Abstract
Candida tropicalis ranks between third and fourth among Candida species most commonly isolated from clinical specimens. Invasive candidiasis and candidemia are treated with amphotericin B or echinocandins as first-line therapy, with extended-spectrum triazoles as acceptable alternatives. Candida tropicalis is usually susceptible to all antifungal agents, although several azole drug-resistant clinical isolates are being reported. However, C. tropicalis resistant to amphotericin B is uncommon, and only a few strains have reliably demonstrated a high level of resistance to this agent. The resistance mechanisms operating in C. tropicalis strains isolated from clinical samples showing resistance to azole drugs alone or with amphotericin B cross-resistance were elucidated. Antifungal drug resistance was related to mutations of the azole target (Erg11p) with or without alterations of the ergosterol biosynthesis pathway. The antifungal drug resistance shown in vitro correlated very well with the results obtained in vivo using the model host Galleria mellonella. Using this panel of strains, the G. mellonella model system was validated as a simple, nonmammalian minihost model that can be used to study in vitro-in vivo correlation of antifungals in C. tropicalis. The development in C. tropicalis of antifungal drug resistance with different mechanisms during antifungal treatment has potential clinical impact and deserves specific prospective studies.
INTRODUCTION
In recent years, an epidemiological change among fungal infections has been observed. Although Candida albicans is still the most common species isolated from patients infected with pathogenic yeasts, the frequency of isolation of non-albicans Candida species has increased (1, 2). The frequency of isolates of non-albicans Candida species differs with geographical location. Candida tropicalis ranks between third and fourth among the species most commonly isolated (3, 4), although it is considered the most frequently isolated species in the Asia-Pacific region (5–7) and the second most frequently isolated species in Brazil and Latin America (20.9% and 13.2%, respectively) (1, 8). Moreover, C. tropicalis is almost always associated with the development of fungal infections, and it is often related to higher mortality than other non-albicans Candida species and C. albicans, particularly in neutropenic and oncology patients (2, 9, 10).
Invasive candidiasis and candidemia are treated with amphotericin B or an echinocandin as first-line therapy, with extended-spectrum triazoles as acceptable alternatives (11, 12). Candida tropicalis is usually susceptible to all antifungal agents, although several cases of fluconazole and azole cross-resistance in clinical isolates have been reported (5, 13, 14), especially in the Asia-Pacific region (1, 15). On the other hand, C. tropicalis resistance to amphotericin B is rare, and very few strains have reliably demonstrated high-level resistance to this agent (16, 17).
Limited specific information is available about the molecular mechanisms of resistance to azoles in C. tropicalis, although they are likely to be similar to those identified in other Candida species. To date, Candida sp. azole resistance has been linked to different molecular mechanisms. (i) Mutations in the azole target, 14α-sterol demethylase (Erg11p), result in a reduction or loss of affinity with azoles or an incapacity to bind azoles. Also, ERG11 overexpression seems to play a role in resistance. (ii) The upregulation of multidrug efflux transporters (ABC [ATP-binding cassette]/MFS [major facilitator superfamily]) leads to decreased drug concentrations within the fungal cell. (iii) A bypass in the ergosterol biosynthesis pathway may develop as a result of mutations in sterol Δ5,6-desaturase (ERG3) (18–20). In C. albicans, the most frequent azole resistance mechanisms reported are increased efflux of the drug (85%) and mutations in the ERG11 sequence (65%) (21). In fact, numerous Erg11p mutations have been described, but only a few have been directly associated with azole resistance (22). Also, the role of amino acid substitutions has been elucidated by heterologous expression of the ERG11 gene (23, 24). In addition, increasing evidence points to a relationship between amino acid substitutions and changes in protein conformation involved in azole resistance (25, 26). For C. tropicalis, overexpression of C. tropicalis ERG11 (CtERG11) associated with missense mutations (Y132F or S154F) has been described as the azole resistance mechanism in clinical isolates (14, 27). Additionally, in vitro development of fluconazole resistance in C. tropicalis linked to the upregulation of two multidrug efflux transporter genes, CtMDR1 and CtCDR1, has been reported (28).
In general, adequate measurement of in vitro antifungal activity in terms of MICs is considered of clinical relevance and often gives useful therapeutic information for patient management. The correlation between in vitro and in vivo data can be explored using different experimental models. A mouse model of systemic candidiasis has been used to evaluate the effects of azole resistance mechanisms on the efficacy of azole treatment (29, 30). Murine models are still considered the gold standard for the measurement of in vivo drug efficacy but have as a main disadvantage that a large number of animals are needed to determine the dose-response relationships of the antifungal. Increasing concern about the use of mammals, i.e., the cost, the number used, and ethical issues related to the pain of the animals during the procedures, has promoted the use of alternative, nonmammalian invertebrate models for testing microbial virulence and for screening the efficacy of antimicrobial agents. Among these, Galleria mellonella presents certain benefits in comparison with other nonmammalian models of infection (31). Previous studies have documented that G. mellonella is a valid model for the evaluation of microbial virulence and/or the efficacies of different antimicrobial agents against infections caused by different bacteria and fungi (32–38).
In this work, we investigated the resistance mechanisms operating in C. tropicalis strains showing azole cross-resistance alone or combined with amphotericin B resistance. We also tested the in vivo antifungal responses of C. tropicalis strains, both susceptible and resistant to amphotericin B, fluconazole, and voriconazole, in the model host G. mellonella in order to correlate the susceptibility phenotypes of those strains with those shown in vitro.
MATERIALS AND METHODS
Strains, media, and growth conditions.
Four Candida tropicalis strains were used in this study: (i) two ATCC strains, ATCC 750 and ATCC 200956, and (ii) two clinical isolates (CL-6835 and TP-13650) recovered from two patients with candidemia who had been treated with antifungal drugs previously (see Table 1). These isolates were identified by morphological features and were confirmed by DNA internal transcribed spacer (ITS) sequencing. Yeast cells were grown overnight in Sabouraud liquid medium (Becton, Dickinson and Company, MD, USA) at 30°C with constant shaking (150 rpm). Cells were harvested by centrifugation and were washed twice with sterile distilled water. Inoculum sizes were adjusted using a hemocytometer chamber. Yeast nitrogen base (YNB) solid medium was used in the metabolic inhibitor assays.
Table 1.
Mutations in ERG11 and ERG3 genes, corresponding amino acid substitutions, and MICs of antifungal drugs for Candida tropicalis isolates
| Isolate | Previous treatmenta | Gene sequence analysisb |
MIC (μg/ml)c |
||||||
|---|---|---|---|---|---|---|---|---|---|
| 14α-Sterol demethylase |
Sterol Δ5,6-desaturase |
||||||||
| ERG11 (bp) | Erg11p (aa) | ERG3 (bp) | Erg3p (aa) | AMB | FLC | VRC | ANF | ||
| ATCC 750 | Unknown | G366A | R122R | 0.25 | 0.5 | 0.03 | 0.03 | ||
| ATCC 200956 | Unknown | Δ132 bp | Δ44 aa | C773T | S258F | 2 | >64 | >8 | 0.03 |
| —d | D275V | ||||||||
| G1362A | K454K | ||||||||
| C1534G | P511A | ||||||||
| T1554C | I517I | ||||||||
| CL-6835 (clinical) | FLC, AMB, and CAS | T906C | I302I | A334G | S113G | 2 | >64 | >8 | 0.03 |
| G1391A | G464D | G366A | R122R | ||||||
| T637C | L212L | ||||||||
| TP-13650 (clinical) | FLC, VRC, and TRB | T225C | C75C | 0.25 | >64 | >8 | 0.03 | ||
| G264A | L88L | ||||||||
| A395T | Y132F | ||||||||
| T783C | I261I | ||||||||
| G1362A | K454K | ||||||||
| T1554C | I517I | ||||||||
CAS, caspofungin; TRB, terbinafine.
Substitutions in amino acid sequences are shown in boldface.
Determined by broth microdilution according to the established method of AFST-EUCAST. AMB, amphotericin B; FLC, fluconazole; VRC, voriconazole; ANF, anidulafungin. EUCAST antifungal clinical breakpoints for C. tropicalis are as follows: for AmB, ≤1 μg/ml for susceptibility and >1 μg/ml for resistance; for FLC, ≤2 μg/ml for susceptibility and >4 μg/ml for resistance; for VRC, ≤0.12 μg/ml for susceptibility and >0.12 μg/ml for resistance; and for ANF, ≤0.06 μg/ml for susceptibility and >0.06 μg/ml for resistance.
—, 132-bp deletion, between nucleotides 824 and 945, results in a 44-amino-acid deletion, between aa 276 and 319. The change in the open reading frame is the cause of the D275V amino acid substitution.
Antifungal drugs.
Amphotericin B (AMB) (Sigma-Aldrich Quimica, Madrid, Spain), fluconazole (FLC) (Pfizer SA, Madrid, Spain), voriconazole (VRC) (Pfizer SA), and anidulafungin (ANF) (Merck & Company, Inc., NJ, USA) were used for the determination of MICs and for assessment of their efficacies against C. tropicalis infection in the Galleria mellonella model. The antifungal doses were calculated by taking into account the therapeutic dose used for humans and the MIC values for the strains studied (11). Stock solutions of voriconazole, fluconazole, amphotericin B, and anidulafungin were first prepared in dimethyl sulfoxide (DMSO), and then dilutions in water for injection were made to obtain the following concentrations: 400 and 80 μg/ml of voriconazole; 800, 360, and 80 μg/ml of fluconazole; 120 and 80 μg/ml of amphotericin B; and 400 μg/ml of anidulafungin.
Antifungal susceptibility testing.
