Abstract
Antimicrobial peptides (AMPs) can cause lysis of target bacteria by directly inserting themselves into the lipid bilayer. This killing mechanism confounds the identification of the intracellular targets of AMPs. To circumvent this, we used a shuttle vector containing the inducible expression of a human cathelicidin-related AMP, LL-37, to examine its effect on Escherichia coli TOP10 under aerobic and anaerobic growth conditions. Induction of LL-37 caused growth inhibition and alteration in cell morphology to a filamentous phenotype. Further examination of the E. coli cell division protein FtsZ revealed that LL-37 did not interact with FtsZ. Moreover, intracellular expression of LL-37 results in the enhanced production of reactive oxygen species (ROS), causing lethal membrane depolarization under aerobic conditions. Additionally, the membrane permeability was increased after intracellular expression of LL37 under both aerobic and anaerobic conditions. Transcriptomic analysis revealed that intracellular LL-37 mainly affected the expression of genes related to energy production and carbohydrate metabolism. More specifically, genes related to oxidative phosphorylation under both aerobic and anaerobic growth conditions were affected. Collectively, our current study demonstrates that intracellular expression of LL-37 in E. coli can inhibit growth under aerobic and anaerobic conditions. While we confirmed that the generation of ROS is a bactericidal mechanism for LL-37 under aerobic growth conditions, we also found that the intracellular accumulation of cationic LL-37 influences the redox and ion status of the cells under both growth conditions. These data suggest that there is a new AMP-mediated bacterial killing mechanism that targets energy metabolism.
INTRODUCTION
The response of Escherichia coli to antimicrobial peptides (AMPs) has been studied extensively, but previous studies have focused mainly on incubation of the test strains with extracellular AMPs under aerobic growth conditions (1). It is generally believed that the mechanism by which AMPs kill bacteria involves three steps. The first is attraction to the bacterial surface through electrostatic binding between the cationic peptides and the anionic structure on the surface of the bacterial cell. This happens immediately when the AMPs encounter the surfaces of the bacterial cells. The second step involves the translocation of AMPs by passing through the bacterial capsular polysaccharides before they reach the cytoplasmic membrane. During this process, AMPs interact directly with the outer membrane. This detailed process includes initial aggregation at the membrane surface, followed by entry into the lipid bilayer and eventual pore formation in the outer membrane. Membrane disruption is believed to be the primary mode of action of AMPs, because irreversible membrane damage can cause the expulsion of cell contents, resulting in cell death (2, 3). However, a certain number of AMPs can pass through the cell membrane without rupturing the cells (4). Once the AMPs pass through the outer membrane into the cytoplasm, they can attack intracellular nonmembrane targets (5). Various intracellular targets have been identified, including those involved in the synthesis of DNA, RNA, and proteins (6–10). However, since the various killing mechanisms that have been identified were obtained using incubation methods in the presence of oxygen and the number of AMPs that cross the cell membrane cannot be measured accurately, the internal killing mechanism of AMPs is not fully understood. Therefore, development of a genetic system that intracellularly expresses AMPs may help us to identify and understand the roles of key intracellular targets.
Host defense peptides (HDPs) are important effector molecules. They are the innate host defense chemicals on the mucosal barrier that provide effective protection against a wide range of invading microorganisms. HDPs are cationic peptides. They include cecropins, magainins, bactenecins, protegrins, defensins, and cathelicidins (11). LL-37, which contains 37 amino acids derived from the hydrolysis of an 18-kDa propeptide (human CAP18 [hCAP18]), is an HDP and the only human member of the cathelicidin family (12). LL-37 has been shown to be an integral part of the human innate immune defense against invasive Gram-negative and Gram-positive bacteria (13). The typical α-helical secondary structure present in physiologically relevant solutions, coupled with the positive charges, enables each peptide to exert various physiological effects. So far, LL-37 has been shown to neutralize the lipopolysaccharides (LPS) on Gram-negative bacteria, thereby inhibiting LPS-mediated cytokine production (14). However, enhancement of the membrane permeability through carpeting or toroidal action has been reported to be the major antibacterial mode of action (15). Time-resolved fluorescence microscopy has shown that AMPs also act by binding to the peptidoglycan in the bacterial septal region penetrating into the bacterial cytoplasm (4).
It has previously been established that the free radicals produced during the exposure of bacterial cells to AMPs show strong antibacterial activity. Joly et al. found that human defensins, such as HBD-2 and HBD-3, are less lethal to strict anaerobes than to facultative bacteria (16). The presence of dissolved oxygen in the growth medium can affect the efficacy of human defensins HBD-3 and HBD-2 against E. coli and Bacteroides fragilis (17). Considering the presence of an oxygen gradient in the natural niche of the intestinal tract ranging from a higher oxygen concentration near the surface of the mucosa to strict anaerobic conditions in the center of the lumen, the mode of action of AMPs should be investigated under both aerobic and anaerobic growth conditions.
In the current study, we developed a genetic system for inducing intracellular expression of LL-37 to evaluate its effect on the growth of E. coli under aerobic and anaerobic growth conditions. Although the intracellular expression of LL-37 is an artificial phenomenon, AMPs have been reported to have the capacity to penetrate the bacterial cell membrane and enter the cytoplasm (4). We propose that the intracellular expression of AMPs is an experimental model that enables us to identify potential intracellular targets.
MATERIALS AND METHODS
Bacterial strains, construction of plasmids, and growth conditions.
E. coli strain TOP10 was purchased from TransGen Biotech, Inc. The plasmid pHERD30T, containing the PBAD promoter, was used for gene expression, as previously reported (18). The nucleotide sequence encoding LL-37 was synthesized and used as a template for amplification by PCR with the following primers: LL-37F (5′-CATGCCATGGAAATGCTGCTGGGCGATTTTTTTCGCAAAAGCA-3′; the NcoI site is underlined) and LL-37R (5′-CCGGAATTCCTGCAGTCAGCTTTC-3′; the EcoRI site is underlined). After digestion of the PCR products with NcoI and EcoRI, the corresponding restriction fragment was cloned into pHERD30T at the NcoI/EcoRI sites, giving rise to pHERD30T-LL-37. The completed construct was transformed into E. coli TOP10 cells by a standard electrical transformation procedure. The construct was verified by direct DNA sequencing.
We used E. coli strain EC258, a Δara derivative of MC4100 containing green fluorescent protein (GFP)-labeled FtsZ, to assess the interactions between intracellularly expressed LL-37 and the cell division proteins. Strain EC258 was kindly provided by David S. Weiss at the University of Iowa. pHERD30T-LL-37 and the empty pHERD30T (vector control) were transformed into E. coli strain EC258 via electroporation, and the cell morphology was detected using phase-contrast microscopy following intracellular expression of LL-37.
Analyses of antimicrobial activity.
