Abstract
Acinetobacter baumannii is a nosocomial opportunistic pathogen that can cause severe infections, including hospital-acquired pneumonia, wound infections, and sepsis. Multidrug-resistant (MDR) strains are prevalent, further complicating patient treatment. Due to the increase in MDR strains, the cationic antimicrobial peptide colistin has been used to treat A. baumannii infections. Colistin-resistant strains of A. baumannii with alterations to the lipid A component of lipopolysaccharide (LPS) have been reported; specifically, the lipid A structure was shown to be hepta-acylated with a phosphoethanolamine (pEtN) modification present on one of the terminal phosphate residues. Using a tandem mass spectrometry platform, we provide definitive evidence that the lipid A isolated from colistin-resistant A. baumannii MAC204 LPS contains a novel structure corresponding to a diphosphoryl hepta-acylated lipid A structure with both pEtN and galactosamine (GalN) modifications. To correlate our structural studies with clinically relevant samples, we characterized colistin-susceptible and -resistant isolates obtained from patients. These results demonstrated that the clinical colistin-resistant isolate had the same pEtN and GalN modifications as those seen in the laboratory-adapted A. baumannii strain MAC204. In summary, this work has shown complete structure characterization including the accurate assignment of acylation, phosphorylation, and glycosylation of lipid A from A. baumannii, which are important for resistance to colistin.
INTRODUCTION
The opportunistic pathogen Acinetobacter baumannii is a Gram-negative aerobic coccobacillus and is a leading cause of nosocomial infections globally (1–4). Infections include hospital and community-acquired pneumonia, wound infections, and sepsis, leading to increased mortality. Additionally, A. baumannii has emerged as a major pathogen in U.S. military personnel in field hospitals in Iraq and Afghanistan (5, 6). A. baumannii strains have developed antimicrobial resistance, including resistance to the cationic microbial peptide (CAMP) colistin (polymyxin E), complicating patient treatment and furthering the cause for the development of new antimicrobial therapies. Thus, A. baumannii has emerged as a pathogen of great clinical concern.
Initial research on A. baumannii pathogenesis focused on defining the genes and mechanisms responsible for antimicrobial resistance. The capsular polysaccharide and lipopolysaccharide (LPS), the major component of the Gram-negative bacterial cell wall, act in concert to block access of complement to the cell wall, inhibiting bacterial membrane lysis. LPS is located in the outer leaflet of the outer membrane of Gram-negative bacteria and consists of lipid A, the core oligosaccharide, and the O-specific antigen. Lipid A is the bioactive component of LPS and is responsible for activating the innate immune system via toll-like receptor 4 (TLR4), which potentially initiates a cascade of inflammatory cytokine production that, if unchecked, can lead to septic shock. Modifications of lipid A can drastically alter its immunostimulatory ability as well as resistance to antibiotics. For example, the addition of positively charged residues including ethanolamine, aminoarabinose, and glucosamine to lipid A modulates CAMP resistance (Fig. 1) (7–9).
Fig 1.
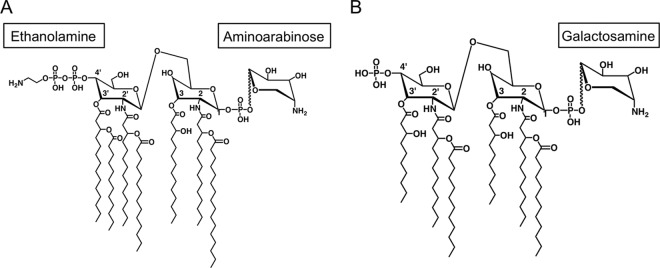
Modification of the lipid A component of lipopolysaccharide by positively charged residues, including ethanolamine, aminoarabinose, and glucosamine, alters resistance to CAMPs. (A) Salmonella Typhimurium; (B) Francisella tularensis.
A previous report shows that a colistin-resistant (Colr) strain of A. baumannii contained a pEtN addition with suggested acyl chain positioning for the hepta-acylated lipid A structure (10, 11). Using a tandem mass spectrometry platform and the laboratory-adapted MAC204 colistin-resistant strain, we confirmed the addition of pEtN and identified a novel second amino sugar modification, GalN. To correlate our structural observations with clinically relevant samples, we analyzed lipid A extracted from matched colistin-susceptible (Cols) and -resistant A. baumannii isolates from individual patients before and after colistin treatment. Using a multifaceted mass spectrometric platform, we observed similar lipid A structures with phosphoethanolamine (pEtN) and galactosamine (GalN) additions that were present only in resistant strains from patients treated with colistin. Taken together, the pattern and location of lipid A acylation, phosphorylation, and glycosylation potentially underpin a critical role in the overall ability of A. baumannii to present resistance to colistin.
MATERIALS AND METHODS
Bacteria.
Acinetobacter baumannii colistin-resistant strain MAC204 was provided by Mark Adams, Case Western University, Cleveland, OH. Strain ATCC 17978 was obtained from the ATCC (American Type Culture Collection, Manassas, VA, USA). Strain MAC204 was generated by inducing spontaneous mutants of the wild-type A. baumannii strain MAC203 (12), a strain that was isolated from late-exponential-phase cultures by selection on Lysogeny Broth (LB; Difco) plates containing 1.5% agar and 1 μg/ml colistin, resulting in strain ATCC 17978 Colr (MAC201). This strain was used to select for colistin-susceptible revertants by growth without colistin resulting in strain ATCC 17978 Colr_Rev (MAC203). This strain was subsequently selected for colistin resistance as described above, except 2 μg/ml colistin was used in selection, resulting in strain MAC204.
Three pairs of colistin-susceptible and -resistant isolates, 1494/1508 (JA637), 2382/2384 (JA566), and 2949/2949A (JA942), respectively, collected from three individual patients, were provided by Yohei Doi, University of Pittsburgh Medical Center (see Table S1 in the supplemental material) under IRB number PRO12060302. Clinical isolates were grown overnight at 37°C in LB supplemented with 1 mM MgCl2. Susceptibility profiles were determined by Etest (bioMérieux, St. Louis, MO), according to the manufacturer's procedures. The bacteria were grown in LB supplemented with 1 mM MgCl2 and 2 μg/ml colistin at 37°C in a shaking incubator at 250 rpm for 20 h (8).
LPS and lipid isolation and purification.
LPS was extracted using a hot phenol-water method (13). Freeze-dried bacteria were resuspended in endotoxin-free water at a concentration of 10 mg/ml. A 12.5-ml volume of 90% phenol was added, and the resultant mixture was vortexed and incubated in a hybridization oven at 65°C. The mixture was cooled on ice and centrifuged at 10,000 rpm at room temperature for 30 min. The aqueous phase was collected, and an equal volume of endotoxin-free water was added to the organic phase. The sample was treated as above, and the aqueous phases were combined, dialyzed against Milli-Q purified water to remove residual phenol, frozen, and then freeze-dried. The resultant pellet was resuspended at a concentration of 10 mg/ml in endotoxin-free water, treated with DNase (Sigma, St. Louis, MO) at 100 μg/ml and RNase A (Sigma) at 25 μg/ml, and incubated at 37°C for 1 h in a water bath. Proteinase K (Sigma) was added to a final concentration of 100 μg/ml and incubated for 1 h in a 37°C water bath (14). The solution was then extracted with an equal volume of water-saturated phenol. The aqueous phase was collected and dialyzed against Milli-Q purified water and freeze-dried as above. The LPS was further purified by the addition of chloroform-methanol (2:1, vol/vol) to remove membrane phospholipids (15) and further purified by an additional water-saturated phenol extraction and 75% ethanol precipitation to remove contaminating lipoproteins (16). For mass spectrometry (MS) structural analysis, 1 mg of purified LPS was converted to lipid A by mild-acid hydrolysis with 1% sodium dodecyl sulfate (SDS) at pH 4.5 as described previously (17). Resulting lipid A was analyzed using mass spectrometry as described below.
