Abstract
We report here the affinity and antibacterial activity of a structurally similar class of neomycin dimers. The affinity of the dimer library for rRNA was established by using a screen that measures the displacement of fluorescein-neomycin (F-neo) probe from RNA. A rapid growth inhibition assay using a single drug concentration was used to examine the antibacterial activity. The structure-activity relationship data were then rapidly analyzed using a two-dimensional ribosomal binding-bacterial inhibition plot analysis.
INTRODUCTION
Aminoglycoside antibiotics are successful chemotherapeutics and have been used as effective, broad-spectrum treatments for a number of life-threatening bacterial diseases. The emergence of antibiotic-resistant bacterial strains associated with common aminoglycosides has challenged the clinical use of these antibiotics (1–4). In response to the emergence of these resistant strains, libraries of modified aminoglycosides are being developed.
Although a number of binding sites have been noted (5–7), the primary site at which neomycin and other aminoglycosides bind is the aminoacyl tRNA recognition region, the A-site, of the ribosome (8–10). Several studies have characterized the antibiotic class of aminoglycosides using a 27-base oligonucleotide that models the Escherichia coli ribosomal A-site (9, 11–13).
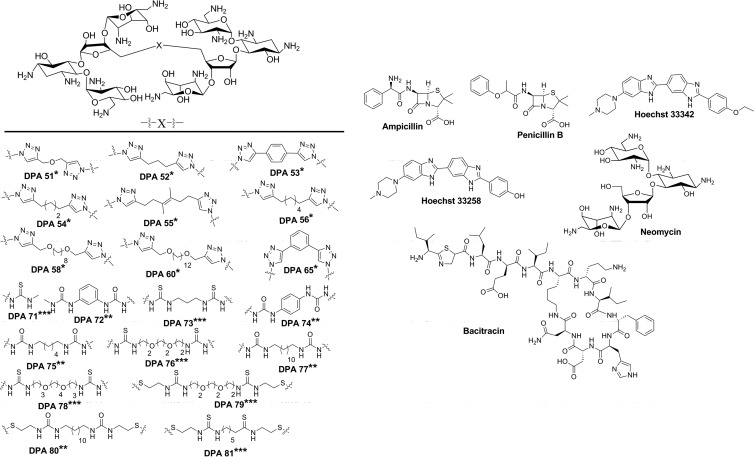
We describe here the screening of a library of structurally similar neomycin dimer compounds that vary in class by linker type (Fig. 1). The majority of recent previous studies of new aminoglycoside compounds have focused on low-throughput MIC calculations assuming the mechanism of aminoglycoside action is A-site binding but with no validation of this mechanism. Several of these recent aminoglycoside modifications have been reported that do not provide any information on the binding of compounds to the RNA A-site (14–19), although previous reports by Mobashery and Wong labs have studied RNA binding and antibacterial activity of modified aminoglycosides using low-throughput methods (20–29). In the present study, each compound was screened using high-throughput capable methods for both binding to a model ribosomal A-site and its ability to inhibit bacterial growth of a variety of bacterial strains in order to correlate the model A-site rRNA binding of each compound to the antibacterial activity.
Fig 1.
Structures of neomycin dimers and commercially available compounds included in the compound library. The neomycin dimers are grouped by linkage type: *, triazole based; **, urea based; and ***, thiourea based.
The A-site binding assay used here was recently developed by Watkins et al. (30). This high-throughput fluorescence-based competition assay uses a 27-base RNA model of the ribosomal A-site and a fluorescent reporter molecule, F-neo (Fig. 2a). This assay is capable of measuring the relative binding affinity of a drug library compared to neomycin, and the assay was validated for A-site binding by accurately ranking the relative binding affinities of known aminoglycosides with previously established results. The binding of F-neo to the A-site results in a quenching of the fluorescence of the probe. The displacement of the probe from the model A-site results in an increase in the fluorescence of the probe. The relative binding affinity is associated with the difference in the fluorescence in the absence of the drug and in the presence of the drug. This study represents the first practical application of this assay to a structurally similar class of aminoglycoside library for which no A-site binding data or antibacterial activity data exists.
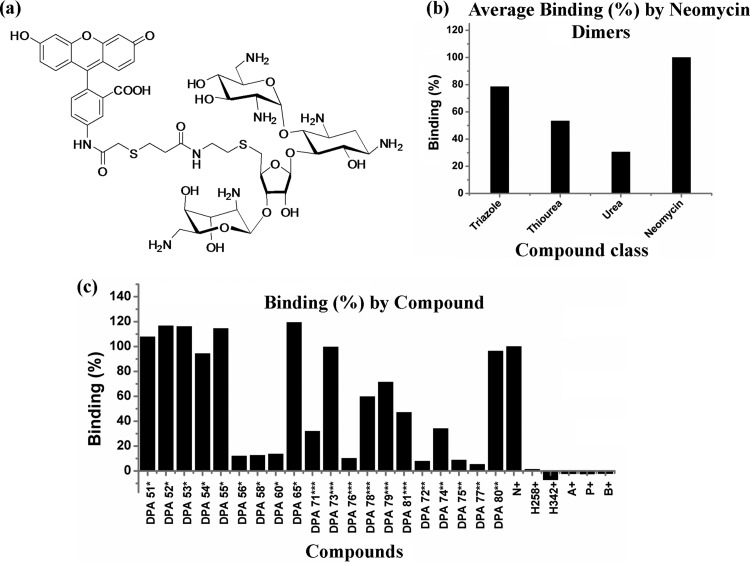
Fig 2.
F-neo competitive binding screen. (a) Structure of F-neo probe. (b) rRNA binding determined by measuring the change in florescence (F-Fo) due to displacement of F-neo by A-site binding compounds. (c) Percent binding by compound. A displacement assay was performed with each compound of interest to F-neo:A-site complex using an excitation/emission of 485/535 nm. All experiments were performed in 10 mM MOPSO (pH 7.0), 50 mM NaCl, and 0.4 mM EDTA. N, neomycin; H258, Hoechst 33258; H342, Hoechst 33342; A, ampicillin; P, penicillin; B, bacitracin.
The neomycin dimer library was screened for antibacterial activity using a single concentration high throughput assay developed by De La Fuente et al. This method has been previously used as an initial antibacterial screening technique in order to identify promising classes of compounds in a small-molecule library (31). We have adapted this screen to be used with our compound library to provide a high-throughput method for identifying compounds with antibacterial activity. In order to verify this single concentration screen as a valid estimate of antibacterial activity, the MIC was determined for each compound.
Finally, we describe a combination of these two screens into a ribosomal binding-bacterial inhibition plot (RB-BIP) analysis, of a compound library of known antibacterial compounds and a neomycin dimer library. Previous work has used compound screening assays and cell-based assays to allow the correlation of mechanism with action within the cell (24, 32–34); however, this RB-BIP data analysis allows the direct cross-correlation of two high-throughput screens to allow for the rapid correlation and interpretation of rRNA binding and antibiotic activity. The analysis allows the compounds to be quickly characterized into four categories, indicating that a drug (i) binds its rRNA target and does not inhibit growth (due to resistance or reduced uptake), (ii) is both effective at binding its target and inhibits growth, (iii) does not bind its target and does not inhibit growth (ineffective compound due to predicted mechanism), or (iv) does not bind its target and inhibits growth. RB-BIP analysis provides rapid insight into the structure-activity relationship (SAR) for the dimer library and may be adaptable to other systems to provide similar information.