Antifungal susceptibility was determined by broth microdilution (BMD). MICs were determined by following the recommendations of the Antifungal Susceptibility Testing Subcommittee of the European Committee on Antibiotic Susceptibility Testing (AFST-EUCAST) (39). For amphotericin B, the breakpoints were defined based on the distribution of MICs for wild-type strains, determined by a EUCAST method (susceptibility breakpoint, ≤1.0 μg/ml) (40). For fluconazole, voriconazole, and anidulafungin, breakpoints proposed by the AFST-EUCAST were used to interpret susceptibility results (41).
Metabolic inhibitor susceptibility assays.
The susceptibilities of the strains to different classes of metabolic inhibitors were determined by a visual spotting assay on YNB medium with different drug concentrations (42). Strains were grown in Sabouraud liquid medium as described above, and cell concentrations were adjusted for a starting solution containing 105 cells per ml. Then 10-fold serial dilutions were carried out, and 5 μl of each dilution was spotted onto agar plates containing either 2 μg/ml amphotericin B, 10 μg/ml brefeldin A (Sigma-Aldrich Quimica SA, Madrid, Spain), 200 μg/ml hygromycin B (Sigma-Aldrich Quimica SA), or 2 μg/ml terbinafine (Novartis, Basel, Switzerland). Drug-free YNB medium was included as a control. The plates were incubated for 48 h at 37°C before reading.
Amplification and sequencing of ergosterol biosynthesis genes.
The ERG11 and ERG3 genes were PCR amplified and sequenced using a panel of oligonucleotide primers (synthesized by Sigma Genosys, Madrid, Spain) (see Table S1 in the supplemental material). Genomic DNA was extracted using the DNeasy Plant minikit (Qiagen Inc., Valencia, CA, USA) and was used as the template for PCR amplification. Amplification conditions consisted of 5 min of denaturation at 94°C, followed by 30 cycles of 30 s at 94°C for denaturation, 40 s at 50°C for annealing, and 50 s at 72°C for elongation, and by a final elongation step of 10 min at 72°C. PCR products were purified with the High Pure PCR product purification kit (Roche Diagnostics, Madrid, Spain) according to the manufacturer's recommendations. PCR products were semiquantified by agarose gel electrophoresis and were used as the templates for sequencing, which was performed with the Dye Terminator cycle sequencing quick-start kit (Beckman Coulter Inc., Fullerton, CA). The sequencing PCR products were then purified through G50 Sepharose columns (Amersham Biosciences) and, finally, were sequenced on a CEQ8000 DNA analysis system (Beckman Coulter) with the forward and reverse primers used for PCR amplification (see Table S1 in the supplemental material).
The amino acid sequences of the genes encoding putative 14α-sterol demethylase (ERG11) and sterol Δ5,6-desaturase (ERG3) were deduced from nucleotide sequences and were analyzed using the MegAlign software package (DNAStar, Inc., Lasergene, Madison, WI, USA). The amino acid alignments were derived by CLUSTAL analysis (43).
Sterol analysis.
Sterols were extracted from C. tropicalis after 18 h of growth in Sabouraud liquid medium at 37°C with shaking (150 rpm), using the protocol described previously (44, 45). Briefly, cells were harvested by centrifugation, washed with sterile water, lyophilized, and weighed to determine the dry weight (in grams) of the fungus. About 150 to 200 mg was used for total-sterol extraction. A 3-ml volume of a 25% alcoholic potassium hydroxide solution (methanol-ethanol, 3:2) was added to the dried pellets and was mixed by vortexing for 1 min. The mixture was incubated at 85°C in a water bath for 1 h. Neutral lipids were extracted twice by addition of a mixture of 1 ml of water and 1.5 ml of hexane, followed by vigorous vortex mixing for 3 min. The upper hexane layer was transferred to a clean glass tube, evaporated, and stored at −20°C until analysis. One hundred micrograms of androstanol (Sigma) was added to the samples as an internal standard for sterol recovery and quantification. Sterols were converted into their trimethylsilyl ethers by reaction with a 1:1 solution of N,O-bis-(trimethylsilyl)trifluoroacetamide (85°C, 60 min) in toluene. Neutral lipid fractions were analyzed on a Trace gas chromatograph coupled to a quadrupole mass analyzer (Thermo Fisher Scientific, Manchester, United Kingdom). The GC program and MS operation conditions were the same as those described previously (45, 46). Sterol identification was based on interpretation and comparison of mass spectra and retention time data with previous data (45, 47). The amount of each sterol (in micrograms) relative to the dry weight (in grams) of the fungus was calculated from three biological replicates analyzed in triplicate by GC-MS. The relative sterol composition was expressed as the ratios of individual sterol concentrations to the total sterol content. The significance of differences in sterol composition (P < 0.0001) was determined by a nonparametric Mann-Whitney test.
Homology modeling of Erg11p.
Homology models of wild-type C. tropicalis Erg11p and mutated sequences containing amino acid substitutions G464D, G464S, Y132H, and Y132F were constructed using the SWISS-MODEL platform (http://swissmodel.expasy.org) (48–50). The crystal structure of human lanosterol 14α-demethylase (HS-CYP51) in complex with ketoconazole, deposited in the Protein Data Bank (http://www.rcsb.org) with code 3LD6 (51), was used as the template. The amino acid sequence identity between this template and C. tropicalis Erg11p was 39%. The N-terminal region of C. tropicalis Erg11p (residues 1 to 50), defined to comprise the membrane-spanning domains, and therefore without counterparts in the template protein, was deleted prior to protein modeling.
Analysis of R6G uptake and efflux.
The activity of efflux pumps in azole- and/or amphotericin B-susceptible and -resistant C. tropicalis strains (ATCC 200956, ATCC 750, CL-6835, and TP-13650) was evaluated using rhodamine 6G (R6G) according to the protocol previously described by Vandeputte et al. in 2005 (14) and by Maesaki et al. in 1999 (52). Accumulation of R6G was measured by flow cytometry in the FL2 channel (collecting the emission signal at 585 ± 42 nm). A total of 10,000 cells were recorded using a FACSCalibur cytometer (FL2 channel; CellQuest; BD Biosciences), and the fluorescence intensity of R6G-stained cells was determined. Unstained cells were included as a negative control in all cases. Data were processed using FlowJo software (version 7.1.6; TreeStar Inc., Ashland, OR, USA). Experiments were performed at least twice with each strain on different days. Results were expressed as mean fluorescence intensities (MIF) and standard deviations (SD).
Evaluation of antifungal efficacy in Galleria mellonella infected with C. tropicalis.
Galleria mellonella caterpillars in the final instar (stage of larval development) (R.J. Mous Live Bait, the Netherlands) were used. Larvae were stored in the dark at 25°C prior to use. Ten caterpillars (body weight, 400 ± 100 mg) were employed per group. Candida tropicalis strains were grown in liquid Sabouraud medium as described above. Preliminary experiments were performed with all the strains to define an appropriate range of inoculum concentrations that would cause 80 to 100% mortality at 72 to 96 h postinfection. The inocula were determined by counting in a hemocytometer chamber and were adjusted in phosphate-buffered saline (PBS) to 2 × 108 cells per ml for ATCC 750, CL-6855, and TP-13650 and to 4 × 108 cells per ml for ATCC 200956. Viable counts were performed to confirm the correct size of the inocula.
A 10-μl Hamilton syringe (Fisher Scientific, Madrid, Spain) was used to infect the larvae by injecting 10 μl of the inoculum suspension into the hemocoel of each G. mellonella through the last left proleg. Within 2 h of the infection, 10 μl of an antifungal solution (either 400 or 80 μg/ml voriconazole, 800, 360, or 80 μg/ml of fluconazole, 120 or 80 μg/ml of amphotericin B, or 400 μg/ml of anidulafungin) was injected into a different proleg using the same technique. After antifungal treatment, caterpillars were incubated at 37°C for 7 days. Larva survival was monitored daily, and larvae were considered dead when they did not respond to physical pressure. The following control groups were included: untouched larvae, pierced larvae, larvae receiving PBS, and larvae given toxic doses of antifungals (400 μg/ml of voriconazole, 800 μg/ml of fluconazole, 120 μg/ml of amphotericin B, or 400 μg/ml of anidulafungin). Each experiment was performed at least three times. and the results are reported as mean values. To simplify some of the figures, the PBS control group results are not presented.
The survival rate of caterpillars was plotted against time, and P values were calculated by the log rank (Mantel-Cox) test using GraphPad Prism, version 5. P values of <0.01 were considered significant.
Nucleotide sequence accession numbers.
The full nucleotide sequence of the ERG11 gene of C. tropicalis ATCC 750 was used as the reference sequence (GenBank accession number M23673). The full nucleotide sequences of the ERG11 genes of ATCC 750, ATCC 200956, and the two C. tropicalis clinical isolates, CL-6835 and TP-13650, and their deduced amino acid sequences have been deposited in the GenBank database under accession numbers KC542323, KC542324, KC542325, and KC542326, respectively. The full nucleotide sequence of the ERG3 gene (encoding sterol Δ5,6-desaturase) of C. tropicalis MYA-3404 was used as the reference sequence for ERG3 (GenBank accession number XM002550136). The full nucleotide sequences of the ERG3 genes of the four strains used in this study (ATCC 750, ATCC 200956, CL-6835, and TP-13650) have been deposited in the GenBank database under accession numbers KC542319, KC542320, KC542321, and KC542322, respectively.
RESULTS
Antifungal susceptibility testing.
The MIC values of fluconazole (FLC), voriconazole (VRC), amphotericin B (AMB), and anidulafungin (ANF) are shown in Table 1. Candida tropicalis strain ATCC 750 was susceptible to all the antifungals tested. Isolate TP-13650 was resistant to fluconazole and voriconazole but susceptible to amphotericin B and anidulafungin. Resistance to all antifungal drugs except anidulafungin was observed for strains ATCC 200956 and CL-6835.
Metabolic inhibitor susceptibility assays.