Small-scale lipid A isolation from whole cells.
A. baumannii lipid A was prepared using an ammonium hydroxide-isobutyric acid-based extraction procedure (18). Briefly, approximately 10 mg of lyophilized material derived from an overnight culture was resuspended in 400 μl of isobutyric acid and 1 M ammonium hydroxide (5:3, vol/vol) and incubated at 100°C for 1 h. After cooling, individual samples were centrifuged for 15 min at 2,000 × g, and supernatants were collected and diluted 1:1 (vol/vol) with endotoxin-free water. The samples were subsequently frozen and lyophilized overnight. The resultant powdered material was then washed twice with 1 ml of methanol, and the insoluble lipid A was extracted in 200 μl of a mixture of chloroform, methanol, and water (3:1:0.25 [vol/vol/vol]). One microliter of this extract was then spotted onto a matrix-assisted laser desorption ionization (MALDI) plate followed by 1 μl of norharmane matrix (Sigma) and air dried.
Fatty acid analysis.
LPS fatty acids were converted to fatty acid methyl esters and analyzed by gas chromatography (GC) as previously described (19, 20). Briefly, 10 mg of lyophilized bacterial cell pellet was incubated at 70°C for 1 h in 500 μl of 90% phenol and 500 μl of water. Samples were then cooled on ice for 5 min and centrifuged at 10,000 rpm for 10 min. The aqueous layer was collected, and 500 μl of water was added to the lower (organic) layer and incubated again. This process was repeated twice more, and all aqueous layers were pooled. Two milliliters of ethyl ether was added to the harvested aqueous layers. This mixture was then vortexed and centrifuged at 3,000 rpm for 5 min. The lower (organic) phase was then collected, and 2 ml of ether was added back to the remaining aqueous phase. This process was carried out twice more. The collected organic layer was then frozen and lyophilized overnight. LPS fatty acids were converted to fatty methyl esters, in the presence of 10 μg pentadecanoic acid (Sigma) as an internal standard, with 2 M methanolic HCl (Alltech, Lexington, KY) at 90°C for 18 h.
Mass spectrometry. (i) ESI-LTQ-FT MS.
Lipid A was analyzed by electrospray ionization-linear ion trap-Fourier transform ion cyclotron resonance mass spectrometry (ESI-LTQ-FT MS) in the negative ion mode on an LTQ-FT linear ion trap Fourier transform ion cyclotron resonance mass spectrometer (Thermo Scientific, San Jose, CA). Samples were diluted to 1 mg/ml in chloroform-methanol (1:1, vol/vol) and infused at a rate of 1.0 ml/min via a fused silica capillary (inner diameter, 75 μm; outer diameter, 360 μm) with a 30-μm spray tip (New Objective, Woburn, MA). Instrument calibration and tuning parameters were optimized using a solution of Ultramark 1621 (Lancaster Pharmaceuticals, PA) in both positive and negative ion modes. For experiments acquired in the ion cyclotron resonance (ICR) cell, the mass resolving power was set to 100,000 and ion populations were held constant by automatic gain control at 1.0 × 106 for mass spectrum (MS) acquisition and at 5.0 × 105 for tandem mass spectra (MSn), respectively. For tandem mass spectra, the precursor ion selection window was set to 4 to 8 Da and the collision energy was set to 30% on the instrument scale. The collision-induced dissociation (CID) MSn analyses in the linear ion trap were acquired with an ion population of 1.0 × 104 and maximum fill time of 200 ms. The subsequent MS2, MS3, and MS4 events had an isolation window of 2 Da with a collision energy of 25%. All spectra were acquired over a time period of 1 to 2 min and averaged. Typically, MS events were mass analyzed in the ICR cell and MS2, MS3, and MS4 events were mass analyzed in the LTQ. Infrared multiphoton dissociation (IRMPD) MS2 events were acquired in the ICR cell using detection parameters similar to the ones described above. Precursor ions were irradiated by IR photons produced by a CO2 laser (Synrad Firestar Series V20, model FSV20SFB 75 W [10.2 to 10.8 μm]) with pulse durations of 20 to 100 ms and pulse power of 20 to 80%. Data were acquired and processed using Xcalibur, version 1.4 (Thermo Scientific), utilizing 7-point Gaussian smoothing.
(ii) MALDI-TOF MS.
Lipid A was analyzed on an AutoFlex Speed MALDI-time of flight (TOF) mass spectrometer (Bruker Daltonics, Billerica, MA). Samples were dissolved in 10 μl of norharmane (20 mg/ml) in chloroform-methanol-water, 4:4:1 (vol/vol/vol), and 0.5 μl of sample was analyzed and spotted directly onto the MALDI target plate. Data were acquired in reflectron negative and positive modes with a Smartbeam laser with a 1-kHz repetition rate, and up to 4,000 shots were accumulated for each spectrum. Instrument calibration and all other tuning parameters were optimized using Agilent Tuning mix (Agilent Technologies, Foster City, CA). Data were acquired and processed using flexControl and flexAnalysis version 3.3 (Bruker Daltonics).
Amino sugar assay chemicals and reagent.
Glucosamine hydrochloride, N-acetylglucosamine, 2-amino-2-methyl-1, and 3-propanediol were from Sigma. 2-Galactoseamine hydrochloride was from Calbiochem (Gibbstown, NJ). The derivatizing reagent, 6-aminoquinoyl-N-hydroxysuccinimidyl carbamate (AQHSC), was purchased from Waters (AccQ-Fluor; Milford, MA). Trifluoroacetic acid (TFA) was from Sigma. All high-pressure liquid chromatography (HPLC) solvents and buffers were of analytical grade.
LPS purification and lipid A isolation.
LPS was isolated using a small-scale isolation method for mass spectrometry analysis as described previously (21). Lipid A was isolated using mild-acid hydrolysis as previously described (17).
Sample preparation.
Standards and biological samples were prepared as described previously (22). Briefly, standards were dissolved in 200 μl 1.0 M TFA, vortexed, and heated at 90°C for 30 min or 24 h. The samples were then frozen, lyophilized, and dissolved in 50 μl 0.2 M borate buffer, pH 8.8, prior to the addition of 50 μl of 1 mg/ml AccQ-Tag derivatizing reagent in acetonitrile. The samples were vortexed, incubated at room temperature for 15 to 30 min, and then evaporated to dryness under nitrogen. Derivatized samples were reconstituted in 100 μl of distilled water, vortexed, and transferred to injection vials for analysis. For analysis of biological samples, 10 μg of lipid A isolated from individual preparations was used. They were then derivatized by the same procedure as the standards as described above.
Instrumentation.
All analyses were performed on a liquid chromatography tandem mass spectrometry (LC-MS/MS) platform.
Liquid chromatography.
The analytes were separated using a Develosil 5u RP-Aqueous C-30 column (150 by 2.1 mm) (Phenomenex, Torrance, CA) using a 10 mM ammonium acetate (pH 7.5)–acetonitrile (ACN) gradient. The initial organic was 2.0% ACN and increased to 5.0% at 0.5 min. Thereafter, there was a linear increase to 10% ACN at 6.5 min and then to 20% ACN at 17 min. The ACN was increased to 80% for a 1-min washout and then dropped back to the initial (2%) ACN and reequilibrated for 8.0 min. The flow was 0.4 ml throughout the run, the temperature was 25°C, and the run time was 30 min. Eluent before 4.5 and after 16 min was diverted from the mass spectrometer.
