Abstract
Quinolones trap the covalent gyrase-DNA complex in Escherichia coli, leading to cell death. Processing activities for trapped covalent complex have not been characterized. A mutant strain lacking SbcCD nuclease activity was examined for both accumulation of gyrase-DNA complex and viability after quinolone treatment. Higher complex levels were found in ΔsbcCD cells than in wild-type cells after incubation with nalidixic acid and ciprofloxacin. However, SbcCD activity protected cells against the bactericidal action of nalidixic acid but not ciprofloxacin.
TEXT
DNA topoisomerases change DNA topology through a DNA breakage and rejoining mechanism in which the DNA breakage step involves the formation of a covalent topoisomerase-DNA complex that is rapidly reversed upon DNA rejoining (1). The quinolones are effective broad-spectrum antimicrobial agents that target bacterial type IIA topoisomerases, DNA gyrase (2), and topoisomerase IV (3, 4) by trapping the covalent topoisomerase-DNA intermediate. Although much is known about the mechanism of action of quinolones, the repair of quinolone-mediated lesions is poorly understood. Repair of lesions caused by quinolones is expected to reduce bacterial cell death. In eukaryotes, a number of repair activities for trapped topoisomerase-DNA complexes have been reported (5–12), suggesting that functional redundancy from multiple repair pathways may also be found in bacteria. One of the most extensively studied eukaryotic repair proteins is the highly conserved Mre11/Rad50 (MR) complex (13). MR complexes consist of a nuclease (Mre11 in humans and Saccharomyces cerevisiae, with the homologues SbcD in Escherichia coli and Gp46 in bacteriophage T4) and a structural, ATP-dependent, coiled-coil protein (Rad50 in humans and yeast, with the homologues SbcC in E. coli and Gp47 in bacteriophage T4) (Fig. 1). In E. coli, previous biochemical studies with purified components demonstrated that the enzymatic activity of the SbcCD nuclease can unblock DNA ends sealed by covalently bound biotin-streptavidin protein complex when the activity of the enzyme was restricted to the DNA duplex termini (14). However, experimental evidence for E. coli SbcCD participation in repair of trapped topoisomerase-DNA complex in cultured cells has not been reported. In order to gain insights into the mechanisms through which bacteria can deal with quinolone-mediated lesions, we investigated a role for the SbcCD nuclease in the repair of quinolone-trapped gyrase-DNA complexes.
Fig 1.
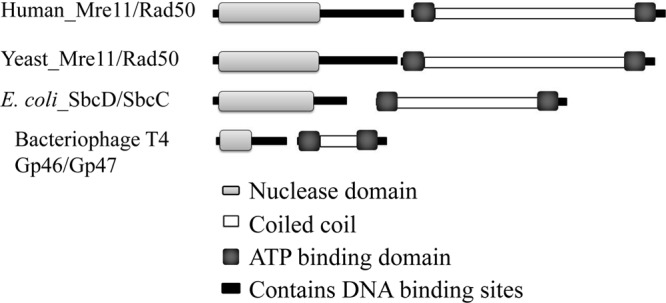
Homologous Mre11 and Rad50 protein domains. Sequence regions corresponding to the nuclease domain and ATPase domain are light and dark gray, respectively. The coiled-coil regions are white. The diagram was drawn approximately to scale using the Conserved Domain Architecture Retrieval Tool (CDART), available at the National Center for Biotechnology Information website (32). There is 27 to 39% identity when homologous domains of yeast Mre11/Rad50 and E. coli SbcCD are aligned.
We previously developed a method for the analysis of covalent topoisomerase-DNA complexes accumulated on chromosomal DNA of E. coli cells in cultures (15). A similar protocol using cesium chloride density gradient sedimentation to detect topoisomerase-DNA complexes has been used to characterize the relevant processing and repair activities in eukaryotes (16–18). In this study, we utilized our previously developed method to investigate repair of quinolone-stabilized gyrase-DNA complexes by the SbcCD protein.