MATERIALS AND METHODS
Bacterial strains and materials.
The bacterial strains Enterobacter cloacae ATCC 13047, E. coli, ATCC 25922, Pseudomonas aeruginosa ATCC 27853, Serratia marcescens ATCC 13880, and methicillin-resistant Staphylococcus aureus (MRSA) ATCC 33591 were purchased from American Type Culture Collection (ATCC; Manassas, VA). Staphylococcus aureus ATCC 25923, Staphylococcus epidermidis ATCC 12228, and Streptococcus pyogenes ATCC 12384 were a generous gift from Tamara McNealy (Clemson University, Clemson, SC). All culture supplies and chemicals were purchased from Thermo Fisher Scientific (Pittsburgh, PA) and Sigma-Aldrich (St. Louis, MO).
Compound library.
Neomycin sulfate, Hoechst stains 33258 and 33342, ampicillin sodium salt, penicillin G sodium salt, and bacitracin were all obtained from commercial sources. Synthesis of DPA 79 and all triazole-linked dimers has been reported previously by us (35–37). The thiourea linkers DPA 71, DPA 73, DPA 76, DPA 78, and DPA 81 and the urea linkers DPA 72, DPA 74, DPA 75, DPA 77, and DPA 80 were synthesized as previously described for the synthesis of DPA 79 (37), and the characterization details will be reported elsewhere.
Single concentration antibacterial screen.
The antibacterial activity of the compound library was assessed by using the eight bacterial strains described above. The screening of the compound library performed here is an adaptation of a turbidometric assay described by De La Fuente et al. (31). Adaptations include the reduction of drug concentration to 5 μM, the use of Mueller-Hinton II cation-adjusted broth, and individual screening of drugs. In addition, manual pipettes (single channel and multichannel) were used rather than robotic liquid handlers. The S. pyogenes screen also included a supplement of 5% (vol/vol) lysed horse blood in accordance with Clinical and Laboratory Standards Institute standards (38). Diluent controls, water, and 2% (vol/vol) dimethyl sulfoxide were included on each plate. The turbidity of each sample was measured by recording the optical density at 595 nm of each sample after incubation at 35°C without agitation for 20 h. Growth inhibition was determined as the percent growth subtracted from 100%. The percent growth was calculated as the ratio of the optical densities of cultures in the presence of test compounds versus the optical densities of cultures grown without compound treatment. In cases where treatment appeared to enhance the growth of the culture, inhibition was scored as zero rather than as negative inhibition.
Determination of MIC.
Confirmation of single concentration antibacterial screen was determined for neomycin and the dimer compounds by determining the MIC in each of the strains according to the standard broth microdilution method. In short, a serial 2-fold dilution assay was set up in 96-well microtiter plates (38). Wells were inoculated with 105 CFU/ml. The CFU/ml was calculated using equation 1 (see reference 39):
| (1) |
Lysed horse blood at a final concentration of 5% (vol/vol) was included for the S. pyogenes studies. The 96-well plates were incubated at 35°C without agitation for 20 h. Optical density measurements at 595 nm were recorded after 20 h, and the MIC was determined.
F-neo competitive binding assay.
The described compound library was screened using an F-neo (Fig. 2a) competitive binding assay as described in detail by Watkins et al. (30). In short, the displacement assay was performed with each test compound at a 0.3 μM concentration to 0.1 μM F-neo:A-site complex using 100 reads per well at an excitation/emission of 485/535 nm. All experiments were performed in 10 mM MOPSO (β-hydroxy-4-morpholinepropanesulfonic acid, 3-morpholino-2-hydroxypropanesulfonic acid; pH 7.0), 50 mM NaCl, and 0.4 mM EDTA. The displacement of F-neo by test compounds was determined from the increase in fluorescence measured after the addition of the test compound to wells of a 96-well plate containing the F-neo:A-site complex compared to the fluorescence of the F-neo:A-site complex alone (ΔF). The percent binding was calculated from the ratio of the change in fluorescence by the addition of the test compound (ΔFdrug) with the change in fluorescence by addition of neomycin (ΔFneomycin) using equation 2:
| (2) |
The binding affinities of the 20 neomycin dimers and the commercially available compounds neomycin, ampicillin, penicillin, bacitracin, Hoechst 33258, and Hoechst 33342 were calculated as a percentage of neomycin binding in the RB-BIP analysis.
RESULTS
Bacterial growth inhibition assays.
The single-concentration (5 μM) growth inhibition screen of neomycin dimers was performed on five neomycin-susceptible strains and three neomycin-resistant strains with the compound library. The average growth inhibition of all neomycin dimers within a linker class indicates that compounds with the thiourea linker are the broadest class of antibiotics within this library (Fig. 3a). The thiourea compounds have an average inhibition of >80% across all neomycin-susceptible strains, S. aureus (ATCC 25923), E. coli, S. marcescens, E. cloacae, and S. epidermidis (S. epidermidis is not shown in Fig. 3). Compounds DPA 73, DPA 78, DPA 79, and DPA 81 of the thiourea linker class all inhibit growth of all susceptible strains by ≥80%, and DPA 71 inhibits the growth of all susceptible strains by ≥40% (Fig. 3b). The outlier within the thiourea class appears to be DPA 76, which only inhibits S. aureus (ATCC 25923) by >40%.
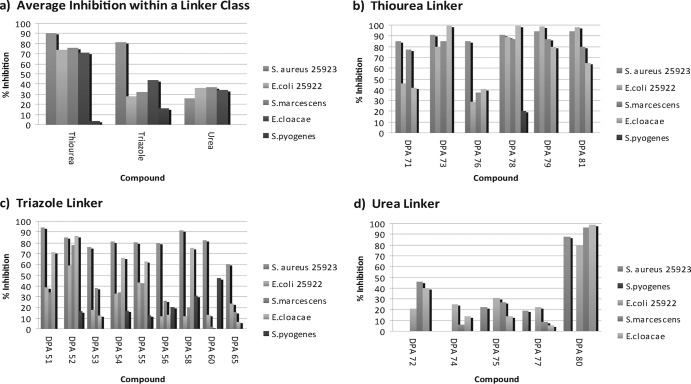
Fig 3.
Representative data from an initial antibacterial screen using S. aureus (ATCC 25923) and E. coli (ATCC 25922) showing the average percent inhibitions (a) and the percent inhibitions with thiourea (b), triazole (c), and urea (d) linkers. Cultures at an optical density of 0.005 at 595 nm were treated with compound at a final concentration of 5 μM in cation-adjusted Mueller-Hinton Broth II. Cultures were incubated for 20 h at 35°C. The optical density at 595 nm was recorded, and the percent inhibition was calculated. N, neomycin; H258, Hoechst 33258; H342. Hoechst 33342; A, ampicillin; P, penicillin; B, bacitracin.
