Abstract
Candida albicans is a prevailing fungal pathogen with a diploid genome that can adapt to environmental stresses by losing or gaining an entire chromosome or a large portion of a chromosome. We have previously found that the loss of one copy of chromosome 5 (Ch5) allows for adaptation to the toxic sugar l-sorbose. l-Sorbose is similar to caspofungin and other antifungals from the echinocandins class, in that it represses synthesis of cell wall glucan in fungi. Here, we extended the study of the phenotypes controlled by Ch5 copy number. We examined 57 strains, either disomic or monosomic for Ch5 and representing five different genetic backgrounds, and found that the monosomy of Ch5 causes elevated levels of chitin and repressed levels of 1,3-β-glucan components of the cell wall, as well as diminished cellular ergosterol. Increased deposition of chitin in the cell wall could be explained, at least partially, by a 2-fold downregulation of CHT2 on the monosomic Ch5 that encodes chitinase and a 1.5-fold upregulation of CHS7 on Ch1 that encodes the protein required for wild-type chitin synthase III activity. Other important outcomes of Ch5 monosomy consist of susceptibility changes to agents representing four major classes of antifungals. Susceptibility to caspofungin increased or decreased and susceptibility to 5-fluorocytosine decreased, whereas susceptibility to fluconazole and amphotericin B increased. Our results suggest that Ch5 monosomy represents an unrecognized C. albicans regulatory strategy that impinges on multiple stress response pathways.
INTRODUCTION
Candida albicans, a unicellular budding fungus, is a part of normal flora of the gut and genitalia. In healthy individuals, C. albicans causes superficial infection. However, in severely immunocompromised patients, C. albicans can cause systemic infections that lead to lethality, making this microbe an important opportunistic pathogen.
C. albicans' diploid genome of approximately 14.6 Mbp is organized into eight pairs of chromosomes that are known for their instability. The occurrence of aneuploid chromosomes is well documented in vitro, establishing that any chromosome can spontaneously become aneuploid upon culture in rich standard medium, that any kind of aneuploidy can occur, including loss or gain of an entire chromosome or a large portion of chromosome, and that aneuploidy is well tolerated (1–5). In addition, a growing number of studies report the occurrence of various chromosome alterations in animal models or human patients (2, 6–8). Reported aneuploidies include trisomies or duplication of the left arm of Ch5 (8–10).
Laboratory studies provided insight into these chromosome alterations and demonstrated that in lethal or limiting environments, C. albicans can use reversible aneuploidy for survival and adaptation. For example, upon culture in media in which glucose was replaced by the toxic sugar l-sorbose, cells that do not utilize sorbose (Sou−) survive, predominantly due to the loss of one chromosome 5 (Ch5), and acquire the ability to grow on sorbose (Sou+) (2, 11, 12). Our long-term study of regulation by Ch5 monosomy revealed association of this regulation with an unanticipated complexity. For example, Ch5 carries multiple unique regions for negative control of growth on sorbose, with each region containing at least one unique negative controlling element, called CSU (control of sorbose utilization). The regions are scattered along Ch5, and the final number of regions is yet to be established. The monosomic condition of Ch5 downregulates, as expected, at least CSU51 (orf19.1105.2) and CSU53 (orf19.3931) from Ch5 and also upregulates SOU1 (sorbose utilization) from Ch4 that encodes sorbose reductase, which catalyzes the first step in the catabolic pathway of l-sorbose (11, 13, 14, and E. Rustchenko, unpublished data). Furthermore, antisense regulation of CSUs is involved, as at least CSU51 and CSU53 produce, in addition to sense transcripts, long noncoding antisense transcripts, designated ASUs (activation of sorbose utilization). ASU and CSU transcripts are inversely related, and ASU elements act by enhancing growth on sorbose, i.e., counteracting CSU (14). Adding to the complexity, transcription from monosomic Ch5 is under the control of various mechanisms. Approximately 34% of transcripts, including known CSUs, are controlled directly by the 2-fold decrease of template DNA. Approximately 9% of transcripts are upregulated by 2-fold or more above the disomic level. Strikingly, however, 58% of genes on Ch5 exhibit dosage compensation to counteract monosomy and keep transcripts at or close to the disomic level (15).
Sorbose kills fungi by interfering with cell wall synthesis (16–19). This is similar to the action of the antifungal drugs from the echinocandin class (20–22). The main purpose of the present study was to investigate the relationship between Ch5 copy number and susceptibility to a major antifungal agent of this class, caspofungin (CAS), as well as other major antifungals. We report here that adaptation-related Ch5 monosomy induces a complex regulatory network that results in remodeling of the structure of the cell wall and cellular membranes. As a result, the cell wall 1,3-β-glucan and chitin content decreased and increased, respectively, whereas the content of cellular ergosterol decreased. These changes result in phenotypes that include alteration of susceptibility to at least four major antifungals, caspofungin, 5-fluorocytosine (5FC), fluconazole (FLU), and amphotericin B (AmB), and they are likely to be involved in the generation of drug-resistant strains.
MATERIALS AND METHODS
Strains, media, growth conditions, and primers.
The strains JRCT1, RO29R4, and RO20G1 were randomly chosen from two collections of clinical isolates containing either strains from AIDS patients or strains collected from mothers and prematurely born children at Strong Memorial Hospital of Rochester. Collections are stored in frozen vials at −70°C, a procedure which interrupts cellular metabolism, preventing induction and propagation of genetic instability (4). The C. albicans laboratory strains SC5314 and 3153A were stored at −70°C upon arrival in our laboratory. These strains were previously extensively characterized for their electrophoretic karyotypes (4, 5).