E. coli strain TOP10 containing pHERD30T-LL-37 and the empty plasmid pHERD30T (vector control) were induced using 0.2% arabinose and grown aerobically in Luria-Bertani (LB) broth (1% Bacto tryptone, 0.5% yeast extract, and 0.5% NaCl) at 37°C for the indicated periods. Gentamicin (13 μg/ml) was added to the culture media as appropriate. Overnight cultures were diluted to a starting optical density at 600 nm (OD600) of 0.1 and inoculated at 1-in-10 (vol/vol) proportions into the LB medium in 15-milliliter conical tubes and incubated at 37°C with vigorous shaking. A 0.5-ml aliquot was removed every hour to monitor the growth curve by measuring the OD600. Empty control plasmids were run in parallel with all growth experiments. For anaerobic growth experiments, overnight cultures were inoculated into the anaerobic LB medium containing 0.2% arabinose and incubated without shaking in an AW 500TG anaerobic chamber (Electrotek, England) for 48 h. Five-hundred-microliter aliquots were removed at 2-h intervals to monitor bacterial growth. For determinations of the viable-cell counts (CFU), 100-μl aliquots were removed after induction of expression of LL-37 for 16 h under aerobic growth conditions and spotted by 10-fold serial dilution in four replicates onto LB agar plates. The plates were incubated for 18 h at 37°C, and the bacterial populations were enumerated. For the CFU counts under anaerobic growth conditions, 100-μl-aliquot cultures were collected after 24 h of LL-37 expression. Viable bacteria were counted using the previously described 10-fold dilution method.
Microscopy and measurement of cell length.
E. coli strain TOP10 containing either pHERD30T-LL-37 or pHERD30T was grown in LB broth supplemented with 0.2% arabinose and 13 μg/ml gentamicin under aerobic conditions for 13 h and under anaerobic conditions for 24 h. The cells were then harvested and fixed on a glass slide. The cells were incubated in the dark at room temperature with a mixture of 10 μl of 20 μg/ml DAPI (4′-6-diamidino-2-phenylindole; Sigma, USA) in 1 ml phosphate-buffered saline (PBS) buffer for 5 to 10 min, washed 2 or 3 times with PBS buffer, and examined using laser confocal microscopy (LSM 510; Zeiss, Germany) at a magnification of ×630. To differentiate the integrities of the cell membrane and the nucleoid, the cells were stained with N-(3-triethylammonium-propyl)-4-(6-(4-(diethylamino)-phenyl) (hexatrienyl) pyridinium dibromide (FM 4-64) (Sigma, USA; 10 mg/ml) for 10 min and examined using confocal microscopy (LSM 510; Zeiss, Germany). FM 4-64 stains the lipid membranes with red fluorescence (excitation and emission, 515 and 640 nm), and DAPI stains the nucleoid with blue fluorescence (excitation and emission, 350 and 470 nm).
To determine the effects of the arabinose concentration on the morphological changes in E. coli cells, cells were collected at 0, 3, 13, and 24 h after the addition of 0.2% arabinose. The cells were then stained with DAPI and visualized using an epifluorescence microscope (CX-51; Olympus, Japan) with a DM 400 filter. All images were acquired using Image-Pro Plus software and measured using Image-Pro Plus 6.0 software (Media Cybernetics, Inc., USA) to determine changes in the lengths of the cells as a function of time. The average lengths of 60 to 70 cells were analyzed to determine the bacterial length.
To determine whether extracellular LL-37 can permeate E. coli cells and affect cell morphology, E. coli TOP10 cells and EC258 were mixed with 15 μM LL-37 in 200 μl M9 medium and incubated at 37°C for 24 h, as previously described by Rosenberger et al. (19). Cell morphology was examined every 3 h for 24 h using an epifluorescence microscope.
Determination of membrane permeability and integrity.
The effect of intracellular expression of LL-37 on membrane depolarization was measured using the voltage-sensitive dye DiBAC4 (3) and flow cytometric analysis (20). In brief, samples consisting of at least 100,000 cells were collected at 2-h intervals from LB medium containing 13 μg/ml gentamicin and resuspended in 1 ml of 1× PBS containing 10 μg/ml DiBAC4 (3). Samples were then incubated in the dark for 15 min and analyzed using the Guava EasyCyte flow cytometer (Guava Technologies, Millipore, Billerica, MA, USA). Cell integrity was determined using propidium iodide (PI) combined with Syto9 according to the manufacturer's instructions (Invitrogen, Carlsbad, CA, USA).
Detection of free radicals.
Samples were collected at 2-h intervals from LB medium with 13 μg/ml gentamicin and resuspended in 500 μl of 1× PBS, pH 7.2, containing 10 μM HPF [3′-(phydroxyphenyl) fluorescein] (Invitrogen, Carlsbad, CA, USA). Samples were incubated in the dark for 15 min, and the cell pellet was centrifuged. The cells were resuspended in 1 ml sterile, filtered 1× PBS for flow cytometric analysis using a Guava EasyCyte.
To measure the concentration of total reactive oxygen species (ROS) after intracellular expression of LL-37 in E. coli, samples were grown in 100 ml LB medium at 37°C. The bacterial cells were collected at 2-h intervals from LB medium and resuspended in 500 μl of 1× PBS after centrifugation. The samples were incubated with the cell-permeable fluorogenic probe 2′,7′-dichlorodihydrofluorescein diacetate (DCFH-DA) at a final concentration of 50 mM for 20 min. Intracellular ROS levels were analyzed using Guava EasyCyte flow cytometry.
RNA isolation, cDNA library construction, and sequencing.
Total RNA was isolated from E. coli TOP10 cells containing pHERD30T-LL-37 with or without intracellular expression of LL-37 using TRIzol (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. DNA was removed from the RNA extracts by incubation with RNase-free DNase I (New England BioLabs, Beverly, MA, USA) for 30 min at 37°C. The quality of total RNA was assessed by using a 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA) and checked using agarose gel electrophoresis. rRNAs were then removed from the total mRNA in accordance with the instructions included with the Ribo-Zero rRNA Removal Kit (Epicentre, Madison, WI, USA). In brief, after addition of the fragmentation buffer, the mRNA was broken down into short fragments with an average length of 200 bp. The first-strand cDNA was then synthesized using a random-hexamer primer with the mRNA fragments as templates. DNA polymerase I buffer, deoxynucleoside triphosphates (dNTPs), RNase H, and DNA polymerase I were added to synthesize the second strand. Double-stranded cDNA was purified using a QiaQuick PCR Extraction Kit (Qiagen, Hilden, Germany) and washed with EB buffer (10 mM Tris-Cl, pH 8.5) for end repair and the addition of the single nucleotide A (adenine). Finally, the sequencing adaptors were attached to the fragments. After gel purification and PCR amplification, the cDNA products were analyzed with an Illumina HiSeq 2000 (Illumina, USA). RNA sequencing was performed by staff at Zhejiang Tiank (Hangzhou, China).
Bioinformatics analyses.
The reference genome sequence for E. coli K-12 was downloaded from GenBank (GI 49175990). The clean reads were then aligned with the E. coli DH10B reference genome (GI 170079663) using CLC Genomics Workbench (CLC bio) with default parameters. Gene coverage was equal to the ratio of the number of bases in the gene in question that were covered by unique reads to the total number of bases in that gene.
The RPKM (reads per kilobase per million mapped reads) method was used to measure gene expression levels (21). The changes in gene expression (log2 ratio) were estimated according to the normalized gene expression value in each sample. In this study, we used an absolute value of the log2 ratio of >1 as the threshold to determine the differences in gene expression. Functional annotation and classification of the commonly regulated genes under both aerobic and anaerobic growth conditions were conducted using the following databases: the NR protein database (NCBI), the gene ontology (GO) database, the Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.genome.jp/kegg/expression/), and the Clusters of Orthologous Groups (COG) database (http://www.ncbi.nlm.nih.gov/COG/). All the unique sequences were classified into different functional groups based on their COG codes, and pathways were carried out by KEGG.