The thiourea linker class compounds of the neomycin library are largely ineffective on the resistant strains S. pyogenes, MRSA, and P. aeruginosa (Fig. 3a and b [the MRSA and P. aeruginosa data are not shown]; data are available in Fig. 5 for all strains [see below]). The inhibition of DPA 78 by 20% is the only measurable inhibition by any of the thiourea compounds of S. pyogenes at 5 μM. No thiourea compounds inhibit the growth of any resistant strain by >30%.
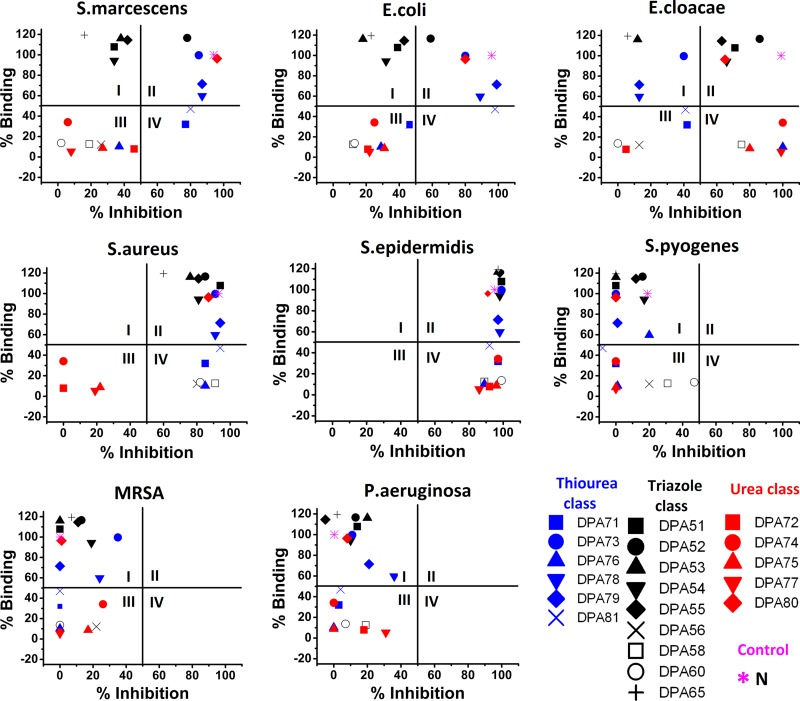
Fig 5.
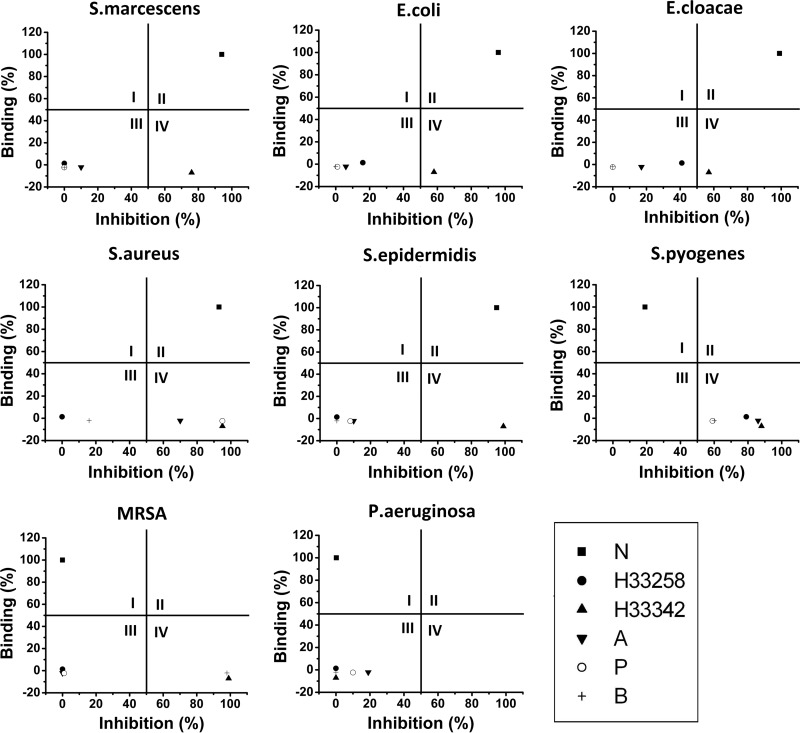
RB-BIP comparing F-neo displacement assay and antibacterial screen results using novel neomycin dimers. Percent binding is the displacement of F-neo by neomycin. Percent inhibition is the percent inhibition of growth by the compound of interest compared to the untreated cultures in the designated strain. Results for S. marcescens, E. coli, E. cloacae, S. aureus, S. epidermidis, S. pyogenes, MRSA, and P. aeruginosa are shown. Quadrants are indicated by Roman numerals.
The triazole linker class of neomycin dimers also has broad antibiotic activity across susceptible strains of the bacteria (Fig. 3a). Compound DPA 52 was the most effective compound, inhibiting all neomycin-susceptible strains growth by >50% (Fig. 3c). Compounds DPA 51, DPA 54, and DPA 55 have a somewhat broad range, inhibiting S. aureus (ATCC 25923) and E. cloacae by >60% and E. coli and S. marcescens by ≥40%. Compounds DPA 53, DPA 56, and DPA 65 are the least effective compounds in the triazole class of linkers, significantly inhibiting the growth of S. aureus (ATCC 25923) only.
The triazole linker class is the most effective compound class against the resistant strain S. pyogenes. DPA 60 inhibits the growth of this neomycin-resistant strain by >50%. In addition, DPA 58 inhibits the growth of S. pyogenes by >30%. The triazole linker compounds are the only class of neomycin dimers in the library with multiple compounds that inhibit a resistant strain by >20%, and half of the compounds in the class have measurable inhibition, i.e., >10%, against all resistant strains (Fig. 3c, MRSA and P. aeruginosa data not shown). Data are available in Fig. 5 for all strains (see below).
The neomycin dimers with the urea linker class are the most ineffective compounds across all strains in the library (Fig. 3a). Only two compounds in the urea class inhibit the growth of a bacterial strain other than S. epidermis, a strain inhibited by all compounds within the library, by >40%. An exception in the urea class of neomycin dimer is DPA 80, which inhibits the growth of all susceptible strains by ≥80% (Fig. 3d). In fact, if this compound is removed from the average inhibition of the urea class of compounds, no strain has an average inhibition of >25% for the urea class, and most inhibition falls at or below 20%.
The single concentration screen results are confirmed by the MIC using a broth microdilution assay.