Mass spectrometry data.
All mass spectra were acquired using positive electrospray ionization mode. Selected reaction monitoring (SRM) studies were performed on a Thermo TSQ Quantum Ultra Triple Stage quadrupole mass spectrometer. The scans of the derivatives universally showed an intense signal at (analyte MW + 170 + H)+ with minimal fragmentation. This is equivalent to m/z 350 (GalA and GluA). The observed signals were consistent with the formation of the N-quinoyl-N′-aminosaccharide urea as the derivative. The SRM transitions for both GalN and GlcN were 350-171. GalN and GlcN had the same transition and were differentiated by chromatographic retention times. The ionization conditions used in SRM were as follows: spray voltage, 3,000 V; capillary temperature, 40°C; capillary offset, 10 V; vaporizer temperature, 40°C; sheath gas pressure, 5 lb/in2; auxiliary gas pressure, 5 lb/in2; ion sweep gas pressure, 0.5 lb/in2.
RESULTS
Lipid A structure from a laboratory-adapted colistin-resistant strain of A. baumannii.
As colistin-resistant strains of A. baumannii are an increasing problem in clinical settings, we hypothesized that generating strains with increased resistance to colistin would result in lipid A modifications specific to these colistin-resistant strains. To determine the presence of colistin-induced lipid A modifications resulting in the A. baumannii MAC204 laboratory strain, we generated an A. baumannii colistin-resistant laboratory-adapted strain. We now describe the structural determination of the chemical structure of lipid A from A. baumannii LPS using MALDI-TOF MS along with ESI MS and MSn mass spectra. Particular emphasis will be devoted to determining acyl chain positioning, phosphoethanolamine (pEtN) addition, and glycan (GalN) modification (both location and identification).
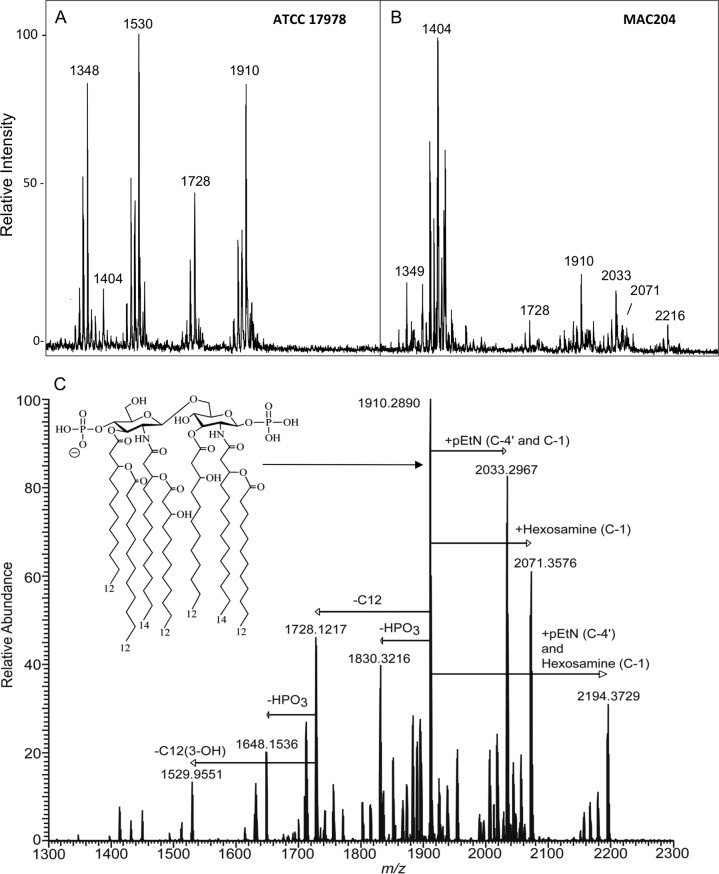
After overnight growth at 37°C in rich medium (LB), lipid A was isolated from the colistin-resistant A. baumannii strain MAC204 and its colistin-susceptible parent strain ATCC 17978 and analyzed by MALDI-TOF mass spectrometry in the negative ion mode. Analysis of lipid A isolated from strain ATCC 17978 (Fig. 2A) showed that an abundant [M-H]− ion was at m/z 1,910, which was tentatively identified as a singly deprotonated lipid A structure that contained two phosphate groups and seven acyl chains (i.e., diphosphoryl hepta-acylated lipid A). Other abundant [M-H]− ions were associated with different acylation or phosphorylation configurations. Of note, ions at m/z 2,033 and 2,071 were absent from these spectra. In contrast, lipid A isolated from the colistin-resistant strain MAC204 (Fig. 2B) displayed not only the [M-H]− at m/z 1,910 but also ions at m/z 2,033 and m/z 2,071, consistent with pEtN and hexosamine additions, respectively. In this strain, we also saw an [M-H]− ion at m/z 2,216, corresponding to a pEtN plus a hexosamine addition along with a sodium adduct. Previous reports have shown A. baumannii lipid A to be modified with pEtN, but to the best of our knowledge, hexosamine has not been previously reported for colistin-resistant A. baumannii strains.
Fig 2.
Negative ion mode MALDI-TOF MS mass spectra of A. baumannii ATCC 17978 (A) and MAC204 (B). (C) Negative ion mode ESI FT-ICR mass spectrum of lipid A isolated from A. baumannii LPS. All ions are singly charged. The inset structure is a proposed configuration for bis-phosphoryl, hepta-acylated lipid A from A. baumannii. pEtN, phosphoethanolamine; C12, lauric acid; C12(3-OH), 3-hydroxy lauric acid; HPO3, phosphate; C-1, C-1 position phosphate houses the modification; C-4′, C-4′ position phosphate houses the modification.
To definitively identify the lipid A additions detected by MALDI-TOF MS analysis, we performed high-order negative ion mode ESI LTQ-FT mass spectrometry of lipid A isolated from A. baumannii strain MAC204, with results shown in Fig. 2C. The most abundant ion at m/z 1,910 from Fig. 2C corresponded to the structure identified by MALDI-TOF MS. Hereinafter, all initial structure characterization using precursor ion m/z values were supported by elemental composition based on accurate mass measurements and literature when available. Several other prominent singly charged ions were recorded at m/z values greater than 2,000 Da. These ions corresponded to lipid A structures containing the addition of pEtN (m/z 2,033), that of hexosamine (m/z 2,071), and both modifications (m/z 2,194).
Determination of acyl chain positioning of lipid A isolated from A. baumannii LPS using tandem mass spectrometry.
Tandem mass spectrometric experiments were carried out to confirm the location of the seven fatty acids. Initial characterization based on elemental composition from accurate mass measurements of the hepta-acylated lipid A anion, m/z 1,910, revealed the presence of four primary fatty acids, i.e., two 3-hydroxylauric acid [C12(3-OH)] acyl chains and two 3-hydroxymyristic acid [C14(3-OH)] acyl chains and of three secondary fatty acids, i.e., one C12(3-OH) acyl chain and two lauric acid (C12) acyl chains. A similar acylation pattern has been reported previously, yet conclusive evidence for assignment of each fatty acid to a specific location was not determined (22, 23).
