Abstract
Rilpivirine is a nonnucleoside reverse transcriptase inhibitor used to treat HIV-1. In the present study, the pathways responsible for the biotransformation of rilpivirine were defined. Using human liver microsomes, the formation of two mono- and two dioxygenated metabolites were detected via ultra high-performance liquid chromatography-tandem mass spectrometry (UHPLC-MS/MS). Mass spectral analysis of the products suggested that these metabolites resulted from oxygenation of the 2,6-dimethylphenyl ring and methyl groups of rilpivirine. Chemical inhibition studies and cDNA-expressed cytochrome P450 (CYP) assays indicated that oxygenations were catalyzed primarily by CYP3A4 and CYP3A5. Glucuronide conjugates of rilpivirine and a monomethylhydroxylated metabolite of rilpivirine were also detected and were found to be formed by UDP-glucuronosyltransferases (UGTs) UGT1A4 and UGT1A1, respectively. All metabolites that were identified in vitro were detectable in vivo. Further, targeted UHPLC-MS/MS-based in vivo metabolomics screening revealed that rilpivirine treatment versus efavirenz treatment may result in differential levels of endogenous metabolites, including tyrosine, homocysteine, and adenosine. Rilpivirine biotransformation was also assessed across species using liver microsomes isolated from a range of mammals, and the metabolite profile identified using human liver microsomes was largely conserved for both oxidative and glucuronide metabolite formation. These studies provide novel insight into the metabolism of rilpivirine and the potential differential effects of rilpivirine- and efavirenz-containing antiretroviral regimens on the endogenous metabolome.
INTRODUCTION
Rilpivirine (RPV; Edurant), 4-[[4-[[4-[(1E)-2-cyanoethenyl]-2,6-dimethylphenyl]amino]-2-pyrimidinyl]amino]benzonitrile, is a more recently developed nonnucleoside reverse transcriptase inhibitor that has been FDA approved for oral administration in combination therapy for the treatment of drug-naive individuals infected with human immunodeficiency virus 1 (HIV-1) (1). RPV is a cyanovinyl diarylpyrimidine and as such has inherent molecular flexibility, allowing for multiple modes of allosteric binding within the hydrophobic binding pocket of the HIV reverse transcriptase; therefore, RPV has a higher genetic barrier to resistance than the initially developed nonnucleoside reverse transcriptase inhibitors, efavirenz and nevirapine (2, 3). In addition, because of its antiviral efficacy, RPV is being developed as an injectable, long-acting formulation for potential use in HIV preexposure prophylaxis (4, 5). ECHO and THRIVE clinical trials, comparing RPV-tenofovir-emtricitabine (Complera) treatment to that with efavirenz-tenofovir-emtricitabine (Atripla), demonstrated that the RPV coformulation has more potent antiviral activity and fewer adverse side effects than does a daily antiretroviral efavirenz-based regimen. In these studies, the most noted adverse events associated with the efavirenz-containing regimen were psychiatric symptoms and rash; however, the molecular mechanism(s) that underlie the differences in the safety profiles of RPV-containing versus efavirenz-containing regimens is unknown (6, 7). While we recently demonstrated that the primary metabolite of efavirenz, 8-hydroxyefavirenz, may play a causal role in efavirenz-mediated toxicities, including hepatic and neuronal toxicity, this type of analysis is not yet possible for RPV, as the metabolites of this drug have yet to be defined (8, 9).
The cytochrome P450 (CYP) superfamily of heme-containing monooxygenases plays a major role in the metabolism of a variety of clinically relevant drugs, including many antiretrovirals. The package inserts for Edurant and Complera state that RPV is primarily metabolized in the liver by CYP3A4 (10, 11); however, primary data detailing the identities of the RPV metabolites and the relative contributions of individual CYP isozymes to the formation of these metabolites have yet to be reported. In addition, a potential role for conjugative metabolism of RPV by phase II drug-metabolizing enzymes, including UDP-glucuronosyltransferases (UGTs), has yet to be demonstrated. Since RPV is taken in combination with other therapeutic agents, defining the routes of RPV metabolism should help to facilitate a more robust and reliable prediction of potential drug-drug interactions involving RPV. Further, elucidation of the pathways of RPV metabolism may lend mechanistic insight to the understanding of potential interindividual differences in RPV exposure and clearance.
The objectives of this study were to (i) comprehensively characterize the biotransformation of RPV in vitro through the utilization of human liver microsomes, cDNA-expressed CYP and UGT isozymes, and primary human hepatocytes analyzed by ultrahigh-performance liquid chromatography-tandem mass spectrometry (UHPLC-MS/MS) and (ii) gain an understanding of the profiles of endogenous small molecules that regulate key cellular processes in individuals receiving Complera versus Atripla in order to spur the elucidation of the molecular mechanism(s) that may underlie the observed differences in the safety profiles of RPV and efavirenz clinically. Collectively, the data lend novel insight into the routes of RPV metabolism which we anticipate will help to facilitate the prediction of drug-drug interactions involving RPV. Further, these studies provide a foundation for a mechanistic understanding of the pathways that modulate RPV exposure in vivo.
MATERIALS AND METHODS
Materials.
Rilpivirine (RPV) was obtained through the National Institutes of Health AIDS Research and Reference Reagent Program (Germantown, MD). Dimethyl sulfoxide (DMSO) was used as the vehicle for all reagents and was purchased from Mediatech Inc. (Manassas, VA). (+)-N-3-Benzylnirvanol, furafylline, ketoconazole, sulfaphenazole, and quinidine were purchased from Sigma-Aldrich Co. (St. Louis, MO), and 2-phenyl-2-(1-piperidinyl)propane (PPP) was purchased from Santa Cruz Biotechnology Inc. (Santa Cruz, CA).
RPV metabolism by liver microsomes, cDNA-expressed CYPs, and cDNA-expressed UGTs.
RPV (20 μM) was incubated with liver microsomes (Xenotech LLC, Lenexa, KS) from the following animals at a concentration of 2 mg/ml: human (mixed sex, pool of 50), beagle dog (male, pool of 8), golden (Syrian) (GS) hamster (male, pool of 100), Hartley albino (HA) guinea pig (male, pool of 50), Sinclair minipig (male, pool of 3), cynomolgus (CYN) monkey (male, pool of 10), rhesus monkey (male, pool of 6), BALB/c mouse (male, pool of 800), CD1 mouse (male, pool of 1,000), New Zealand (NZ) rabbit (male, pool of 8), and Sprague-Dawley (SD) rat (male, pool of 433). For CYP and UGT individual contribution experiments, 20 μM RPV was incubated with the following cDNA-expressed enzymes (Supersomes, BD Biosciences): CYP1A2, -2B6, -2C8, -2C9, -2C19, -2D6, -3A4, and -3A5 and UGT1A1, -1A3, -1A4, -1A6, -1A7, -1A8, -1A9, -1A10, -2B4, -2B7, -2B15, and -2B17 at concentrations of 10 pmol/ml and 0.2 mg/ml, respectively.
Liver microsomes (2 mg/ml) were preincubated with RPV at 37°C for 5 min in 100 mM potassium phosphate buffer, pH 7.4, prior to the addition of an NADPH-regenerating system (BD Biosciences). RPV incubations with human liver microsomes and liver microsomes from other species proceeded at 37°C for a total of 30 min. Formation of glucuronidated metabolites in the liver microsome preparations required the simultaneous addition of NADPH and a UGT reaction mix (BD Biosciences) containing 25 mM uridine 5′-diphosphoglucuronic acid (UDPGA) and alamethicin, and reactions proceeded at 37°C for 60 min.
cDNA-expressed CYPs (10 pmol/ml) were preincubated with RPV at 37°C for 5 min in 100 mM potassium phosphate buffer, pH 7.4, prior to the addition of an NADPH-regenerating system from which the reaction proceeded at 37°C for a total of 30 min. cDNA-expressed UGT assays, however, required a modified reaction preparation. RPV was preincubated for 30 min at 37°C in 100 mM potassium phosphate buffer, pH 7.4, with 10 pmol/ml of CYP3A4 and an NADPH-regenerating system in order to allow for hydroxylated-metabolite formation, to which the UDPGA- and alamethicin-containing reaction mix was added. Glucuronide metabolite formation was then initiated by the addition of 0.2 mg/ml of each individual UGT isozyme, and reactions proceeded at 37°C for an additional 60 min.
Chemical inhibition of CYPs was performed by preincubating 2 mg/ml of human liver microsomes for 5 min in a reaction mixture containing an NADPH-regenerating system, 100 mM potassium phosphate buffer, pH 7.4, and the following concentrations of an inhibitor or vehicle: 20 μM furafylline (CYP1A2 inhibitor), 30 μM PPP (CYP2B6 inhibitor), 20 μM sulfaphenazole (CYP2C9 inhibitor), 10 μM (+)-N-3-benzylnirvanol (CYP2C19 inhibitor), 1 μM quinidine (CYP2D6 inhibitor), 1 μM ketoconazole (CYP3A4 and -3A5 inhibitor), and 0.2% DMSO. These final concentrations of reagents were based on previously reported studies (12–14). Following the 5-min preincubation at 37°C, RPV was added to each inhibitor or DMSO control treatment, and the reactions were allowed to proceed at 37°C for a total of 30 min.