In order to confirm the screen results, broth microdilution was completed with neomycin and the 20 neomycin dimer compounds using the eight strains included here (Table 1). Initial MIC determinations of small sample of randomly selected dimers with various bacteria strains indicated that compounds inhibiting growth by ≥50% at 5 μM would inhibit growth completely at a concentration of 10 to 15 μM. The single concentration hit, inhibiting growth by ≥50% at 5 μM, was considered a false positive if the MIC is >15 μM. The compound was considered false negative if the MIC was determined to be ≤15 μM and was not determined to be a hit, as defined by bacterial growth inhibition by 50% at 5 μM in the single point screen.
Table 1.
Antibacterial activities presented as drug MICs at which no visible growth was observed
| Compound | Mol wt | Drug MICs (μg/ml or μM) for various strainsa: |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
S. aureus |
S. epidermidis |
MRSA |
S. pyogenes |
E. coli |
E. cloacae |
S. marcescens |
P. aeruginosa |
||||||||||
| μg/ml | μM | μg/ml | μM | μg/ml | μM | μg/ml | μM | μg/ml | μM | μg/ml | μM | μg/ml | μM | μg/ml | μM | ||
| Neomycinb | 908.88 | 0.57 | 0.63 | 0.1 | 0.1 | 145 | 159.5 | >1,160 | >1,280 | 5 | 5.5 | 2 | 2.2 | 2 | 2.2 | 582 | 640.4 |
| DPA 51 | 1,810.4 | 4.5–9.1 | 2.5–5 | 1.25 | 0.69 | >80 | >44.2 | 20–40 | 11–22† | 10–20 | 5.5–11† | 20 | 11.04 | 20–40 | 11–22† | 20–40 | 11–22† |
| DPA 52 | 1,802.42 | 2.5 | 1.4 | 1.25 | 0.69 | >80 | >44.4 | 40–80 | 22–44 | 20 | 11 | 20 | 11.1 | 20–40 | 11–22* | 40–80 | 22–44 |
| DPA 53 | 1,843 | 36.9 | 20* | 9 | 5 | >80 | >43.4 | >80 | >43.4 | 40 | 21.7 | 80 | 43.4 | 80 | 43.4 | 40 | 21.7 |
| DPA 54 | 1,823.01 | 5–10 | 2.7–5.5 | 2.5 | 1.4 | >80 | >43.9 | >80 | >43.9 | 80 | 43.9 | 80 | 43.9* | 20–40 | 11–22† | >80 | >43.9 |
| DPA 55 | 1,877.1 | 5 | 2.5 | 1.25 | 0.66 | >80 | >42.6 | 40–80 | 21–42 | 40 | 21.3 | 80 | 42.6* | 20–40 | 11–22† | >80 | >42.6 |
| DPA 56 | 1,851.06 | 2.3–4.6 | 1.25–2.5 | 1.1 | 0.63 | 80 | 43.2 | >80 | >43.2 | 20 | 10.3† | 19–37 | 10–20† | 37 | 20 | >80 | >43.2 |
| DPA 58 | 1,939.57 | 2.4 | 1.25 | 1.2 | 0.63 | >80 | >41.2 | >80 | >41.2 | 80 | 41.2 | 20 | 9.8 | 40 | 20.1 | >80 | >41.2 |
| DPA 60 | 1,995.27 | 5–10 | 2.5–5 | 1.2 | 0.63 | 40 | 20.1 | >80 | >40.1 | 20–40 | 10–20† | >80 | >40.1 | >80 | >40.1 | >80 | >40.1 |
| DPA 65 | 1,843 | 2 | 1 | 1.2 | 0.63 | 37 | 20.1 | >80 | >43.4 | 37 | 20 | >80 | >43.4 | >80 | >43.4 | 37 | 20 |
| DPA 71 | 1,706.91 | 10 | 6 | 1.25–2.5 | 0.7–1.5 | >80 | >46.9 | >80 | >46.9 | 40–80 | 23.5–47 | 20–40 | 12–24† | 20–40 | 12–24* | >80 | >46.9 |
| DPA 72 | 1,824.98 | >80 | >43.8 | 20 | 11 | >80 | >43.8 | >80 | >43.8 | >80 | >43.8 | >80 | >43.8 | 20–40 | 11–22† | >80 | >43.8 |
| DPA 73 | 1,823.09 | 2.3 | 1.25 | 1.1 | 0.63 | 80 | 43.9 | >80 | >43.9 | 9 | 5 | 10 | 4.9 | 9–18 | 5–10 | >80 | >43.9 |
| DPA 74 | 1,824.98 | 40 | 22 | 5 | 3 | >80 | >43.8 | >80 | >43.8 | >80 | >43.8 | 80 | 43.8 | >80 | >43.8 | >80 | >43.8 |
| DPA 75 | 1,833.04 | 2.5–5 | 1.4–2.7† | 1.25 | 0.68 | >80 | >43.6 | 40–80 | 22–44 | 20–40 | 11–22† | 40 | 21.8 | 20–40 | 11–22† | >80 | >43.6 |
| DPA 76 | 1,897.17 | 10–20 | 5.3–10.5 | 2.5–5 | 1.3–2.6 | >80 | >42.2 | >80 | >42.2 | 40 | 21.1 | 40 | 21.1 | 20–40 | 11–22† | >80 | >42.2 |
| DPA 77 | 1,917.2 | 19.2–38.3 | 10–20† | 2.3–4.8 | 1.25–2.5 | >80 | >41.7 | >80 | >41.7 | 40 | 20.8 | 38 | 19.8 | 40 | 20.9 | >80 | >41.7 |
| DPA 78 | 1,953.28 | 20 | 10† | 4.9 | 2.5 | 39 | 20 | >80 | >41.0 | 5–10 | 2.5–5 | 5 | 2.5 | 2.4–4.9 | 1.25–2.5 | >80 | >41.0 |
| DPA 79 | 2,017.41 | 10 | 5 | 2.5 | 1.25 | >80 | >39.7 | >80 | >39.7 | >80 | >39.7* | >80 | >39.7* | >80 | >39.7* | >80 | >39.7 |
| DPA 80 | 2,037.44 | 1.25–2.5 | 0.6–1.2 | 1.25 | 0.61 | 40 | 19.7 | 40–80 | 20–40 | 20 | 9.8 | 40 | 19.6* | >80 | >39.3* | >80 | >39.3 |
| DPA 81 | 2,934.88 | 1.25–2.5 | 0.4–0.9 | 2.5 | 0.85 | >80 | >27.3 | >80 | >27.3 | 20 | 6.8 | 80 | 27.3* | 20–40 | 7–14 | >80 | >27.3 |
Strains: S. aureus ATCC 25923, S. epidermidis ATCC 12228, methicillin-resistant S. aureus ATCC 33591, S. pyogenes ATCC 12384, E. coli ATCC 25922, E. cloacae ATCC 13047, S. marcescens ATCC 13880, and P. aeruginosa ATCC 27853. MICs for which the screening results were not clearly confirmed: *, ≥50% inhibition; and †, <50% inhibition in a single point screen.
That is, neomycin sulfate.