Microarray data accession number.
All mRNA sequences from this study were deposited in GEO (http://www.ncbi.nlm.nih.gov/geo/) with accession number GSE43408.
RESULTS
Effect of intracellular expression of LL-37 on the growth of E. coli.
In order to determine the intracellular effect of cathelicidin peptide on the growth of E. coli, we first synthesized the LL-37 gene and inserted it into pHERD30T containing a PBAD promoter (pHERD30T-LL-37) (18). pHERD30T-LL-37 was then transformed into E. coli strain TOP10 cells via electrical transformation. Control strains were constructed through electrical transformation of the empty vector pHERD30T. The effect of intracellular expression of LL-37 on the growth of E. coli was tested after the addition of 0.2% arabinose to the LB broth at the beginning of the growth experiment. As shown in Fig. 1A, the growth of E. coli was inhibited after the addition of arabinose to the growth medium, but growth was not affected in the control strain containing the empty vector. To confirm the inhibitory effect of intracellular expression of LL-37 on the growth of E. coli, 0.2% arabinose was added when the E. coli cells reached OD600s of 0.5 and 1.2. In those cases, bacterial growth declined after 4 h of induction, suggesting that the intracellular expression of LL-37 directly affected E. coli growth (see Fig. S1 in the supplemental material). Consistent with these results, intracellular expression of LL-37 also caused a decline in the growth of E. coli under anaerobic growth conditions (Fig. 1B). The inhibitory activity of intracellular expression of LL-37 was further confirmed by plating (CFU and CFU/ml). After the expression of LL-37, bacterial populations decreased from 6.81 log10 CFU/ml to 5.22 log10 CFU/ml in cultures grown under aerobic conditions for 13 h and from 6.42 log10 CFU/ml to 5.48 log10 CFU/ml under anaerobic conditions for 24 h (see Fig. S2 in the supplemental material).
Fig 1.
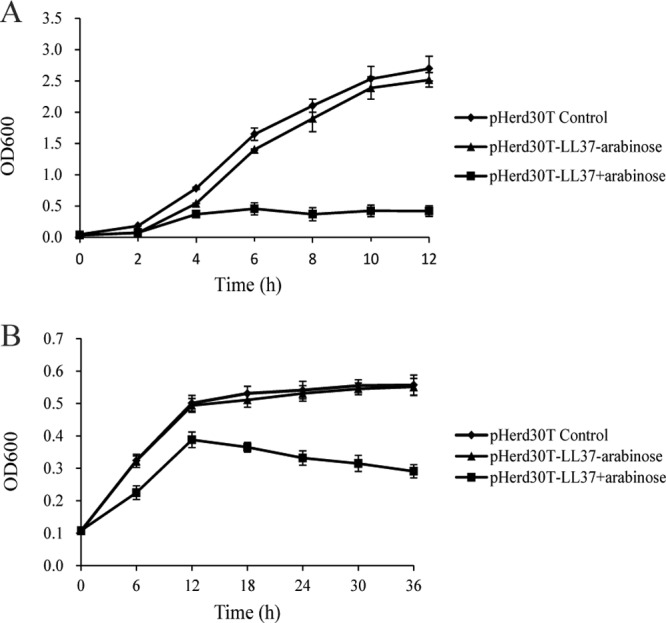
Effects of intracellular expression of LL-37 on the growth of E. coli under aerobic (A) and anaerobic (B) growth conditions. E. coli strain TOP10 harboring pHERD30T-LL37 was grown in LB under aerobic and anaerobic growth conditions. Bacterial growth was monitored by the optical density at 600 nm after induction with 0.2% arabinose. The results shown are means ± standard deviations (SD) of biological triplicates.
Cell morphology after intracellular and extracellular exposure to LL-37.
The cell morphology of E. coli following exposure to intracellular expression of LL-37 was examined by light microscopy. As shown in Fig. 2A, the cell morphology changed from the typical rod-shaped phenotype to a highly elongated filamentous phenotype following intracellular expression of LL-37. No change was observed in those cells containing the empty plasmid (see Fig. S3A in the supplemental material). The cell membrane was observed under confocal microscopy using a lipophilic probe, FM 4-64. A uniform membrane with clear boundaries was observed in the control cells and cells after intracellular expression of LL-37 for 3 h (see Fig. S3B and C in the supplemental material); however, the cell membrane exhibited a patched pattern after intracellular expression of LL-37 for 13 h under aerobic growth conditions (Fig. 2C). A septum was not observed between the cells, and DAPI staining showed segregated nucleoids unevenly distributed inside the cytoplasm (Fig. 2E). These data suggest that the intracellular expression of LL-37 affects the formation of the bacterial septa. The cell membranes maintained their integrity and showed clear boundaries after 24 h of intracellular expression of LL-37 under anaerobic growth conditions (Fig. 2D and F).
Fig 2.
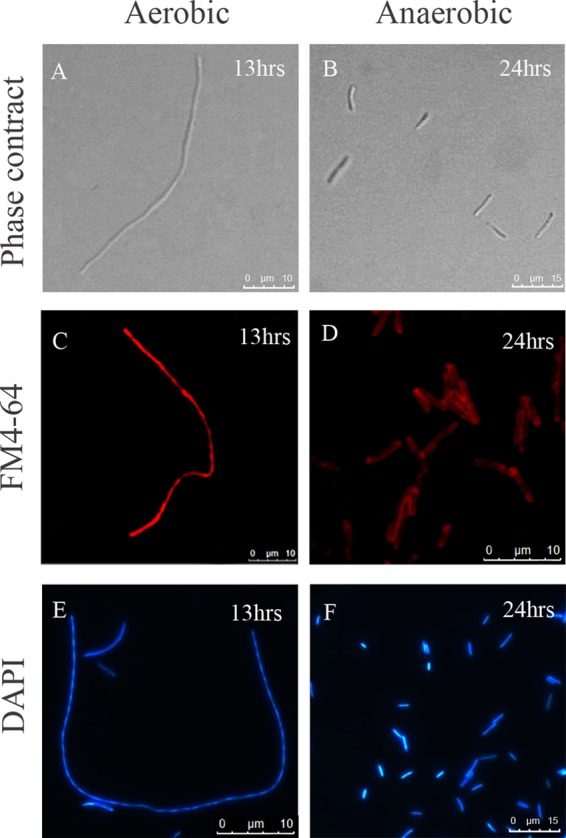
Cell morphology of E. coli strain TOP10 after induction of LL-37 expression under aerobic and anaerobic growth conditions. The cell morphologies of E. coli strains harboring pHERD30T-LL37 were detected by phase-contrast microscopy after induction with 0.2% arabinose under aerobic growth for 13 h (A) and anaerobic growth for 24 h (B). Cell membranes (C and D) were stained with FM 4-64 and DNA (E and F) with DAPI and visualized by epifluorescence microscopy.