The susceptibility of C. tropicalis strains to different metabolic inhibitors was used as a screening method to identify those yeasts with alterations in membrane fluidity. Candida tropicalis strains with cross-resistance to azole drugs and amphotericin B were more susceptible to metabolic inhibitors (brefeldin A or hygromycin B) and to terbinafine than the wild-type strain or the strain with azole drug resistance only. Changes in membrane fluidity were clearly detectable when brefeldin A was used (see Fig. S2 in the supplemental material). These results suggest that amphotericin B-resistant isolates may have an alteration of the membrane that increases its permeability, and therefore, these compounds could pass through the membrane relatively easily, resulting in the hypersusceptible phenotype observed in vitro.
ERG11 and ERG3 gene sequence analyses. (i) ERG11 gene sequence.
No changes were observed between the ERG11 sequence of strain ATCC 750 determined here and that deposited in GenBank (accession number M23673). Comparison of the ERG11 gene sequence of strain ATCC 200956 with the available corresponding sequence for strain ATCC 750 revealed a deletion of 132 bp that corresponds to the absence of 44 amino acids within the I helix (Δ276–319) of the protein (Table 1; see also Fig. S1B in the supplemental material) and an amino acid substitution (D275V) with restoration of the open reading frame. PCR amplification using two primer combinations with one of the oligonucleotides designed within the deleted sequence clearly showed that the deletion had occurred in the two alleles (see Fig. S4 in the supplemental material). Therefore, strain ATCC 200956 was homozygous for the ERG11 gene deletion. Analysis of the ERG11 gene sequences of strains CL-6835 and TP-13650 revealed unique missense mutations for each strain corresponding to amino acid substitutions G464D and Y132F, respectively (see Fig. S1C and A in the supplemental material). Silent mutations were also observed in these strains (Table 1).
(ii) ERG3 gene sequence.
The full nucleotide sequence of the ERG3 gene (encoding sterol Δ5,6-desaturase) of C. tropicalis MYA-3404 was used as a reference sequence for ERG3 (GenBank accession number XM002550136). Sequencing of the ERG3 gene showed single missense mutations, C773T and A334G, in the ERG3 sequences of ATCC 200956 and CL-6835, respectively, resulting in the S258F and S113G amino acid substitutions. In addition, CL-6835 presented two point mutations in the ERG3 gene (T637C and G366A) that did not affect the amino acid sequence. Neither ATCC 750 nor isolate TP-13650 presented any missense mutations in the entire ERG3 gene sequence (Table 1).
Sterol analysis.
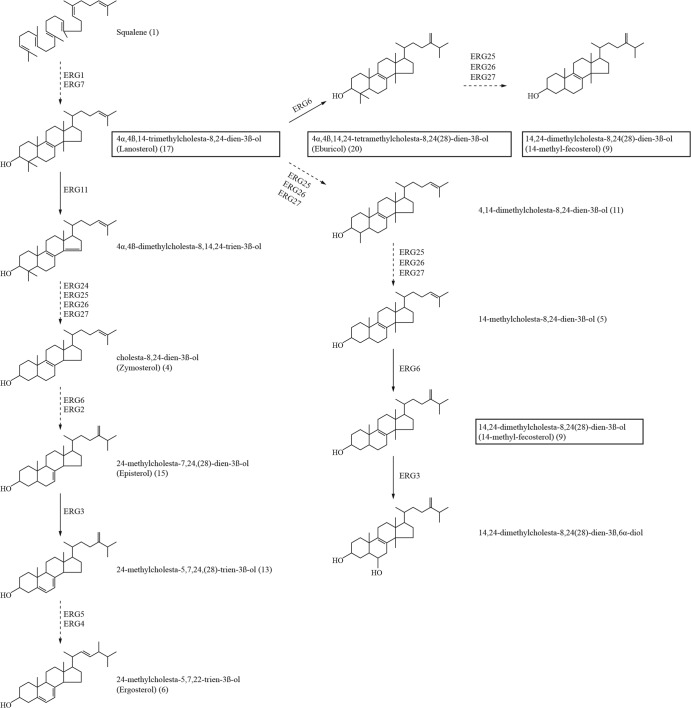
To investigate the contribution of membrane sterol composition to antifungal drug resistance, the sterol profiles of susceptible and resistant strains were analyzed by gas chromatography (GC)–mass spectrometry (MS). Sterols were classified according to their GC retention times and mass spectra, and a total of 22 sterols were detected (Table 2). The relative sterol composition for each strain is given in Table 2 based on the retention times of the sterols. Ergosterol was found to be the major sterol in strains ATCC 750 and TP-13650. In contrast, no ergosterol was detected in either ATCC 200956 or CL-6835. This absence of ergosterol suggested possible alterations in the sterol contents of these two strains. We then grouped these metabolites according to the presence of the methyl group on carbon atom 14. Figure S3 in the supplemental material shows statistically significant accumulations of C-14-methylated sterols in ATCC 200956 and CL-6835 that were not observed in strains ATCC 750 and TP-13650. C-14-methylated sterols in ATCC 200956 and CL-6835 comprised 14-methyl fecosterol, 14-methylcholesta-8,24-dien-3β-ol, 4,14-dimethylcholesta-8,24-dien-3β-ol, lanosterol, and eburicol.
Table 2.
Sterol compositions of Candida tropicalis strains
| Compound no. | Sterol |
% of total sterolsa in strain: |
||||
|---|---|---|---|---|---|---|
| Systematic nameb | Common name | ATCC 750 | ATCC 200956 | CL-6835 | TP-13650 | |
| 1 | Squalene | 8.54 | 0.17 | 1.45 | ND | |
| 2 | 24-Methylcholesta-5,7,9(11),22-tetraen-3β-ol | 0.15 | ND | ND | ND | |
| 3 | S28:3 | 0.08 | ND | ND | ND | |
| 4 | Cholesta-8,24-dien-3β-ol | Zymosterol | 8.97 | ND | ND | 20.60 |
| 5 | 14-Methylcholesta-8,24-dien-3β-ol | ND | 0.11 | 0.07 | ND | |
| 6 | 24-Methylcholesta-5,7,22-trien-3β-ol | Ergosterol | 64.96 | ND | ND | 60.84 |
| 7 | 24-Methylcholesta-7,22-dien-3β-ol | 0.20 | ND | ND | 0.51 | |
| 8 | 24-Methylcholesta-5,7,22,24(28)-tetraen-3β-ol | 0.81 | ND | ND | 5.20 | |
| 9 | 14,24-Dimethylcholesta-8,24(28)-dien-3β-ol | 14-Methyl fecosterol | ND | 61.08 | 52.67 | ND |
| 10 | 24-Methylcholesta-8,24(28)-dien-3β-ol | Fecosterol | 1.91 | ND | ND | 2.31 |
| 11 | 4,14-Dimethylcholesta-8,24-dien-3β-ol | 0.12 | 1.64 | 0.63 | ND | |
| 12 | 4α-S28:2 | ND | ND | ND | 0.21 | |
| 13 | 24-Methylcholesta-5,7,24(28)-trien-3β-ol | 2.56 | ND | ND | 3.43 | |
| 14 | 24-Methylcholesta-5,7-dien-3β-ol | 4.11 | ND | ND | 2.23 | |
| 15 | 24-Methylcholesta-7,24(28)-dien-3β-ol | Episterol | 4.45 | ND | ND | 2.06 |
| 16 | 24-Methylcholest-7-en-3β-ol | ND | ND | ND | 0.03 | |
| 17 | 4α,4β,14-Trimethylcholesta-8,24-dien-3β-ol | Lanosterol | 2.32 | 25.80 | 27.75 | 1.83 |
| 18 | 4α,24-Dimethylcholesta-8,24(28)-dien-3β-ol | 0.11 | ND | ND | ND | |
| 19 | 4α,4β-Dimethylcholesta-8,24(28)-dien-3β-ol | 0.53 | ND | ND | 0.65 | |
| 20 | 4α,4β,14,24-Tetramethylcholesta-8,24(28)-dien-3β-ol | Eburicol | 0.15 | 8.68 | 17.42 | 0.10 |
| 21 | 4α,4β,24-Trimethylcholesta-8,24(28)-dien-3β-ol | 0.03 | ND | ND | ND | |
| 22 | Unidentified | ND | 2.51 | ND | ND | |
ND, not detected.
S28:3 and 4α-S28:2 refer to sterols with 28 carbon atoms and 3 or 2 double bonds, respectively.
Analysis of R6G uptake and efflux.
Rhodamine 6G (R6G) is a fluorescent substrate of the multidrug transporter superfamily (conferring multidrug resistance [MDR]) that has already been used to demonstrate the activity of these proteins in azole-resistant isolates of Candida albicans. In fact, this dye is exported from cells by the same types of transporters as those used for azoles in yeasts (52). There were no remarkable differences among the R6G uptake/efflux ratios of the different C. tropicalis strains, despite their different antifungal drug susceptibility profiles (see Fig. S5 in the supplemental material). Levels of retention of R6G, after washing with PBS and incubation in fresh YPD medium (15 min), were similar for all strains. The percentages of retention of the total R6G quantified initially ranged from 58 to 83%. Some differences in the initial mean fluorescence intensities (MIF) were observed. For strains ATCC 750 and TP-13650, both susceptible to amphotericin B, the MIF were 239 ± 28 and 365 ± 196, respectively. In contrast, for the amphotericin B-resistant strains ATCC 200956 and CL-6835, the intensities of fluorescence were higher (1,881 ± 28 and 949 ± 4, respectively).
Homology modeling of Erg11p.