Of the 168 broth microdilution studies completed, 93% (156 studies) of the MIC results confirmed the results found in the single point screen, either the compound inhibited growth by <50% and the MIC was >15 μM or the compound inhibited growth by >50% and the MIC was <15 μM (Fig. 4 and Table 1).
Fig 4.
RB-BIP validation comparing F-neo displacement assay and antibacterial screen results using known drug controls. Percent binding is the displacement of F-neo by neomycin. Percent inhibition is the percent inhibition of growth by the compound of interest compared to the untreated cultures in the designated strain. Results for S. marcescens, E. coli, E. cloacae, S. aureus, S. epidermidis, S. pyogenes, MRSA, and P. aeruginosa are shown. Quadrants are indicated by Roman numerals.
In this comparison of single point screen results and MIC values, only 7% of the MIC values did not confirm the single point screen results. The false positives from the screen appear to be dependent on the cell strain, with four of the nine false positives distributed evenly between the neomycin dimer classes in the E. cloacae strain, two false negatives in S. marcescens, and one false positive in each of E. coli and S. aureus (Table 1, designated by “*”). Only three false negatives occur in the single concentration screen. One occurs in the S. aureus, while the other two false negatives occur in the E. coli strain (Table 1, designated by “†”). In addition, the MIC values of the false negatives within the E. coli strain fall only slightly below the cutoff for the being classified as effective compounds by the MIC.
F-neo competitive binding screen.
The F-neo displacement assay used here detects the binding ability of a compound to the model ribosomal A-site. The aminoglycoside neomycin binds with high affinity to the model A-site. Neomycin causes a large change in fluorescence (ΔF), due to the displacement of F-neo probe from the model ribosomal A-site (Fig. 2b). Known antibacterial drugs that do not bind to the ribosomal A-site (Hoechst 33258, Hoechst 33342, ampicillin, penicillin, and bacitracin) showed no change in the fluorescence, indicating that they do not bind with a comparable affinity to neomycin to the model RNA A-site (Fig. 2b).
The ΔF due to the displacement of F-neo varied considerably among the neomycin dimers. Neomycin dimer DPA 73 from the thiourea class had a high binding affinity compared to neomycin. Two thiourea dimers, DPA 78 and DPA 79, bind the A-site with 60% neomycin affinity or greater. Compound DPA 71 and DPA 81 bind to the model A-site moderately with <50% of the binding affinity of neomycin. The weakest binding molecule of the thiourea class of linkers was DPA 73, binding with only 10% of the relative binding affinity of neomycin.
The triazole linker class had six compounds—DPA 51, DPA 52, DPA 53, DPA 54, DPA 55, and DPA 65–that bind the A-site with similar or greater affinity compared to neomycin. The remaining three dimers from the triazole class—DPA 56, DPA 58, and DPA 60—bind weakly to the model RNA A-site with <15% of the binding affinity of neomycin.
As a group, the urea linker class of dimers bind the model A-site weakly. However, DPA 80 has an affinity for the model A-site that approaches that of neomycin. Only DPA 74, of the remaining urea dimers—DPA 72, DPA 74, DPA 75, and DPA 77—displaces F-neo from the A-site by >10% relative to neomycin, with close to 34% displacement.
RB-BIP analysis.
In order to obtain SAR for a structurally similar compound's ability to bind to the model ribosomal A-site and inhibit bacterial growth, the data from the two previously mentioned screens were plotted on ribosomal binding-bacterial inhibition plots (RB-BIPs; Fig. 4 and 5). RB-BIPs outline the percent ribosomal binding of the compound compared to neomycin on the y axis and the percent bacterial inhibition compared to untreated cultures on the x axis. The RB-BIP can then be divided into quadrants: I, II, III, and IV. Compounds plotted in quadrant I exhibit strong binding to the model ribosomal A-site but weakly inhibit bacterial growth. Compounds plotted in quadrant II display both high binding to the ribosomal A-site and the inhibition of bacterial growth. Quadrant III of the RB-BIP contains compounds with low ribosomal binding and inhibition of bacterial growth. The final quadrant, quadrant IV, shows compounds exhibiting inhibition of bacterial growth with weak binding to the model A-site.
RB-BIP analysis: validation with known drug controls.
(i) RB-BIP analysis was performed with a standard compound library with known A-site binding affinity on the five neomycin-susceptible strains (Fig. 4). As expected, neomycin graphs into quadrant II, indicating that the inhibition of the neomycin-susceptible strains follow the predicted mechanism of ribosomal A-site binding. The control compounds Hoechst 33258, ampicillin, penicillin, and bacitracin appear in quadrant III in four of the neomycin-susceptible strains. These control compounds have low binding affinity for the ribosomal A-site and limited inhibition of growth in the three Gram-negative bacteria strains and the Gram-positive strain, S. epidermidis. The inhibition of growth in S. aureus (ATCC 25923) by Hoechst 33258 and bacitracin is also low, since these compounds appear in quadrant III. However, ampicillin and penicillin do inhibit the growth of S. aureus (ATCC 25923), as noted by their presence in quadrant IV for this strain. Hoechst 33342 consistently appears in quadrant IV of the RB-BIP. Hoechst 33342 is a known non-A-site binding control but is capable of inhibiting many bacterial strains by an alternate mechanism, as indicated by the presence of Hoechst 33342 in quadrant IV.
(ii) RB-BIP analysis was performed with a standard compound library with known A-site binding affinity on the three neomycin-resistant strains (Fig. 4). As expected, neomycin shifts from quadrant II to quadrant I in the RB-BIP analysis of the neomycin-resistant strains. The established antibiotic compounds that work by alternate mechanisms were more successful against MRSA and S. pyogenes, as indicated by their presence in quadrant IV, with Hoechst 33342 and bacitracin inhibiting growth in both strains. Penicillin, ampicillin, and Hoechst 33258 inhibited the growth of S. pyogenes. As expected, all compounds were found in either quadrant I or quadrant III in the RB-BIP analysis with P. aeruginosa due to the lack of inhibition of growth regardless of the mechanism of action.
RB-BIP analysis of dimers with neomycin-susceptible strains.
(i) RB-BIP analysis of the neomycin dimer library was performed on three Gram-negative neomycin-susceptible strains: S. marcescens, E. coli, and E. cloacae (Fig. 5). One triazole linker class compound (DPA 52), three thiourea class compounds (DPA 73, DPA 78, and DPA 79), and one urea class compound (DPA 80) appear in quadrant II, indicating that binding to the ribosomal A-site is the mechanism of growth inhibition by these compounds for in S. marcescens, E. coli, and E. cloacae. In addition to these five compounds, three triazole class dimers—DPA 51, DPA 54, and DPA 55—approach quadrant II from quadrant I. Conversely, the triazole class dimers, DPA 53 and DPA 65, appear more entrenched in quadrant I. The greater inhibition of growth may indicate that DPA 54 and DPA 55 are better candidates for antibiotics than DPA 53 or DPA 65, despite the higher affinity for the ribosomal A-site observed in DPA 53 and DPA 65.