The resulting 100-μl reaction volumes from the previously described metabolism assays were quenched with 100 μl of acetonitrile at various times and incubated on ice for 10 min prior to centrifugation at 10,000 × g for 10 min at 4°C. Following centrifugation, supernatants of these assays were dried under vacuum before reconstitution in 100 μl of methanol, from which 1 μl was injected for UHPLC-MS/MS analysis.
UHPLC-MS/MS analysis of RPV oxygenated and glucuronide metabolites.
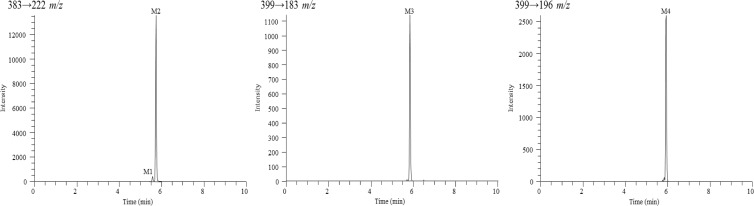
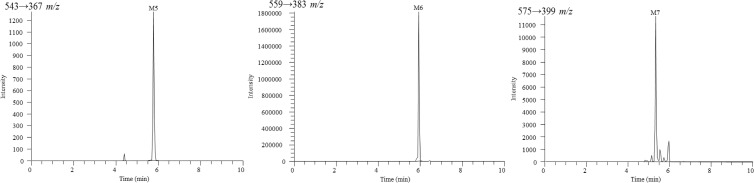
A UHPLC-MS/MS method was developed and optimized for the detection of RPV metabolites using a Dionex UltiMate 3000 UHPLC system (Thermo Scientific, Pittsburgh, PA) coupled to a Thermo Scientific TSQ Vantage triple-stage quadrupole mass spectrometer. Samples were resolved using a Polaris C18-A column (5 μm, 100 by 2.0 mm; Agilent Technologies, Santa Clara, CA) at a flow rate of 0.4 ml/min. A multistep gradient was implemented using mobile phases A (water, 0.1% formic acid) and B (acetonitrile, 0.1% formic acid): 10% B for 0.0 to 1.0 min, increased from 10% B to 55% B over 1.0 to 8.0 min, held at 100% B from 8.0 to 8.3 min, and then decreased to 10% B for 8.3 to 10 min for column reequilibration. The electrospray ionization interface was set to positive ion mode, and the following instrument parameters were used: ion transfer capillary temperature, 300°C; spray voltage, 5,500 V; sheath and auxillary nitrogen gas pressures, 60 and 15, respectively; and collision energy, 30 V for RPV and oxygenated metabolites and 60 V for glucuronidated metabolites. Metabolite identification was initially performed in product ion (MS/MS) mode, and selected reaction monitoring (SRM) mode was used for the detection of the relative abundance levels of metabolites. Metabolites were identified as those ions with a signal-to-noise ratio of five or greater that exhibited NADPH and/or UDPGA dependence. The following transitions in SRM mode were monitored (parent mass, Q1→product ion, Q3): 367→195 m/z (RPV), 383→222 m/z (monohydroxylated RPV; M1 and M2), 399→183 m/z (dihydroxylated RPV; M3), 399→196 m/z (dihydroxylated RPV; M4), 543→367 m/z (RPV glucuronide conjugate; M5), 559→383 m/z (monohydroxylated RPV glucuronide conjugate; M6), and 575→399 m/z (dihydroxylated RPV glucuronide conjugate; M7). The approximate retention times for metabolites M1 through M7 were 5.51, 5.66, 5.78, 5.88, 5.74, 5.90, and 5.32 min, respectively, and 6.48 min for RPV.
RPV treatment of primary human hepatocytes.
Six- and twelve-well plates of primary human hepatocytes with Matrigel overlay were purchased from Xenotech, LLC; these hepatocytes had been isolated from three individual donors (lots 1155, 1157, and 1158). The ages (in years) and sexes of the donors were as follows: 43 and female, 59 and female, and 36 and male. Cell viabilities were reported to be 74.7%, 77.9%, and 74.9%, respectively. Hepatocytes were incubated overnight in Williams' medium E (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum, 1% penicillin-streptomycin, and 1% l-glutamine at 37°C in a 5% CO2 humidified environment. Prior to treatment, hepatocytes were placed into fresh medium to which either 0.1% DMSO (vehicle) or 10 μM RPV was added for 6-, 12-, and 24-h incubations. Medium from each DMSO and RPV time point was collected, dried under vacuum, and reconstituted in 100 μl of methanol, of which 1 μl was injected onto the UHPLC-MS/MS system for analysis.
Human plasma and urine sample preparation for UHPLC-MS/MS analysis.
Deidentified plasma and urine samples were obtained from participants (a total of six) who were already on an antiretroviral drug regimen and had been recruited by the Drug Development Unit of the Johns Hopkins University School of Medicine Division of Clinical Pharmacology after providing written informed consent for participation in a protocol approved by the institutional review board of the Johns Hopkins Medical Institutions. Blood and urine samples were obtained from three HIV-1-infected individuals receiving Complera, a coformulation of RPV-tenofovir-emtricitabine, and from three HIV-1-infected individuals receiving Atripla, a coformulation of efavirenz-tenofovir-emtricitabine. The ages (in years) and sexes of participants being treated with Complera were as follows: 29 and male, 45 and female, and 42 and male. The ages (in years) and sexes of participants being treated with Atripla were as follows: 50 and male, 58 and female, and 40 and male. Whole blood was centrifuged for 10 min at 1,100 × g in EDTA-coated tubes, from which the resulting supernatant was collected as plasma and live virus was heat inactivated for 10 min at 60°C. Urine was centrifuged for 10 min at 12,000 × g, and the resulting supernatant was collected. For UHPLC-MS/MS analysis, 100 μl of plasma or urine was extracted with 500 μl of acetonitrile and incubated on ice for 10 min, followed by centrifugation at 10,000 × g for 10 min. For RPV metabolite detection, Complera clinical samples were dried under vacuum and then six individual extracts of either plasma or urine were pooled and reconstituted in a total of 100 μl of methanol, of which 5 μl was injected on the UHPLC-MS/MS system for analysis.
UHPLC-MS/MS-based targeted screen of plasma and urine endogenous metabolomic profiles.
Plasma and urine from the previously described Complera clinical samples were utilized in a targeted screen of endogenous small-molecule metabolites. The metabolic profiles of these samples were further analyzed in comparison to plasma and urine of HIV-infected individuals receiving therapeutic treatment with Atripla. Noninfected, non-antiretroviral-treated human plasma was prepared by the manufacturer using an EDTA anticoagulant (lots BRH642964, BRH642965, and BRH642967) as the experimental samples were, and urine (lots BRH720679, BRH720681, and BRH720682) that was purchased from Bioreclamation, LLC (Westbury, NY), was processed in the same manner as the experimental samples. Separate positive ion and negative ion mode UHPLC-MS/MS methods were developed and optimized for the detection of a range of metabolites using a Dionex UltiMate 3000 UHPLC system coupled to a Thermo Scientific TSQ Vantage triple-stage quadrupole mass spectrometer using the same instrument parameters as defined above. Samples were resolved using a Polaris C18-A column (5 μm, 100 by 2.0 mm; Agilent Technologies). The positive ion mode method used a multistep gradient of mobile phases A (water, 0.5% acetic acid) and B (methanol, 0.5% acetic acid): 5% B for 0.0 to 1.1 min at 0.3 ml/min, increased from 5 to 95% B for 1.1 to 4.5 min at 0.3 ml/min, held at 95% B for 4.5 to 5.0 min at 0.4 ml/min and then 95% B for 5.0 to 5.1 min at 0.3 ml/min; the column was reequilibrated with 5% B from 5.1 to 6 min at 0.3 ml/min. The negative ion mode method used a flow gradient at 0.35 ml/min of mobile phases A (water, 0.1% formic acid) and B (acetonitrile, 0.1% formic acid): 10% B for 0.0 to 0.9 min, increased from 10 to 90% B for 0.9 to 4.0 min; the column was reequilibrated with 10% B for 4.0 to 5 min. Extracted plasma and urine samples as previously described were reconstituted in either 50 μl of mobile phase A (water, 0.1% formic acid) for analysis by positive ion mode or 50 μl of a 1:1 solution of mobile phase A to methanol for negative ion mode analysis, of which 5 μl was injected for both UHPLC-MS/MS analyses. SRM mode transitions were developed for the detection and determination of relative abundance levels of each endogenous metabolite via infusion of synthetic standards for each molecule. The optimized parent mass-to-product ion transitions and collision energies used for each metabolite in this targeted screen, as well as their approximate retention times, are listed in Tables S1 and S2 in the supplemental material for detection in the positive ion and negative ion modes, respectively.