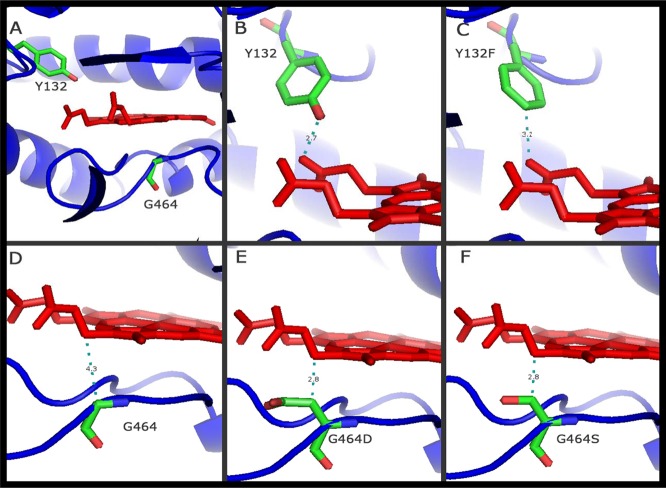
The crystal structure of human lanosterol 14α-demethylase (HS-CYP51) was used to generate a 3-dimensional (3D) homology model of the amino acid sequence of wild-type C. tropicalis Erg11p. The heme cofactor was extracted from the HS-CYP51 structure and was merged into the models obtained. Alignment of the two sequences revealed that there were no insertions or deletions in any of the secondary-structure regions of the protein. The fungus-specific insertion in the C-terminal part of the sequence was modeled as a loop and was supported by the ANOLEA prediction (53), which reported negative energy values for most of the residues of this section. Moreover, none of the residues studied in this paper were located within this region of the model; rather, they were located in conserved regions that have been intensively studied, as discussed below. Therefore, models for Erg11p with the G464S or Y132F substitution were generated using the same approach. Modeling of the partially deleted Erg11p from strain ATCC 200956 was not possible because of the lack of sufficient identity with the available crystal structures of proteins that could be useful as templates. Therefore, this model is not shown but is discussed below. Figure 1A shows the positions of the mutations analyzed, both close to the heme group. The different models revealed that the amino acid substitutions affected the heme environment in different ways. The wild-type strain shows a tyrosine at position 132 (Fig. 1B), which interacts directly with the heme through the hydroxyl group. Two mutations were analyzed for this site, Y132F and Y132H, but only the former (the one found in this work) is shown. Interaction of the protein with the heme group would be avoided because of the absence in the F residue of the hydroxyl group present in the Y residue, positioned in the 3-Å environment of the heme group (Fig. 1C). Another two mutations were analyzed for the amino acid at position 464. In contrast with Y132, glycine at position 464 (Fig. 1D) is close to the heme group but not within the 3-Å environment and probably does not interact with it. In contrast, the substitution G464S or G464D might affect the environment of the heme group, changing the interaction, due not only to the nucleophilic character of serine (Fig. 1F) or the acidic character of aspartic acid (Fig. 1E) but also to the fact that the side chains of these two amino acids are larger than that of glycine.
Fig 1.
Positions of the point mutations of Candida tropicalis Erg11p analyzed according to the homology models generated in this study. (A, B, and D) Wild-type Candida tropicalis Erg11p (Y132 and G464). (C, E and F) Positions of the mutations analyzed in the protein: Y132F, G464D, and G464S, respectively. The residues studied are shown in stick representation (green, carbon atoms; red, oxygen atoms; blue, nitrogen atoms). Heme is represented in red. The distance between the heme and the closest atom of the amino acid studied is indicated by a dashed green line and a number giving the distance in daltons. The crystal structure of human lanosterol 14α-demethylase (HS-CYP51) in complex with ketoconazole (Protein Data Bank code 3LD6) was used as a template. Y, tyrosine; F, phenylalanine; G, glycine; D, aspartic acid; S, serine.
Galleria mellonella in vivo infection model and antifungal response.
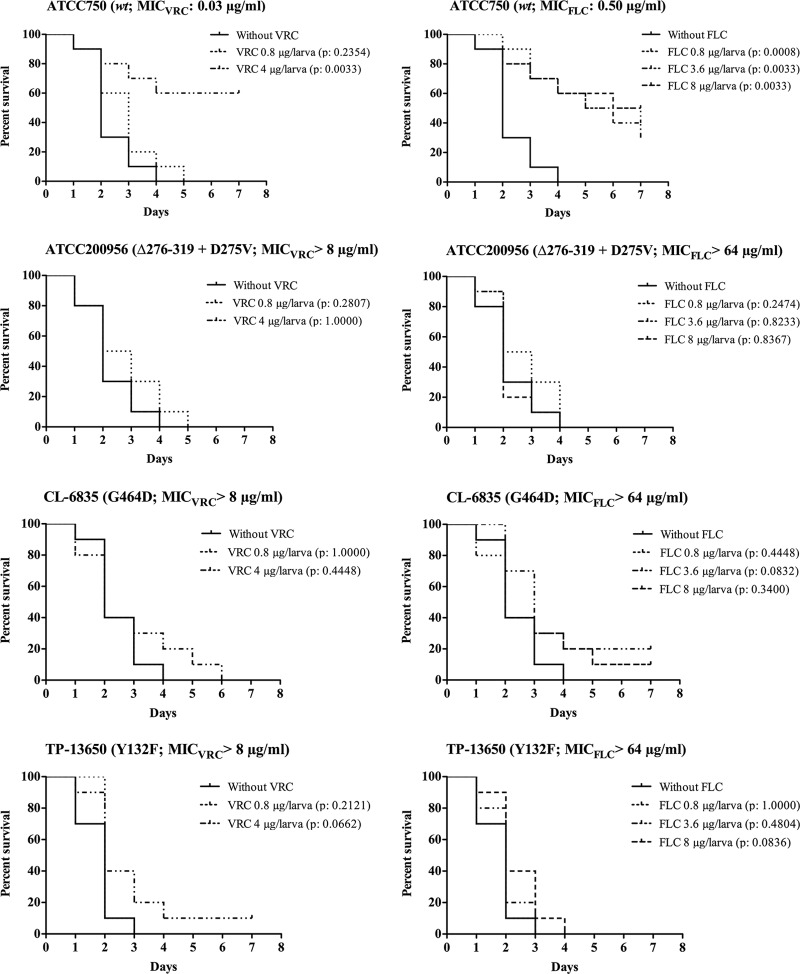
Galleria mellonella larvae were infected with an inoculum of 2 × 106 CFU/larva when strain ATCC 750, CL-6835, or TP-13650 was used. An inoculum of 4 × 106 CFU/larva was used for ATCC 200956, since this strain shows reduced virulence in this model (37). In all cases, 80% of infected and untreated larvae died between the 3rd and 4th day postinfection. For each strain inoculated, the survival of the larvae was dependent on both the antifungal dose administered and the in vitro MIC (Fig. 2 to 4).
Fig 2.
Efficacies of fluconazole (FLC) and voriconazole (VRC) for Galleria mellonella infected with different strains of Candida tropicalis. Ten larvae per group were infected with 2 × 106 cells of C. tropicalis ATCC 750, CL-6835, or TP-13650. For C. tropicalis ATCC 200956, an inoculum of 4 × 106 cells was used. After inoculation, larvae were treated with 0.8 or 4 μg of VRC/larva (subtherapeutic and therapeutic doses, respectively) or with 0.8, 3.6, or 8 μg of FLC/larva (subtherapeutic and therapeutic doses and twice the therapeutic dose, respectively). An untreated control group was included. Larva survival was monitored daily; the larvae are considered dead when they do not respond to touch.
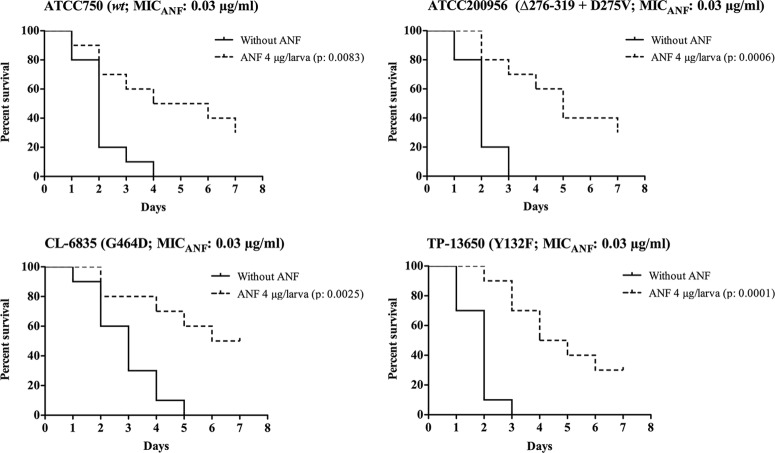
Fig 4.
Efficacy of anidulafungin (ANF) in Galleria mellonella infected with different strains of Candida tropicalis. Ten larvae per group were infected with 2 × 106 cells of C. tropicalis ATCC 750, CL-6835, or TP-13650. For C. tropicalis ATCC 200956, an inoculum of 4 × 106 cells was used. After inoculation, larvae were treated with 4 μg of ANF/larva. An untreated control group was included. Larva survival was monitored daily; the larvae are considered dead when they do not respond to touch.
The results showed that at therapeutic doses of azole drugs (10 mg of voriconazole/kg of body weight/day, equivalent to 4 μg/larva, and 9 mg of fluconazole/kg of body weight/day, equivalent to 3.6 μg/larva), larvae infected with azole-susceptible strain ATCC 750 showed significantly improved survival relative to that of their untreated control groups (P, 0.0033 for both VRC and FLC) (Fig. 2). In contrast, when G. mellonella larvae were infected with an azole-resistant strain (ATCC 200956, TP-13650, or CL-6835) and treated with a therapeutic (10 mg of voriconazole/kg/day, equivalent to 4 μg/larva, or 9 mg of fluconazole/kg/day, equivalent to 3.6 μg/larva) or subtherapeutic (2 mg/kg/day, equivalent to 0.8 μg/larva) dose of an azole, the percentages of survival of these groups did not differ significantly from those of their respective untreated control groups (Fig. 2). In addition, an increase in the fluconazole dosage to 20 mg/kg/day (equivalent to 8 μg/larva) failed to improve survival when larvae were infected with these strains (Fig. 2).