In order to investigate a potential role for SbcCD nuclease in the repair of quinolone-stabilized gyrase-DNA complexes, a mutant strain with an sbcCD deletion was used to determine the effect of the mutation on viability after quinolone treatment. The ΔsbcCD strain was constructed by P1clr100 transduction selecting for kanamycin resistance using E. coli strains BW27784 [Δ(araD-araB)567 ΔlacZ4787(::rrnB-3) λ− Δ(araH-araF)570(::FRT) ΔaraEp-532::FRT ϕPcp18araE533 Δ(rhaD-rhaB)568 hsdR514] (19) and PFG947 [F-ara Δ(gpt-lac)5 thi Rifr (Pro−) ΔsbcCD::Kan] (20) as the recipient and donor, respectively. Following transduction, the kanamycin resistance marker was removed as described previously (21). Transduction of the sbcCD::Kan allele and removal of the kanamycin marker were confirmed by PCR with the primers5′-CCCTCTGTATTCATTATCCTGCTG-3′ and 5′-TCTGTTTGGGTATAATCGCGCCCATGC-3′.
Nalidixic acid was added to exponentially growing cultures (optical density at 600 nm [OD600] = 0.4) of the E. coli parental strain BW27784 and the ΔsbcCD mutant derivative at 8 μg/ml (2× MIC). Following addition of the drug, cultures were further incubated for 30 to 90 min before dilution and plating on drug-free LB agar plates. Colonies were counted following overnight incubation of the agar plates. As shown in Fig. 2A, the ΔsbcCD deletion mutant showed an ∼6-fold decrease in viable CFU/ml following 90 min incubation with 8 μg/ml nalidixic acid compared to the viable counts at time zero. In contrast, only growth inhibition, not a decrease in viable CFU/ml, was seen for the parental strain at this 2× MIC nalidixic acid concentration. Measurement of MIC by the broth macrodilution method showed that the ΔsbcCD mutation did not change the MIC for nalidixic acid, which correlates with inhibition of DNA replication (22). The ΔsbcCD mutation had no significant effect on growth in the absence of quinolone, as seen in a spotting assay and in a growth curve monitored by absorption at 600 nm (see Fig. S1 in the supplemental material). Collectively, these data support the contention that for quinolones, blocking growth and killing cells are distinct events.
Fig 2.
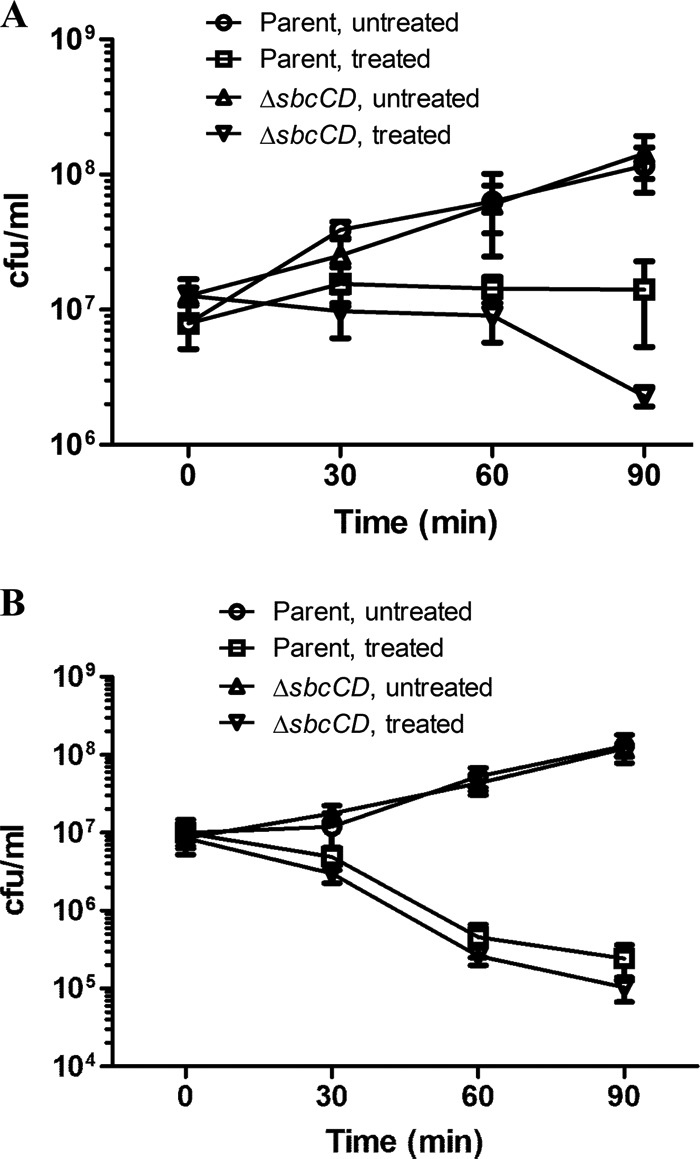
Effect of ΔsbcCD mutation on viability following nalidixic acid and ciprofloxacin treatments. (A) Exponentially growing cultures of E. coli strain BW27784 (parental strain) and its mutant derivative ΔsbcCD were incubated for 30, 60, or 90 min with 8 μg/ml of nalidixic acid corresponding to 2× MIC. Aliquots of treated culture after incubation with the drug were removed, diluted, and plated on LB agar for overnight incubation to determine the number of viable colonies. (B) Parental and ΔsbcCD strains were grown to exponential phase and incubated with 0.062 μg/ml ciprofloxacin (2× MIC) for the indicated times. The data are averages and standard errors from at least 3 independent experiments.