Two thiourea dimers, DPA 71 and DPA 81, appear to be approaching quadrant II from quadrants III and IV in all of the neomycin-susceptible Gram-negative strains, with growth inhibition of >50% in S. marcescens (Fig. 5) and approaching 50% in E. coli and E. cloacae (Fig. 4). Both of these compounds have an A-site binding percentage approaching 50% compared to neomycin.
The least effective compounds in both rRNA binding and inhibition are located exclusively in quadrant III. This quadrant houses four triazole class dimers (DPA 52, DPA 56, DPA 58, and DPA 60), one thiourea compound (DPA 76), and four urea compounds (DPA 72, DPA 74, DPA 75, and DPA 77), which all have low binding affinity for the ribosomal A-site and limited inhibition of growth in these three Gram-negative bacterial strains.
(ii) RB-BIP analysis of the neomycin dimer library was performed on two Gram-positive neomycin-susceptible strains: S. aureus and S. epidermidis (Fig. 5). The neomycin-susceptible Gram-positive bacteria tested here appear to be much more susceptible to the dimer compounds than the neomycin-susceptible Gram-negative strains to A-site binding antibiotics. All compounds within quadrant I of the Gram-negative S. marcescens, E. coli, and E. cloacae shift to quadrant II of the neomycin-susceptible Gram-positive strains. This trend continues with two other thiourea dimers (DPA 71 and DPA 81) which appeared in both quadrants III and IV of the Gram-negative neomycin-susceptible strains and are exclusively in quadrant IV of the Gram-positive neomycin-susceptible strains, indicating that the Gram-positive strains are more susceptible to even relatively weak A-site binders. S. epidermidis was highly susceptible to almost all compounds tested either through ribosomal A-site binding compounds (as shown in quadrant II) or by binding to an rRNA different from the model A-site used here (as shown in quadrant IV). This strain of bacteria was the only strain susceptible to inhibition by the poor A-site binding neomycin urea-linked dimers DPA 72, DPA 74, DPA 75, and DPA 77, which shift to quadrant IV in the S. epidermidis.
RB-BIP neomycin-resistant strains.
RB-BIP analysis of the neomycin dimer library was performed on strains resistant to neomycin: S. pyogenes, S. aureus (MRSA, ATCC 33591), and P. aeruginosa (Fig. 5). All compounds that bind to the model ribosomal A-site shift from quadrant II to quadrant I, indicating that the mechanism of resistance for neomycin is effective in the resistance to the dimer compounds that bind to the A-site.
All neomycin dimer compounds that previously inhibited growth despite showing poor binding to the model rRNA A-site were also unable to significantly inhibit growth of any of the neomycin-resistant strains. However, DPA 60 does approach quadrant IV from quadrant III in S. pyogenes, indicating that this compound may be useful as an agent against this strain and this activity does not likely stem from binding to the model A-site rRNA used here.
DISCUSSION
Single concentration bacterial inhibition assay as a rapid estimation of compound efficacy.
A single concentration screen for bacterial inhibition studies was adapted from previous work to determine a compounds ability to inhibit bacterial growth in a rapid high-throughput format. De la Fuente et al. established this screen using P. aeruginosa and E. coli to identify classes of small-molecule antimicrobials (31). The single concentration of 5 μM used to estimate the inhibition of a compound was chosen by analyzing the effective concentration of neomycin (control) to inhibit the growth of the susceptible strains (S. aureus, S. epidermidis, E. coli, S. marcescens, and E. cloacae) by >95%.
In order to validate the single point screen as an effective method for estimating an effective compound for future development, the MIC was determined for each compound examined for each strain. In the single concentration bacterial inhibition screen, a compound was considered a hit if the compound inhibited the growth of the bacterial strain by >50% at 5 μM. Because the MIC curves are nonlinear, the single concentration screen could possibly over- or underestimate the efficacy of the compound. However, the MIC determination of all neomycin dimer compounds within this library across all eight strains included here confirmed the results of the single concentration assay in 93% of the screen.
Description and construction of RB-BIPs.
High-throughput screening allows for the rapid testing of large compound libraries for target affinity and/or biological effect. Because large amounts of data are obtained rapidly from this screening, a high-throughput method of data analysis should be part of the strategy in library screening. We combined the data analysis of two screening methodologies of target screening for ribosomal A-site binding and antibacterial activity in a variety of bacteria strains into a single two-dimensional plot. RB-BIP analysis allows the interpretation of two separate techniques simultaneously. The analysis is more powerful than the single interpretation alone because it allows correlations to be drawn as to possible cause and effect. Below we identify several observations taken directly from the plots of our data that are useful guides for SAR studies of structurally similar libraries, such as those used here.
RB-BIP analysis of neomycin dimers.
(i) The mechanisms of inhibition of growth of a strain can be directly correlated with a proposed pharmacodynamic model of binding. The use of quadrants within the plots allows for the rapid interpretation of data showing compounds that inhibit bacterial growth with the binding to the model target ribosomal A-site. In RB-BIP, quadrant II is correlated with A-site binding and inhibition of growth. The presence of neomycin within this quadrant in all strains susceptible to this antibiotic supports this interpretation. In the present analysis, quadrant II is defined as 50% inhibition of growth and 50% A-site binding compared to neomycin. This quadrant acts as the hit window for the screening analysis. Here, the hit window was defined by sectioning the plot at 50% binding and 50% inhibition. Fifty percent was chosen because of the commonality of finding concentrations at which 50% inhibition occurs (i.e., IC50, MIC50, etc.). The size of the quadrant and the number of compounds within the quadrant can be adjusted by changing either parameter of percent growth inhibition or binding, as appropriate for library size, limiting the number of compounds for downstream testing, or minimal needed effect.
(ii) The transition from quadrant II to quadrant I provides insight into the mechanism of resistance from the susceptible strains to the resistant strains. Using RB-BIP analysis, this resistance information is readily available through rapid interpretation of the plots of multiple strains. The same analysis can be used when the transition from quadrant IV to quadrant III is observed with compounds that do not bind to the model A-site rRNA. The RB-BIP analysis provides rapid information on SAR of compounds for binding to a proposed pharmacodynamic model.
(iii) The triazole based linker compounds can be divided into two groups: compounds that bind to A-site and compounds that do not bind to the A-site. Although all of the triazole-linked dimers included here are effective at inhibiting the growth of the Gram-positive neomycin-susceptible strains, the analysis indicates that the binding to the model A-site rRNA may not be the primary binding target. Some of the triazole dimers appear to inhibit growth by binding to the A-site, as indicated by their presence in quadrant II; the other triazole dimers appear to inhibit growth but do not bind to the model A-site, as indicated by their presence in quadrant IV. In addition, only the subset of triazole compounds that bind to the A-site are somewhat effective against the neomycin-susceptible Gram-negative strains. The difference in the compounds appears to be a maximal linker length and the rigidity of the linker introduced by a double bond within the linker. Using the RB-BIP, an obvious difference in the binding and inhibition is apparent when the linker length is extended to include six carbon atoms.