The preparation of yeast-peptone-dextrose (YPD), sorbitol, and l-sorbose media has been described previously (15, 23–25). In order to prepare solid medium, 2% (wt/vol) agar was added. Cells were routinely incubated at 37°C. Care was taken to grow and maintain cells in a way that prevents induction of chromosome instability (for details, see references 4 and 14).
Primers used in this study are presented in Table 1.
Table 1.
List of primers
| Primer purpose and genea | Primer sequence |
|---|---|
| Amplification of the MTL locus | |
| MTLa1 (orf19.3201) | F, TACAATTGAAGCGTGAGAGGCAGGAGA |
| R, TCATCATCCATCTGGTCGCTTACTTCA | |
| MTLα1 (orf19.10712) | F, GCGATGCTCCAAGAAGAGACACAAGAG |
| R, TAATCCAAAGCCTCGCATAACCAGTCC | |
| Amplification of HS1 or HS2 regions of GSC1 (orf19.2929) | |
| HS1-A | F, CGGTGCTCAACATTTGAGTCGTCGTAT |
| HS1-A | R, TTGATTTCCATTTCCGTGGTAGCTAAA |
| HS2-A | F, TGCTGGTATGAATGCCATGATGAGAGG |
| HS2-A | R, GGTGCTTGCCAATGAGAAACTGTACCA |
| Sequencing of HS1 and HS2 regions of GSC1 (orf19.2929) | |
| HS1-S | F, CGGCATATGCTGTGTCGATTGT |
| HS1-S | R, TGAACGACCAATGGAGAAGA |
| HS2-S | F, TTGGTGCTGGTATGGGAGAACA |
| HS2-S | R, GCACCACCAACGGTCAAATCA |
| Semiquantitative RT-PCR | |
| CHS7 (orf19.2444) | F, TGTTGATTGTGGTGTTAGTCCTCCA |
| R, AACATAAACGGCCAATATGACAGCA | |
| POM152* (orf19.2081) | F, GGCCGATCCAGCAACCAAACATT |
| R, TCACAGCACACGAATTGATTCCAGA | |
| CHT2 (orf19.3895) | F, CATCAGCTTTGGCCAGTGCCTCT |
| R, GCAGAAGCAACATCGCTGAAACCA | |
| orf19.2113* | F, CAAAGGTCAGCAGCAGTGGCACA |
| R, GCATCATCGGCAGCATTGGGTAA | |
| TAC1 (orf19.3188) | F, AACCATGCGGAATTCACGTCCAA |
| R, TGTTGCTGGTGAACGACCTGTGC | |
| DAD2* (orf19.3551) | F, AGAGACGATGAATGGAGGAACTGC |
| R, CATTTGATTGTCCAACACGCACTC | |
| ERG11 (orf19.922) | F, TGGGATACTGCTGCTGCCAAAGCTA |
| R, TCCCAAATGATTTCTGCTGGTTCA | |
| WBP1* (orf19.2298) | F, CTTGGCCCAGTGAGTGCCATCAG |
| R, TTCATCGTTTGCGAAGTGTCTCACG | |
| CDR1 (orf19.6000) | F, GCAAGTGAGGTATGGTGTTGCGAGA |
| R, CCAAGGCATCAGCTGAAGGACGA | |
| RPL17B* (orf19.4490) | F, TCTGCCCGTGGTTCTTACTTGAGAG |
| R, TGACAGATTTAGCAGGCCATCTAGC | |
| CDR2* (orf19.5958) | F, TGCTACTGCCATGTCACTCTCCACA |
| R, CGGTACCTTGGACAACTGTGCTTCC | |
| orf19.6866* | F, CCCACCAGAGAAACGGTCAACCA |
| R, GGCCAAACGTTTCAAGTTTGCTTCG | |
| MDR1 (orf19.5604) | F, TGGCCACTGGTGGTGCTAGTGTTG |
| R, TCTGTCGTTACCGGTGATGGCTCTC | |
| orf19.2804* | F, CAACCTGCATCCCAAATTCCAA |
| R, TTGCGTCGTTGCATCAATTGTC | |
| FUR1 (orf19.2640) | F, GAACCAATTACCAGTTGAAGAAGCA |
| R, TCCTCCTGTGGCCAACATTGGAT | |
| orf19.2484* | F, TCTCAGATATCATTGCGCCAGTT |
| R, TCCTGATCCATCGTCGTCAGCAC | |
| GSC1 (orf19.2929) | F, TTGCTTCGTCAAGATGGGCTGCT |
| R, CCAATGGCATGACGGCAAAGAAT | |
| GAR1* (orf19.1165) | F, AAACCACCAAGCGTGGGTCCAAA |
| R, TACTGTTTCCACGGGCACCTCCA |
An asterisk indicates a reference gene that was placed below the corresponding genes studied.
Spot assay.
The spot dilution assay was performed on solid medium (26). Briefly, cells from a −70°C stock were streaked for independent colonies on YPD plates and incubated at 37°C until young colonies of the approximate size of 1 × 105 to 3 × 105 cells/colony grew up. Colonies then were collected, and serial 10-fold dilutions were prepared in sterile distilled water with the aid of a hemacytometer. The corresponding suspensions were plated at 104, 103, 102, and 101 CFU per spot. The plates were incubated for 2 to 8 days at 37°C and photographed with a Molecular Imager Gel Doc XR+ system (Bio-Rad).