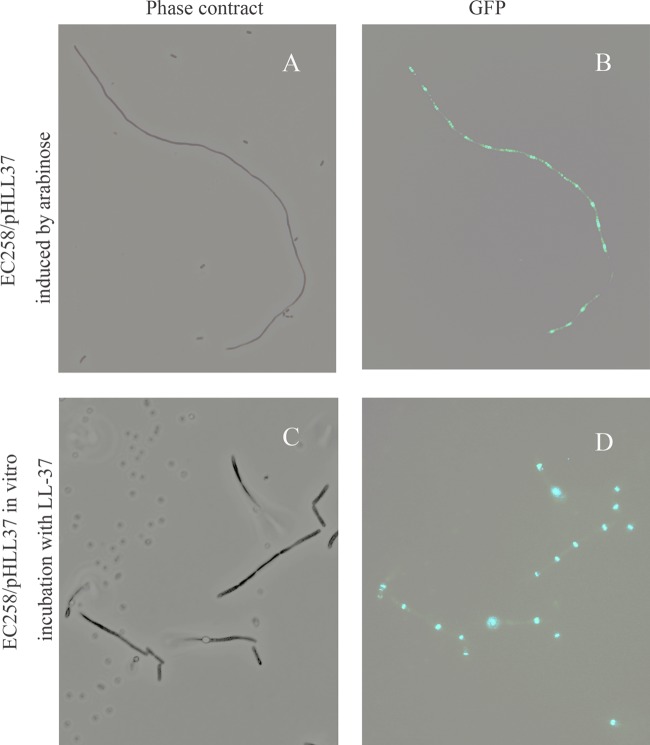
To further determine the impact of intracellular LL-37 on cell division, pHERD30T-LL-37 was transformed into E. coli strain EC528, which contains GFP-labeled FtsZ. After expressing LL-37 by adding 0.2% arabinose, multiple Z rings were clearly observed. These Z rings were regularly located with the filamentous cell, indicating that intracellular LL-37 may not affect cell division directly (Fig. 3A and B). A regular single Z ring was noted in the control cells. We next tested whether the extracellular LL-37 also affected the shape of bacterial cells. After in vitro incubation for 24 h in M9 medium containing a sublethal concentration of LL-37, a filamentous phenotype was observed in both the E. coli TOP10 strain (see Fig. S4 in the supplemental material) and EC258 (Fig. 3C). Interestingly, regular Z rings were also identified on the elongated cells (Fig. 3D).
Fig 3.
(A and B) Cell morphology and FtsZ ring position of E. coli EC528 harboring pHERD30T-LL37 after induction of LL37 by adding 0.2% arabinose for 13 h. (C and D) Cells were externally treated with 15 μM LL37 in M9 medium and incubated for 24 h. Z rings were observed using fluorescence microscopy.
The effect of the arabinose concentration on the lengths of bacterial cells was investigated under aerobic and anaerobic growth conditions. The lengths of E. coli cells gradually increased as induction time increased. The average length reached 68 ± 8.5 μm when 1% arabinose was added. In contrast, the average length of the control cells was 2.54 ± 1.14 μm. As demonstrated in Fig. 4A, the concentration of arabinose showed a positive correlation with the length of E. coli cells. The longest cells were detected in the presence of 1% arabinose in the medium. The induction of LL-37 expression also affected the lengths of the E. coli cells under anaerobic conditions. However, cell length showed little correlation with the concentration of the inducer arabinose (Fig. 4B). The average cell length was recorded at 4.46 ± 0.9 μm after induction with 0.2% arabinose for 24 h, whereas the lengths of the control cells were comparable to the cell lengths under aerobic growth conditions (Fig. 4B).
Fig 4.
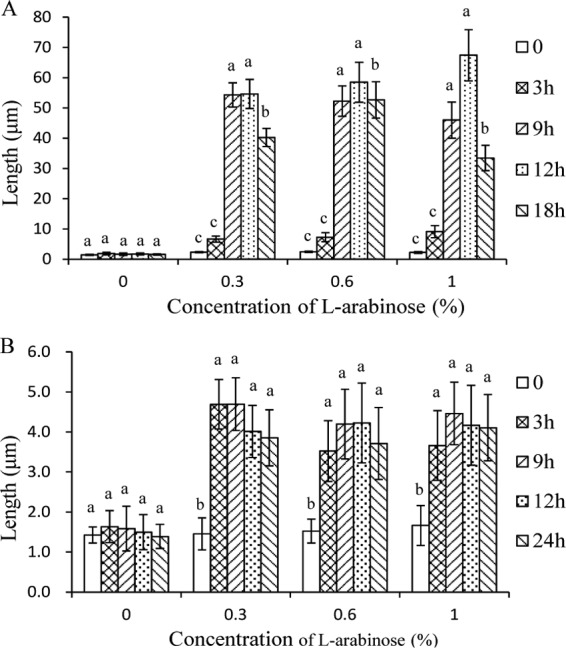
Effects of arabinose concentrations on the lengths of E. coli cells harboring pHERD30T-LL37 under aerobic (A) and anaerobic (B) growth conditions. Cells were grown in LB medium, and the images were acquired by using fluorescence microscopy. The average lengths of 60 to 70 cells were measured using Image-Pro Plus 6.0 software. Statistical analysis was performed by one-way analysis of variance (ANOVA) followed by Duncan's test. The data are presented as means ± SD of 30 to 40 independent biological samples. The lowercase letters indicate statistically significant differences (P < 0.05) in length between l-arabinose-treated and untreated groups at different sampling time points.
Intracellular expression of LL-37 affects membrane integrity.
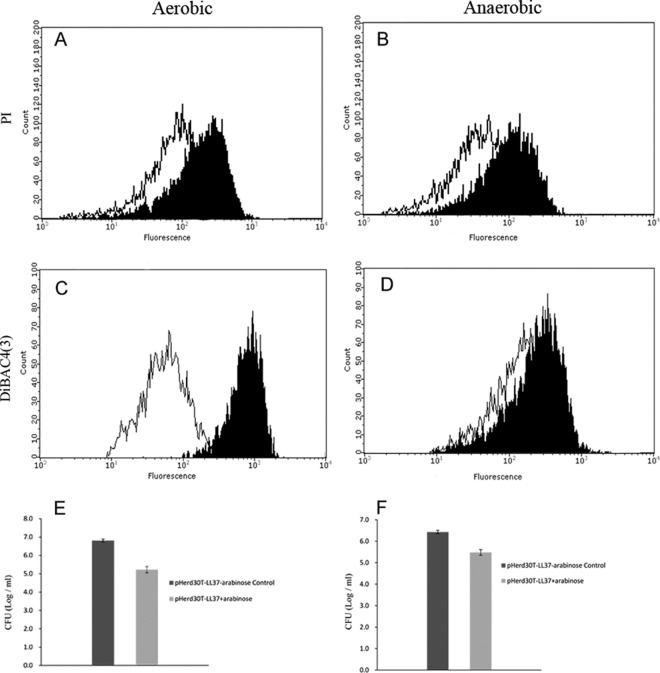
The specific fluorescent dyes DiBAC4 (3) and PI were used to monitor changes in bacterial cell membrane potential and cell membrane integrity via flow cytometry. The membrane permeability and polarization were determined for E. coli cells grown under both aerobic and anaerobic conditions. To test membrane permeability, a membrane-impermeable DNA-staining dye, PI, was used. A one-dimensional gate was created, and cells in these regions were sorted using the high-throughput mode. As shown in Fig. 5A, the fluorescence intensity increased in the cells in comparison with the control cells after 13 h of intracellular expression of LL-37 under both aerobic and anaerobic growth conditions. These data suggest that the presence of intracellular LL-37 affects the permeability of the bacterial cell membrane regardless of the presence of oxygen (Fig. 5A and B).