Statistical analysis.
Data analysis was performed using GraphPad Prism (version 6; GraphPad Software Inc., San Diego, CA). Statistical significance was determined by Student's t test and is depicted in figures as follows: *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
RESULTS
In vitro detection of RPV oxygenated and glucuronide metabolites.
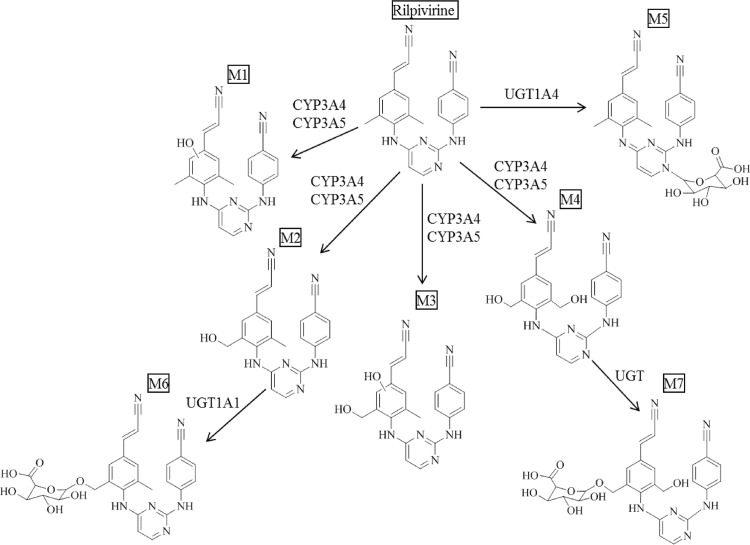
RPV metabolites were initially characterized using a human liver microsome assay and analyzed by UHPLC-MS/MS. Tandem mass spectrometry was used for metabolite detection and provided structural information of metabolites based on the resulting fragment ions; however, SRM mode allowed for greater sensitivity in detecting metabolites and was, therefore, utilized in determining the relative abundance for all metabolites. Since the metabolites of RPV were previously unreported, synthetic standards of these compounds could not be obtained. Thus, the assays that were developed in this study for the purpose of metabolite identification are solely qualitative. A total of seven RPV metabolites were detected using the human liver microsome in vitro assay and are indicated as metabolite 1 (M1) through metabolite 7 (M7). Two monohydroxylated metabolites, M1 and M2 (383 m/z), and two dihydroxylated metabolites, M3 and M4 (399 m/z), resulted from oxygen insertion into RPV (Fig. 1). Three glucuronides resulted from direct glucuronidation of RPV (543 m/z) and glucuronidation of mono- and dioxygenated RPV metabolites (559 m/z and 575 m/z, respectively) (Fig. 2). Hydroxylated metabolites were monitored via SRM mode using the following transitions (parent mass, Q1→product ion, Q3): 383→222 m/z (M1 and M2), 399→183 m/z (M3), and 399→196 m/z (M4). From a 30-min human liver microsomal metabolism assay, M2 was observed to be approximately 13.0-fold more abundant than M1, and M4 was approximately 2.3-fold more abundant than M3. The following transitions were used for monitoring glucuronidated metabolites: 543→367 m/z (M5), 559→383 m/z (M6), and 575→399 m/z (M7). From a 60-min human liver microsomal metabolism assay in the presence of UDPGA, the abundance of glucuronide M6 was observed to be approximately 1,500.0-fold and 163.3-fold greater than the relative abundance of M5 and M7 glucuronides, respectively. Other phase II metabolites, such as sulfated metabolites, were not detected under these experimental conditions.
Fig 1.
Extracted ion chromatograms for RPV monohydroxylated metabolites, M1 and M2, and dihydroxylated metabolites, M3 and M4. Human liver microsomes (2 mg/ml) were incubated with 20 μM RPV for 30 min at 37°C in the presence of an NADPH-regenerating system. RPV metabolites were detected by UHPLC-MS/MS in SRM mode using the following transitions: 383→222 m/z (M1 and M2), 399→183 m/z (M3), and 399→196 m/z (M4). Chromatograms are representative of three individual experiments. It should be noted that the y axes differ for each metabolite due to the observed intensity of the ions.
Fig 2.
Extracted ion chromatograms for RPV glucuronidated metabolites, M5, M6, and M7. Peaks not indicated as metabolites in these chromatograms were also present in the minus-NADPH and UDPGA controls and resulted in intensities below a signal-to-noise ratio of five. Human liver microsomes (2 mg/ml) were incubated with 20 μM RPV for 60 min at 37°C in the presence of an NADPH-regenerating system and UDPGA. Metabolites were detected by UHPLC-MS/MS in SRM mode using the following transitions: 543→367 m/z (M5), 559→383 m/z (M6), and 575→399 m/z (M7). Chromatograms are representative of three individual experiments. It should be noted that the y axes differ for each metabolite due to the observed intensity of the ions.
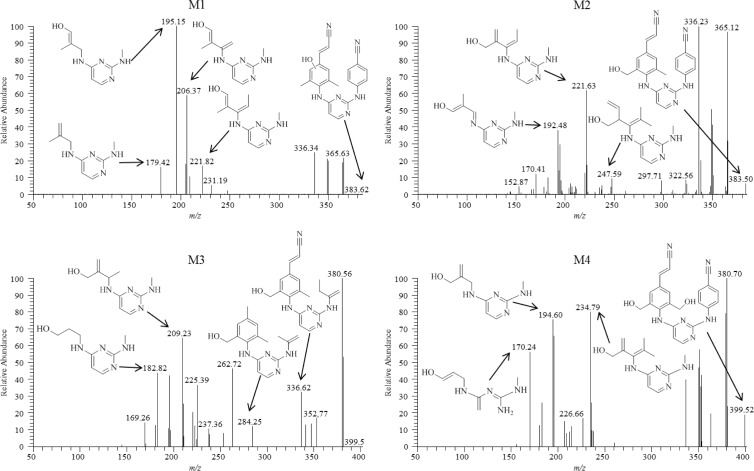
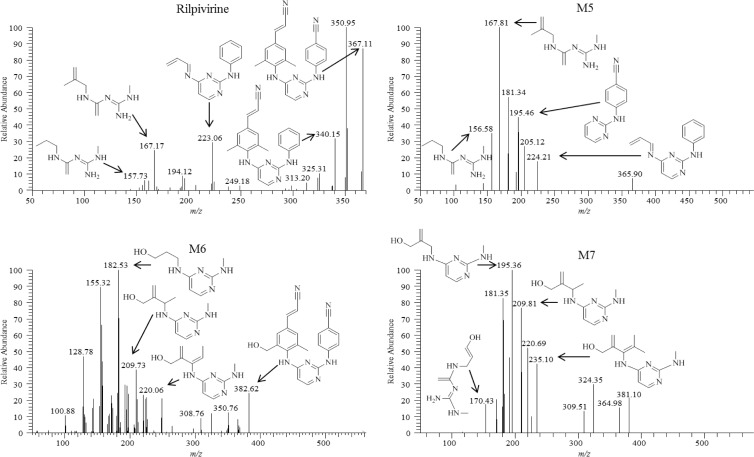
Chemical structures of these metabolites were proposed through use of individual fragmentation spectra observed in MS/MS mode. For monohydroxylated metabolite M1, it is proposed that fragment ions 179.42, 195.15, 206.37, and 221.82 m/z correspond to the loss of C9H14N4, C9H14N4O, C10H14N4O, and C11H16N4O, respectively. The observed fragment ions 192.48, 221.63, and 247.59 m/z for the more abundant monohydroxylated metabolite M2 are proposed to correspond to the loss of C9H12N4O, C11H16N4O, and C13H20N4O, respectively (Fig. 3). For dihydroxylated metabolite M3, the fragment ions 182.82, 209.23, 284.25, and 336.62 m/z are proposed to be consistent with the loss of C8H14N4O, C10H16N4O, C16H20N4O, and C19H21N5O, respectively (Fig. 3). Dihydroxylated metabolite M4 fragment ions, 170.24, 194.60, and 234.79 m/z, were predicted to result from the loss of C7H14N4O, C9H14N4O, and C12H18N4O, respectively (Fig. 3). The structures of glucuronidated metabolites were elucidated in a similar fashion, and the resulting fragmentation spectra were compared to those observed for RPV or for the predicted corresponding oxidative metabolite. Glucuronide M5 is proposed to result from direct N-linked glucuronidation of RPV on the pyrimidine ring, as evidenced by the fact that human liver microsomes could form this metabolite in the absence of an NADPH-regenerating system (required for cytochrome P450-dependent activity). In addition, the mass-to-charge ratio of this metabolite as well as the fragmentation pattern was consistent with direct conjugation of glucuronic acid to RPV itself (Fig. 4). The most abundant 167.81 m/z fragment ion of M5 was commensurate with the loss of C8H16N4 and was also observed to be a predominant fragment ion for RPV (Fig. 4). Comparison of the M6 fragmentation spectrum to that of the monohydroxymethyl metabolite M2 suggested O-linked glucuronidation occurring after initial oxidative metabolism, with the fragment ion 382.62 m/z corresponding to loss of M2 itself, C22H18N6O, and the fragment ion 220.06 m/z corresponding to the loss of C11H16N4O as was similarly observed for the fragmentation of M2 (Fig. 4). Furthermore, comparison of the fragment ions of M7 to those of the dihydroxymethyl metabolite M4 also indicated O-linked glucuronidation after initial oxidative metabolism, with the fragment ions 170.43, 195.36, and 235.10 m/z proposed to correlate to the loss of C7H14N4O, C9H14N4O, and C12H18N4O, respectively, as was similarly observed for the fragmentation of M4 (Fig. 4). From the predicted modes of fragmentation for these metabolites, we propose that M1 results from oxygen insertion on the 2,6-dimethylphenyl ring, M2 results from oxygen insertion on either methyl group, M3 results from oxygen insertion on a methyl group as well as the dimethylphenyl ring, and M4 results from oxygen insertion on both methyl groups.