Broth microdilution assay for susceptibility testing.
MICs were determined in accordance with the CLSI reference M27-A3 broth microdilution method (27). An inoculum of 2 × 103 cells/ml was prepared in RPMI 1640 medium. Sterile polystyrene 96-well microtiter plates were prepared with serial 2-fold dilutions of the drugs, and then 50 μl of the inoculum was added to each well, resulting in a final concentration of 1 × 103 cells/ml in a total volume of 100 μl per well. Inoculum-free, as well as drug-free, control wells were included. Each strain was tested in duplicate on the plates. Modifications included incubating the plates on a rotating shaker at 150 rpm at 35°C for 48 h and recording growth by reading the plates with a microplate reader (Spectra Max M5; Molecular Devices Corp.) at 600 nm. MICs were determined as the lowest concentration of drug that caused a significant diminution (>50%) of growth compared to growth in the drug-free control wells.
Determination of chitin content in the cell wall.
The chitin content was determined by measuring the absorbance of glucosamine released by acid hydrolysis of the purified cell wall according to reference 28. Briefly, approximately 3,000 CFU was placed on YPD plates and incubated at 37°C until young colonies of the approximate size of 1 × 105 to 3 × 105 cells/colony grew up. Colonies then were washed from the surface of plates with sterile distilled water and collected by centrifugation. Cells were washed three times with sterile distilled water, suspended in sterile distilled water, and disrupted with 0.5-mm glass beads (11079105; BioSpec Products, Inc.) using a Mini-Beadbeater (BioSpec Products, Bartlesville, OK). Cell debris was washed five times with 1 M NaCl. Cell wall was extracted in SDS-MerOH extraction buffer (50 mM Tris, 2% sodium dodecyl sulfate, 0.3 M β-mercaptoethanol, 1 mM EDTA, pH 8.0) at 100°C for 10 min. The extracts were washed three times in sterile distilled water and then dried with a SpeedVac concentrator (Phoenix Equipment Inc.). Approximately 4 mg dry weight of cell wall extracts was hydrolyzed in 1 ml of 6 M HCl at 100°C for 17 h. After evaporation at 65°C, 1 ml of sterile distilled water was added. A volume of 0.1 ml of the sample was mixed with 0.1 ml of 1.5 M Na2CO3 in 4% acetyl acetone. The mixture was incubated at 100°C for 20 min, and then 0.7 ml of 96% ethanol and 0.1 ml of 1.6 g of p-dimethylaminobenzaldehyde in 30 ml of concentrated HCl and 30 ml of ethanol were added. One h later, the absorbance at 520 nm was measured with a plate reader (Spectra Max M5; Molecular Devices Corp.). The glucosamine concentration in each sample was determined from a standard curve of 0 to 0.5 mg/ml of glucosamine (Sigma-Aldrich, St. Louis, MO). Subsequently, chitin content was calculated as a percentage of the cell wall dry weight.
Determination of 1,3-β-glucan level in the cell wall.
The 1,3-β-glucan content was measured with the aniline blue method according to reference 29. Briefly, cells were prepared as described above, washed twice with TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0), suspended in 250 μl of TE buffer, and then adjusted with a spectrophotometer (Beckman DU-64 spectrophotometer) to an optical density at 600 nm (OD600) of 0.2. A total of 0.5 ml of the cell suspension was mixed with 0.1 ml of 6 M NaOH. 1,3-β-Glucan was solubilized by incubation in a water bath at 80°C for 30 min, followed by addition of 2.1 ml of aniline blue mix consisting of 0.03% aniline blue, 0.18 M HCl, and 0.49 M glycine-NaOH, pH 9.5. The tubes were vortexed briefly and then incubated for 30 min at 50°C and for an additional 30 min at room temperature. Fluorescence of 1,3-β-glucan was measured with a fluorescence plate reader (Spectra Max M5; Molecular Devices Corp.). An excitation wavelength of 400 nm/slit and an emission wavelength of 460 nm/slit with a cutoff of 455 nm were used to measure fluorescence. Fluorescence intensities were compared.
Determination of the cellular content of ergosterol.
Total cellular sterols were extracted by the alcoholic KOH method, and the ergosterol content was determined as a percentage of the wet weight of the cells according to references 30 and 31. Briefly, cells were prepared as described above and boiled at 85°C for 1 h in 25% alcoholic KOH (25 g of KOH and 35 ml of sterile water, brought to 100 ml with 100% ethanol). One ml of sterile water and 3 ml of n-heptane (J. T. Baker, Stehelin AG, Basel, Switzerland) were added, followed by vigorous vortexing for 3 min. The heptane layer was diluted 5-fold in 100% ethanol and measured by a spectrophotometer (Spectra Max M5; Molecular Devices Corp.) at both 281.5 and 230 nm. Ergosterol content was calculated as % ergosterol = [(A281.5/290 × F/pellet weight] − [A230/518 × F/pellet weight], where F is the factor for dilution in ethanol.
Determination of resistance to zymolyase.
Cells were prepared as described above and adjusted with a spectrophotometer (Beckman DU-64) to an OD600 of 0.5 in the solution of 10 mM Tris-HCl, pH 7.5, containing 50 μg/ml of zymolyase 20T (U.S. Biological, Swampscott, MA). Cells were incubated at 37°C for 4 h. The decrease in optical density was monitored at a time interval of 0.5 h with the spectrophotometer.