In order to determine the positioning of the seven fatty acids, we conducted a series of tandem mass spectrometric experiments aimed at highlighting diagnostic cross-ring and glycosidic cleavage product ions that provided evidence for pinpointing acyl chain positions. Detailed descriptions of the tandem mass spectra (MSn) experiments, the corresponding rationale for fatty acid assignment, and the MSn spectra are given in Fig. S1 and S2 in the supplemental material. The hepta-acylated lipid A structure was determined to have the following configuration: the C-2 position contained a primary amide-linked C14(3-OH) and a secondary ester-linked C12, the C-3 position contained a primary ester-linked C12(3-OH), the C-2′ position contained a primary amide-linked C14(3-OH) and a secondary ester-linked C12(3-OH), and the C-3′ position contained a primary ester-linked C12(3-OH) and a secondary ester-linked C12. The acyl chain configuration as outlined above held true for all hepta-acylated lipid A extracted from A. baumannii. The proposed structure of hepta-acylated lipid A extracted from A. baumannii is shown in the inset in Fig. 2.
Finally, to determine the amount of total fatty acids in lipid A, A. baumannii strain ATCC_ C2B was grown at 37°C overnight, lipid A was extracted, and the fatty acids were analyzed by capillary gas chromatography using flame ionization detection (FID). As shown in Fig. 3, this analysis yielded percentages of total fatty acids of 24.6% ± 0.7% C12, 9.3% ± 0.1% 2-OH C12, 29.9% ± 0.1% 3-OH C12, and 36.2% ± 0.5% 3-OH C14, confirming the structural characterization by MS above.
Fig 3.
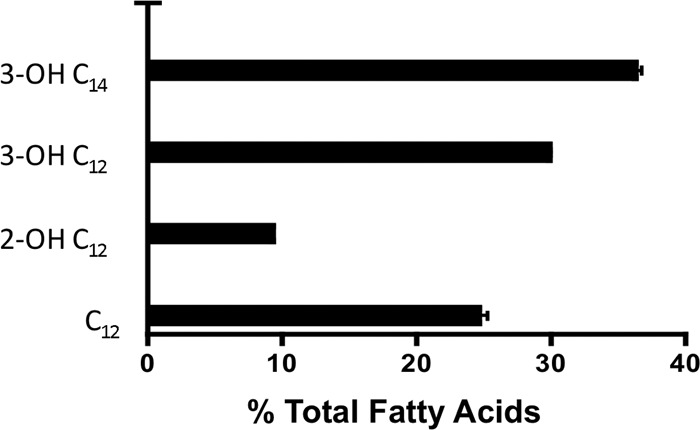
Fatty acid analysis via GC-FID displayed via percentage of fatty acid present to total fatty acids.
Identification and localization of pEtN addition to lipid A isolated from A. baumannii LPS using tandem mass spectrometry.
Tandem mass spectrometric experiments on the precursor ion at m/z 2,033, which corresponded to the addition of a pEtN substituent to hepta-acylated lipid A, were carried out in turn to correctly identify and localize the pEtN modification. Accurate mass measurement of the hepta-acylated structure (m/z 1,910) plus the 123 mass unit modification precursor ion at m/z 2,033 revealed the elemental composition of the 123 mass unit modification to be C2H6NPO3, which corresponded to pEtN (C2H6NPO3; accurate mass, 123.0085; experimental mass, 123.0161; 7.6 millimass units [mmu]). The pEtN modification has been reported previously, yet evidence for localizing the pEtN group was suggestive (22, 23). We determined the location of the pEtN modification via a series of tandem mass spectrometric experiments aimed at highlighting diagnostic product ions that provided convincing evidence for pEtN localization.
Gas phase dissociation of the precursor ion at m/z 2,033 resulting in the generation of an MS2 mass spectrum is highlighted in Fig. S3 in the supplemental material. The overwhelming dissociation pathway from the precursor ion at m/z 2,033 involved the neutral loss of the pEtN group at m/z 1,910. The ion at m/z 1,910 corresponded to diphosphoryl hepta-acylated lipid A (see above for acyl chain assignments). In addition to the abundant ion at m/z 1,910, a series of more modestly abundant product ions resulting from competitive and consecutive neutral loss(es) from pEtN, fatty acids, and phosphate were present. These ions provided insight regarding the acyl chain and phosphate configuration; however, they did not yield conclusive evidence for pEtN assignment. pEtN localization was achieved via examination of product ions resulting from cross-ring and glycosidic cleavages.
Diagnostic glycosidic and cross-ring product ions allowed us to assign the pEtN modification to both the reducing and the nonreducing ends of the lipid A disaccharide backbone. The MSn spectra and detailed analysis are shown in Fig. S3 in the supplemental material. Moreover, the pEtN group was found to be attached to the lipid A backbone via a phosphodiester bond at either the C-4′ monophosphate or the C-1 monophosphate position. Of particular note, the phosphorylation pattern for lipid A extracted from A. baumannii LPS was determined to be both bisphosphorylated and pyrophosphorylated (see Fig. S3 in the supplemental material). At this level of MS analysis, we did not observe pEtN attached directly to a pyrophosphate group or pEtN attached without linkage to the C-4′ or C-1 monophosphate group. The presence of pyrophosphate in the structure corresponding to m/z 2,033 was confirmed in the IRMPD (infrared multiphoton disassociation) MS2 mass spectrum of the ion at m/z 2,033 (data not shown). A previous report (24) estimated the abundance of pyrophosphorylated lipid A extracted from Yersinia pestis LPS to be near 5%. Equally apparent from this report was the vast majority (∼95%) of diphosphorylated lipid A with a bisphosphorylated configuration. Therefore, assuming that A. baumannii lipid A is predominately composed of bisphosphate and due to the inability to sufficiently characterize pyrophosphorylated lipid A from A. baumannii, we focused our efforts on characterizing the PEtN modification in the context of bisphosphorylated A. baumannii lipid A.
Localization of the glycan addition to lipid A isolated from A. baumannii LPS using tandem mass spectrometry.
Tandem mass spectrometric experiments on the precursor ion at m/z 2,071, which corresponded to the addition of a hexosamine group to hepta-acylated lipid A, were carried out in order to localize the glycan modification. Accurate mass measurement of the hepta-acylated plus the 161 mass unit modification precursor ion at m/z 2,071 revealed the elemental composition of the 161 mass unit modification to be a hexosamine group (C6H11NO4; accurate mass, 161.0688; experimental mass, 161.0598; mass difference, 9.0 mmu). The glycan modification has not been reported previously. A series of tandem mass spectrometric experiments were conducted to highlight diagnostic product ions that provided convincing evidence for localizing the glycan modification.
Gas phase dissociation of the precursor ion at m/z 2,071 resulting in the generation of an MS2 mass spectrum is highlighted in Fig. S4 in the supplemental material. The overwhelming dissociation pathway from the precursor ion at m/z 2,071 involved the neutral loss of the hexosamine group at m/z 1,910. The ion at m/z 1,910 corresponded to diphosphoryl hepta-acylated lipid A (see above for phosphate and acyl chain assignments). Similar to structure determination as described above, the glycan localization was achieved via examination of diagnostic product ions resulting from cross-ring and glycosidic cleavages.
We identified several glycosidic cleavages that allowed us to assign the glycan modification to the reducing end of the lipid A disaccharide (see Fig. S3 in the supplemental material). Furthermore, the glycan group was found to be attached to the lipid A backbone via a phosphodiester bond at the C-1 monophosphate position. At this level of MS sophistication, we did not observe the glycan attached directly to a pyrophosphate group or attached without linkage to the C-1 monophosphate group. Once again, it was assumed, as outlined previously, that A. baumannii lipid A is predominately composed of bisphosphate lipid A, and therefore we focused our efforts on characterizing the hexosamine modification in context with bisphosphorylated A. baumannii lipid A.
Assignment of the phosphoethanolamine (pEtN) and glycan modifications to lipid A isolated from A. baumannii LPS using tandem mass spectrometry.