Fig 3.
Fragmentation spectra of monohydryoxylated metabolites M1 and M2 (top) and dihydroxylated metabolites M3 and M4 (bottom). Proposed modes of fragmentation are indicated by the fragment ion corresponding to the appropriate mass-to-charge ratio. Human liver microsomes (2 mg/ml) were incubated with 20 μM RPV for 30 min at 37°C in the presence of an NADPH-regenerating system. Metabolites were detected in MS/MS mode using parent masses of 383 m/z for M1 and M2 and 399 m/z for M3 and M4. Spectra are representative of three individual experiments.
Fig 4.
Fragmentation spectra of RPV (top) and glucuronidated metabolites M5 (top) and M6 and M7 (bottom). Proposed modes of fragmentation are indicated by the fragment ion corresponding to the appropriate mass-to-charge ratio. Human liver microsomes (2 mg/ml) were incubated with 20 μM RPV for 60 min at 37°C in the presence of an NADPH-regenerating system and UDPGA. Metabolites were detected in MS/MS mode using parent masses of 367, 543, 559, and 575 m/z for RPV, M5, M6, and M7, respectively. Spectra are representative of three individual experiments.
CYP3A4 and CYP3A5 are primarily responsible for the oxidative metabolism of RPV.
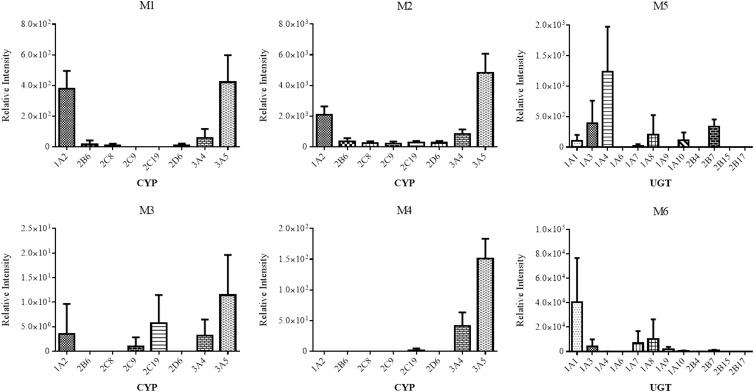
Following identification of RPV metabolites, cDNA-expressed CYPs were employed in order to determine which CYP isozymes had the ability to catalyze the formation of RPV metabolites. The cDNA-expressed CYPs used in these experiments were CYP1A2, -2B6, -2C8, -2C9, -2C19, -2D6, -3A4, and -3A5 and are representative of the principal drug-metabolizing CYPs expressed in human liver (15). CYP3A5 was found to metabolize RPV to M1, M2, M3, and M4 at the greatest abundances, followed by CYP1A2 and CYP3A4 (Fig. 5). CYP2C19 also displayed the ability to catalyze the formation of M3, while relatively minor formation of M3 by CYP2C9 was also observed.
Fig 5.
Relative contributions of individual drug-metabolizing CYPs and UGTs to RPV metabolite formation. cDNA-expressed CYPs (10 pmol/ml) were incubated with 20 μM RPV for 30 min at 37°C in the presence of an NADPH-regenerating system. For UGT assays, cDNA-expressed CYP3A4 (10 pmol/ml) was preincubated with 20 μM RPV, an NADPH-regenerating system, and UDPGA from 30 min at 37°C, to which 0.2 mg/ml of cDNA-expressed UGTs was added for an additional 60-min incubation. Metabolite formation was detected by UHPLC-MS/MS in SRM mode using the following transitions: 383→222 m/z (M1 and M2), 399→183 m/z (M3), 399→196 m/z (M4), 543→367 m/z (M5), and 559→383 m/z (M6). Glucuronide M7 was not detected under the UGT assay conditions. Data are representative of the mean relative peak intensities of three experiments ± standard deviations.
Chemical inhibition experiments were performed in an effort to probe the role of each CYP in the biotransformation of RPV in a system in which all of the above-mentioned isozymes were present. Human liver microsomes were preincubated for 5 min with the DMSO vehicle or CYP inhibitor prior to the addition of RPV. Treatment of microsomes with 1 μM ketoconazole, which is an inhibitor of both CYP3A4 and CYP3A5 due to their high amino acid sequence similarities, resulted in the most significantly decreased formation of M1, M2, M3, and M4 relative to the DMSO control, with approximate decreases of 33.3% ± 6.3%, 30.5% ± 11.1%, 4.6% ± 2.3%, and 8.1% ± 2.8%, respectively (means ± standard deviations of three replicates). In addition, M1 formation was inhibited by 30 μM PPP (CYP2B6 inhibitor), 20 μM sulfaphenazole (CYP2C9 inhibitor), and 10 μM (+)-N-3-benzylnirvanol (CYP2C19 inhibitor), with decreases of 58.1% ± 17.2%, 64.8% ± 16.7%, and 49.1% ± 17.4%, respectively, relative to the DMSO control. (+)-N-3-Benzylnirvanol additionally inhibited M2 formation, with a decrease of 51.8% ± 17.2% compared to control incubations. PPP and (+)-N-3-benzylnirvanol inhibited M3 formation as well, with decreases of 59.5% ± 14.2% and 53.4% ± 14.3%, respectively, relative to the control samples. Pretreatment with furafylline (20 μM; CYP1A2 inhibitor) and 1 μM quinidine (CYP2D6 inhibitor) did not result in abrogation of oxidative metabolite formation.
Since RPV and the structurally similar antiretroviral drug etravirine were previously demonstrated to increase CYP3A4 mRNA expression (16–18), we tested whether RPV had the ability to induce its own metabolism using primary human hepatocytes. Hepatocytes isolated from three donors were treated with either the DMSO vehicle or 10 μM RPV for 6, 12, or 24 h. Hepatocyte medium was collected after each time point for UHPLC-MS/MS analysis (see Fig. S1 in the supplemental material). Hydroxylated metabolites M1, M2, and M4 were detectable at all time points; M3 was observed only at 6 h and 24 h. Glucuronides M5 and M6 were present at each time point examined following RPV treatment; however, glucuronide M7 was not detected in the medium of any of the three hepatocyte preparations. While M1 and M2 exhibited a trend toward an increase in abundance over 24 h of RPV treatment, their levels did not reach statistical significance; interestingly, an increase in the mRNA expression of CYP3A4 over time was indeed observed, with changes of 7.4-fold (range, 2.3- to 17.4-fold), 8.8-fold (range, 3.05- to 13.6-fold), and 19.5-fold (range, 3.02- to 28.4-fold) relative to the DMSO control for 6, 12, and 24 h, respectively.
UGT1A1 and UGT1A4 contribute to glucuronidation of RPV and monooxygenated metabolite M2.
In order to determine which individual UGT isozymes contribute to the formation of glucuronidated metabolites of RPV, indicated here as M5, M6, and M7, assays were performed using cDNA-expressed UGTs. M5 was formed by direct N-linked glucuronidation of RPV on the pyrimidine ring in a non-NADPH-dependent manner as determined by incubation with human liver microsomes. The formation of glucuronides M6 and M7 was observed to be NADPH dependent, suggesting initial oxygenation of RPV prior to UGT-mediated conjugation with glucuronic acid. Therefore, CYP3A4 was used for coincubations with individual cDNA-expressed UGT isozymes in the presence of RPV, an NADPH-regenerating system, and UDPGA in order to ensure that the oxygenated metabolites were formed at levels that would allow for the facile detection of the subsequent glucuronide-conjugated metabolites M6 and M7. The cDNA-expressed UGT isozymes used for these assays were UGT1A1, -1A3, -1A4, -1A6, -1A7, -1A8, -1A9, -1A10, -2B4, -2B7, -2B15, and -2B17, all of which are expressed in human liver (19). UGT1A4 was found to be primarily responsible for the formation of M5, and UGT1A1 was found to be primarily responsible for the formation of M6 (Fig. 5). Glucuronide M7 was not formed under these experimental conditions using any of the UGTs tested.