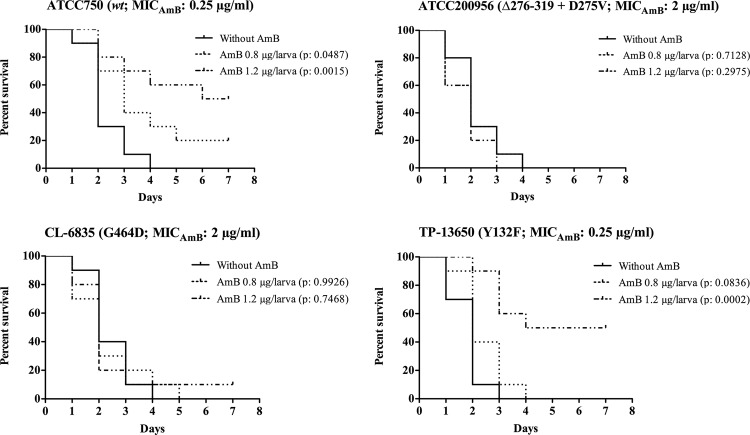
Amphotericin B treatment (1.2 μg/larva, equivalent to a therapeutic dose of 3 mg/kg/day) improved the survival of larvae infected with strain ATCC 750 (P, 0.0015) or TP-13650 (P, 0.0002) but not that of those infected with a strain (ATCC 200956 or CL-6835) cross-resistant to azole and amphotericin B (Fig. 3).
Fig 3.
Efficacy of amphotericin B (AMB) in Galleria mellonella infected with different strains of Candida tropicalis. Ten larvae per group were infected with 2 × 106 cells of C. tropicalis ATCC 750, CL-6835, or TP-13650. For C. tropicalis ATCC 200956, an inoculum of 4 × 106 cells was used. After inoculation, larvae were treated with 0.8 or 1.2 μg of AMB/larva (subtherapeutic and therapeutic doses, respectively). An untreated control group was included. Larva survival was monitored daily; the larvae are considered dead when they do not respond to touch.
In contrast, anidulafungin was the most active antifungal drug against all strains regardless of their azole/amphotericin B susceptibility profiles, with a larval survival range between 30 and 50% at the end of the experiment (Fig. 4). None of the drugs were toxic to the larvae at any of the concentrations used (data not shown).
DISCUSSION
Candida tropicalis is usually susceptible to all antifungal agents. However, antifungals have been used extensively for the prophylaxis or treatment of candidiasis, and there are increasing reports of azole resistance for this species (13, 14, 27, 54). In fact, the azole-resistant clinical isolates of C. tropicalis studied in this work were recovered from two patients suffering from candidemia and treated with different antifungals (Table 1). The precise mechanisms responsible for azole drug resistance in Candida species have been extensively studied and reviewed (18–20, 55). In Candida albicans, numerous ERG11 mutations have been described, but only a few of them have been associated with amino acid substitutions and azole resistance (22). The most frequently reported modifications are the substitutions Y132H, D278E, S405F, G464S, and R467K (14, 22). However, little information is available concerning the molecular mechanisms leading to azole resistance in the pathogenic yeast C. tropicalis. Acquired resistance to azole drugs in clinical isolates of C. tropicalis due to overexpression of a CtERG11 gene associated with missense mutations (Y132F and S145F) has been reported (13, 26).
In this study, we intensively investigated the molecular bases for C. tropicalis strains presenting a pattern of resistance to azole drugs, alone or in combination with resistance to amphotericin B. Cross-resistance to azoles and amphotericin B drugs was observed in two of the C. tropicalis strains analyzed (ATCC 200956 and CL-6835), while strain TP-13650 was resistant only to azole drugs. We also confirmed this in vitro phenotype in vivo by using an alternative minihost model of candidiasis.
Enhanced drug efflux activation is one of the major mechanisms causing azole resistance in yeast, although it could be accompanied by other resistance mechanisms. This mechanism is a consequence of transcriptional activation of genes encoding drug efflux pump proteins belonging to the ABC (encoded by Candida drug resistance [CDR] genes) and major facilitator (encoded by MDR genes) superfamilies of transporters (20). In order to search for concomitant resistance mechanisms related to increased efflux pump activity, we tested this possibility using rhodamine 6G (R6G). This compound is a fluorescent substrate of the multidrug transporter superfamily and has been used to demonstrate the activity of these proteins in azole-resistant isolates of C. albicans, because this dye uses the same transporters as azoles in yeasts (52). Our results showed that all strains captured R6G, although amphotericin B-resistant strains showed higher uptake/efflux ratios than amphotericin B-susceptible strains at time zero. This phenomenon could be explained by higher membrane permeability due to changes in the sterol composition of the membrane. However, analysis of the uptake and efflux of R6G for all azole-resistant or wild-type C. tropicalis strains shows that the efflux pumps do not seem to play a fundamental role in azole resistance in C. tropicalis, in agreement with previous results (14, 27).
The antifungal susceptibility phenotype of strains ATCC 200956 and CL-6835 resembled that of C. albicans ERG11/ERG11 double mutant strains showing resistance to azoles as well as to amphotericin B on either the wild-type or the ERG3/ERG3 mutant background (42). Also, our data were consistent with those for the C. albicans ERG11-null mutation when the strains were tested against different metabolic inhibitors (see Fig. S2 in the supplemental material). These data suggest that the susceptibilities of both antifungal cross-resistant C. tropicalis strains to these metabolic compounds might be related to changes in the composition and permeability of the fungal membrane and are likely associated with the functionality of Erg11p and alterations in ergosterol biosynthesis (42, 56). Sterol analysis confirmed the absence of ergosterol (Table 2) and the accumulation of 14-methyl sterols in C. tropicalis strains ATCC 200956 and CL-6835 (Fig. 5; see also Fig. S3 in the supplemental material). In fact, this sterol profile was similar to those of different Candida species with ERG11 gene deletions (42, 56). Sequencing and structural analysis of the proteins showed that strain ATCC 200956 has a deletion of 44 amino acids in Erg11p that corresponds to the excision of the “I” helix in the azole target. That deletion would presumably render the protein nonfunctional, which would explain both the lack of ergosterol and the accumulation of 14α-methylated sterols, and therefore the resistance to amphotericin B. In fact, during the revision of this work, an identical azole/amphotericin B resistance mechanism was described in a clinical isolate of C. tropicalis isolated in Tunisia (54).
Fig 5.
Ergosterol biosynthetic pathway in Candida tropicalis. Major sterol intermediates that accumulate in C. tropicalis strains with nonfunctional Erg11p are boxed. Broken arrows indicate multiple enzymatic steps; solid arrows indicate single enzymatic steps. The numbers in parentheses refer to the sterols described in Table 2.
Examination of the C. albicans Erg11p 3D protein models in combination with azole drugs suggests that the portion of the protein that is lacking (helix I) is likely to be the cause of the lack of interaction either with the substrate (lanosterol) or with azole drugs (26, 57). In fact, in Candida glabrata and Saccharomyces cerevisiae, a single mutation at that level is enough to nullify the function of native Erg11p (56, 58).
The effects of Erg11p mutations at G464 and Y132 on azole susceptibility have been reported previously in different yeasts and filamentous fungi (59–63). The Y132H and G464S modifications conferred azole resistance on C. albicans (59). Also, the effects of different ERG11 mutations have been explored by heterologous expression in S. cerevisiae (24, 64). In addition, several studies performed with microsomal fractions and purified enzyme preparations have been used to analyze the relationship between Erg11p amino acid substitutions and reductions in the level of azole binding (22, 65). Those authors concluded that the azole nitrogen could not interact with the iron of the heme of C. albicans Erg11p (CaErg11p) with Y132H or G464S, as normally occurs to produce inhibition. In contrast, the altered protein retained the ability to metabolize the substrate, comparably to the ERG11 wild-type strain, allowing resistant mutants to produce ergosterol (23, 64).
That is the case in strain TP-13650; the presence of phenylalanine at position 132 produces the loss of a bridge of hydrogen that is normally formed between heme and tyrosine. This modification in the environment of heme would alter the binding to azoles without modifying the activity of the enzyme. This difference is marked by a wild-type sterol profile, resistance to metabolic inhibitors (due to a normal cytoplasmic membrane), and susceptibility to amphotericin B. The modeling of wild-type protein and a variant with equivalent residues mutated in fungal plant pathogens, such as Y137F in Mycosphaerella graminicola, has provided similar results (65), which are consistent with the resistance recorded for other plant pathogens (Y136F in Mycosphaerella fijiensis, Y136F in Erysiphe graminis, and Y136F in Uncinula necator [66–68]).
On the other hand, although G464 cannot interact with fluconazole directly, this position might be important for the conformation of the heme environment. Changing glycine to other residues would be expected to decrease the flexibility required for interdomain conformational changes upon inhibitor or substrate binding (57). In strain CL-6835, the absence of ergosterol and a pattern of sterol metabolites similar to that of strain ATCC 200956 would suggest that the Erg11p amino acid substitution G464D (Fig. 1E), located at the catalytic heme binding domain, will affect equally the affinity of the enzyme for the azole and its affinity for the substrate. This could be due to the negative charge or the larger size of aspartic acid, producing a great disturbance in the environment of the heme and preventing the binding of the substrate/inhibitor. In fact, in C. albicans Erg11p with the G464S substitution, although no difference in the affinity of the substrate was observed between the mutant and the wild-type protein, reduced sterol demethylase catalytic activity was observed (63). The sterol profile confirmed that possibility, since this strain has a complete lack of ergosterol and an accumulation of 14α-methylated sterols, thus showing cross-resistance to azole and amphotericin B, as was the case with strain ATCC 200956. Interestingly, strain CL-6835 is the first reported isolate that carries an aspartic acid substitution at position G464.