Colony PCR.
Cells were prepared as described above for colony PCR. An individual colony was picked and suspended in 50 μl of sterile water in a 250-μl microcentrifuge tube, and PCR was performed according to methods from reference 32.
DNA profiling by aCGH.
Chromosome ploidy was determined by array comparative genome hybridization (aCGH) according to reference 15 with tiling microarrays that were custom designed by Roche NimbleGen Inc. Each of the microarrays contains 710,907 probes covering the C. albicans genome four times, with approximately 20,000 probes covering Ch5 (15).
RNA profiling by expression arrays.
Expression data of several genes of interest were extracted from raw data available at GEO under the accession numbers GSM455127 to GSM455132. These data were obtained with custom-designed CustomArray expression microarrays from CombiMatrix Corporation (Mukilteo, WA). Arrays are based on 6,321 open reading frames (ORF) found in assembly 19 of the C. albicans genome sequence. Each such microarray contains oligonucleotides that were designed to correspond to the 5′ and 3′ regions of the ORFs. Each chip contains 11,898 probes that spread across 12,000 spots (see reference 15 for details).
RT-PCR and semiquantitative analysis.
RNA isolation, reverse transcription-PCR (RT-PCR), and semiquantitative RT-PCR analysis were conducted as previously described by us (14, 15).
Miscellaneous.
DNA sequencing was performed in the Core Facility at the University of Rochester using a BigDye Terminator v3.1 cycle sequencing kit on an ABI 3730 Prism genetic analyzer. The PCR method was previously described by Kravets et al. (15). Separation of chromosomes was previously described by Ahmad et al. (4).
Microarray data accession number.
Raw data were deposited in the Gene Expression Omnibus (GEO), http://www.ncbi.nlm.nih.gov/geo, under accession number GSE48093.
RESULTS
Generation of sequential mutants having monosomic or reduplicated condition of Ch5.
To study the effect of Ch5 copy number, we used a large group of related strains having either one or two copies of Ch5. We took advantage of previously generated mutants with one Ch5 copy that were derived from the laboratory strain 3153A: parental disomy of Ch5→Ch5 monosomy due to loss of either one of two copies of Ch5. We also took advantage of a previously constructed sequential series also derived from 3153A: parental disomy of Ch5→Ch5 monosomy→duplication of remaining Ch5 (11; reviewed in reference 2), as observed, for example, in the strains 3153A→Sor122→Sor122-R (Table 2). Monosomic mutants of 3153A were generated by a well-established procedure of passing strains through medium containing toxic l-sorbose as the sole source of carbon. While most of the Sou− parental cells died on sorbose plates, Ch5 monosomy occurred with relatively high frequency in the surviving cells and rendered the Sou+ phenotype (11, 12). Each Sou+ mutant was generated from independently grown cultures and resulted from an independent mutational event. Derivatives with duplicated remaining Ch5 copies were obtained from the monosomic mutants in the absence of selection for sorbose. The chromosome condition of monosomic mutants and duplicated derivatives was determined by precise separation of chromosomes with pulsed-field gel electrophoresis (PFGE) (11, 12). In addition, the chromosome condition of the mutant Sor125(55), which was previously published under the name Sor55 (11), has been analyzed by the aCGH approach (15).
Table 2.
Summary of phenotypes in strains alternating between 1 and 2 Ch5 copiesa
| Ch5 condition | Strain | MTL locus | Resistance phenotype |
|||
|---|---|---|---|---|---|---|
| CAS | 5FC | FLU | AmB | |||
| Parent | RO29R4 | a/α | ||||
| Monosomic | Sor322, Sor323, Sor324, Sor325, Sor326 | a | S | R | S | S |
| Sor321 | α | P | R | S | S | |
| Duplicated | Sor322-R, Sor321-R | a/a or α/α | P | P | P | P |
| Parent | JRCT1 | a/α | ||||
| Monosomic | Sor91, Sor92, Sor93 | α | R | R | S | S |
| Duplicated | Sor91-R, Sor92-R, Sor93-R | α/α | P | R | P | P |
| Parent | RO20G1 | a/α | ||||
| Monosomic | Sor341, Sor343, Sor344, Sor345, Sor346 | a or α | S | R | S | S |
| Parent | SC5314 | a/α | ||||
| Monosomic | Sor71, Sor72, Sor73 | a or α | R | R | S | S |
| Duplicated | Sor71-R, Sor72-R, Sor73-R | a/a or α/α | P | P | P | P |
| Parent | 3153A | a/α | ||||
| Monosomic | Sor122, Sor123 | α | S | R | S | S |
| Sor41, Sor43, Sor1210(60) | a | R | R | S | P | |
| Sor126, Sor128 | α | P | R | S | P | |
| Sor125(55) | α | S | R | S | P | |
| Duplicated | Sor122-R, Sor123-R | α/α | P | P | P | P |
| Sor125(55)-R | α/α | P | P | P | P | |
| Control derived from 3153Ac | ||||||
| Monosomic | F6-RA, RB, RC, RDb | a | S | R | S | S |
| Disomic | Sor5 | a/α | P | P | P | P |
| m5, m6 | a/α | P | P | P | P | |
| m15 | a/α | S | P | P | P | |
| m16 | a/α | P | P | P | P | |
| m2, m8 | a/α | S | P | S | S | |
| m7 | a/α | S | R | S | P | |
Derivatives with the same genetic background are shown in sequential order from top to bottom. See Results for further explanation. CAS, 5FC, FLU, and AmB stand for caspofungin, 5-fluorocytosin, fluconazole, and amphotericin B, respectively. S, R, and P denote increased susceptibility or sensitivity, decreased susceptibility, resistance, and no susceptibility change or parental, respectively. Two prominent patterns of susceptibility to antifungals are indicated by boldface. Note that Sor125(55) and Sor1210(60) were previously published under the names Sor55 and Sor60, respectively (15).