Tandem mass spectrometric experiments on the precursor ion at m/z 2,194, which corresponded to the addition of both pEtN and hexosamine substituents to hepta-acylated lipid A were performed to correctly identify and localize these substituents. Accurate mass measurement of the hepta-acylated plus the 123 and 161 mass unit modifications at m/z 2,194 revealed the elemental composition of the 123 mass unit modification to be pEtN (C2H6NPO3; accurate mass, 123.0085; experimental mass, 123.0001; 8.4 mmu) and the 161 mass unit modification to be hexosamine (C6H11NO4; accurate mass, 161.0688; experimental mass, 161.0620; mass difference, 6.8 mmu). The combined pEtN and hexosamine modifications have not been reported previously.
Gas-phase dissociation of the precursor ion at m/z 2,194 resulting in the generation of a MS2 mass spectrum is highlighted in Fig. S5 in the supplemental material. The overwhelming dissociation pathway from the precursor ion at m/z 2,194 involved the neutral loss of both the pEtN and the hexosamine groups at m/z 1,910. The ion at m/z 1,910 corresponded to diphosphoryl hepta-acylated lipid A (without pEtN and hexosamine). pEtN and hexosamine localization was achieved via examination of product ions resulting from cross-ring and glycosidic cleavages.
Diagnostic product ions (see Fig. S4 and the supplemental material for detailed analysis) allowed us to confidently assign the pEtN modification to the nonreducing end and the hexosamine modification to the reducing end of the lipid A disaccharide backbone. Moreover, both modifications were found to be attached to the lipid A backbone via a phosphodiester bond at the C-4′ monophosphate position for pEtN and at the C-1 monophosphate position for hexosamine. The structure assignment was in context to the predominant bisphosphate lipid A configuration.
Identification of the hexosamine modification to lipid A extracted from LPS of an A. baumannii clinical isolate via LC-MS/MS.
In order to confirm the identity of the hexosamine sugar modification to A. baumannii lipid A, we utilized an LC-MS/MS platform previously developed in our laboratory for detection and quantitation of amino sugars isolated from lipid A in Francisella novicida (25–27). This study demonstrated the detection of the GalN modification to lipid A. The GalN modification resulted in increased resistance to polymyxin B. For this study, we used the Colr clinical isolate 1508, which is highly resistant to colistin (MIC [MIC], 24 μg/ml by Etest) to assay for the presence of amino sugars (Table 1).
Table 1.
Summary of lipid A structural modifications observed by MALDI-TOF in A. baumannii clinical isolates and MIC data
| Clinical strain | Sample source | Colistin Etest MIC (μg/ml) | Colistin therapy | pEtN addition | GalN addition |
|---|---|---|---|---|---|
| 1494 | BALa | 0.06 | No | − | − |
| 1508 | Sputum | 24 | Yes | + | + |
| 2382 | BAL | 0.02 | No | − | − |
| 2384 | BAL | 1.5 | Yes | + | + |
| 2949 | BAL | 1 | No | − | − |
| 2949A | BAL | 48 | Yes | + | + |
BAL, bronchoalveolar lavage fluid.
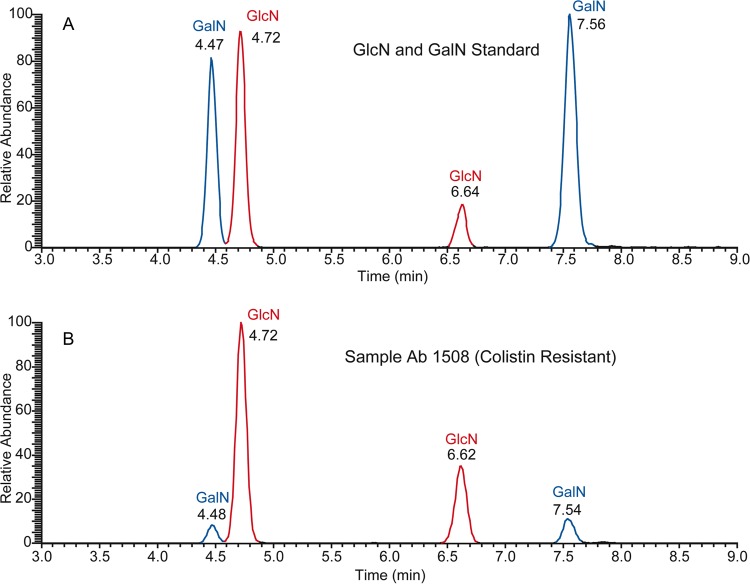
We hypothesized that the presence of GalN, along with pEtN, could be responsible for this phenotype. Indeed, as shown in Fig. 4, we provide evidence for the addition of GalN to A baumannii lipid A. Figure 5A depicts an extracted ion chromatogram for glucosamine (GlcN) and GalN standards. Four peaks are observed and corresponded to two isomers each for GlcN and GalN. Figure 5B corresponds to the Colr clinical isolate 1508.
Fig 4.
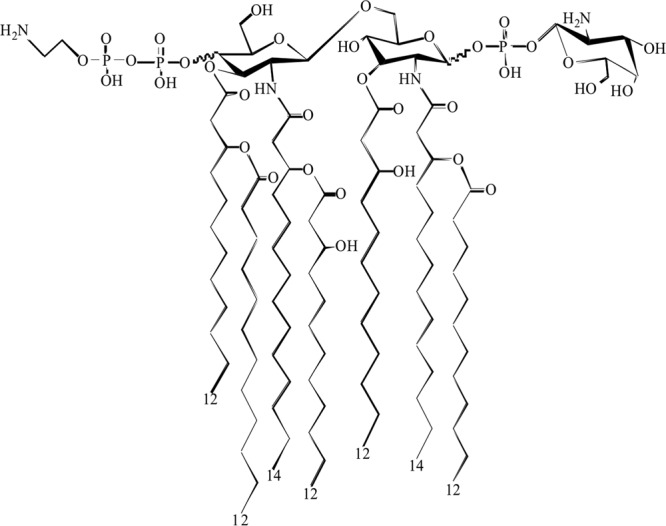
Proposed structure for lipid A extracted from A. baumannii LPS with both the pEtN and GalN modifications.
Fig 5.
LC-MS/MS selected reaction monitoring (SRM)-extracted ion chromatogram for galactosamine standard (A) and lipid A extracted from Colr A. baumannii 1508 LPS (B). Standard concentrations used were 1, 7, and 25 ng/ml.
Analysis of the extracted ion chromatogram for isolate 1508 (Fig. 4B) clearly demonstrates that the hexosamine modification was GalN. The percent relative abundance of lipid A modified with the GalN of clinical isolate 1508 was approximately 30%, showing a substantial presence of this substituent.
Lipid A profiles of A. baumannii clinical strains.
Colistin resistance in A. baumannii has been linked to pEtN modification of lipid A. To determine if these modifications are relevant and detectable in multiple patients' samples, we extracted lipid A from cross-sectional samples of three individual Cols clinical strains, collected from patients before colistin therapy at the University of Pittsburgh Medical Center. After growth at 37°C in rich medium (LB), lipid A was isolated and analyzed by MALDI-TOF mass spectrometry.
