar]
- 25.Hall-Stoodley L, Costerton JW, Stoodley P. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2:95–108 [DOI] [PubMed] [Google Scholar]
- 26.Hawkey PM. 2008. The growing burden of antimicrobial resistance. J. Antimicrob. Chemother. 62(Suppl 1):i1–i9 [DOI] [PubMed] [Google Scholar]
- 27.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. 2009. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 48:1–12 [DOI] [PubMed] [Google Scholar]
- 28.Peschel A, Sahl HG. 2006. The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat. Rev. Microbiol. 4:529–536 [DOI] [PubMed] [Google Scholar]
- 29.Falagas ME, Kasiakou SK. 2006. Toxicity of polymyxins: a systematic review of the evidence from old and recent studies. Crit. Care 10:R27. 10.1186/cc3995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kapoor R, Wadman MW, Dohm MT, Czyzewski AM, Spormann AM, Barron AE. 2011. Antimicrobial peptoids are effective against Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 55:3054–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hou S, Liu Z, Young AW, Mark SL, Kallenbach NR, Ren D. 2010. Effects of Trp- and Arg-containing antimicrobial-peptide structure on inhibition of Escherichia coli planktonic growth and biofilm formation. Appl. Environ. Microbiol. 76:1967–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hannig C, Follo M, Hellwig E, Al-Ahmad A. 2010. Visualization of adherent micro-organisms using different techniques. J. Med. Microbiol. 59:1–7 [DOI] [PubMed] [Google Scholar]
- 33.Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711–745 [DOI] [PubMed] [Google Scholar]
- 34.Travis SM, Anderson NN, Forsyth WR, Espiritu C, Conway BD, Greenberg EP, McCray PB, Jr, Lehrer RI, Welsh MJ, Tack BF. 2000. Bactericidal activity of mammalian cathelicidin-derived peptides. Infect. Immun. 68:2748–2755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goldman MJ, Anderson GM, Stolzenberg ED, Kari UP, Zasloff M, Wilson JM. 1997. Human beta-defensin-1 is a salt-sensitive antibiotic in lung that is inactivated in cystic fibrosis. Cell 88:553–560 [DOI] [PubMed] [Google Scholar]