F6-RA, F6-RB, F6-RC, and F6-RD acquired Ch5 monosomy spontaneously (26).
Sor5 is an exceptional Sou+ mutant having a deletion of one Ch5 copy instead of Ch5 monosomy (36). Spontaneous m mutants have Ch5 lost and reduplicated (m5 and m6), as well as a single (m15) or multiple (m2, m7, m8, and m16) trisomic chromosomes or other chromosomal rearrangements outside Ch5 (2, 23, 37).
We also examined the 3153A Ch5 monosomic mutants F6-RA, F6-RB, F6-RC, and F6-RD that were derived from mutants rescued in the presence of toxic 5-fluoroorotic acid by the amplification of an unknown portion of one copy of Ch5. The mutants F6-RA, F6-RB, F6-RC, and F6-RD spontaneously lost an enlarged Ch5 and retained a normal Ch5, acquiring Ch5 monosomy in the absence of sorbose exposure. The chromosome condition of those strains was also determined by precise separation of chromosomes with PFGE (26).
In this work, Ch5 monosomy of each 3153A mutant was additionally confirmed by demonstrating the loss of heterozygosity at the MTL locus carried on Ch5. For this analysis we used the colony PCR method with primers specific for the MTLa or MTLα locus (Table 1), as exemplified by the representative mutant Sor41 in Fig. 1.
Fig 1.
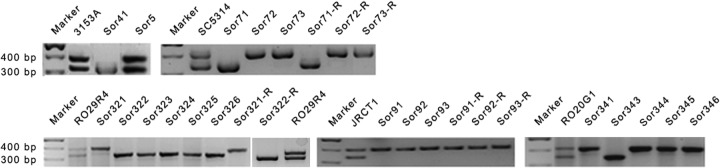
Products of PCR amplification obtained with total DNA of each strain and primers specific for the MTLa or MTLα locus. Presented are parental strains RO29R4, JRCT1, RO20G1, and SC5314 and their mutants, as indicated. Also presented are control amplifications with 3153A and its representative mutant Sor41, monosomic for Ch5 (2), as well as its exceptional mutant, Sor5, that became resistant to sorbose due to a large deletion on one of two Ch5 copies (36). Note that two bands, the upper band for MTLα on Ch5b and the lower band for MTLa on Ch5a, are amplified from DNA of parental strains or the exceptional Sor5, while one band of either kind is amplified from the mutants. See Table 2 for the relationships between strains.
Finally, using the approaches described above, we also generated Ch5 monosomic mutants from three clinical isolates, RO29R4, JRCT1, and RO20G1 (Materials and Methods), as well as from the sequencing strain SC5314 (http://www.candidagenome.org/) (Table 2). Each newly derived mutant was confirmed for the loss of heterozygosity at the MTL locus on Ch5 as described above (Fig. 1). In four representative mutants, Sor91, Sor321, Sor322, and Sor343, having three different genetic backgrounds (Table 2), Ch5 monosomy was confirmed as the only aneuploid change with the aCGH approach (Fig. 2). A total of 8 monosomic mutants, derivatives of the strains RO29R4, JRCT1, and SC5314, were used to create multiple series of related strains, parental→Ch5 monosomy→Ch5 reduplicated, similar to the 3153A series (Table 2).
Fig 2.
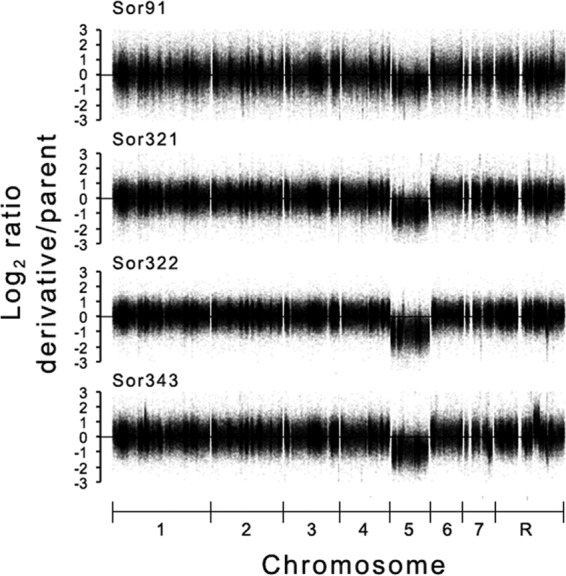
DNA profiling of the monosomic Ch5 by tiling aCGH. The aCGH log2 of the derivative/parental ratio for each of the tiling probes on each chromosome is plotted according to the chromosomal position of the probe. Thus, every point of the plot corresponds to a probe. The horizontal line at position 0 on the ordinate corresponds to no DNA change. Note the global diminution of the DNA on Ch5b but not on the other chromosomes in the derivatives Sor91, Sor321, Sor322, and Sor343 (Table 2).