In vivo detection of RPV metabolites in plasma and urine of individuals receiving Complera.
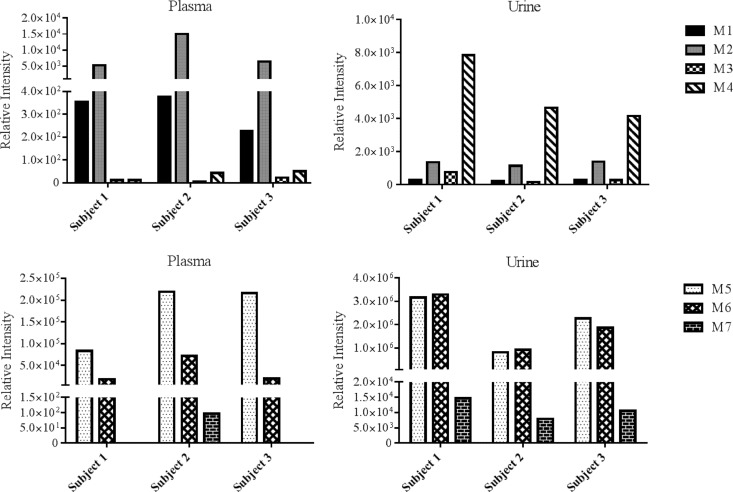
In an effort to determine whether the metabolites that were identified through our in vitro studies were also formed in vivo, RPV metabolites were measured in plasma and urine collected from individuals receiving the RPV-containing drug Complera, a coformulation of RPV-tenofovir-emtricitabine. The UHPLC-MS/MS SRM transitions developed for the analysis of in vitro metabolite formation by human liver microsomes and cDNA-expressed CYP and UGT isozymes were employed in detecting oxygenated and glucuronidated RPV metabolites in these clinical samples. While all of the metabolites that were identified in vitro were indeed detectable in vivo, variation in the abundance of metabolites was observed across the three subjects (Fig. 6). Further, although M7 was detectable in all of the urine samples that were analyzed, this metabolite was present in the plasma of only one subject. Monohydroxylated metabolites, M1 and M2, were more abundant in plasma than dihydroxylated metabolites, M3 and M4, while M4 was the most abundant oxygenated metabolite detected in urine. M5 was the most abundant glucuronide in plasma, while M5 and M6 were observed to be similarly more abundant than glucuronide M7 in urine. Relative plasma RPV levels were determined to be approximately 70% of the combined plasma nonmetabolized drug and metabolite levels in these subjects. Glucuronide M5 accounted for 24 to 34% of this total, followed by glucuronide M6, with 5 to 10%, and monooxygenated M2, with 1 to 2%. M1, M3, and M4 accounted for <1% of the total. In contrast, RPV was found to be a relatively minor component of the urine collected, as it accounted for only approximately <1% of the collective nonmetabolized drug and metabolite levels in urine. The urine glucuronide levels for M5 and M6 were approximately 50% of total detectable drug and RPV metabolites, and M1, M2, M3, M4, and M7 accounted for <1%.
Fig 6.
In vivo detection of RPV metabolites, M1 through M4 (top) and M5 through M7 (bottom), in human plasma and urine. Samples were collected from clinical subjects on an RPV-based antiretroviral regimen. Plasma and urine were extracted with acetonitrile in preparation for UHPLC-MS/MS analysis. Metabolites were detected in SRM mode using the following transitions: 383→222 m/z (M1 and M2), 399→183 m/z (M3), 399→196 m/z (M4), 543→367 m/z (M5), 559→383 m/z (M6), and 575→399 m/z (M7). Data shown are representative of three individual participants.
Comparison of Complera and Atripla endogenous metabolomic profiles using a UHPLC-MS/MS targeted screen.
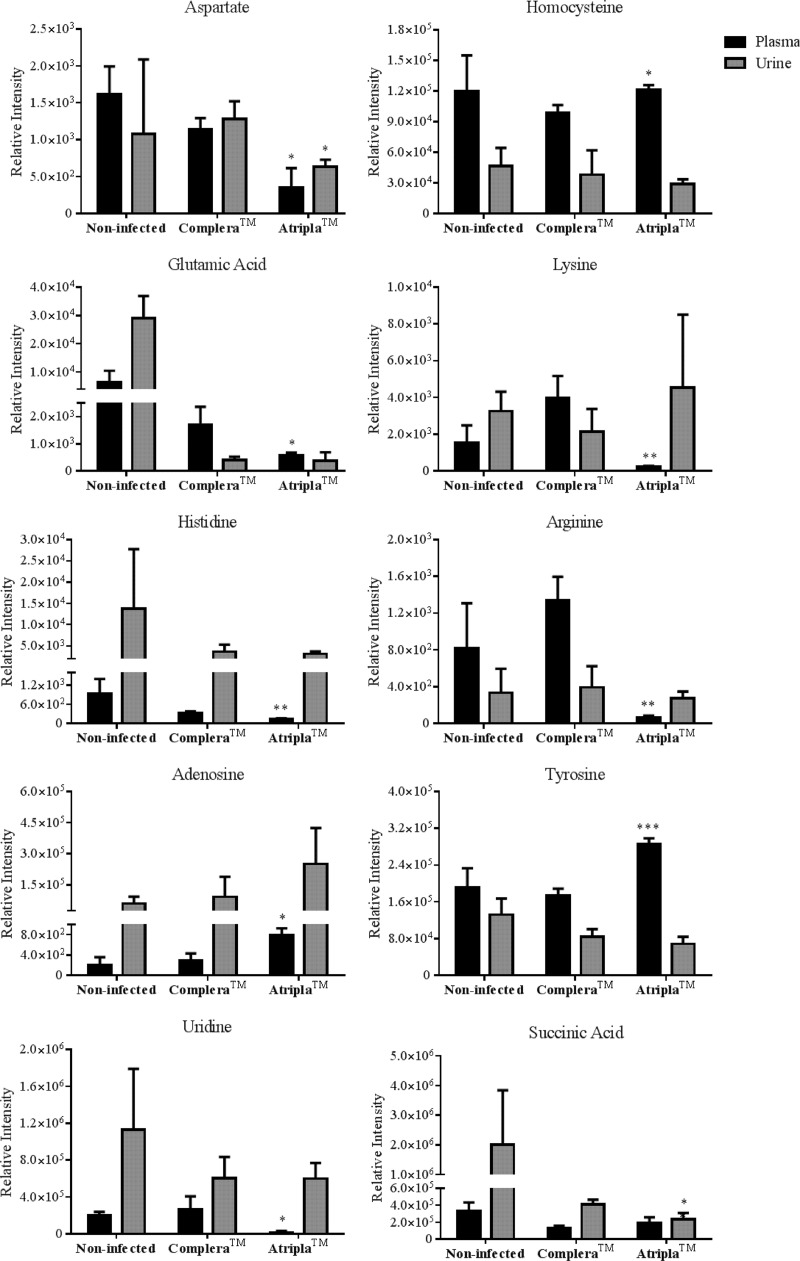
A novel UHPLC-MS/MS-based metabolomics screen was developed in an effort to characterize differences in the endogenous metabolomes of HIV-infected individuals receiving Complera (RPV-tenofovir-emtricitabine) or Atripla (efavirenz-tenofovir-emtricitabine). While the purpose of the analysis was to probe for variation in the levels of endogenous small molecules that participate in critical cellular processes in subjects being treated with each of these drugs, plasma and urine samples from HIV-negative individuals that had not received Complera or Atripla were also measured. The latter samples were used to provide a context for understanding levels of these molecules in human subjects that have not previously been treated with either drug. In this screen, 19 total metabolites, including essential and nonessential amino acids, nucleotides, and key intermediates of the citric acid cycle, methionine-homocysteine cycle, and urea cycle, were targeted for detection by UHPLC-MS/MS (see Tables S1 and S2 in the supplemental material) in the plasma and urine collected from both drug-treated groups as well as the noninfected, non-antiretroviral-treated group. Of the detectable endogenous metabolites, 10 exhibited significant differences in relative abundance levels in the comparison of plasma and urine samples from the two treatment groups (Fig. 7). Specifically, aspartate, glutamic acid, histidine, uridine, lysine, and arginine were observed at lower levels in the plasma of those subjects treated with Atripla. In contrast, levels of adenosine, homocysteine, and tyrosine in plasma were higher in these subjects than in subjects receiving Complera. Finally, significantly lower levels of aspartate and succinic acid were also observed in the urine of the individuals on Atripla relative to those treated with Complera.
Fig 7.