It has been reported that disruption of ergosterol biosynthesis at the Erg11p level results in toxic metabolites. Cell growth arrest correlates with the accumulation of 14α-methyl-ergosta-8,24(28)-dien-3β,6α-diol in yeasts with a sterol 14α-demethylase gene disruption. In this situation, 14α-methylated sterols are converted to toxic 3,6-diol derivatives by Δ5,6 desaturation performed by the enzyme Erg3p (Fig. 5). However, cells can overcome the effect of such a block by a suppressor mutation in sterol C5,6 desaturation and can thus acquire azole resistance (69). In agreement with this observation, azole-resistant fungal isolates with nonfunctional ERG3 alleles have been described for C. albicans and Candida dubliniensis (70–72). Both strains with a putative block at the Erg11p level (CL-6835 and ATCC 200956) carried ERG3 mutations (Table 1) and showed an accumulation of 24-methylcholesta-5,7,24(28)-trien-3-ol (Table 2), suggesting that these two strains might have evolved to avoid the production of toxic sterols like 14,24-dimethylcholesta-8,24(28)-3β,6α-diol (Fig. 5). On the other hand, it is possible that changes in overall membrane composition could compensate for perturbations in sterol content, a scenario consistent with reports of other ergosterol-deficient yeasts with dysfunctional Erg11p that remain aerobically viable (56). Although our results agree with those of other studies regarding activity alterations of Erg11p and Erg3p (54), future work would include functional characterization of these enzymes by gene deletion or other genetic approaches, such as heterologous expression and site-directed mutagenesis, in order to fully understand the enzymatic activities of these proteins in these particular clinical isolates.
Since two of the strains, CL-6835 and TP-13652, were isolated from patients with candidiasis refractory to antifungal treatment, we attempted to determine the therapeutic effects of azoles in an in vivo model. Due to the strong structural and functional similarities between the innate immune responses of insects and mammals (73), several minihosts are now being employed to study alterations in microbial virulence and the efficacy of antimicrobial agents. Among them, Galleria mellonella presents certain benefits in comparison with other nonmammalian models of infection (31). For example, the larvae can be maintained at a temperature range of 25°C to 37°C, thus reproducing both at the temperature at which fungi exist in the environment and at the temperature at which they are capable of producing invasive infections in humans. In addition, an accurate inoculum of the pathogen can be delivered directly into the host's body (34). Finally, the number of larvae used and the ease of handling also represent important advantages. Previous studies have documented that G. mellonella is a valid model for the evaluation of microbial virulence and/or the efficacies of different antimicrobial agents in infections caused by different bacteria (32–38) and fungi, such as Cryptococcus neoformans (34), Candida spp. (38), Fusarium spp. (35), and Aspergillus fumigatus (36). Recently, Mesa-Arango et al. (37) demonstrated that C. tropicalis is able to infect and kill G. mellonella. The lethal effect of C. tropicalis depends on both the yeast dose and the incubation temperature. In larvae infected with antifungal-susceptible C. tropicalis, the fungal burdens increased in a time-dependent manner, and treatment with either amphotericin B or fluconazole reduced the number of CFU in the worm. Also, antifungal treatment was associated with the formation of cell aggregates around infected areas. Taking this into account, along with other published works (34, 35), we used the G. mellonella model to evaluate the efficacies of different antifungal drugs against C. tropicalis infection. The in vivo response correlates very well with the antifungal susceptibility phenotype shown in vitro. Fluconazole or voriconazole treatment is capable of improving the survival of larvae infected with azole-susceptible C. tropicalis (ATCC 750). These drugs improve survival in a dose-dependent manner. In fact, voriconazole and fluconazole are effective when the doses recommended for humans to treat invasive candidiasis or candidemia are used (11). On the other hand, these antifungal drugs are ineffective for larvae infected with azole-resistant strains (ATCC 200956, CL-6835, and TP-13650). However, amphotericin B treatment at therapeutic doses (1.2 μg/larva) improves larval survival when strains ATCC 750 and TP-13650 (amphotericin B susceptible) are used for infection but fail to improve the survival of larvae infected with strain CL-6835 or ATCC 200956 (amphotericin B resistant). The response to anidulafungin was used as a control for drug efficacy and to evaluate if the model was able to reproduce the in vitro phenotype of susceptibility to echinocandin drugs for all the strains. Anidulafungin treatment showed a decrease in the mortality of larvae, independently of the strain. We confirmed that the G. mellonella host is very useful for evaluation of the in vivo efficacies of antifungal agents in an experimental model of candidiasis.
In summary, we report the study of different mechanisms responsible for single and combined azole and amphotericin B resistance in C. tropicalis clinical isolates. Although increased efflux or missense mutations in ERG11 are the most frequent mechanisms involved in azole resistance, the decreased susceptibility of these isolates to azoles seemed to be due to an ERG11 gene deletion/amino acid substitution in an area of the protein affecting the site for both substrate binding and azole binding. The interruption of the ergosterol biosynthesis pathway would, in turn, be responsible for amphotericin B resistance in strains lacking functional Erg11p. The results of our study elucidate important characteristics and potential mechanisms of fluconazole and amphotericin B resistance in C. tropicalis. In addition, we validated G. mellonella as a simple nonmammalian model system that can be used to study in vitro-in vivo correlation of antifungals. Galleria mellonella larvae infected with Candida tropicalis strains showing in vitro resistance to amphotericin B, fluconazole, and voriconazole failed treatment with these antifungals but responded to echinocandins.
Supplementary Material
ACKNOWLEDGMENTS
E.M. was supported by the European Science Foundation (Fuminomics 06-RNP-132), the Research Projects from the Spanish Ministry of Science and Innovation (ERA-NET Pathogenomics [7th FP], BFU2008-04709-E/BMC), and the Fondo de Investigacion Sanitaria (PI12_02376, MPY1003/13). L.A.-F. is funded by the Spanish Fondo de Investigación Sanitaria with a Miguel Servet fellowship (FIS ref. CP11/00026). A.F. is funded by a fellowship from the Agencia Española de Cooperación Internacional para el Desarrollo (MAEC-AECID, Convocatoria 2011–2012, 0000557290). A.A.-I. and L.B.-M. have a research contract from the Spanish Network for Research in Infectious Diseases (REIPI RD06/0008). A.C.M.-A. is funded by a grant from the Fundacion Carolina and the University of Antioquia, Medellin, Colombia.
We thank Franco Forastiero for computer drawing assistance and Gema del Rio for technical support.
All authors declare that this research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
ADDENDUM
During the revision of this article, a related work, on the same resistance mechanism, by Eddouzi et al. (54) was accepted by Antimicrobial Agents and Chemotherapy.
Footnotes
Published ahead of print 22 July 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00477-13.
REFERENCES
- 1.Pfaller MA, Diekema DJ, Gibbs DL, Newell VA, Ellis D, Tullio V, Rodloff A, Fu W, Ling TA. 2010. Results from the ARTEMIS DISK Global Antifungal Surveillance Study, 1997 to 2007: a 10.5-year analysis of susceptibilities of Candida species to fluconazole and voriconazole as determined by CLSI standardized disk diffusion. J. Clin. Microbiol. 48:1366–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Negri M, Silva S, Henriques M, Oliveira R. 2012. Insights into Candida tropicalis nosocomial infections and virulence factors. Eur. J. Clin. Microbiol. Infect. Dis. 31:1399–1412 [DOI] [PubMed] [Google Scholar]
- 3.Pfaller MA, Messer SA, Moet GJ, Jones RN, Castanheira M. 2011. Candida bloodstream infections: comparison of species distribution and resistance to echinocandin and azole antifungal agents in intensive care unit (ICU) and non-ICU settings in the SENTRY Antimicrobial Surveillance Program (2008–2009). Int. J. Antimicrob. Agents 38:65–69 [DOI] [PubMed] [Google Scholar]
- 4.Pemán J, Cantón E, Quindós G, Eraso E, Alcoba J, Guinea J, Merino P, Ruiz-Pérez-de-Pipaon MT, Pérez del Molino L, Linares-Sicilia MJ, Marco F, García J, Roselló EM, Gómez-Garcia-de-la-Pedrosa E, Borrell N, Porras A, Yagüe G, on behalf of the FUNGEMYCA Study Group 2012. Epidemiology, species distribution and in vitro antifungal susceptibility of fungaemia in a Spanish multicentre prospective survey. J. Antimicrob. Chemother. 67:1181–1187 [DOI] [PubMed] [Google Scholar]