Overall, 57 strains were used in this study, including 5 parental strains with two normal Ch5 copies representing 5 different genetic backgrounds, 29 strains with Ch5 monosomy, 11 strains with reduplicated Ch5, and 8 control strains that spontaneously acquired various chromosomal alterations. Of the 29 monosomic strains, 14 were derived from 3 clinical isolates and 15 were derived from 2 laboratory strains (Table 2).
Ch5 copy number controls susceptibility to caspofungin, 5-fluorocytosine, fluconazole, and amphotericin B.
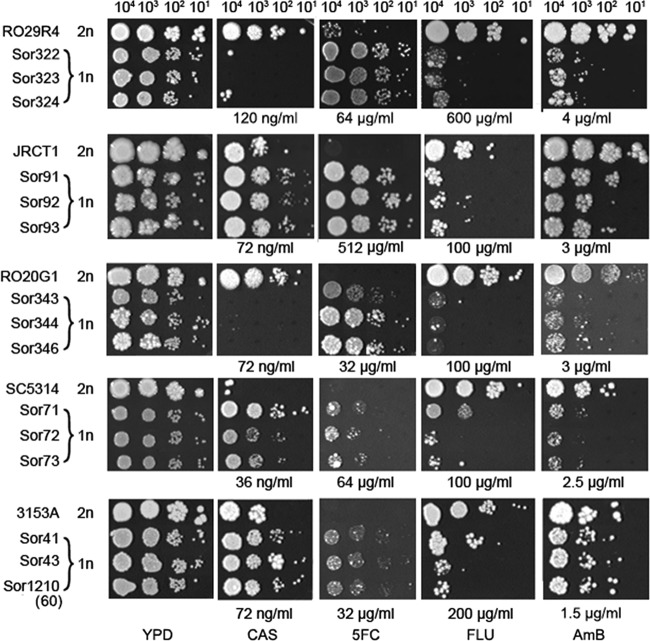
As expected from our previously published studies (2, 11) and as described above, all sorbose-generated Sou+ mutants became monosomic for Ch5, in contrast to Sou− parentals disomic for Ch5. The strains with remaining duplicated Ch5 reverted, as expected, to Sou−. All strains either monosomic or disomic for Ch5 (Table 2) were tested for the ability to grow in the presence of the antifungals caspofungin, 5-fluorocytosine, fluconazole, and amphotericin B. The test was performed with a plate spot assay, as shown in Fig. 3. Each measurement was repeated up to four times. Because different strains have different intrinsic susceptibilities for the same drug, we conducted pilot experiments with a range of concentrations for each drug that started from close to 0 and gradually increased. For each drug, we identified several optimal drug concentrations that revealed growth differences between each parental strain and its derivatives based on the number of growing spots (see Fig. S1 to S4 in the supplemental material). For example, no growth in any of the four spots on the plate (Fig. 3, Sor322 versus RO29R4 in the presence of caspofungin), or greatly reduced growth in first two spots from left to right and no growth in the two remaining spots (Fig. 3, Sor71 versus SC5314 in the presence of fluconazole), is considered suppressed growth compared to that of the parentals. The presence of a few colonies, instead of confluent growth, in the two leftmost spots was interpreted as resulting from mutations that confer resistance; the presence of faster-growing papillae over the confluent growth was interpreted as resulting from compensatory mutations allowing growth. The growth difference between a parental strain and its derivative was attributed to a difference in drug susceptibility (Table 2). The agar-based spot assay has been reported to be a precise and reproducible method for determining relative caspofungin susceptibility (33). Our experiments also confirmed the spot assay as a precise and reproducible method to determine relative caspofungin susceptibility as well as other relative antifungal susceptibilities (see Fig. S1 to S4 in the supplemental material).
Fig 3.
Examples of susceptibility of the mutants with monosomic Ch5 to antifungal agents compared to parental strains. Shown is the spot assay for growth on control YPD medium as well as on YPD medium in the presence of CAS, 5FC, FLU, or AmB. Drug concentrations are indicated. From left to right, 104, 103, 102, and 101 cells were spotted on each plate and incubated at 37°C for up to 8 days. Control YPD plates were incubated for 2 days. Strains are indicated on the left. Names of the derivatives are shown under the names of the corresponding parentals, RO29R4, JRCT1, RO20G1, SC5314, and 3153A (Table 2). The monosomy, 1n, or disomy, 2n, condition of Ch5 is indicated. Larger papillae are interpreted as arising from mutations compensating for growth, as these appeared on supplemented as well as control media. Note that Ch5 monosomic mutants demonstrated less growth and smaller colonies than their normal parents on the control YPD medium. However, this pattern is reversed in the presence of toxic agent CAS or 5FC or enhanced in the presence of toxic agent FLU or AmB.
We found that monosomy of either one of two Ch5 copies was closely correlated with growth changes in the presence of multiple drugs compared to the parental strain. Specifically, Ch5 monosomy decreased susceptibility to caspofungin in 9 out of 29 of the tested mutants, increased susceptibility in 17 mutants, and did not affect susceptibility in 3 mutants, as shown in Fig. 3 and summarized in Table 2. For more results, see Fig. S1 to S4 in the supplemental material. All monosomic mutants showed increased resistance (indicated in Table 2 as R) to 5-fluorocytosine. Most of the monosomic mutants showed increased sensitivity (S) to both fluconazole and amphotericin B, except 6 out of 8 of the 3153A mutants that retained parental (P)-like growth in the presence of amphotericin B. The MTLa or MTLα locus, each carried on one Ch5 copy, did not seem to segregate with the fluconazole phenotype, as previously debated (8, 34, 35). A total of 22 out of 29 mutants, or 76%, revealed two prominent patterns of susceptibility, R/R/S/S or S/R/S/S, referring to caspofungin/5-fluorocytosine/fluconazole/amphotericin B susceptibilities (Table 2).