To demonstrate SbcCD-mediated processing of quinolone-stabilized gyrase-DNA complexes in cell cultures, the level of gyrase-DNA covalent complexes in ΔsbcCD chromosomal DNA was compared to that in the parental E. coli strain following quinolone treatment. The previously described method based on CsCl gradient centrifugation (15) was utilized to separate free gyrase from DNA-bound gyrase based on their different densities. Representative immunoblots, probed with antibodies against GyrA, from experiments repeated at least three times are shown. As expected, accumulation of gyrase covalent complexes in chromosomal DNA was observed by immunoblotting following 30 min of incubation with 1× (4 μg/ml) and 2× (8 μg/ml) MIC of nalidixic acid in both the parental and ΔsbcCD mutant strains (Fig. 3A, fraction 3). However, 1.7-fold ± 0.4-fold-higher and 2.0-fold ± 0.1-fold-higher covalent complex levels were observed in the ΔsbcCD than in the parental strain at 1× MIC and 2× MIC, respectively (means ± standard errors of the means [SEM]) (Fig. 3A). This difference in covalent complex accumulation was not due to a difference in gyrase expression, as shown in blots of fractions containing free proteins (Fig. 3A, fractions 8 and 9) and Western blots of the total GyrA subunit (data not shown). Because quinolone inhibition of DNA synthesis (22–24) and gyrase covalent complex formation (15, 24) takes place within minutes following addition of the drug to the medium, we speculate that potential repair activities for trapped gyrase-DNA complexes may act within the first minutes of incubation with quinolones as well. Therefore, the accumulation of gyrase-DNA covalent complexes in ΔsbcCD mutant was investigated at earlier time points (5 and 15 min). Levels of gyrase-DNA complex in the ΔsbcCD mutant were 3.7-fold ± 1.4-fold and 4.1-fold ± 2.7-fold (means ± SEM) higher than those in the parental strain at 5 and 15 min, respectively, after treatment with 8 μg/ml nalidixic acid (2× MIC) (Fig. 3B).
Fig 3.
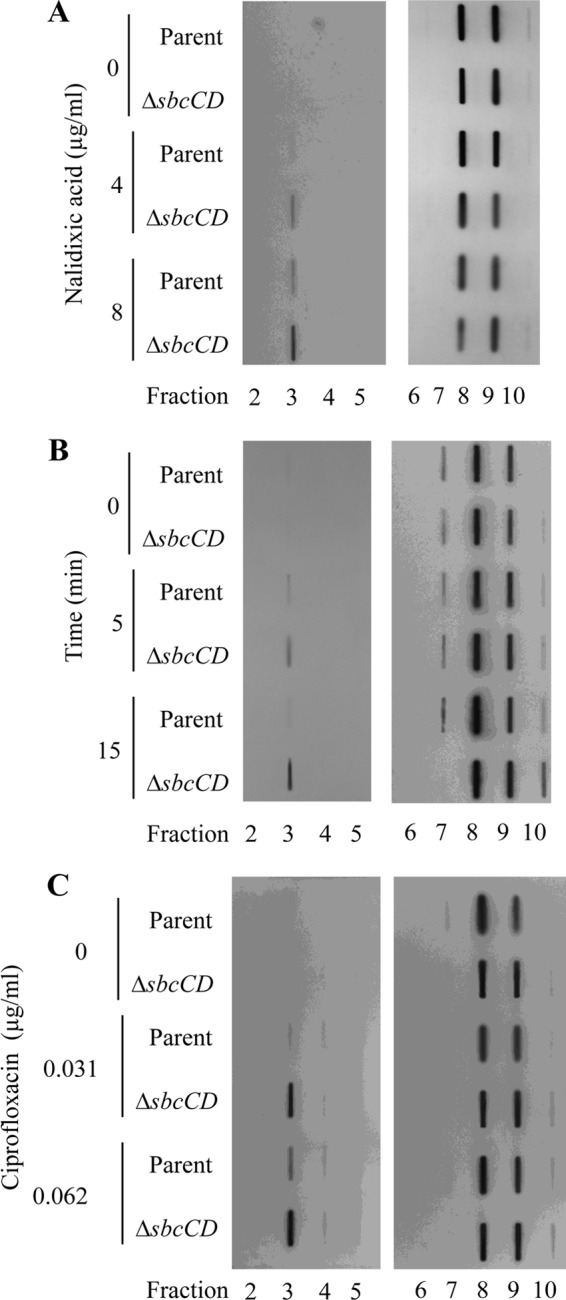
Increased levels of covalently bound gyrase in the ΔsbcCD mutant compared to parental strain. (A) Exponentially growing cultures of E. coli parental strain BW27784 and its ΔsbcCD mutant derivative were incubated for 30 min in the absence or presence of 4 μg/ml (1× MIC) or 8 μg/ml (2× MIC) of nalidixic acid. Cells were then lysed in the presence of a detergent, and gyrase-DNA complexes were separated from free gyrase by sedimentation in a CsCl density gradient. Gradients were fractionated