Fig 5.
Intracellular expression of LL37 causes membrane depolarization under aerobic, but not anaerobic, conditions. (A to D) E. coli strain TOP10 harboring pHERD30T-LL37 was grown in LB medium and induced by adding 0.2% arabinose under aerobic growth for 13 h (A and C) or anaerobic growth for 24 h (B and D). Samples consisting of at least 100,000 cells were stained with HPF (PI) (A and B) and DiBAC4(3), which stained the depolarized cells, where it binds to intracellular proteins (C and D). Black line, uninduced; filled histogram, induced with arabinose. (E and F) Log change in CFU per milliliter under aerobic growth for 13 h (E) and anaerobic growth conditions for 24 h (F). CFU data are presented as log10. The error bars represent standard deviations of triplicate samples. The experiments were repeated at least three times with similar results.
In parallel, changes in cell membrane potential were evident in the E. coli cells after intracellular expression of LL-37 under aerobic and anaerobic growth conditions. DiBAC4 (3) fluorescence shifted significantly toward the higher intensity with the cells after intracellular expression of LL-37 under aerobic growth conditions (Fig. 5C). In contrast, the DiBAC4 (3) fluorescence changed to a much smaller extent in the anaerobic culture after intracellular expression of LL-37, suggesting that the change in cell membrane potential is less significant (Fig. 5D).
Formation of free radicals after intracellular expression of LL-37.
The difference in the depolarization of bacterial cell membranes under aerobic and anaerobic growth conditions implies that the presence of oxygen in the growth medium may act as one of the effectors of the killing action of LL-37. Previous studies have suggested that free radicals (ROS) can be generated through leakage of the respiration chain after exposure to AMPs. This might affect the bacterial cell morphology and membrane depolarization. In this way, the total concentrations of ROS and hydroxyl radicals were determined using flow cytometry coupled with DCFH-DA and HPF, respectively. As shown in Fig. 6A, the level of total ROS showed significant elevation after 4 h of intracellular expression of LL-37. In contrast, the fluorescence intensity in the E. coli cells stained with HPF after the expression of LL-37 remained relatively stable in comparison to control cells, suggesting that OH· was not the major free radical produced under the stress of AMPs (Fig. 6B).
Fig 6.
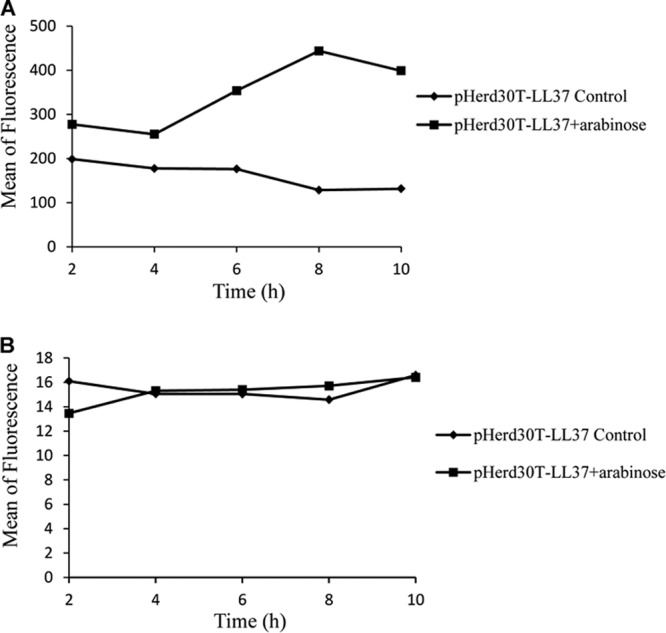
Effects of intracellular expression of LL-37 on ROS (A) and the formation of hydroxyl radicals (B). E. coli strain TOP10 harboring pHERD30T-LL37 was grown in LB medium and induced by 0.2% arabinose under aerobic growth conditions. Samples consisting of at least 100,000 cells were collected at different intervals and stained with DCFH-DA, an indicator of intracellular activity of ROS, and HPF.
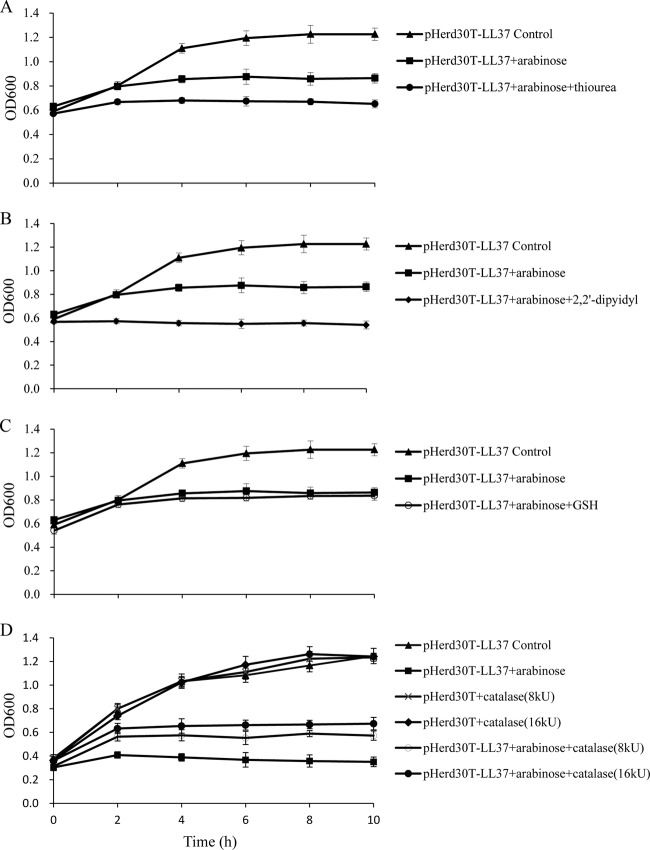
Since the intracellular expression of LL-37 results in the accumulation of ROS intermediates, we tested the effects of antioxidants, such as 2,2′-dipyidyl, thiourea, glutathione (GSH), and catalase, on bacterial survival after intracellular expression of LL-37 under aerobic growth conditions. Consistent with our previous results, the ion chelator 2,2′-dipyidyl, the OH· scavenger thiourea, and GSH showed no protective effects (Fig. 7A, B, and C), but the presence of catalase (8 kU and 16 kU) in LB medium increased the level of survival of E. coli after intracellular expression of LL-37 for 2 h (Fig. 7D).
Fig 7.
Exogenous addition of catalase partially neutralizes the growth-inhibitory effects of LL-37 in E. coli TOP10 cells. Shown are growth curves of E. coli expressing LL-37 in LB medium in the absence and presence of 0.1% l-arabinose; 150 μM thiourea (A), 500 μM 2,2′-dipyidyl (B), and 4 mM glutathione (C) were added to the medium, and catalase was included at 8 kU and 16 kU final concentrations (D).
Transcriptional response to the intracellular expression of LL-37.