Metabolic profiles of plasma and urine collected from HIV-infected individuals receiving either Complera (RPV-tenofovir-emtricitabine) or Atripla (efavirenz-tenofovir-emtricitabine). The observed relative abundances of each analyte are shown alongside those in plasma and urine collected from noninfected, non-antiretroviral-treated individuals. The data for the 10 endogenous metabolites, of the 19 monitored, that were determined to be significantly different in either the plasma, urine, or both between the drug-treated groups are shown. Plasma and urine were extracted with acetonitrile in preparation for UHPLC-MS/MS analysis. The relative abundance of each metabolite was detected in SRM mode using either positive or negative ionization. The transitions that were used for each analyte are listed in Tables S1 and S2 in the supplemental material. Data are representative of the mean relative intensities for three subjects per treatment ± standard deviations. Statistically significant differences of Atripla treatment from Complera are indicated as follows: *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
RPV oxidative and glucuronide metabolite formation across species.
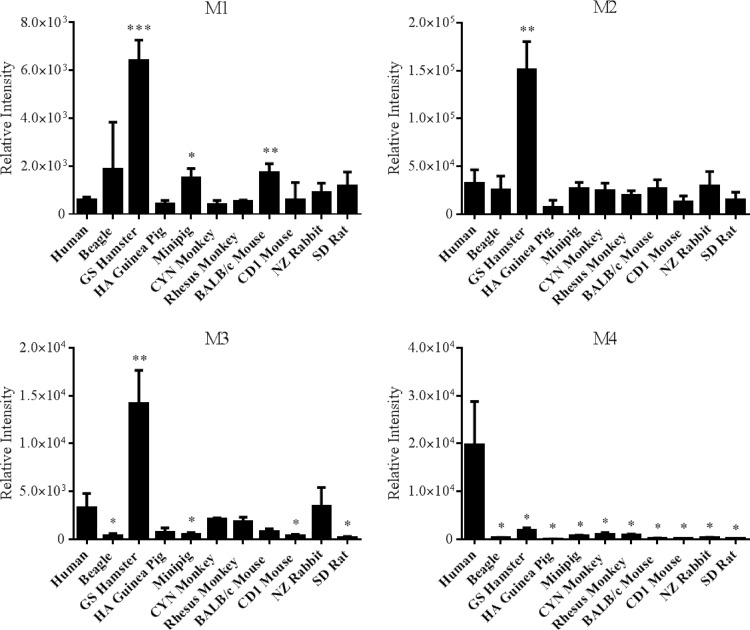
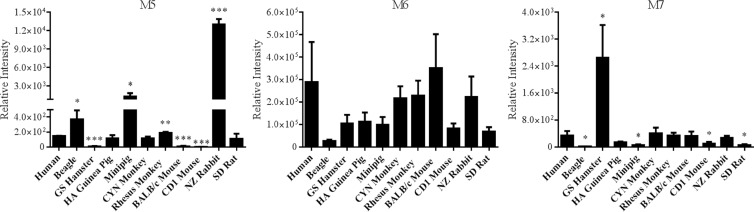
The metabolism of RPV by a range of species used in preclinical drug development was investigated in order to determine whether biotransformation of RPV by any of these animals paralleled what we observed in humans. Using liver microsomes, monohydroxylated metabolites M1 and M2 and dihydroxylated metabolites M3 and M4 were found to be formed at various abundances by each species of the 10 mammals that were investigated in these studies (Fig. 8). Interestingly, the formation of M1, M2, and M3 by golden (Syrian) hamster liver microsomes occurred at levels approximately 11.0-fold, 4.6-fold, and 4.3-fold, respectively, greater than production by human liver microsomes. However, all of these species formed M4 at lower levels than observed using human liver microsomes. RPV glucuronide formation was observed to also be rather conserved across species (Fig. 9). The abundance of glucuronide M5, which we have previously identified in human plasma to be the predominant in vivo-glucuronidated metabolite, was greatest in the beagle, Sinclair minipig, rhesus monkey, and New Zealand rabbit liver microsomal incubations. The lowest levels of M5 production were observed when golden (Syrian) hamster, BALB/c mouse, and CD1 mouse liver microsomes were employed. In addition, the formation of the glucuronidated metabolite M7 was greater using golden (Syrian) hamster liver microsomes than human liver microsomes.
Fig 8.
Comparison of RPV oxidative metabolism across species. Liver microsomes (2 mg/ml) were collected from the following species: human, beagle, GS hamster, HA guinea pig, Sinclair minipig, CYN monkey, rhesus monkey, BALB/c mouse, CD1 mouse, NZ rabbit, and SD rat. The microsomes were incubated with 20 μM RPV for 30 min at 37°C in the presence of an NADPH-regenerating system. RPV metabolites were detected by UHPLC-MS/MS in SRM mode using the following transitions: 383→222 m/z (M1 and M2), 399→183 m/z (M3), and 399→196 m/z (M4). Data are representative of the mean relative intensities for three experiments ± standard deviations. Statistically significant differences from human liver microsome metabolite formation are indicated as follows: *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
Fig 9.
Comparison of RPV phase II metabolism across species. Liver microsomes (2 mg/ml) were collected from the following species: human, beagle, GS hamster, HA guinea pig, Sinclair minipig, CYN monkey, rhesus monkey, BALB/c mouse, CD1 mouse, NZ rabbit, and SD rat. The microsomes were incubated with 20 μM RPV for 60 min at 37°C in the presence of an NADPH-regenerating system and UDPGA. Metabolites were detected by UHPLC-MS/MS in SRM mode using the following transitions: 543→367 m/z (M5), 559→383 m/z (M6), and 575→399 m/z (M7). Data are representative of the mean relative intensities for three experiments ± standard deviations. Statistically significant differences from human liver microsome glucuronide formation are indicated as follows: *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
DISCUSSION
In this study, the routes of RPV metabolism were systematically investigated in order to identify the metabolites of RPV as well as the enzymes that contribute to the formation of these products (Fig. 10). In total, seven RPV metabolites were characterized. Tandem mass spectral analysis revealed that the oxidative metabolites, M1 through M4, resulted from mono- or dioxygenation of the 2,6-dimethylphenyl ring itself or either of the two methyl groups located on this phenyl ring. Even though we did indeed observe distinct fragments for M1 and M2, there were a number of similarities in the fragmentation spectra of these metabolites that made it difficult to definitively assign oxygen insertion onto the methyl group or the 2,6-dimethylphenyl ring for either of these metabolites; however, due to the fact that the methyl group of the 2,6-dimethylphenyl ring may be more readily accessible for oxygenation than for direct oxygen insertion on one of the carbon atoms within the phenyl ring, we propose that it is reasonable for M2 to result from methylhydroxylation of RPV, since it was observed at an apparently greater abundance than M1. Glucuronide M5 resulted from direct N-linked glucuronidation of RPV, and glucuronides M6 and M7 were proposed to be produced via O-linked glucuronidation of mono- and dihydroxymethyl metabolites M2 and M4, respectively. Fecal excretion has been previously demonstrated to be the primary route of RPV clearance (20). Although we did not analyze fecal samples in the present study, using plasma and urine from individuals receiving a RPV-containing regimen, we were able to demonstrate that all of the metabolites that were identified in this study in vitro were indeed formed in vivo. Complementary liver microsomal metabolism studies were performed in order to examine the conservation of RPV biotransformation across species. While the assay developed here to monitor metabolite formation was not quantitative, it facilitated the identification of RPV metabolites and perhaps lays the foundation for the development of a quantitative assay for the measurement of RPV metabolites. Synthetic standards are required in order to definitively determine the levels of these molecules, since differences in ionization could contribute to the observed variability in relative abundances. Therefore, the results presented here can be used to guide the synthesis of RPV metabolite standards for this purpose. Interestingly, each of the species investigated in these studies was able to form all of the metabolites of RPV that were observed in humans, with golden (Syrian) hamster liver microsomes being the only to produce several metabolites of RPV to greater abundance than human liver microsomes. Further, no additional metabolites beyond those that were identified through the human liver microsomal metabolism assays were produced in the incubations performed using liver microsomes isolated from the nonhuman animals. These data suggest that several of these animals may have utility as preclinical species for use in studies to gain an increased understanding of RPV exposure in vivo.
Fig 10.
Schematic summarizing the phase I and phase II metabolism of RPV. Monohydroxylated metabolites, M1 and M2, and dihydroxylated metabolites, M3 and M4, result from oxygenation of RPV by CYP3A4 and CYP3A5. M5 results from direct glucuronic acid conjugation to RPV by UGT1A4. M6 results from glucuronidation of monooxygenated metabolite M1 by UGT1A1, and M7 results from glucuronidation of dioxygenated metabolite M4; however, the UGT responsible for this conjugation was unable to be determined from our studies.