In 11 analyzed derivatives with reduplicated Ch5, the susceptibility levels predominantly reverted to the parental levels. Sor125(55)-R acquired a caspofungin phenotype somewhere between the parental and monosomic mutant phenotype, whereas Sor321-R acquired better growth than the parental strain grown in the presence of caspofungin.
To further understand how Ch5 copy number controls drug susceptibilities, we used an exceptional Sou+ mutant, Sor5, that arose from 3153A via a large deletion on one Ch5 copy instead of Ch5 monosomy (see Fig. 1 in reference 36) yet showed no susceptibility changes (Table 2; also see Fig. S1 to S4 in the supplemental material). These results suggest that exposure to sorbose alone or lack of the portion of the chromosome described above is not sufficient to induce susceptibility changes; rather, Ch5 monosomy or a larger deletion is required. Consistently, the Ch5 monosomic mutants F6-RA, F6-RB, F6-RC, and F6-RD, which were derived from 3153A without exposure to sorbose, showed an S/R/S/S pattern that is more characteristic of 12 other monosomic sorbose-generated mutants (Table 2). These findings also suggest that the method of obtaining Ch5 monosomy is irrelevant for phenotypes controlled by Ch5 copy number. We next extended our antifungal test to a total of 7 control mutants (m2, m5, m6, m7, m8, m15, and m16) that were derived spontaneously from 3153A during cultivation in rich liquid medium and showed various chromosome rearrangements, as demonstrated with PFGE (2, 23, 37). Mutants m5 and m6, which lost one copy of Ch5 and reduplicated the remaining Ch5, showed the P/P/P/P phenotype, similar to sorbose-generated monosomic mutants with reduplicated Ch5. Mutant m16, which lost one copy of Ch5 and reduplicated the remaining copy, gained an uncharacterized extra chromosome, and a shortened one, chromosome R, also showed the P/P/P/P phenotype. Mutant m15 with trisomy presumably of Ch7, showed strong repression of growth in the presence of caspofungin, i.e., the S/P/P/P phenotype. Mutant m2, with poorly characterized rearrangement of chromosomes, and mutant m8, with putative trisomy of Ch3, Ch6, and Ch7, both showed strong repression of growth in the presence of caspofungin or fluconazole and no change of growth in the presence of other antifungals, i.e., the S/P/S/P phenotype. Finally, m7, with putative monosomy of Ch7, trisomy of Ch3 and Ch4, and trisomy of an unknown chromosome, showed strong repression of growth in the presence of caspofungin or fluconazole as well as decreased susceptibility to 5-fluorocytosine, i.e., the S/R/S/P phenotype. A characteristic feature of some of those mutants is strong repression of growth in the presence of some antifungals. This response seems to be different from that of monosomic mutants. It also seems, based on the example of m7, that an increased number of rearranged chromosomes results in an increase of phenotypic disturbances.
We set out to confirm the results with an agar-based spot assay by a quantitative broth microdilution method that allows determination and comparison of MICs (Materials and Methods). We used a total of three sequential series of strains, parental→Ch5 monosomy→Ch5 reduplication, with each series representing a different genetic background, RO29R4, JRCT1, and SC5314 (Table 3). We also used the strain 3153A and its four Ch5 monosomic derivatives (Table 3). Each strain was tested for growth in the presence of caspofungin, 5-fluorocytosine, fluconazole, and amphotericin B, and results are presented as growth inhibition curves (see Fig. S5 in the supplemented material). MICs were determined and are presented in Table 3. We found that the spot assays were confirmed in every case.
Table 3.
Summary of MICs in series of successive strains alternating between 1 and 2 Ch5 copiesa
| Ch5 condition | Strain | MIC (μg/ml) |
|||
|---|---|---|---|---|---|
| CAS | 5FC | FLU | AmB | ||
| Parental | RO29R4 | 1 | 0.5 | 0.125 | 0.5 |
| Monosomic | Sor322 | 0.125 | 2 | 0.06 | 0.125 |
| Monosomic | Sor321 | 1 | 2 | 0.03 | 0.125 |
| Duplicated | Sor322-R, 321-R | 1 | 0.5 | 0.125 | 0.5 |
| Parental | JRCT1 | 1 | 1 | 0.125 | 0.25 |
| Monosomic | Sor91 | 2 | 8 | 0.06 | 0.125 |
| Duplicated | Sor91-R | 1 | 8 | 0.125 | 0.25 |
| Parental | SC5314 | 0.5 | 0.125 | 0.125 | 0.5 |
| Monosomic | Sor71 | 2 | 0.25 | 0.06 | 0.125 |
| Duplicated | Sor71-R | 0.5 | 0.125 | 0.125 | 0.5 |
| Parental | 3153A | 1 | 0.125 | 0.25 | 0.25 |
| Monosomic | Sor126, Sor128 | 1 | 0.5 | 0.03 | 0.25 |
| Monosomic | Sor41, Sor43 | 2 | 0.5 | 0.03 | 0.25 |
See Table 2 for spot assay results of the strains and for abbreviations.
Ch5 copy number controls susceptibility to cell wall-damaging agents.