from bottom to top and loaded on a slot blot. Slot blots were probed with an anti-GyrA antibody. (Left) CsCl density gradient fractions 2 to 5, collected from the bottom portion of the gradient (higher density). (Right) CsCl density gradient fractions 6 to 10, collected from the top portion of the gradient (lower density). (B) The parent and the ΔsbcCD mutant were grown to exponential phase, and an aliquot was collected immediately before the addition of nalidixic acid to a final concentration of 8 μg/ml (2× MIC) (0 min). After 5 and 15 min of incubation with nalidixic acid, more aliquots were taken. Gyrase-DNA complexes were isolated as described for panel A. (C) The parent and the ΔsbcCD mutant were incubated for 5 min in the absence or presence of 0.031 μg/ml (1× MIC) or 0.062 μg/ml (2× MIC) of ciprofloxacin. Gyrase-DNA complexes were isolated as described for panel A.
Previous studies showed that the cell death pathway triggered by narrow-spectrum quinolones, such as nalidixic and oxolinic acids, is protein synthesis dependent, since nalidixic acid-mediated chromosome fragmentation and cell killing are blocked by chloramphenicol, a protein synthesis inhibitor (25, 26). A second quinolone-mediated cell death pathway that can be induced by expanded-spectrum fluoroquinolones, including ciprofloxacin, is independent of protein synthesis (25–27). In order to investigate whether the repair activity of SbcCD protein has an effect on ciprofloxacin-mediated loss of viability, we compared survival of the ΔsbcCD E. coli strain and the parental strain following 30, 60, and 90 min of incubation with ciprofloxacin at 0.062 μg/ml (2× MIC). Cell death was observed for both the parental strain and the mutant at this ciprofloxacin concentration. No significant difference in loss of viability was observed between the parental strain and the mutant (Fig. 2B). These results suggest that the repair activity of SbcCD has little effect on the protein synthesis-independent cell death pathway induced by ciprofloxacin and thereby provide independent support for the existence of that pathway. Nevertheless, 2.0-fold ± 0.3-fold (mean ± SEM)-greater complex accumulation was seen in the ΔsbcCD mutant than in the parental strain after 5 min incubation with 1× (0.031 μg/ml) or 2× (0.062 μg/ml) MIC of ciprofloxacin (Fig. 3C). This discordance between complex number and killing raises the interesting possibility that lethal complexes differ from bacteriostatic ones, and killing may be a function of the drug being bound in the complex.