To better understand the complete cellular response to LL-37, a global transcriptional profile of E. coli after intracellular expression of LL-37 was compared to that of a control strain under both aerobic and anaerobic growth conditions. Two separate experiments were conducted individually. A total of 5,872,273 and 16,744,564 raw reads, with average lengths of 49 bp and 101 bp, were obtained from the test strain (E. coli TOP10 pHERD30T-LL-37) under aerobic and anaerobic conditions, respectively. Among them, 5,867,603 (99.92% of the total sequences from aerobic growth conditions) and 16,699,993 (99.73% of the total sequences from anaerobic growth conditions) good-quality reads were obtained following filtration of all low-quality reads. Additionally, the numbers of clean reads from the control strain (E. coli TOP10 pHERD30T) were 6,175,414 (99.94%) and 27,517,769 (99.76%) for aerobic and anaerobic growth conditions, respectively. A total of 57.67% (under aerobic conditions) and 44.62% (under anaerobic conditions) of transcripts from the test strain (E. coli TOP10 pHERD30T-LL-37) can be mapped to the E. coli reference genome, in comparison with 70.36% and 44.80% in the control strain (E. coli TOP10 pHERD30T). The transcript abundance was measured using the RPKM method. The transcripts differentially expressed between the test and control strains under aerobic and anaerobic conditions were identified using an algorithm developed by Audic and Claverie (22) and then those differentially expressed genes were analyzed using the COG database. A total of 22 COG functional categories were identified (see Table S1 in the supplemental material). In general, the number of downregulated genes was higher than that of upregulated genes across all categories. There were 301 upregulated and 391 downregulated genes under aerobic conditions, while 185 upregulated and 190 downregulated genes were detected under anaerobic growth conditions (see Fig. S2 in the supplemental material). It is interesting that the largest proportion of altered genes either cannot be classified or grouped into the categories unknown (S) and general function predicted only (R) (see Table S1 in the supplemental material). Apart from that, the categories amino acid transport and metabolism (E), carbohydrate transport and metabolism (G), and energy production and conversion (C) contained the most regulated genes.
We then detected the commonly regulated genes caused by the intracellular expression of LL-37 under both aerobic and anaerobic growth conditions. In total, 51 genes were detected, and among them, 13 genes were found to be upregulated and 38 genes were found to be downregulated (Table 1; see also Fig. S3 in the supplemental material). Interestingly, 6 out of the 13 commonly upregulated genes encoded membrane-associated proteins (AraJ, YbcI, YebE, Rtn, YfcP, and EmrA). The cytoplasmic proteins included RecX and ChpA. The former is responsible for DNA repair, while overexpression of the latter led to programmed cell death. In contrast, the downregulated transcripts were in various locations. Among them, 13 out of 38 proteins were membrane associated (WrbA, NarJ, IhfB, CspD, GalT, YeeL, YohC, NuoA, FrdC, OmpW, SdhD, YcjY, and YnhG), and the rest were located in the cytoplasm or unknown locations.
Table 1.
Genes regulated commonly under aerobic and anaerobic growth conditions
| COG | Downregulated gene | Annotation | Upregulated gene | Annotation |
|---|---|---|---|---|
| Unclassified | bssR | Biofilm formation regulatory protein BssR | ||
| ychH | Hypothetical protein | |||
| yebV | Hypothetical protein | |||
| yohC | Inner membrane protein | |||
| yffR | CPZ-55 prophage; hypothetical protein | |||
| Energy production and conversion (C) | sdhD | Succinate dehydrogenase cytochrome b556 small membrane subunit | ybcI | Inner membrane protein |
| galT | Galactose-1-phosphate uridylyltransferase | araJ | MFS transport protein AraJ | |
| narJ | Molybdenum-cofactor-assembly chaperone subunit (delta subunit) of nitrate reductase 1 | |||
| nuoA | NADH dehydrogenase subunit A | |||
| frdC | Fumarate reductase subunit C | |||
| Carbohydrate transport and metabolism | fucR | DNA-binding transcriptional activator FucR | ECDH10B_1331 | Replication protein 15 (O) |
| gatY | Tagatose-bisphosphate aldolase | |||
| glpF | Glycerol facilitator | |||
| rbsB | d-Ribose transporter subunit RbsB | |||
| yagF | Hypothetical protein | |||
| yegT | Nucleoside transporter | |||
| yegV | Kinase | |||
| yniA | Phosphotransferase/kinase | |||
| Transcription (K) | cspD | Stationary-phase/starvation-inducible regulatory protein CspD | ||
| yhcO | Barnase inhibitor | |||
| Replication and repair (L) | ihfB | Integration host factor subunit beta | ||
| sbmC | DNA gyrase inhibitor | |||
| Cell wall/membrane/envelope biogenesis | ompW | Outer membrane protein W CP4-6 | ECDH10B_1333 | Hypothetical protein |
| yagE | Prophage; lyase/synthase | yfgH | Outer membrane lipoprotein | |
| Cell motility (U) | yfcP | Fimbria-like adhesin protein | ||
| Posttranslational modification, protein turnover, and chaperones (O) | htpX | Heat shock protein HtpX | ||
| Inorganic ion transport and metabolism (P) | dps | DNA starvation/stationary-phase protection protein Dps | ygaP | Inner membrane protein with hydrolase activity |
| yibN | Rhodanese-related sulfurtransferase | |||
| nirD | Nitrite reductase small subunit | |||
| Secondary metabolite biosynthesis, transport, and catabolism (Q) | ycaC | Hydrolase | ||
| ybaW | Hypothetical protein | |||
| wrbA | TrpR binding protein WrbA | recX | Recombination regulator RecX | |
| General function prediction only (R) | ycjY | Hydrolase | ||
| ygaF | Toxin ChpA | |||
| aphA | Acid phosphatase/phosphotransferase | |||
| ynhG | Hypothetical protein | |||
| Function unknown (S) | yeeI | Hypothetical protein | yebE | Hypothetical protein |
| ygiW | Hypothetical protein | |||
| yjbQ | Hypothetical protein | |||
| Signal transduction mechanisms (T) | uspA | Universal stress global response regulator | rtn | Hypothetical protein |
| Defense mechanisms (V) | chpA | Toxin ChpA | ||
| emrA | Multidrug efflux system |
The genes commonly regulated by intracellularly expressed LL-37 were further grouped by COG analysis (Table 1). Consistent with observations from the differentially expressed genes, a large proportion of commonly regulated genes were either unable to be classified or were classified into categories of unknown function (R and S). There were 7 genes involved in energy production and conversion (C). Among them, 5 genes were downregulated and 2 genes were upregulated. It is interesting that genes for fumarate reductase (frdC), NADH dehydrogenase (nuoA), molybdenum cofactor assembly chaperone of nitrate reductase (narJ), succinate dehydrogenase (sdhD), and galactose-1-phosphate uridylyltransferase (galT) are particularly associated with the function of oxidative phosphorylation. In addition, 8 downregulated genes were identified, yagF, yniA, gatY, yegT, yegV, fucR, rbsR, and glpF, belonging to carbohydrate transport and metabolism (G). In addition, there were 3 genes (dps, yibN, and nirD) involved in inorganic ion transport and metabolism (P).