The present study is the first to report primary data detailing the routes of RPV metabolism. As demonstrated through the use of cDNA-expressed CYP isozymes, CYP1A2, -3A4, and -3A5 were able to catalyze the oxygenation of RPV to products M1 through M4. Subsequent chemical inhibition experiments revealed that CYP3A4 and CYP3A5 were predominantly responsible for mono- and dioxygenation of RPV, as the most significant reduction in metabolite formation was observed with the dual CYP3A4/5 inhibitor ketoconazole. Treatments with furafylline, a mechanism-based inactivator of CYP1A2 (21), did not block metabolite formation. This could be the result of compensation of the loss of CYP1A2 activity by another CYP isozyme, for instance, CYP3A4 and/or CYP3A5, resulting in a lack of a significant decrease in RPV metabolism following chemical inhibition of CYP1A2. We observed statistically significant inhibition of M1 formation following incubation of liver microsomes with chemical inhibitors of CYP2B6 (PPP), CYP2C9 (sulfaphenazole), and CYP2C19 [(+)-N-3-benzylnirvanol]; however, RPV was not metabolized to M1 by cDNA-expressed CYP2B6, CYP2C9, or CYP2C19. This variability in the catalytic activities of cDNA-expressed isozymes compared to human liver microsomes could be the result of differences in the protein concentrations of each CYP in these two systems in addition to the fact that in human liver microsomes RPV was being concurrently metabolized to all of the oxygenated products. Therefore, the overall kinetics of formation differ in these two assays. Still, the studies using both cDNA-expressed isozymes and human liver microsomes were congruent and demonstrated a primary role for CYP3A4 and CYP3A5 in RPV oxidative metabolism.
A recent report demonstrated that RPV, similar to the nonnucleoside reverse transcriptase inhibitors etravirine and efavirenz, was able to activate the pregnane X receptor in primary human hepatocytes (17). The pregnane X receptor is a key regulator of drug-metabolizing enzymes, including CYP3A and UGT1A isozymes (22–24). In our study, although we were able to recapitulate the increase in CYP3A4 mRNA expression following incubation of primary human hepatocytes with RPV, we did not observe an increase in the abundance of RPV metabolites via UHPLC-MS/MS analysis of the hepatocyte medium. Oxidative metabolism did not appear to be shunted to glucuronic acid conjugation, as the glucuronides that were detectable in the hepatocyte medium, namely, metabolites M5 and M6, also did not exhibit an increase in abundance over the 24 h of incubation. Although we have previously reported that autoinduction of etravirine metabolism is readily detectable following 24 h of incubation with primary human hepatocytes (17), longer treatment times may be required to observe this effect using RPV. For instance, CYP3A4 protein may not have yet been elevated over the time points that we measured. Further, the metabolites may not be stable in culture medium, thereby resulting in our inability to detect increases in metabolite levels over time. This would need to be tested using synthetic standards for each of these molecules.
Assays performed using cDNA-expressed UGT isozymes demonstrated that UGT1A1 and UGT1A4 were able to catalyze the conjugation of glucuronic acid to moonoxygenated metabolite M2 and RPV, respectively. These results correlated well with our proposed glucuronide metabolite structures, as UGT1A1 is known to facilitate alkyl- and aryl-O-glucuronidation, while UGT1A4 has been demonstrated to participate in the N-glucuronidation of tertiary amines (25). UGT1A1 and UGT1A4 are members of the UGT1A gene superfamily and are most abundantly expressed in the human liver. Of note, glucuronide M7 was not formed by any of the cDNA-expressed UGT isozymes tested under our experimental conditions, nor was this product detectable in hepatocyte medium following up to 24 h of incubation with RPV; however, M7 was detectable in the urine of all three subjects receiving Complera that were analyzed in this study, in addition to being present in the plasma of one of the subjects. Therefore, this metabolite that was initially identified through the use of human liver microsomes appears to indeed be formed in vivo. M7 may be produced by a UGT isozyme beyond those tested, and the lack of formation in the primary human hepatocytes may be due to interindividual variability (the human liver microsomes are isolated from a pool of 50 donors) and/or lower levels of expression of drug-metabolizing enzyme as a result of the isolation or culturing of the hepatocytes. Since this metabolite was observed to be the more minor of the glucuronides detected in vivo, its formation may be more sensitive to the aforementioned variables.
We sought to probe for potential differential physiological effects of an RPV-containing therapy versus an efavirenz-containing therapy through analysis of the plasma and urine endogenous metabolomes. Since analysis at the level of the metabolome can generate large volumes of data, we opted to prioritize the pathways that we investigated. To this end, we developed a targeted UHPLC-MS/MS-based method to detect 32 intracellular metabolites that have been demonstrated to play a role in maintaining cellular homeostasis. Further, while global metabolomics provides relative quantitation of intracellular small molecules, a targeted approach allows for markedly improved detection of less abundant chemical species. Plasma tyrosine levels were observed to be higher in Atripla-treated subjects. In a reaction that predominantly occurs in the liver, the essential amino acid phenylalanine is converted to tyrosine via phenylalanine hydroxylase (26). Plasma phenylalanine levels, however, were comparable between subject groups, suggesting a potential increase in phenylalanine hydroxylase activity with an efavirenz-containing treatment compared to a RPV-based regimen, thus resulting in the increased synthesis of tyrosine by-product. Plasma homocysteine and adenosine levels were also greater with Atripla drug therapy. The S-adenosylmethionine-dependent transmethylation pathway, which is extensively active in the liver, is responsible for the synthesis of homocysteine and adenosine through the hydrolysis of S-adenosylhomocysteine via S-adenosylhomocysteine hydrolase (27). Interestingly, plasma methionine levels were comparable between treatments. As part of the transmethylation cycle, the conversion of homocysteine to methionine is regulated by methionine synthase (27); therefore, we would anticipate observing greater methionine levels in the plasma samples from Atripla-treated individuals. This, however, did not occur, suggesting possible impairment of methionine synthase activity with Atripla treatment. Moreover, it is also plausible that plasma methionine levels remain unchanged as a result of immediate conversion to S-adenosylmethionine. The small molecules S-adenosylhomocysteine and S-adenosylmethionine were initially included in this metabolomics screen, yet neither was consistently detectable in these biological samples.
Aspartate and arginine are two examples of metabolites that were observed to be in lower abundance in the plasma collected from the Atripla drug treatment group relative to those receiving Complera. Interestingly, lower plasma arginine levels have been observed to parallel decreasing HIV RNA levels with an established antiretroviral drug regimen (28). Moreover, arginine and aspartate are key components of the urea cycle, in which argininosuccinate synthase catalyzes the conversion of aspartate to argininosuccinate, which is further hydrolyzed into arginine and fumarate via argininosuccinate lyase, ultimately producing urea (29). Lower arginine and aspartate levels would suggest an increase in the activity of both argininosuccinate synthase and argininosuccinate lyase enzymes. Separate from their role in the urea cycle, lower plasma aspartate levels can result from increased aspartate aminotransferase activity, which catalyzes the conversion of the amino acid and α-ketoglutarate to oxaloacetate and glutamate, and have served as a biomarker for diagnosing fibrosis and cirrhosis of the liver in individuals coinfected with HIV and hepatitis C virus (30). This proposed mechanism is challenging to fully assess, since α-ketoglutarate and oxaloacetate were not detectable in these samples by UHPLC-MS/MS analysis and plasma glutamic acid levels were also observed to be lower in those receiving Atripla. Potential confounding factors for our analyses include the fact that the subjects involved in this study could have had comorbidities or been concomitantly taking medications that might also have had an impact on their plasma and urine metabolomes. In addition, due to the small sample size, we cannot rule out potential effects of the mean age difference between the subjects receiving Atripla or Complera. However, the levels of the endogenous small molecules that we measured were consistent among individuals within each of the drug treatment groups, so much so that the standard deviations were less than 5%, indicating that there may indeed be distinct endogenous small-molecule signatures that can be attributed to RPV- and efavirenz-containing regimens. Further, these data demonstrate for the first time that endogenous metabolites can be readily detected in plasma and urine of individuals receiving these drugs, and we provide an assay that can be applied broadly in measuring these molecules in biological matrices. Overall, these endogenous metabolomic studies need to be further extended to a larger population of individuals for both Complera and Atripla subject groups; however, the present findings suggest possible biochemical pathways that can be tested for modulation by Complera and Atripla.
In summary, this study has defined the in vitro routes of phase I (primarily CYP3A4/5) and phase II (glucuronic acid conjugation) RPV metabolism. Additionally, RPV metabolism was observed to be conserved across a range of mammals, and these data should inform the selection of preclinical animal models for subsequent investigation of RPV pharmacology and/or toxicology in vivo. Taken together, these data provide novel insight into the biotransformation of RPV.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the HIV Prevention Trials Network (NIH U01 AI068613) and NIH R01 GM103853 awarded to N.N.B.
We kindly acknowledge Craig Hendrix and the Drug Development Unit of the Johns Hopkins University School of Medicine Division of Clinical Pharmacology for the recruitment of participants for this study.
Footnotes
Published ahead of print 5 August 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01401-13.