Our results show that the activity of SbcCD helps to prevent cell death from the protein synthesis-dependent cell death pathway associated with nalidixic acid. However, the protein synthesis-independent pathway induced by ciprofloxacin appears not to be impeded by the SbcCD processing activity, possibly because this pathway is triggered more rapidly or irreversibly by the lethal complexes following treatment with ciprofloxacin. There may be multiple processing mechanisms for topoisomerase-DNA complexes, as has been shown for the processing of yeast and human topoisomerase complexes (5, 8, 28–31), which remain to be characterized for E. coli.
Supplementary Material
ACKNOWLEDGMENTS
We thank P. L. Foster, Indiana University, Bloomington, IN, for E. coli strain PFG947.
This research was supported by award R01AI069313 to Y.-C.T.-D. from the National Institute of Allergy and Infectious Diseases.
Footnotes
Published ahead of print 5 August 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00130-13.
REFERENCES
- 1.Wang JC. 2002. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 3:430–440 [DOI] [PubMed] [Google Scholar]
- 2.Gellert M, Mizuuchi K, O'Dea MH, Itoh T, Tomizawa JL. 1977. Nalidixic acid resistance: a second genetic character involved in DNA gyrase activity. Proc. Natl. Acad. Sci. U. S. A. 74:4772–4776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khodursky AB, Zechiedrich EL, Cozzarelli NR. 1995. Topoisomerase IV is a target of quinolones in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 92:11801–11805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soussy CJ, Wolfson JS, Ng EY, Hooper DC. 1993. Limitations of plasmid complementation test for determination of quinolone resistance due to changes in the gyrase A protein and identification of conditional quinolone resistance locus. Antimicrob. Agents Chemother. 37:2588–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cortes Ledesma F, El Khamisy SF, Zuma MC, Osborn K, Caldecott KW. 2009. A human 5′-tyrosyl DNA phosphodiesterase that repairs topoisomerase-mediated DNA damage. Nature 461:674–678 [DOI] [PubMed] [Google Scholar]
- 6.Liu C, Pouliot JJ, Nash HA. 2002. Repair of topoisomerase I covalent complexes in the absence of the tyrosyl-DNA phosphodiesterase Tdp1. Proc. Natl. Acad. Sci. U. S. A. 99:14970–14975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pouliot JJ, Yao KC, Robertson CA, Nash HA. 1999. Yeast gene for a Tyr-DNA phosphodiesterase that repairs topoisomerase I complexes. Science 286:552–555 [DOI] [PubMed] [Google Scholar]
- 8.Sacho EJ, Maizels N. 2011. DNA repair factor MRE11/RAD50 cleaves 3′-phosphotyrosyl bonds and resects DNA to repair damage caused by topoisomerase 1 poisons. J. Biol. Chem. 286:44945–44951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beidler DR, Cheng YC. 1995. Camptothecin induction of a time- and concentration-dependent decrease of topoisomerase I and its implication in camptothecin activity. Mol. Pharmacol. 47:907–914 [PubMed] [Google Scholar]
- 10.Zhang A, Lyu YL, Lin CP, Zhou N, Azarova AM, Wood LM, Liu LF. 2006. A protease pathway for the repair of topoisomerase II-DNA covalent complexes. J. Biol. Chem. 281:35997–36003 [DOI] [PubMed] [Google Scholar]
- 11.Hartsuiker E, Neale MJ, Carr AM. 2009. Distinct requirements for the Rad32(Mre11) nuclease and Ctp1(CtIP) in the removal of covalently bound topoisomerase I and II from DNA. Mol. Cell 33:117–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hong G, Kreuzer KN. 2003. Endonuclease cleavage of blocked replication forks: an indirect pathway of DNA damage from antitumor drug-topoisomerase complexes. Proc. Natl. Acad. Sci. U. S. A. 100:5046–5051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharples GJ, Leach DR. 1995. Structural and functional similarities between the SbcCD proteins of Escherichia coli and the RAD50 and MRE11 (RAD32) recombination and repair proteins of yeast. Mol. Microbiol. 17:1215–1217 [DOI] [PubMed] [Google Scholar]
- 14.Connelly JC, de Leau ES, Leach DR. 2003. Nucleolytic processing of a protein-bound DNA end by the E. coli SbcCD (MR) complex. DNA Repair (Amst.) 2:795–807 [DOI] [PubMed] [Google Scholar]