(iv) A-site binding and bacterial inhibition is favored by thiourea linkages over urea linkages. The change from a thiourea linker to a urea linker would appear to be a conservative change. By analyzing the RB-BIP, we found that this change is detrimental to the model A-site binding and bacterial growth inhibition, as shown by a transition from quadrant II to quadrant III in most strains. This finding may not be readily predictable by simple analysis of the compounds, but RB-BIP analysis allows a correlation to be drawn among the change in linker composition, A-site binding, and bacterial growth inhibition.
(v) The importance of the composition of the linker extends beyond the primary functional groups. The presence of DPA 80 in quadrant II identifies this compound as one of the most effective compounds for A-site binding and bacterial inhibition. DPA 80 is chemically similar to the urea-linked compounds, particularly DPA 77, that are ineffective. However, the presence of a sulfide linkage at the end of the linker and/or the extension of the linker by six atoms compared to DPA 77 appears to compensate for the absence of the thiourea groups within the linker. The RB-BIP analysis highlights how this slight modification in linker composition changes this compound's effectiveness compared to other dimers linked via urea bonds.
In contrast, DPA 76 is chemically similar to the thiourea-linked dimers that are effective, particularly DPA 78. However, DPA 76 only binds weakly to the A-site and only inhibits the growth of E. cloacae and the Gram-positive neomycin-susceptible bacterial strains. The structural difference in the linker of DPA 76 compared to other compounds in this group appears to reduce its ability to inhibit growth in E. coli and S. marcescens and also changes its binding to the model A-site. Further work is under way to investigate the precise role of linker atoms and will be reported in due course.
Conclusion.
The present study is the first application of a novel, A-site binding assay as a screening method for a group of structurally similar dimer compounds. In addition, the validation and use of an antibacterial screen is also presented using this library of neomycin dimers. This SAR study of the neomycin dimers demonstrates the effect of changes within the linker of the molecule on the activity of the compound. A relatively small change to the composition of the linker results in alterations in both the binding properties and the antibacterial properties of the drug.
Historically, the binding assay is typically treated independently from the bacterial inhibition assay and little direct correlation between the bacterial inhibition and the mechanism of inhibition is established. We linked the screening data of two assays into an RB-BIP using a two-dimensional plot. This treatment of the data gives a rapidly interpretable correlation between antibacterial effects and a possible mechanism of action of binding to the proposed model rRNA A-site. The results for our targeted library indicate that the most effective linker class is triazole > thiourea > urea. However, outliers in the thiourea and urea class, DPA 76 and DPA 80, indicate that both linker length and additional functional groups in the linker alone are capable of completely changing the binding to the model rRNA and antibiotic properties of the compound.
RB-BIP analysis brings together the data from both assays into a single plot, so that analysis is rapid and correlations can be drawn quickly. RB-BIP analysis is a two-dimensional first approximation assay that allows for the rapid correlation of a structurally similar set of compound library's efficacy and mechanism. The value of the analysis could be increased by the addition of other dimensions. The addition of other dimensions, such as cellular uptake and different methods of acquired resistance, could be incorporated directly into the plots as three-dimensional mapping (40) (41). The RB-BIP analysis described here could be adapted and expanded to other forms of analysis for SAR studies of structurally similar compounds where screening methodologies may be in place for the determination of drug potency and proposed inhibition mechanisms.
ACKNOWLEDGMENT
We acknowledge the National Institutes of Health (GM097917) for financial support. D.P.A. has ownership interest in NUBAD, LLC.
We thank Tamara McNealy for bacterial strains and for technical advice.
This manuscript was written through contributions from all of the authors. All authors gave approval to the final version of the manuscript.
Footnotes
Published ahead of print 15 July 2013
REFERENCES
- 1.Willis B, Arya DP. 2006. An expanding view of aminoglycoside-nucleic acid recognition. Adv. Carbohydr. Chem. Biochem. 60:251–302 [DOI] [PubMed] [Google Scholar]
- 2.Livermore DM. 2012. Current epidemiology and growing resistance of gram-negative pathogens. Korean J. Intern. Med. 27:128–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Livermore DM. 2003. Bacterial resistance: origins, epidemiology, and impact. Clin. Infect. Dis. 36:S11–S23 [DOI] [PubMed] [Google Scholar]
- 4.Walsh C. 2000. Molecular mechanisms that confer antibacterial drug resistance. Nature 406:775–781 [DOI] [PubMed] [Google Scholar]
- 5.Scheunemann AE, Graham WD, Vendeix FA, Agris PF. 2010. Binding of aminoglycoside antibiotics to helix 69 of 23S rRNA. Nucleic Acids Res. 38:3094–3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dibrov SM, Parsons J, Hermann T. 2010. A model for the study of ligand binding to the rRNA helix h44. Nucleic Acids Res. 38:4458–4465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yonath A. 2005. Antibiotics targeting ribosomes: resistance, selectivity, synergism, and cellular regulation. Annu. Rev. Biochem. 74:649–679 [DOI] [PubMed] [Google Scholar]
- 8.Vaheri A, Pagano JS. 1965. Infectious poliovirus RNA: a sensitive method of assay. Virology 27:434–436 [DOI] [PubMed] [Google Scholar]
- 9.Vicens Q, Westhof E. 2001. Crystal structure of paromomycin docked into the eubacterial ribosomal decoding A-site. Structure 9:647–658 [DOI] [PubMed] [Google Scholar]
- 10.Vicens Q, Westhof E. 2002. Crystal structure of a complex between the aminoglycoside tobramycin and an oligonucleotide containing the ribosomal decoding A-site. Chem. Biol. 9:747–755 [DOI] [PubMed] [Google Scholar]