- 5.Chakrabarti A, Chatterjee SS, Rao KL, Zameer MM, Shivaprakash MR, Singhi S, Singh R, Varma SC. 2009. Recent experience with fungaemia: change in species distribution and azole resistance. Scand. J. Infect. Dis. 41:275–284 [DOI] [PubMed] [Google Scholar]
- 6.Adhikary R, Joshi S. 2011. Species distribution and anti-fungal susceptibility of candidaemia at a multi super-speciality center in Southern India. Indian J. Med. Microbiol. 29:309–311 [DOI] [PubMed] [Google Scholar]
- 7.Kothavade RJ, Kura MM, Valand AG, Panthaki MH. 2010. Candida tropicalis: its prevalence, pathogenicity and increasing resistance to fluconazole. J. Med. Microbiol. 59:873–880 [DOI] [PubMed] [Google Scholar]
- 8.Colombo AL, Nucci M, Park BJ, Nouér SA, Arthington-Skaggs B, da Matta DA, Warnock D, Morgan J. 2006. Epidemiology of candidemia in Brazil: a nationwide sentinel surveillance of candidemia in eleven medical centers. J. Clin. Microbiol. 44:2816–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kontoyiannis DP, Vaziri I, Hanna HA, Boktour M, Thornby J, Hachem R, Bodey GP, Raad II. 2001. Risk factors for Candida tropicalis fungemia in patients with cancer. Clin. Infect. Dis. 33:1676–1681 [DOI] [PubMed] [Google Scholar]
- 10.Chai LY, Denning DW, Warn P. 2010. Candida tropicalis in human disease. Crit. Rev. Microbiol. 36:282–298 [DOI] [PubMed] [Google Scholar]
- 11.Pappas PG, Kauffman CA, Andes D, Benjamin DK, Jr, Calandra TF, Edwards JE, Jr, Filler SG, Fisher JF, Kullberg BJ, Ostrosky-Zeichner L, Reboli AC, Rex JH, Walsh TJ, Sobel JD. 2009. Clinical practice guidelines for the management of candidiasis: 2009 update by the Infectious Diseases Society of America. Clin. Infect. Dis. 48:503–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ullmann AJ, Cornely OA, Donnelly JP, Akova M, Arendrup MC, Arikan-Akdagli S, Bassetti M, Bille J, Calandra T, Castagnola E, Garbino J, Groll AH, Herbrecht R, Hope WW, Jensen HE, Kullberg BJ, Lass-Flörl C, Lortholary O, Meersseman W, Petrikkos G, Richardson MD, Roilides E, Verweij PE, Viscoli C, Cuenca-Estrella M, ESCMID Fungal Infection Study Group 2012. ESCMID guideline for the diagnosis and management of Candida diseases 2012: developing European guidelines in clinical microbiology and infectious diseases. Clin. Microbiol. Infect. 18(Suppl 7):1–8 [DOI] [PubMed] [Google Scholar]
- 13.Myoken Y, Kyo T, Fujihara M, Sugata T, Mikami Y. 2004. Clinical significance of breakthrough fungemia caused by azole-resistant Candida tropicalis in patients with hematologic malignancies. Haematologica 89:378–380 [PubMed] [Google Scholar]
- 14.Vandeputte P, Larcher G, Bergés T, Renier G, Chabasse D, Bouchara JP. 2005. Mechanisms of azole resistance in a clinical isolate of Candida tropicalis. Antimicrob. Agents Chemother. 49:4608–4615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang YL, Wang AH, Wang CW, Cheng WT, Li SY, Lo HJ; TSARY Hospitals 2008. Susceptibilities to amphotericin B and fluconazole of Candida species in Taiwan Surveillance of Antimicrobial Resistance of Yeasts 2006. Diagn. Microbiol. Infect. Dis. 61:175–180 [DOI] [PubMed] [Google Scholar]
- 16.Rex JH, Cooper CR, Merz WG, Jr, Galgiani JN, Anaissie EJ. 1995. Detection of amphotericin B-resistant Candida isolates in a broth-based system. Antimicrob. Agents Chemother. 39:906–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cantón E, Pemán J, Gobernado M, Viudes A, Espinel-Ingroff A. 2004. Patterns of amphotericin B killing kinetics against seven Candida species. Antimicrob. Agents Chemother. 48:2477–2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lupetti A, Danesi R, Campa M, Del Tacca M, Kelly S. 2002. Molecular basis of resistance to azole antifungals. Trends Mol. Med. 8:76–81 [DOI] [PubMed] [Google Scholar]
- 19.Sanglard D, Odds FC. 2002. Resistance of Candida species to antifungal agents: molecular mechanisms and clinical consequences. Lancet Infect. Dis. 2:73–85 [DOI] [PubMed] [Google Scholar]
- 20.Pfaller MA. 2012. Antifungal drug resistance: mechanisms, epidemiology, and consequences for treatment. Am. J. Med. 125:S3–S13 [DOI] [PubMed] [Google Scholar]
- 21.Perea S, López-Ribot JL, Kirkpatrick WR, McAtee RK, Santillán RA, Martínez M, Calabrese D, Sanglard D, Patterson TF. 2001. Prevalence of molecular mechanisms of resistance to azole antifungal agents in Candida albicans strains displaying high-level fluconazole resistance isolated from human immunodeficiency virus-infected patients. Antimicrob. Agents Chemother. 45:2676–2684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morio F, Loge C, Besse B, Hennequin C, Le Pape P. 2010. Screening for amino acid substitutions in the Candida albicans Erg11 protein of azole-susceptible and azole-resistant clinical isolates: new substitutions and a review of the literature. Diagn. Microbiol. Infect. Dis. 66:373–384 [DOI] [PubMed] [Google Scholar]
- 23.Kelly SL, Lamb DC, Kelly DE. 1999. Y132H substitution in Candida albicans sterol 14α-demethylase confers fluconazole resistance by preventing binding to haem. FEMS Microbiol. Lett. 180:171–175 [DOI] [PubMed] [Google Scholar]
- 24.Sanglard D, Ischer F, Koymans L, Bille J. 1998. Amino acid substitutions in the cytochrome P-450 lanosterol 14α-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contribute to resistance to azole antifungal agents. Antimicrob. Agents Chemother. 42:241–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Becher R, Wirsel SG. 2012. Fungal cytochrome P450 sterol 14α-demethylase (CYP51) and azole resistance in plant and human pathogens. Appl. Microbiol. Biotechnol. 95:825–840 [DOI] [PubMed] [Google Scholar]
- 26.Xiao L, Madison V, Chau AS, Loebenberg D, Palermo RE, McNicholas PM. 2004. Three-dimensional models of wild-type and mutated forms of cytochrome P450 14α-sterol demethylases from Aspergillus fumigatus and Candida albicans provide insights into posaconazole binding. Antimicrob. Agents Chemother. 48:568–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang C, Dong D, Yu B, Cai G, Wang X, Ji Y, Peng Y. 2013. Mechanisms of azole resistance in 52 clinical isolates of Candida tropicalis in China. J. Antimicrob. Chemother. 68:778–785 [DOI] [PubMed] [Google Scholar]
- 28.Barchiesi F, Calabrese D, Sanglard D, Falcone Di Francesco L, Caselli F, Giannini D, Giacometti A, Gavaudan S, Scalise G. 2000. Experimental induction of fluconazole resistance in Candida tropicalis ATCC 750. Antimicrob. Agents Chemother. 44:1578–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacCallum DM, Coste A, Ischer F, Jacobsen MD, Odds FC, Sanglard D. 2010. Genetic dissection of azole resistance mechanisms in Candida albicans and their validation in a mouse model of disseminated infection. Antimicrob. Agents Chemother. 54:1476–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyazaki T, Miyazaki Y, Izumikawa K, Kakeya H, Miyakoshi S, Bennett JE, Kohno S. 2006. Fluconazole treatment is effective against a Candida albicans erg3/erg3 mutant in vivo despite in vitro resistance. Antimicrob. Agents Chemother. 50:580–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Desalermos A, Fuchs BB, Mylonakis E. 2012. Selecting an invertebrate model host for the study of fungal pathogenesis. PLoS Pathog. 8:e1002451. 10.1371/journal.ppat.1002451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Desbois AP, Coote PJ. 2011. Wax moth larva (Galleria mellonella): an in vivo model for assessing the efficacy of antistaphylococcal agents. J. Antimicrob. Chemother. 66:1785–1790 [DOI] [PubMed] [Google Scholar]
- 33.Harding CR, Schroeder GN, Reynolds S, Kosta A, Collins JW, Mousnier A, Frankel G. 2012. Legionella pneumophila pathogenesis in the Galleria mellonella infection model. Infect. Immun. 80:2780–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mylonakis E, Moreno R, El Khoury JB, Idnurm A, Heitman J, Calderwood SB, Ausubel FM, Diener A. 2005. Galleria mellonella as a model system to study Cryptococcus neoformans pathogenesis. Infect. Immun. 73:3842–3850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coleman JJ, Muhammed M, Kasperkovitz PV, Vyas JM, Mylonakis E. 2011. Fusarium pathogenesis investigated using Galleria mellonella as a heterologous host. Fungal Biol. 115:1279–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Slater JL, Gregson L, Denning DW, Warn PA. 2011. Pathogenicity of Aspergillus fumigatus mutants assessed in Galleria mellonella matches that in mice. Med. Mycol. 49:S107–S113 [DOI] [PubMed] [Google Scholar]
- 37.Mesa-Arango AC, Forastiero A, Bernal-Martínez L, Cuenca-Estrella M, Mellado E, Zaragoza O. 2013. The non-mammalian host Galleria mellonella can be used to study the virulence of the fungal pathogen Candida tropicalis and the efficacy of antifungal drugs during infection by this pathogenic yeast. Med. Mycol. 51:461–472 [DOI] [PubMed] [Google Scholar]
- 38.Scorzoni L, de Lucas MP, Mesa-Arango AC, Fusco-Almeida AM, Lozano E, Cuenca-Estrella M, Mendes-Giannini MJ, Zaragoza O. 2013. Antifungal efficacy during Candida krusei infection in non-conventional models correlates with the in vitro susceptibility profile. PLoS One 8:e60047. 10.1371/journal.pone.0060047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arendrup MC, Cuenca-Estrella M, Lass-Flörl C, Hope W. EUCAST-AFST 2012. EUCAST technical note on the EUCAST definitive document EDef 7.2: method for the determination of broth dilution minimum inhibitory concentrations of antifungal agents for yeasts EDef 7.2 (EUCAST-AFST). Clin. Microbiol. Infect. 18:E246–E247 [DOI] [PubMed] [Google Scholar]