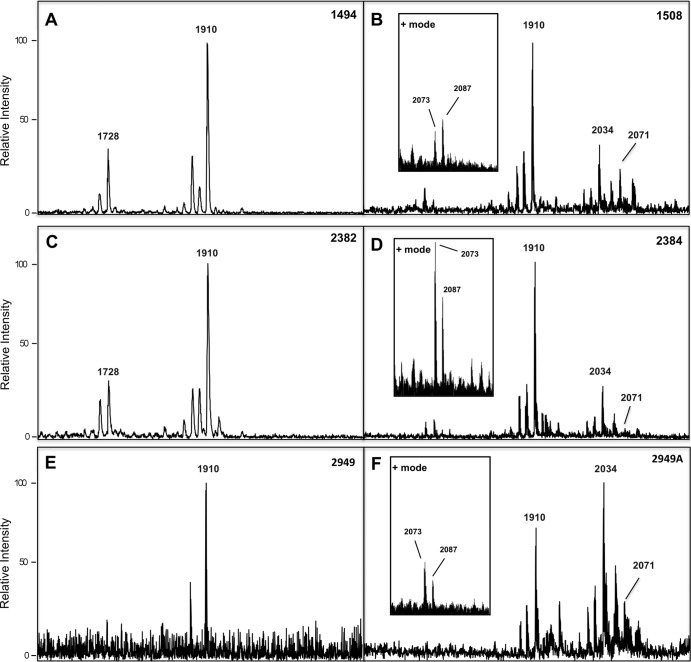
Analysis of lipid A isolated from our cross-sectional samples of Cols clinical strains (Fig. 6A, C, and E) showed that the most abundant [M-H]− ion was at m/z 1,910. Also of note, ions at m/z values corresponding to pEtN (m/z 2,033) and GalN (m/z 2,071) were absent from these spectra. The [M-H]− ion at m/z 1,910 corresponding to a diphosphorylated hepta-acylated lipid A was observed in all mass spectra (Fig. 6). In contrast, lipid A isolated from Colr clinical strains from the same patients after colistin therapy (Fig. 6B, D, and F) displayed not only the abundant [M-H]− at m/z 1,910 but also ions at m/z 2,033 and m/z 2,071, consistent with pEtN and GalN additions, respectively. While A. baumannii lipid A modified with pEtN has been reported in previous studies, GalN has not been previously reported for A. baumannii clinical strains.
Fig 6.
Negative ion mode MALDI-TOF mass spectra of A. baumannii clinical strains matched isolates Cols 1494 and Colr 1508 (A and B), matched isolates Cols 2382 and Colr 2384 (C and D), and matched isolates Cols 2949 and Colr 2949A (E and F). Positive ion mode MALDI-TOF mass spectra are shown in insets in panels B, D, and F. At m/z 2,069 is the GalN ion detected in positive mode, corresponding to the m/z 2,071 GalN ion detected in negative mode.
All Colr clinical strains (Table 1) contained the [M-H]− ions corresponding to pEtN and GalN modifications. To corroborate the detection of the GalN modifications observed in the negative ion mode, we performed positive ion mode [M-H]+ MALDI-TOF MS using lipid A isolated from the same Colr clinical strains shown in Fig. 6B, D, and F (refer to the “+ mode” insets in these panels). We hypothesized that the GalN moiety may be easier to detect in positive mode. The resulting spectra showed detection of GalN (m/z 2,073) in these samples due to the overall difference in net charge in positive mode. GalN should have an m/z of 2073 in positive ion mode rather than 2,071, as the analyte is doubly protonated. Also detected is a peak at m/z 2,087 that is a methylated variant of the GalN peak detected at m/z 2,073. There were also novel peaks detected in positive mode not seen in the negative mode analysis, and these ions are currently being defined. These results clearly demonstrate that lipid A isolated from these clinical strains contain lipid A structures that included the addition of pEtN and GalN substituents in response to colistin treatment. In summary, we show evidence that A. baumannii Colr clinical isolates show the same lipid A modifications as in the Colr laboratory-adapted strain and using both negative- and positive-mode MALDI-TOF MS.
DISCUSSION
In the setting of multidrug-resistant A. baumannii, tigecycline and especially colistin are the agents of last resort to combat this problematic pathogen. Despite previous reports of toxicity concerns, colistin has reemerged as the agent of choice to treat severe systemic and pulmonary A. baumannii infections (2, 28–30). Colistin resistance has been linked to modifications of the lipid A of the LPS moiety from many bacterial species, including A. baumannii strains (10, 11). Indeed, in this study we deduced the lipid A structure from LPS from the Colr A. baumannii MAC204 laboratory-adapted strain, and the resulting lipid A structure contains important modifications for colistin resistance. Using electrospray ionization (ESI) tandem mass spectrometry, we demonstrate that lipid A is hepta-acylated with secondary C12 and C14 acyl chains, bis- and pyrophosphorylated, along with a novel hexosamine addition shown to be GalN at the 1-phosphate position and a pEtN addition at the 4′ phosphate position. Using GC-FID analysis, we were able to quantify the fatty acid content of a Colr A. baumannii isolate. Finally, we used a set of clinical isolates from the same patient, showing that the modifications seen in our structural analysis from the MAC204 Colr laboratory-adapted strain are identical to the lipid A modifications in the Colr clinical isolates. The ability of these modifications to translate to clinical strains is important, as colistin is one of the last lines of defense for treatment against MDR A. baumannii strains. Thus, these important lipid A modifications could play a critical role in colistin resistance and subsequent patient outcome.
Lipid A structural analysis from the genus Acinetobacter was first delineated from the A. radioresistens strain S13 and showed that the major lipid A structure was a di-phosphoryl hepta-acylated structure with an m/z of 1,838.18 (31). Recent work has shown that modification of lipid A is a strategy employed not only in A. baumannii strains but in many other Gram-negative bacteria. Recent reports have shown Shigella flexneri lipid A to be modified by pEtN groups and involved in resistance to extreme acidity (32). Loss of pEtN addition in lptA null strains (lptA encodes the pEtN transferase for the 4′ position of lipid A) of Neisseria gonorrhoeae modulates resistance to complement killing and polymyxin resistance (33). Neisseria commensal strains that lack lptA and pEtN addition to lipid A were more susceptible to polymyxin B and showed an increased inflammatory response via TLR4 (34).
In the flagellated bacterium Campylobacter jejuni, lipid A is decorated with pEtN mediated by EptC, which serves a dual role as a pEtN transferase modulating CAMP resistance and as a modifier of the flagellar rod protein FlgG (35, 36). Helicobacter pylori modifies its lipid A in a two-step process whereby the removal of the 1-phosphate group by a lipid A phosphatase LpxE is followed by the addition of a pEtN residue catalyzed by EptA, resulting in increased resistance to polymyxin (37).
PmrA-LpxT-dependent pEtN modification of lipid A in Salmonella enterica serovar Typhimurium and Escherichia coli strains has been well documented (38) as well as a PmrC-dependent pEtN addition to the lipid A of E. coli O157:H7 (39). Surprisingly, the canine pathogen Capnocytophaga canimorsus and plant pathogens Xanthomonas campestris and Xanthomonas axanopodis have lipid A modified with pEtN, also altering their innate immune responses and disease manifestations. Thus, modification of lipid A by pEtN as a means to evade the effects of antimicrobial agents by altering the electrostatic charge on the bacterial membrane is prevalent (40, 41).
Additionally we have also shown a novel GalN addition to a Colr laboratory-adapted strain and Colr clinical strains as shown by high-order MS along with LC-MS/MS. This modification is also seen on the lipid A from Francisella novicida as published previously by our laboratory (25, 27) and others in the Francisella field (42, 43). In the setting of F. novicida, the enzyme FlmK adds a GalN and a mannose residue to the 1 and 4′ positions of lipid A whereas the FlmF2 enzyme is responsible for the addition of the GalN residue. GalN additions have also been shown to modulate resistance to antimicrobial agents (25), demonstrating the importance of elucidating this novel addition to the A. baumannii lipid A structure. In this context, a GalN modification could play a role in colistin resistance similar to that of aminoarabinose modifications, which have been shown to modulate antimicrobial resistance by altering the electrostatic charge on the bacterial membrane (along with pEtN modifications). Thus, these lipid A modifications have clinical ramifications in the setting of A. baumannii treatment.