DISCUSSION
The goal of this study was to determine the mechanism by which intracellularly expressed LL-37 kills the Gram-negative bacterium E. coli under aerobic and anaerobic growth conditions. Extensive studies have shown that AMPs kill bacteria through multiple mechanisms, including binding to cell wall materials, such as bacterial LPS and peptidoglycan, using electrostatic reactions; disturbing the bilayer structure of the membrane by penetrating into the phospholipid monolayer; and directly attacking intracellular targets, such as enzymes and nucleic acids (5). However, most of these previous studies have elucidated the bactericidal mechanisms of AMPs through in vitro extracellular systems under aerobic growth conditions (23–28). In contrast, we developed a genetic system to intracellularly express LL-37 and examine its impact on the growth of E. coli. Growth curve experiments and CFU detection indicated that intracellular expression of LL-37 was able to inhibit the growth of E. coli under both aerobic and anaerobic conditions. This is consistent with the activity of LL-37 from outside the bacterial cell (29, 30). Interestingly, intracellular production of LL-37, like extracellular production, altered the cell morphology to exhibit an elongated filamentous phenotype. Moreover, this profound phenomenon was observed only under aerobic growth conditions. We initially hypothesized that LL-37 directly interfered with cell division proteins, resulting in a malfunction during cell division. Further microscopic observation with GFP-labeled FtsZ showed that Z rings formed normally, suggesting that the filamentous morphology after expression of LL-37 is not attributable to direct interactions between LL-37 and bacterial cell division proteins. This leads to the formation of multiple Z rings without normal septa in the filamentous phenotype in the presence of oxygen. This suggests that multiple killing mechanisms of LL-37 exist and that the bacterial growth conditions also contribute to growth inhibition.
AMPs commonly use the inhibition of cell division as a mechanism of bacterial killing (5). Rosenberger et al. reported that in vitro incubation of Salmonella enterica serovar Typhimurium with sublethal levels of mouse cathelicidin-related antimicrobial peptide (CRAMP) causes the bacterial cell to become filamentous in shape with arrested septum formation (19). Li et al. also found that the peptide-treated cells are unable to undergo cell division (31). In the current study, we found that, solely under aerobic growth conditions, the lengths of bacterial cells increased proportionally to the concentration of arabinose, which corresponds to the levels of intracellular expression of LL-37.This suggests that the increased cell length is particularly associated with the presence of free radicals, which are produced by intracellular LL-37 in the presence of oxygen. Indeed, the enhanced production of ROS was detected in the current study after intracellular expression of LL-37. RNA-sequencing analysis further confirmed that no single gene was up- or downregulated in the cell division pathway after intracellular expression of LL-37. Therefore, cell elongation is possibly a secondary effect resulting from the production of ROS.
The enhanced production of ROS by the intracellular expression of LL-37 has been demonstrated in the current study. As a consequence, ROS causes a significant change in the cell membrane potential, but not in membrane permeability. It has been established that alteration of cell membrane potential eventually accelerates cell death. Generation of ROS, particularly the highly deleterious hydroxyl radicals, is a common mechanism causing bacterial death in most classes of bactericidal antibiotics (20). However, intracellular expression of LL-37 does not stimulate the production of hydroxyl radicals. Instead, we observed elevated production of total ROS. The protective effect observed from catalase, but not from the iron chelator 2,2′-dipyidyl and the OH· scavenger thiourea, implies that hydrogen peroxide may be the major component of the ROS generated by intracellular exposure to LL-37. This suggests that a different mode of antimicrobial activity may exist between the bactericidal antibiotics and AMPs. Interestingly, the profound membrane depolarization happens only under aerobic growth conditions, while membrane permeability occurred under both aerobic and anaerobic conditions. Therefore, the former is most likely a consequence of ROS production and the latter may be due to the interference of intracellular LL-37. Nuding et al. (17) found that profound membrane depolarization for obligate anaerobes, such as Bacteroides vulgatus 484142B and B. fragilis 484143G, occurs only in the presence of oxygen, which is consistent with our current findings. In fact, the degree of membrane depolarization has been used as an assessing index for AMP efficacy (28). Since LL-37 is a cationic peptide, disruption of the cell membrane has been proposed as the primary mode of action for positively charged AMPs from outside (31, 32). Membrane permeability under anaerobic growth conditions indicates that interference with the bacterial cell membrane is also the major killing mechanism of intracellular LL-37.
Intracellular expression of LL-37 provokes an inhibitory-like response pattern in transcription levels. It has been known that decreases in carbohydrate metabolism and respiration are common phenomena for both antibiotics and AMPs and that supplementation of metabolites, such as glucose, fructose, and mannose, can stimulate the generation of proton-motive force (33). In the current study, genes related to carbohydrate metabolism and energy production were downregulated. Interestingly, the genes involved in oxidative phosphorylation, such as NADH dehydrogenase I (nuoA), succinate dehydrogenase (sdhD), fumarate reductase (frdC), and nitrate reductase (nirD) genes, are downregulated under both aerobic and anaerobic growth conditions. Since genes involved in oxidative phosphorylation always contain redox cofactors, such as menaquinone, quinone, or iron-sulfur clusters, it is proposed that intracellular positively charged LL-37 may affect the redox status or electron transfer. Indeed, when the iron chelator 2,2′-dipyidyl was added to the growth medium, the inhibition effect of intracellular LL-37 on the growth of bacterial cells was enhanced (Fig. 7). Moreover, four genes related to the pathway of inorganic ion transport and metabolism were altered. On the other hand, downregulated NADH dehydrogenase I may contribute significantly to the change in membrane potential, and the reduced respiration may lead to a decline in energy consumption. Therefore, the cationic property of LL-37 may also contribute to the bacterial killing effect inside the cell by interference with the intracellular redox potential or ion strength.
In summary, we have demonstrated that intracellular LL-37 can inhibit the growth of E. coli under both aerobic and anaerobic growth conditions. Additionally, the cell morphology changes to a filamentous phenotype after intracellular expression of LL-37. Physiological experiments, coupled with transcriptional profiling, suggest that LL-37 does not directly interfere with cell division. However, the presence of oxygen, which subsequently induces the production of ROS, significantly affects cell morphology and membrane potential. It seems that the positively charged LL-37 not only affects the membrane integrity of bacterial cells extracellularly, but also influences membrane permeability intracellularly through interference with redox and ion status.
Supplementary Material
ACKNOWLEDGMENTS
The research was supported by grants from the National Basic Research Program of China (973; no. 2012CB721006) and the International S and T Cooperational Project of China (2008DFA32080). H.D.Y. was supported by the Cystic Fibrosis Foundation (CFF-YU11G0) and by NIH P20RR016477 and P20GM103434 to the West Virginia IDeA Network for Biomedical Research Excellence.
Footnotes
Published ahead of print 15 July 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00825-13.