- 15.Aedo S, Tse-Dinh YC. 2012. Isolation and quantitation of topoisomerase complexes accumulated on Escherichia coli chromosomal DNA. Antimicrob. Agents Chemother. 56:5458–5464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adachi N, So S, Koyama H. 2004. Loss of nonhomologous end joining confers camptothecin resistance in DT40 cells. Implications for the repair of topoisomerase I-mediated DNA damage. J. Biol. Chem. 279:37343–37348 [DOI] [PubMed] [Google Scholar]
- 17.Interthal H, Champoux JJ. 2011. Effects of DNA and protein size on substrate cleavage by human tyrosyl-DNA phosphodiesterase 1. Biochem. J. 436:559–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang YW, Regairaz M, Seiler JA, Agama KK, Doroshow JH, Pommier Y. 2011. Poly(ADP-ribose) polymerase and XPF-ERCC1 participate in distinct pathways for the repair of topoisomerase I-induced DNA damage in mammalian cells. Nucleic Acids Res. 39:3607–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khlebnikov A, Datsenko KA, Skaug T, Wanner BL, Keasling JD. 2001. Homogeneous expression of the P(BAD) promoter in Escherichia coli by constitutive expression of the low-affinity high-capacity AraE transporter. Microbiology 147:3241–3247 [DOI] [PubMed] [Google Scholar]
- 20.Storvik KA, Foster PL. 2011. The SMC-like protein complex SbcCD enhances DNA polymerase IV-dependent spontaneous mutation in Escherichia coli. J. Bacteriol. 193:660–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cherepanov PP, Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9–14 [DOI] [PubMed] [Google Scholar]
- 22.Chow RT, Dougherty TJ, Fraimow HS, Bellin EY, Miller MH. 1988. Association between early inhibition of DNA synthesis and the MICs and MBCs of carboxyquinolone antimicrobial agents for wild-type and mutant [gyrA nfxB(ompF) acrA] Escherichia coli K-12. Antimicrob. Agents Chemother. 32:1113–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Snyder M, Drlica K. 1979. DNA gyrase on the bacterial chromosome: DNA cleavage induced by oxolinic acid. J. Mol. Biol. 131:287–302 [DOI] [PubMed] [Google Scholar]
- 24.Chen CR, Malik M, Snyder M, Drlica K. 1996. DNA gyrase and topoisomerase IV on the bacterial chromosome: quinolone-induced DNA cleavage. J. Mol. Biol. 258:627–637 [DOI] [PubMed] [Google Scholar]
- 25.Malik M, Zhao X, Drlica K. 2006. Lethal fragmentation of bacterial chromosomes mediated by DNA gyrase and quinolones. Mol. Microbiol. 61:810–825 [DOI] [PubMed] [Google Scholar]
- 26.Malik M, Hussain S, Drlica K. 2007. Effect of anaerobic growth on quinolone lethality with Escherichia coli. Antimicrob. Agents Chemother. 51:28–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malik M, Nitiss JL. 2004. DNA repair functions that control sensitivity to topoisomerase-targeting drugs. Eukaryot. Cell 3:82–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vance JR, Wilson TE. 2002. Yeast Tdp1 and Rad1-Rad10 function as redundant pathways for repairing Top1 replicative damage. Proc. Natl. Acad. Sci. U. S. A. 99:13669–13674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang SW, Burgin AB, Jr, Huizenga BN, Robertson CA, Yao KC, Nash HA. 1996. A eukaryotic enzyme that can disjoin dead-end covalent complexes between DNA and type I topoisomerases. Proc. Natl. Acad. Sci. U. S. A. 93:11534–11539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee KC, Padget K, Curtis H, Cowell IG, Moiani D, Sondka Z, Morris NJ, Jackson GH, Cockell SJ, Tainer JA, Austin CA. 2012. MRE11 facilitates the removal of human topoisomerase II complexes from genomic DNA. Biol. Open 1:863–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Collignon P, Powers JH, Chiller TM, Aidara-Kane A, Aarestrup FM. 2009. World Health Organization ranking of antimicrobials according to their importance in human medicine: a critical step for developing risk management strategies for the use of antimicrobials in food production animals. Clin. Infect. Dis. 49:132–141 [DOI] [PubMed] [Google Scholar]
- 32.Geer LY, Domrachev M, Lipman DJ, Bryant SH. 2002. CDART: protein homology by domain architecture. Genome Res. 12:1619–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