Of interest is that these observations indicate that the presence or absence of pEtN and GalN on lipid A extracted from A. baumannii isolates may be used as novel biomarkers to be exploited for diagnosis of colistin resistance. Currently, the reliability and accuracy of susceptibility testing methods such as disk diffusion and Etest are compromised by poor agar diffusion of colistin due to its molecular size and poor concordance at low concentration, respectively (44–46). Therefore, further confirmation with different methods is required. Taken together, the potential to determine the presence or absence of modified lipid A with pEtN or GalN on bacterial clinical isolates by mass spectrometry could serve as an alternative diagnostic test to predict the resistance of bacteria to polymyxins and potentially guide therapeutic choices that will improve clinical outcomes.
Supplementary Material
ACKNOWLEDGMENTS
Y.D. was supported in part by a career development award from the NIH (K22AI080584). D.V.Z is funded via grants from the Military Infectious Diseases Research Program (MIDRP) and the Defense Medical Research and Development Program (DMRDP). This material is based upon work supported by, or in part by, the U.S. Army Research Laboratory and the U.S. Army Research Office under Contract/Grant no. W911NF-11-1-0274 to K.R.O.H and R.K.E.
The findings and opinions expressed herein belong to the authors and do not necessarily reflect the official views of the WRAIR, the U.S. Army, or the Department of Defense.
Footnotes
Published ahead of print 22 July 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00865-13.
REFERENCES
- 1.Perez F, Hujer AM, Hujer KM, Decker BK, Rather PN, Bonomo RA. 2007. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 51:3471–3484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Queenan AM, Pillar CM, Deane J, Sahm DF, Lynch AS, Flamm RK, Peterson J, Davies TA. 2012. Multidrug resistance among Acinetobacter spp. in the USA and activity profile of key agents: results from CAPITAL Surveillance 2010. Diagn. Microbiol. Infect. Dis. 73:267–270 [DOI] [PubMed] [Google Scholar]
- 3.Scott P, Deye G, Srinivasan A, Murray C, Moran K, Hulten E, Fishbain J, Craft D, Riddell S, Lindler L, Mancuso J, Milstrey E, Bautista CT, Patel J, Ewell A, Hamilton T, Gaddy C, Tenney M, Christopher G, Petersen K, Endy T, Petruccelli B. 2007. An outbreak of multidrug-resistant Acinetobacter baumannii-calcoaceticus complex infection in the US military health care system associated with military operations in Iraq. Clin. Infect. Dis. 44:1577–1584 [DOI] [PubMed] [Google Scholar]
- 4.Sebeny PJ, Riddle MS, Petersen K. 2008. Acinetobacter baumannii skin and soft-tissue infection associated with war trauma. Clin. Infect. Dis. 47:444–449 [DOI] [PubMed] [Google Scholar]
- 5.Barrette RW, Metwally SA, Rowland JM, Xu L, Zaki SR, Nichol ST, Rollin PE, Towner JS, Shieh WJ, Batten B, Sealy TK, Carrillo C, Moran KE, Bracht AJ, Mayr GA, Sirios-Cruz M, Catbagan DP, Lautner EA, Ksiazek TG, White WR, McIntosh MT. 2009. Discovery of swine as a host for the Reston ebolavirus. Science 325:204–206 [DOI] [PubMed] [Google Scholar]
- 6.Whitman TJ, Qasba SS, Timpone JG, Babel BS, Kasper MR, English JF, Sanders JW, Hujer KM, Hujer AM, Endimiani A, Eshoo MW, Bonomo RA. 2008. Occupational transmission of Acinetobacter baumannii from a United States serviceman wounded in Iraq to a health care worker. Clin. Infect. Dis. 47:439–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Basheer SM, Guiso N, Tirsoaga A, Caroff M, Novikov A. 2011. Structural modifications occurring in lipid A of Bordetella bronchiseptica clinical isolates as demonstrated by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 25:1075–1081 [DOI] [PubMed] [Google Scholar]
- 8.Llobet E, Campos MA, Gimenez P, Moranta D, Bengoechea JA. 2011. Analysis of the networks controlling the antimicrobial-peptide-dependent induction of Klebsiella pneumoniae virulence factors. Infect. Immun. 79:3718–3732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moskowitz SM, Ernst RK, Miller SI. 2004. PmrAB, a two-component regulatory system of Pseudomonas aeruginosa that modulates resistance to cationic antimicrobial peptides and addition of aminoarabinose to lipid A. J. Bacteriol. 186:575–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beceiro A, Llobet E, Aranda J, Bengoechea JA, Doumith M, Hornsey M, Dhanji H, Chart H, Bou G, Livermore DM, Woodford N. 2011. Phosphoethanolamine modification of lipid A in colistin-resistant variants of Acinetobacter baumannii mediated by the pmrAB two-component regulatory system. Antimicrob. Agents Chemother. 55:3370–3379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arroyo LA, Herrera CM, Fernandez L, Hankins JV, Trent MS, Hancock RE. 2011. The pmrCAB operon mediates polymyxin resistance in Acinetobacter baumannii ATCC 17978 and clinical isolates through phosphoethanolamine modification of lipid A. Antimicrob. Agents Chemother. 55:3743–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adams MD, Nickel GC, Bajaksouzian S, Lavender H, Murthy AR, Jacobs MR, Bonomo RA. 2009. Resistance to colistin in Acinetobacter baumannii associated with mutations in the PmrAB two-component system. Antimicrob. Agents Chemother. 53:3628–3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Westphal OJK. 1965. Bacterial lipopolysaccharides: extraction with phenol–water and further applications of the procedure. Methods Carbohydr. Chem. 43:83–91 [Google Scholar]
- 14.Fischer W, Koch HU, Haas R. 1983. Improved preparation of lipoteichoic acids. Eur. J. Biochem. 133:523–530 [DOI] [PubMed] [Google Scholar]
- 15.Folch J, Lees M, Sloane Stanley GH. 1957. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226:497–509 [PubMed] [Google Scholar]
- 16.Hirschfeld M, Ma Y, Weis JH, Vogel SN, Weis JJ. 2000. Cutting edge: repurification of lipopolysaccharide eliminates signaling through both human and murine toll-like receptor 2. J. Immunol. 165:618–622 [DOI] [PubMed] [Google Scholar]
- 17.Caroff M, Tacken A, Szabo L. 1988. Detergent-accelerated hydrolysis of bacterial endotoxins and determination of the anomeric configuration of the glycosyl phosphate present in the “isolated lipid A” fragment of the Bordetella pertussis endotoxin. Carbohydr. Res. 175:273–282 [DOI] [PubMed] [Google Scholar]
- 18.El Hamidi A, Tirsoaga A, Novikov A, Hussein A, Caroff M. 2005. Microextraction of bacterial lipid A: easy and rapid method for mass spectrometric characterization. J. Lipid Res. 46:1773–1778 [DOI] [PubMed] [Google Scholar]