REFERENCES
- 1.Wainberg MA. 2013. Combination therapies, effectiveness, and adherence in patients with HIV infection: clinical utility of a single tablet of emtricitabine, rilpivirine, and tenofovir. HIV AIDS (Auckl.) 5:41–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andries K, Azijn H, Thielemans T, Ludovici D, Kukla M, Heeres J, Janssen P, De Corte B, Vingerhoets J, Pauwels R, de Bethune MP. 2004. TMC125, a novel next-generation nonnucleoside reverse transcriptase inhibitor active against nonnucleoside reverse transcriptase inhibitor-resistant human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 48:4680–4686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Janssen PA, Lewi PJ, Arnold E, Daeyaert F, de Jonge M, Heeres J, Koymans L, Vinkers M, Guillemont J, Pasquier E, Kukla M, Ludovici D, Andries K, de Bethune MP, Pauwels R, Das K, Clark AD, Jr, Frenkel YV, Hughes SH, Medaer B, De Knaep F, Bohets H, De Clerck F, Lampo A, Williams P, Stoffels P. 2005. In search of a novel anti-HIV drug: multidisciplinary coordination in the discovery of 4-[[4-[[4-[(1E)-2-cyanoethenyl]-2,6-dimethylphenyl]amino]-2-pyrimidinyl]amino]benzonitrile (R278474, rilpivirine). J. Med. Chem. 48:1901–1909 [DOI] [PubMed] [Google Scholar]
- 4.Abraham BK, Gulick R. 2012. Next-generation oral preexposure prophylaxis: beyond tenofovir. Curr. Opin. HIV AIDS 7:600–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baert L, van 't Klooster G, Dries W, Francois M, Wouters A, Basstanie E, Iterbeke K, Stappers F, Stevens P, Schueller L, Van Remoortere P, Kraus G, Wigerinck P, Rosier J. 2009. Development of a long-acting injectable formulation with nanoparticles of rilpivirine (TMC278) for HIV treatment. Eur. J. Pharm. Biopharm. 72:502–508 [DOI] [PubMed] [Google Scholar]
- 6.Cohen CJ, Andrade-Villanueva J, Clotet B, Fourie J, Johnson MA, Ruxrungtham K, Wu H, Zorrilla C, Crauwels H, Rimsky LT, Vanveggel S, Boven K. 2011. Rilpivirine versus efavirenz with two background nucleoside or nucleotide reverse transcriptase inhibitors in treatment-naive adults infected with HIV-1 (THRIVE): a phase 3, randomised, non-inferiority trial. Lancet 378:229–237 [DOI] [PubMed] [Google Scholar]
- 7.Molina JM, Cahn P, Grinsztejn B, Lazzarin A, Mills A, Saag M, Supparatpinyo K, Walmsley S, Crauwels H, Rimsky LT, Vanveggel S, Boven K. 2011. Rilpivirine versus efavirenz with tenofovir and emtricitabine in treatment-naive adults infected with HIV-1 (ECHO): a phase 3 randomised double-blind active-controlled trial. Lancet 378:238–246 [DOI] [PubMed] [Google Scholar]
- 8.Bumpus NN. 2011. Efavirenz and 8-hydroxyefavirenz induce cell death via a JNK- and BimEL-dependent mechanism in primary human hepatocytes. Toxicol. Appl. Pharmacol. 257:227–234 [DOI] [PubMed] [Google Scholar]
- 9.Tovar-y-Romo LB, Bumpus NN, Pomerantz D, Avery LB, Sacktor N, McArthur JC, Haughey NJ. 2012. Dendritic spine injury induced by the 8-hydroxy metabolite of efavirenz. J. Pharmacol. Exp. Ther. 343:696–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.TibotecPharmaceuticals 2011. Edurant (rilpivirine) tablets full prescribing information. Tibotec Pharmaceuticals, County Cork, Ireland [Google Scholar]
- 11.Gilead Sciences Inc 2011. Complera highlights of prescribing information. Gilead Sciences Inc., Foster City, CA [Google Scholar]
- 12.Suzuki H, Kneller MB, Haining RL, Trager WF, Rettie AE. 2002. (+)-N-3-Benzyl-nirvanol and (−)-N-3-benzyl-phenobarbital: new potent and selective in vitro inhibitors of CYP2C19. Drug Metab. Dispos. 30:235–239 [DOI] [PubMed] [Google Scholar]
- 13.Walsky RL, Obach RS. 2007. A comparison of 2-phenyl-2-(1-piperidinyl)propane (ppp), 1,1′,1″-phosphinothioylidynetrisaziridine (thioTEPA), clopidogrel, and ticlopidine as selective inactivators of human cytochrome P450 2B6. Drug Metab. Dispos. 35:2053–2059 [DOI] [PubMed] [Google Scholar]
- 14.Ward BA, Gorski JC, Jones DR, Hall SD, Flockhart DA, Desta Z. 2003. The cytochrome P450 2B6 (CYP2B6) is the main catalyst of efavirenz primary and secondary metabolism: implication for HIV/AIDS therapy and utility of efavirenz as a substrate marker of CYP2B6 catalytic activity. J. Pharmacol. Exp. Ther. 306:287–300 [DOI] [PubMed] [Google Scholar]
- 15.Ingelman-Sundberg M, Sim SC. 2010. Pharmacogenetic biomarkers as tools for improved drug therapy; emphasis on the cytochrome P450 system. Biochem. Biophys. Res. Commun. 396:90–94 [DOI] [PubMed] [Google Scholar]
- 16.Sharma D, Lau AJ, Sherman MA, Chang TK. 2013. Agonism of human pregnane X receptor by rilpivirine and etravirine: comparison with first generation non-nucleoside reverse transcriptase inhibitors. Biochem. Pharmacol. 85:1700–1711 [DOI] [PubMed] [Google Scholar]
- 17.Yanakakis LJ, Bumpus NN. 2012. Biotransformation of the antiretroviral drug etravirine: metabolite identification, reaction phenotyping, and characterization of autoinduction of cytochrome P450-dependent metabolism. Drug Metab. Dispos. 40:803–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weiss J, Haefeli WE. 2013. Potential of the novel antiretroviral drug rilpivirine to modulate the expression and function of drug transporters and drug-metabolising enzymes in vitro. Int. J. Antimicrob. Agents 41:484–487 [DOI] [PubMed] [Google Scholar]
- 19.Ohno S, Nakajin S. 2009. Determination of mRNA expression of human UDP-glucuronosyltransferases and application for localization in various human tissues by real-time reverse transcriptase-polymerase chain reaction. Drug Metab. Dispos. 37:32–40 [DOI] [PubMed] [Google Scholar]
- 20.Lachau-Durand S, Mamidi RNVS, Cuyckens F, Michlova V, Mannens G, Raoof A. 2009. Absorption, metabolism and excretion of TMC278, an NNRTI, after a single oral dose of 150mg in healthy male volunteers. HIV Med. 10(Suppl 2):93 [Google Scholar]
- 21.Fairman DA, Collins C, Chapple S. 2007. Progress curve analysis of CYP1A2 inhibition: a more informative approach to the assessment of mechanism-based inactivation? Drug Metab. Dispos. 35:2159–2165 [DOI] [PubMed] [Google Scholar]
- 22.Willson TM, Kliewer SA. 2002. PXR, CAR and drug metabolism. Nat. Rev. Drug Discov. 1:259–266 [DOI] [PubMed] [Google Scholar]
- 23.Moore LB, Maglich JM, McKee DD, Wisely B, Willson TM, Kliewer SA, Lambert MH, Moore JT. 2002. Pregnane X receptor (PXR), constitutive androstane receptor (CAR), and benzoate X receptor (BXR) define three pharmacologically distinct classes of nuclear receptors. Mol. Endocrinol. 16:977–986 [DOI] [PubMed] [Google Scholar]
- 24.Gardner-Stephen D, Heydel JM, Goyal A, Lu Y, Xie W, Lindblom T, Mackenzie P, Radominska-Pandya A. 2004. Human PXR variants and their differential effects on the regulation of human UDP-glucuronosyltransferase gene expression. Drug Metab. Dispos. 32:340–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tukey RH, Strassburg CP. 2000. Human UDP-glucuronosyltransferases: metabolism, expression, and disease. Annu. Rev. Pharmacol. Toxicol. 40:581–616 [DOI] [PubMed] [Google Scholar]
- 26.Hufton SE, Jennings IG, Cotton RG. 1995. Structure and function of the aromatic amino acid hydroxylases. Biochem. J. 311(Part 2):353–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Williams KT, Schalinske KL. 2010. Homocysteine metabolism and its relation to health and disease. Biofactors 36:19–24 [DOI] [PubMed] [Google Scholar]
- 28.Kurz K, Teerlink T, Sarcletti M, Weiss G, Zangerle R, Fuchs D. 2012. Asymmetric dimethylarginine concentrations decrease in patients with HIV infection under antiretroviral therapy. Antivir. Ther. 17:1021–1027 [DOI] [PubMed] [Google Scholar]
- 29.Haines RJ, Pendleton LC, Eichler DC. 2011. Argininosuccinate synthase: at the center of arginine metabolism. Int. J. Biochem. Mol. Biol. 2:8–23 [PMC free article] [PubMed] [Google Scholar]
- 30.Resino S, Sanchez-Conde M, Berenguer J. 2012. Coinfection by human immunodeficiency virus and hepatitis C virus: noninvasive assessment and staging of fibrosis. Curr. Opin. Infect. Dis. 25:564–569 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.