REFERENCES
- 1.Hale JD, Hancock RE. 2007. Alternative mechanisms of action of cationic antimicrobial peptides on bacteria. Expert Rev. Anti Infect. Ther. 5:951–959 [DOI] [PubMed] [Google Scholar]
- 2.Matsuzaki K. 1999. Why and how are peptide-lipid interactions utilized for self-defense? Magainins and tachyplesins as archetypes. Biochim. Biophys. Acta 1462:1–10 [DOI] [PubMed] [Google Scholar]
- 3.Oren Z, Shai Y. 1998. Mode of action of linear amphipathic alpha-helical antimicrobial peptides. Biopolymers 47:451–463 [DOI] [PubMed] [Google Scholar]
- 4.Sochacki KA, Barns KJ, Bucki R, Weisshaar JC. 2011. Real-time attack on single Escherichia coli cells by the human antimicrobial peptide LL-37. Proc. Natl. Acad. Sci. U. S. A. 108:E77–E81. 10.1073/pnas.1101130108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brogden KA. 2005. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 3:238–250 [DOI] [PubMed] [Google Scholar]
- 6.Boman HG, Agerberth B, Boman A. 1993. Mechanisms of action on Escherichia coli of cecropin P1 and PR-39, two antibacterial peptides from pig intestine. Infect. Immun. 61:2978–2984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yonezawa A, Kuwahara J, Fujii N, Sugiura Y. 1992. Binding of tachyplesin I to DNA revealed by footprinting analysis: significant contribution of secondary structure to DNA binding and implication for biological action. Biochemistry 31:2998–3004 [DOI] [PubMed] [Google Scholar]
- 8.Hancock RE, Chapple DS. 1999. Peptide antibiotics. Antimicrob. Agents Chemother. 43:1317–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park CB, Kim HS, Kim SC. 1998. Mechanism of action of the antimicrobial peptide buforin II: buforin II kills microorganisms by penetrating the cell membrane and inhibiting cellular functions. Biochem. Biophys. Res. Commun. 244:253–257 [DOI] [PubMed] [Google Scholar]
- 10.Skerlavaj B, Romeo D, Gennaro R. 1990. Rapid membrane permeabilization and inhibition of vital functions of gram-negative bacteria by bactenecins. Infect. Immun. 58:3724–3730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sass V, Schneider T, Wilmes M, Korner C, Tossi A, Novikova N, Shamova O, Sahl HG. 2010. Human beta-defensin 3 inhibits cell wall biosynthesis in staphylococci. Infect. Immun. 78:2793–2800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gudmundsson GH, Agerberth B, Odeberg J, Bergman T, Olsson B, Salcedo R. 1996. The human gene FALL39 and processing of the cathelin precursor to the antibacterial peptide LL-37 in granulocytes. Eur. J. Biochem. 238:325–332 [DOI] [PubMed] [Google Scholar]
- 13.Bucki R, Leszczynska K, Namiot A, Sokolowski W. 2010. Cathelicidin LL-37: a multitask antimicrobial peptide. Arch. Immunol. Ther. Exp. 58:15–25 [DOI] [PubMed] [Google Scholar]
- 14.Rosenfeld Y, Shai Y. 2006. Lipopolysaccharide (endotoxin)-host defense antibacterial peptides interactions: role in bacterial resistance and prevention of sepsis. Biochim. Biophys. Acta 1758:1513–1522 [DOI] [PubMed] [Google Scholar]
- 15.Lee CC, Sun Y, Qian S, Huang HW. 2011. Transmembrane pores formed by human antimicrobial peptide LL-37. Biophys. J. 100:1688–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joly S, Maze C, McCray PB, Jr, Guthmiller JM. 2004. Human beta-defensins 2 and 3 demonstrate strain-selective activity against oral microorganisms. J. Clin. Microbiol. 42:1024–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nuding S, Zabel LT, Enders C, Porter E, Fellermann K, Wehkamp J, Mueller HA, Stange EF. 2009. Antibacterial activity of human defensins on anaerobic intestinal bacterial species: a major role of HBD-3. Microbes Infect. 11:384–393 [DOI] [PubMed] [Google Scholar]
- 18.Qiu D, Damron FH, Mima T, Schweizer HP, Yu HD. 2008. PBAD-based shuttle vectors for functional analysis of toxic and highly regulated genes in Pseudomonas and Burkholderia spp. and other bacteria. Appl. Environ. Microbiol. 74:7422–7426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosenberger CM, Gallo RL, Finlay BB. 2004. Interplay between antibacterial effectors: a macrophage antimicrobial peptide impairs intracellular Salmonella replication. Proc. Natl. Acad. Sci. U. S. A. 101:2422–2427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kohanski MA, Dwyer DJ, Wierzbowski J, Cottarel G, Collins JJ. 2008. Mistranslation of membrane proteins and two-component system activation trigger antibiotic-mediated cell death. Cell 135:679–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5:621–628 [DOI] [PubMed] [Google Scholar]
- 22.Audic S, Claverie JM. 1997. The significance of digital gene expression profiles. Genome Res. 7:986–995 [DOI] [PubMed] [Google Scholar]
- 23.Hong RW, Shchepetov M, Weiser JN, Axelsen PH. 2003. Transcriptional profile of the Escherichia coli response to the antimicrobial insect peptide cecropin A. Antimicrob. Agents Chemother. 47:1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Otvos L, Jr, de Olivier Inacio V, Wade JD, Cudic P. 2006. Prior antibacterial peptide-mediated inhibition of protein folding in bacteria mutes resistance enzymes. Antimicrob. Agents Chemother. 50:3146–3149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pietiainen M, Francois P, Hyyrylainen HL, Tangomo M, Sass V, Sahl HG, Schrenzel J, Kontinen VP. 2009. Transcriptome analysis of the responses of Staphylococcus aureus to antimicrobial peptides and characterization of the roles of vraDE and vraSR in antimicrobial resistance. BMC Genomics 10:429–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou Y, Chen WN. 2011. iTRAQ-coupled 2-D LC-MS/MS analysis of membrane protein profile in Escherichia coli incubated with apidaecin IB. PLoS One 6:e20442. 10.1371/journal.pone.0020442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou Y, Chen WN. 2011. iTRAQ-coupled 2-D LC-MS/MS analysis of cytoplasmic protein profile in Escherichia coli incubated with apidaecin IB. J. Proteomics 75:511–516 [DOI] [PubMed] [Google Scholar]
- 28.Epand RF, Pollard JE, Wright JO, Savage PB, Epand RM. 2010. Depolarization, bacterial membrane composition, and the antimicrobial action of ceragenins. Antimicrob. Agents Chemother. 54:3708–3713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bals R, Wang X, Zasloff M, Wilson JM. 1998. The peptide antibiotic LL-37/hCAP-18 is expressed in epithelia of the human lung where it has broad antimicrobial activity at the airway surface. Proc. Natl. Acad. Sci. U. S. A. 95:9541–9546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brissette CA, Lukehart SA. 2007. Mechanisms of decreased susceptibility to β-defensins by Treponema denticola. Infect. Immun. 75:2307–2315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li L, Shi Y, Su G, Le G. 2012. Selectivity for and destruction of Salmonella typhimurium via a membrane damage mechanism of a cell-penetrating peptide ppTG20 analogue. Int. J. Antimicrob. Agents 40:337–343 [DOI] [PubMed] [Google Scholar]
- 32.Hartmann M, Berditsch M, Hawecker J, Ardakani MF, Gerthsen D, Ulrich AS. 2010. Damage of the bacterial cell envelope by antimicrobial peptides gramicidin S and PGLa as revealed by transmission and scanning electron microscopy. Antimicrob. Agents Chemother. 54:3132–3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allison KR, Brynildsen MP, Collins JJ. 2011. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature 473:216–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.