- 19.Darveau RP, Cunningham MD, Bailey T, Seachord C, Ratcliffe K, Bainbridge B, Dietsch M, Page RC, Aruffo A. 1995. Ability of bacteria associated with chronic inflammatory disease to stimulate E-selectin expression and promote neutrophil adhesion. Infect. Immun. 63:1311–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Somerville JE, Jr, Cassiano L, Bainbridge B, Cunningham MD, Darveau RP. 1996. A novel Escherichia coli lipid A mutant that produces an antiinflammatory lipopolysaccharide. J. Clin. Invest. 97:359–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yi EC, Hackett M. 2000. Rapid isolation method for lipopolysaccharide and lipid A from gram-negative bacteria. Analyst 125:651–656 [DOI] [PubMed] [Google Scholar]
- 22.Kussak A, Weintraub A. 2002. Quadrupole ion-trap mass spectrometry to locate fatty acids on lipid A from Gram-negative bacteria. Anal. Biochem. 307:131–137 [DOI] [PubMed] [Google Scholar]
- 23.Boue SM, Cole RB. 2000. Confirmation of the structure of lipid A from Enterobacter agglomerans by electrospray ionization tandem mass spectrometry. J. Mass Spectrom. 35:361–368 [DOI] [PubMed] [Google Scholar]
- 24.Chan S, Reinhold VN. 1994. Detailed structural characterization of lipid A: electrospray ionization coupled with tandem mass spectrometry. Anal. Biochem. 218:63–73 [DOI] [PubMed] [Google Scholar]
- 25.Kanistanon D, Hajjar AM, Pelletier MR, Gallagher LA, Kalhorn T, Shaffer SA, Goodlett DR, Rohmer L, Brittnacher MJ, Skerrett SJ, Ernst RK. 2008. A Francisella mutant in lipid A carbohydrate modification elicits protective immunity. PLoS Pathog. 4:e24. 10.1371/journal.ppat.0040024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Domon B, Costello CE. 1988. Systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconjugate 5:397–409 [Google Scholar]
- 27.Kalhorn TF, Kiavand A, Cohen IE, Nelson AK, Ernst RK. 2009. A sensitive liquid chromatography/mass spectrometry-based assay for quantitation of amino-containing moieties in lipid A. Rapid Commun. Mass Spectrom. 23:433–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cai Y, Chai D, Wang R, Liang B, Bai N. 2012. Colistin resistance of Acinetobacter baumannii: clinical reports, mechanisms and antimicrobial strategies. J. Antimicrob. Chemother. 67:1607–1615 [DOI] [PubMed] [Google Scholar]
- 29.Gales AC, Jones RN, Sader HS. 2011. Contemporary activity of colistin and polymyxin B against a worldwide collection of Gram-negative pathogens: results from the SENTRY Antimicrobial Surveillance Program (2006-09). J. Antimicrob. Chemother. 66:2070–2074 [DOI] [PubMed] [Google Scholar]
- 30.Gordon NC, Wareham DW. 2010. Multidrug-resistant Acinetobacter baumannii: mechanisms of virulence and resistance. Int. J. Antimicrob. Agents 35:219–226 [DOI] [PubMed] [Google Scholar]
- 31.Leone S, Sturiale L, Pessione E, Mazzoli R, Giunta C, Lanzetta R, Garozzo D, Molinaro A, Parrilli M. 2007. Detailed characterization of the lipid A fraction from the nonpathogen Acinetobacter radioresistens strain S13. J. Lipid Res. 48:1045–1051 [DOI] [PubMed] [Google Scholar]
- 32.Martinic M, Hoare A, Contreras I, Alvarez SA. 2011. Contribution of the lipopolysaccharide to resistance of Shigella flexneri 2a to extreme acidity. PLoS One 6:e25557. 10.1371/journal.pone.0025557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewis LA, Shafer WM, Dutta Ray T, Ram S, Rice PA. 2013. Phosphoethanolamine residues on the lipid A moiety of Neisseria gonorrhoeae lipooligosaccharide modulate binding of complement inhibitors and resistance to complement killing. Infect. Immun. 81:33–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.John CM, Liu M, Phillips NJ, Yang Z, Funk CR, Zimmerman LI, Griffiss JM, Stein DC, Jarvis GA. 2012. Lack of lipid A pyrophosphorylation and functional lptA reduces inflammation by Neisseria commensals. Infect. Immun. 80:4014–4026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cullen TW, Madsen JA, Ivanov PL, Brodbelt JS, Trent MS. 2012. Characterization of unique modification of flagellar rod protein FlgG by Campylobacter jejuni lipid A phosphoethanolamine transferase, linking bacterial locomotion and antimicrobial peptide resistance. J. Biol. Chem. 287:3326–3336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cullen TW, O'Brien JP, Hendrixson DR, Giles DK, Hobb RI, Thompson SA, Brodbelt JS, Trent MS. 2013. EptC of Campylobacter jejuni mediates phenotypes involved in host interactions and virulence. Infect. Immun. 81:430–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herrera CM, Hankins JV, Trent MS. 2010. Activation of PmrA inhibits LpxT-dependent phosphorylation of lipid A promoting resistance to antimicrobial peptides. Mol. Microbiol. 76:1444–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Falagas ME, Rafailidis PI, Matthaiou DK. 2010. Resistance to polymyxins: mechanisms, frequency and treatment options. Drug Resist. Updat. 13:132–138 [DOI] [PubMed] [Google Scholar]
- 39.Kim SH, Jia W, Parreira VR, Bishop RE, Gyles CL. 2006. Phosphoethanolamine substitution in the lipid A of Escherichia coli O157:H7 and its association with PmrC. Microbiology 152:657–666 [DOI] [PubMed] [Google Scholar]
- 40.Ittig S, Lindner B, Stenta M, Manfredi P, Zdorovenko E, Knirel YA, dal Peraro M, Cornelis GR, Zahringer U. 2012. The lipopolysaccharide from Capnocytophaga canimorsus reveals an unexpected role of the core-oligosaccharide in MD-2 binding. PLoS Pathog. 8:e1002667. 10.1371/journal.ppat.1002667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Silipo A, Sturiale L, Garozzo D, Erbs G, Jensen TT, Lanzetta R, Dow JM, Parrilli M, Newman MA, Molinaro A. 2008. The acylation and phosphorylation pattern of lipid A from Xanthomonas campestris strongly influence its ability to trigger the innate immune response in Arabidopsis. Chembiochem 9:896–904 [DOI] [PubMed] [Google Scholar]
- 42.Llewellyn AC, Zhao J, Song F, Parvathareddy J, Xu Q, Napier BA, Laroui H, Merlin D, Bina JE, Cotter PA, Miller MA, Raetz CR, Weiss DS. 2012. NaxD is a deacetylase required for lipid A modification and Francisella pathogenesis. Mol. Microbiol. 86:611–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beasley AS, Cotter RJ, Vogel SN, Inzana TJ, Qureshi AA, Qureshi N. 2012. A variety of novel lipid A structures obtained from Francisella tularensis live vaccine strain. Innate Immun. 18:268–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Balaji V, Jeremiah SS, Baliga PR. 2011. Polymyxins: antimicrobial susceptibility concerns and therapeutic options. Indian J. Med. Microbiol. 29:230–242 [DOI] [PubMed] [Google Scholar]
- 45.Lo-Ten-Foe JR, de Smet AM, Diederen BM, Kluytmans JA, van Keulen PH. 2007. Comparative evaluation of the VITEK 2, disk diffusion, etest, broth microdilution, and agar dilution susceptibility testing methods for colistin in clinical isolates, including heteroresistant Enterobacter cloacae and Acinetobacter baumannii strains. Antimicrob. Agents Chemother. 51:3726–3730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Galani I, Kontopidou F, Souli M, Rekatsina PD, Koratzanis E, Deliolanis J, Giamarellou H. 2008. Colistin susceptibility testing by Etest and disk diffusion methods. Int. J. Antimicrob. Agents 31:434–439 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.