- 40.Lass-Flörl C, Arendrup MC, Rodriguez-Tudela JL, Cuenca-Estrella M, Donnelly P, Hope W; European Committee on Antimicrobial Susceptibility Testing–Subcommittee on Antifungal Susceptibility Testing 2011. EUCAST technical note on amphotericin B. Clin. Microbiol. Infect. 17:E27–E29 [DOI] [PubMed] [Google Scholar]
- 41.Arendrup MC, Rodriguez-Tudela JL, Lass Flörl C, Cuenca-Estrella M, Donnelly JP, Hope W. EUCAST-AFST 2011. EUCAST technical note on anidulafungin. Clin. Microbiol. Infect. 17:E18–E20 [DOI] [PubMed] [Google Scholar]
- 42.Sanglard D, Ischer F, Parkinson T, Falconer D, Bille J. 2003. Candida albicans mutations in the ergosterol biosynthetic pathway and resistance to several antifungal agents. Antimicrob. Agents Chemother. 47:2404–2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ. 1998. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23:403–405 [DOI] [PubMed] [Google Scholar]
- 44.Arthington-Skaggs BA, Lee-Yang W, Ciblak MA, Frade JP, Brandt ME, Hajjeh RA, Harrison LH, Sofair AN, Warnock DW, for the Candidemia Active Surveillance Group 2002. Comparison of visual and spectrophotometric methods of broth microdilution MIC end point determination and evaluation of a sterol quantitation method for in vitro susceptibility testing of fluconazole and itraconazole against trailing and nontrailing Candida isolates. Antimicrob. Agents Chemother. 46:2477–2481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alcazar-Fuoli L, Mellado E, Garcia-Effron G, Lopez JF, Grimalt JO, Cuenca-Estrella JM, Rodriguez-Tudela JL. 2008. Ergosterol biosynthesis pathway in Aspergillus fumigatus. Steroids 73:339–347 [DOI] [PubMed] [Google Scholar]
- 46.Méjanelle L, Lòpez JF, Gunde-Cimerman N, Grimalt JO. 2001. Ergosterol biosynthesis in novel melanized fungi from hypersaline environments. J. Lipid Res. 42:352–358 [PubMed] [Google Scholar]
- 47.Yasmin S, Alcazar-Fuoli L, Gründlinger M, Puempel T, Cairns T, Blatzer M, Lopez JF, Grimalt JO, Bignell E, Haas H. 2012. Mevalonate governs interdependency of ergosterol and siderophore biosyntheses in the fungal pathogen Aspergillus fumigatus. Proc. Natl. Acad. Sci. U. S. A. 109:E497–E504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arnold K, Bordoli L, Kopp J, Schwede T. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195–201 [DOI] [PubMed] [Google Scholar]
- 49.Kiefer F, Arnold K, Künzli M, Bordoli L, Schwede T. 2009. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 37:D387–D392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peitsch MC. 1995. Protein modeling by E-mail. Bio/Technology 13:658–660 [Google Scholar]
- 51.Strushkevich N, Usanov SA, Park HW. 2010. Structural basis of human CYP51 inhibition by antifungal azoles. J. Mol. Biol. 397:1067–1078 [DOI] [PubMed] [Google Scholar]
- 52.Maesaki S, Marichal P, Vanden Bossche H, Sanglard D, Kohno S. 1999. Rhodamine 6G efflux for the detection of CDR1 overexpressing azole-resistant Candida albicans strains. J. Antimicrob. Chemother. 44:27–31 [DOI] [PubMed] [Google Scholar]
- 53.Melo F, Feytmans E. 1998. Assessing protein structures with a non-local atomic interaction energy. J. Mol. Biol. 277:1141–1152 [DOI] [PubMed] [Google Scholar]
- 54.Eddouzi J, Parker JE, Vale-Silva LA, Coste A, Ischer F, Kelly S, Manai M, Sanglard D. 2013. Molecular mechanisms of drug resistance in clinical Candida species isolated in Tunisian hospitals. Antimicrob. Agents Chemother. 57:3182–3193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marichal P, Koymans L, Willemsens S, Bellens D, Verhasselt P, Luyten W, Borgers M, Ramaekers FC, Odds FC, Bossche HV. 1999. Contribution of mutations in the cytochrome P450 14α-demethylase (Erg11p, Cyp51p) to azole resistance in Candida albicans. Microbiology 145:2701–2713 [DOI] [PubMed] [Google Scholar]
- 56.Hull CM, Parker JE, Bader O, Weig M, Gross U, Warrilow AG, Kelly DE, Kelly SL. 2012. Facultative sterol uptake in an ergosterol-deficient clinical isolate of Candida glabrata harboring a missense mutation in ERG11 and exhibiting cross-resistance to azoles and amphotericin B. Antimicrob. Agents Chemother. 56:4223–4232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Podust LM, Poulos TL, Waterman MR. 2001. Crystal structure of cytochrome P450 14α-sterol demethylase (CYP51) from Mycobacterium tuberculosis in complex with azole inhibitors. Proc. Natl. Acad. Sci. U. S. A. 98:3068–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ishida N, Aoyama Y, Hatanaka R, Oyama Y, Imajo S, Ishiguro M, Oshima T, Nakazato H, Noguchi T, Maitra US, Mohan VP, Sprinson DB, Yoshida Y. 1988. A single amino acid substitution converts cytochrome P450(14DM) to an inactive form, cytochrome P450SG1: complete primary structures deduced from cloned DNAS. Biochem. Biophys. Res. Commun. 155:317–323 [DOI] [PubMed] [Google Scholar]
- 59.Chau AS, Mendrick CA, Sabatelli FJ, Loebenberg D, McNicholas PM. 2004. Application of real-time quantitative PCR to molecular analysis of Candida albicans strains exhibiting reduced susceptibility to azoles. Antimicrob. Agents Chemother. 48:2124–2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sionov E, Chang YC, Garraffo HM, Dolan MA, Ghannoum MA, Kwon-Chung KJ. 2012. Identification of a Cryptococcus neoformans cytochrome P450 lanosterol 14α-demethylase (Erg11) residue critical for differential susceptibility between fluconazole/voriconazole and itraconazole/posaconazole. Antimicrob. Agents Chemother. 56:1162–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rodero L, Mellado E, Rodriguez AC, Salve A, Guelfand L, Cahn P, Cuenca-Estrella M, Davel G, Rodriguez-Tudela JL. 2003. G484S amino acid substitution in lanosterol 14-α demethylase (ERG11) is related to fluconazole resistance in a recurrent Cryptococcus neoformans clinical isolate. Antimicrob. Agents Chemother. 47:3653–3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pelaez T, Gijón P, Bunsow E, Bouza E, Sánchez-Yebra W, Valerio M, Gama B, Cuenca-Estrella M, Mellado E. 2012. Resistance to voriconazole due to a G448S substitution in Aspergillus fumigatus in a patient with cerebral aspergillosis. J. Clin. Microbiol. 50:2531–2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kelly SL, Lamb DC, Loeffler J, Einsele H, Kelly DE. 1999. The G464S amino acid substitution in Candida albicans sterol 14α-demethylase causes fluconazole resistance in the clinic through reduced affinity. Biochem. Biophys. Res. Commun. 262:174–179 [DOI] [PubMed] [Google Scholar]
- 64.Park HG, Lee IS, Chun IJ, Yun CH, Johnston JB, Ortiz de Montellon PR, Kima D. 2011. Heterologous expression and characterization of the sterol 14α-demethylase CYP51F1 from Candida albicans. Arch. Biochem. Biophys. 509:9–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mullins JGL, Parker JE, Cools HJ, Togawa RC, Lucas JA, Fraaije BA, Kelly DE, Kelly S. 2011. Molecular modelling of the emergence of azole resistance in Mycosphaerella graminicola. PLoS One 6:e20973. 10.1371/journal.pone.0020973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Délye C, Laigret F, Corio-Costet MF. 1997. A mutation in the 14α-demethylase gene of Uncinula necator that correlates with resistance to a sterol biosynthesis inhibitor. Appl. Environ. Microbiol. 63:2966–2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cañas-Gutiérrez GP, Angarita-Velásquez MJ, Restrepo-Flórez JM, Rodríguez P, Moreno CX, Arango R. 2009. Analysis of the CYP51 gene and encoded protein in propiconazole-resistant isolates of Mycosphaerella fijiensis. Pest. Manag. Sci. 65:892–899 [DOI] [PubMed] [Google Scholar]
- 68.Délye C, Bousset L, Corio-Costet MF. 1998. PCR cloning and detection of point mutations in the eburicol 14α-demethylase (CYP51) gene from Erysiphe graminis f. sp. hordei, a “recalcitrant” fungus. Curr. Genet. 34:399–403 [DOI] [PubMed] [Google Scholar]
- 69.Kelly SL, Lamb DC, Corran AJ, Baldwin BC, Kelly DE. 1995. Mode of action and resistance to azole antifungals associated with the formation of 14α-methylergosta-8,24(28)-dien-3β,6α-diol. Biochem. Biophys. Res. Commun. 207:910–915 [DOI] [PubMed] [Google Scholar]
- 70.Pinjon E, Moran GP, Jackson CJ, Kelly SL, Sanglard D, Coleman DC, Sullivan DJ. 2003. Molecular mechanisms of itraconazole resistance in Candida dubliniensis. Antimicrob. Agents Chemother. 47:2424–2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Morio F, Pagniez F, Lacroix C, Miegeville M, Le Pape P. 2012. Amino acid substitutions in the Candida albicans sterol Δ5,6-desaturase (Erg3p) confer azole resistance: characterization of two novel mutants with impaired virulence. J. Antimicrob. Chemother. 67:2131–2138 [DOI] [PubMed] [Google Scholar]
- 72.Martel CM, Parker JE, Bader O, Weig M, Gross U, Warrilow AG, Rolley N, Kelly DE, Kelly SL. 2010. Identification and characterization of four azole-resistant erg3 mutants of Candida albicans. Antimicrob. Agents Chemother. 54:4527–4533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kavanagh K, Reeves EP. 2004. Exploiting the potential of insects for in vivo pathogenicity testing of microbial pathogens. FEMS Microbiol. Rev. 28:101–112 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.