- 11.Barbieri CM, Kaul M, Pilch DS. 2007. Use of 2-aminopurine as a fluorescent tool for characterizing antibiotic recognition of the bacterial rRNA A-site. Tetrahedron 63:3567–6574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshizawa S, Fourmy D, Puglisi JD. 1998. Structural origins of gentamicin antibiotic action. EMBO J. 17:6437–6448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barbieri CM, Kaul M, Bozza-Hingos M, Zhao F, Tor Y, Hermann T, Pilch DS. 2007. Defining the molecular forces that determine the impact of neomycin on bacterial protein synthesis: importance of the 2′-amino functionality. Antimicrob. Agents Chemother. 51:1760–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bera S, Dhondikubeer R, Findlay B, Zhanel GG, Schweizer F. 2012. Synthesis and antibacterial activities of amphiphilic neomycin B-based bilipid conjugates and fluorinated neomycin B-based lipids. Molecules 17:9129–9141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dhondikubeer R, Bera S, Zhanel GG, Schweizer F. 2012. Antibacterial activity of amphiphilic tobramycin. J. Antibiot. (Tokyo) 65:495–498 [DOI] [PubMed] [Google Scholar]
- 16.Findlay B, Zhanel GG, Schweizer F. 2012. Neomycin-phenolic conjugates: polycationic amphiphiles with broad-spectrum antibacterial activity, low hemolytic activity and weak serum protein binding. Bioorg. Med. Chem. Lett. 22:1499–1503 [DOI] [PubMed] [Google Scholar]
- 17.Bera S, Zhanel GG, Schweizer F. 2011. Synthesis and antibacterial activity of amphiphilic lysine-ligated neomycin B conjugates. Carbohydr. Res. 346:560–568 [DOI] [PubMed] [Google Scholar]
- 18.Bera S, Zhanel GG, Schweizer F. 2010. Antibacterial activity of guanidinylated neomycin B- and kanamycin A-derived amphiphilic lipid conjugates. J. Antimicrob. Chemother. 65:1224–1227 [DOI] [PubMed] [Google Scholar]
- 19.Zhang J, Keller K, Takemoto JY, Bensaci M, Litke A, Czyryca PG, Chang CW. 2009. Synthesis and combinational antibacterial study of 5″-modified neomycin. J. Antibiot. (Tokyo) 62:539–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kling D, Chow C, Mobashery S. 2007. Binding of antibiotics to the aminoacyl-tRNA site of bacterial ribosome, p 225–233 In Arya DP. (ed), Aminoglycoside antibiotics: from chemical biology to drug discovery. John Wiley & Sons, Inc., Hoboken, NJ [Google Scholar]
- 21.Russell RJ, Murray JB, Lentzen G, Haddad J, Mobashery S. 2003. The complex of a designer antibiotic with a model aminoacyl site of the 30S ribosomal subunit revealed by X-ray crystallography. J. Am. Chem. Soc. 125:3410–3411 [DOI] [PubMed] [Google Scholar]
- 22.Haddad J, Kotra LP, Llano-Sotelo B, Kim C, Azucena EF, Jr, Liu M, Vakulenko SB, Chow CS, Mobashery S. 2002. Design of novel antibiotics that bind to the ribosomal acyltransfer site. J. Am. Chem. Soc. 124:3229–3237 [DOI] [PubMed] [Google Scholar]
- 23.Llano-Sotelo B, Azucena EF, Kotra LP, Mobashery S, Chow CS. 2002. Aminoglycosides modified by resistance enzymes display diminished binding to the bacterial ribosomal aminoacyl-tRNA site. Chem. Biol. 9:455–463 [DOI] [PubMed] [Google Scholar]
- 24.Sucheck SJ, Wong AL, Koeller KM, Boehr DD, Draker K, Sears P, Wright GD, Wong C. 2000. Design of bifunctional antibiotics that target bacterial rRNA and inhibit resistance-causing enzymes. J. Am. Chem. Soc. 122:5230–5231 [Google Scholar]
- 25.Greenberg WA, Priestley ES, Sears PS, Alper PB, Rosenbohm C, Hendrix M, Hung S, Wong C-H. 1999. Design and synthesis of new aminoglycoside antibiotics containing neamine as an optimal core structure: correlation of antibiotic activity with in vitro inhibition of translation. J. Am. Chem. Soc. 121:6527–6541 [Google Scholar]
- 26.Alper PB, Hendrix M, Sears P, Wong C-H. 1998. Probing the specificity of aminoglycoside-rRNA interactions with designed synthetic analogs. J. Am. Chem. Soc. 120:1965–1978 [Google Scholar]
- 27.Wong CH, Hendrix M, Manning DD, Rosenbohm C, Greenberg WA. 1998. A library approach to the discovery of small molecules that recognize RNA: use of a 1,3-hydroxyamine motif as core. J. Am. Chem. Soc. 120:8319–8327 [Google Scholar]
- 28.Wong CH, Hendrix M, Priestley ES, Greenberg WA. 1998. Specificity of aminoglycoside antibiotics for the A-site of the decoding region of rRNA. Chem. Biol. 5:397–406 [DOI] [PubMed] [Google Scholar]
- 29.Hendrix M, Priestley ES, Joyce GF, Wong CH. 1997. Direct observation of aminoglycoside-RNA interactions by surface plasmon resonance. J. Am. Chem. Soc. 119:3641–3648 [DOI] [PubMed] [Google Scholar]
- 30.Watkins D, Norris FA, Kumar S, Arya DP. 2013. A fluorescence-based screen for ribosome binding antibiotics. Anal. Biochem. 434:300–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De La Fuente R, Sonawane ND, Arumainayagam D, Verkman AS. 2006. Small molecules with antimicrobial activity against Escherichia coli and Pseudomonas aeruginosa identified by high-throughput screening. Br. J. Pharmacol. 149:551–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu KC, Fang JM, Jan JT, Cheng TJ, Wang SY, Yang ST, Cheng YS, Wong CH. 2012. Enhanced anti-influenza agents conjugated with anti-inflammatory activity. J. Med. Chem. 55:8493–8501 [DOI] [PubMed] [Google Scholar]
- 33.Su CY, Cheng TJ, Lin MI, Wang SY, Huang WI, Lin-Chu SY, Chen YH, Wu CY, Lai MM, Cheng WC, Wu YT, Tsai MD, Cheng YS, Wong CH. 2010. High-throughput identification of compounds targeting influenza RNA-dependent RNA polymerase activity. Proc. Natl. Acad. Sci. U. S. A. 107:19151–19156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cochrane A, Murley LL, Gao M, Wong R, Clayton K, Brufatto N, Canadien V, Mamelak D, Chen T, Richards D, Zeghouf M, Greenblatt J, Burks C, Frappier L. 2009. Stable complex formation between HIV Rev and the nucleosome assembly protein, NAP1, affects Rev function. Virology 388:103–111 [DOI] [PubMed] [Google Scholar]
- 35.Kumar S, Kellish P, Robinson WE, Wang D, Appella DH, Arya DP. 2012. Click dimers to target HIV TAR RNA conformation. Biochemistry 51:2331–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumar S, Arya DP. 2011. Recognition of HIV TAR RNA by triazole linked neomycin dimers. Bioorg. Med. Chem. Lett. 21:4788–4792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar S, Xue L, Arya DP. 2011. Neomycin-neomycin dimer: an all-carbohydrate scaffold with high affinity for AT-rich DNA duplexes. J. Am. Chem. Soc. 133:7361–7375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.National Committee for Clinical Laboratory Standards 1997. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals; approved standard, 2nd ed. NCCLS document M31-A2. National Committee for Clinical Laboratory Standards, Wayne, PA [Google Scholar]
- 39.Cudic M, Lockatell C, Johnson D, Otvos L. 2003. In vitro and in vivo activity of an antibacterial peptide analog against uropathogens. Peptides 24:807–820 [DOI] [PubMed] [Google Scholar]
- 40.Helander IM, Mattila-Sandholm T. 2000. Fluorometric assessment of gram-negative bacterial permeabilization. J. Appl. Microbiol. 88:213–219 [DOI] [PubMed] [Google Scholar]
- 41.Toth M, Chow JW, Mobashery S, Vakulenko SB. 2009. Source of phosphate in the enzymic reaction as a point of distinction among aminoglycoside 2″-phosphotransferases. J. Biol. Chem. 284:6690–6696 [DOI] [PMC free article] [PubMed] [Google Scholar]