We asked whether Ch5 monosomy affects the structure of the cell wall. For this, monosomic or reduplicated strains were grown, in three independent experiments, in the presence of a cell wall-damaging agent: caffeine, sodium dodecyl sulfate, or hygromycin B. Strains Sor5, m2, m5, m6, m7, m8, m15, and m16 (Table 2), derivatives of 3153A that acquired various aneuploidies, were tested in a single experiment. As shown in Fig. 4, all 29 monosomic mutants tested became more sensitive to caffeine, sodium dodecyl sulfate, or hygromycin B than parental 3153A (complete results are provided in Fig. S6 and Table S1 in the supplemental material). All 11 strains with duplicated Ch5 tested showed no difference from parentals, i.e., the reversed phenotype, with one exception. Some of the control strains showed a mixed response: at least 7 of 15 strains tested showed no growth difference from parental 3153A.
Fig 4.
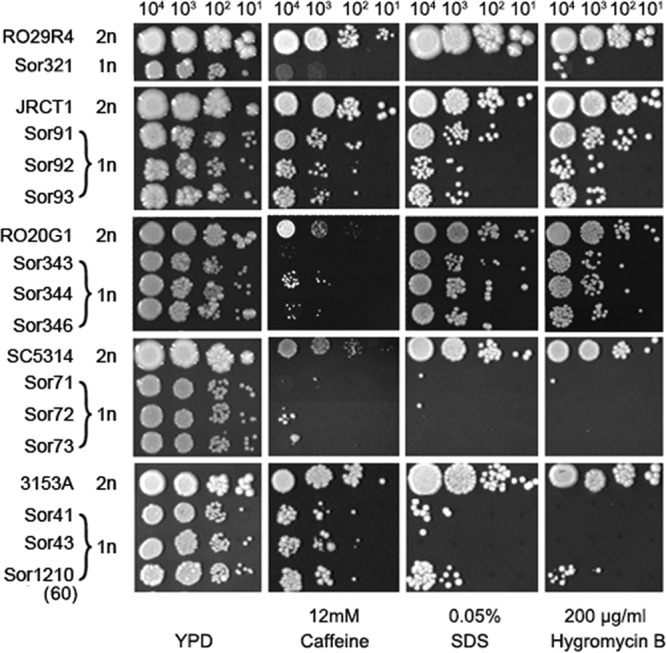
Susceptibilities of the mutants with monosomic Ch5 to cell wall-damaging agents compared to parental strains. Shown is the spot assay for growth on control YPD medium as well as on YPD medium in the presence of caffeine, SDS, hygromycin B, and Congo red. Drug concentrations are indicated. From left to right, 104, 103, 102, and 101 cells were spotted on each plate and incubated at 37°C for 2 to 5 days. Control YPD plates were incubated for 2 days. Strains are indicated on the left (Table 2). The monosomy, 1n, or disomy, 2n, of Ch5 is indicated.
In addition, monosomic mutants, reduplicated strains, or control strains derived from 3153A were tested to determine if their cell wall was resistant to zymolyase, which specifically hydrolyzes 1,3-β bonds in glucan. For this purpose, we conducted a time course experiment. We incubated cell suspensions with 50 μg/ml of zymolyase 20T at 37°C and measured the absorbance at time intervals of 0.5 h with a Spectra Max M5 microplate fluorescence reader. All 16 monosomic mutants tested acquired resistance to zymolyase, while all 10 reduplicated strains tested had no increase in resistance (Fig. 5; also see Table S1 in the supplemental material). Importantly, all of the control strains analyzed, Sor5, m2, m5, m6, m7, m8, m15, and m16, retained a level of zymolyase resistance similar to that of the parental strain. Zymolyase resistance has been suggested to result from changes in the structure of the glucan network (38, 39) or from masking, leading to less zymolase-mediated cleavage of 1,3-β-glucan due to changes in the external mannoprotein layer (40, 41).
Fig 5.
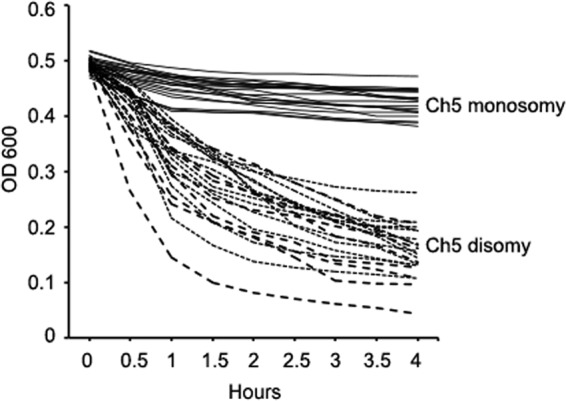
Survival of strains monosomic or disomic for Ch5 in the presence of zymolyase. Ch5 monosomic or disomic strains are indicated with continuous or dotted lines, respectively. The control derivatives of 3153A carrying various chromosome alterations are indicated with dashed lines. Ch5 ploidy is also indicated. The following monosomic mutants are included: Sor321, Sor322, Sor91, Sor92, Sor93, Sor71, Sor72, Sor73, Sor122, Sor123, Sor41, Sor43, Sor1210(60), Sor126, Sor128, and Sor125(55). The following reduplicated strains are included: Sor321-R, Sor91-R, Sor92-R, Sor93-R, Sor71-R, Sor72-R, Sor73-R, Sor125Sor125(55)-R, Sor122-R, and Sor123-R. The following control derivatives of 3153A are included: Sor5, m2, m5, m6, m7, m8, m15, and m16. The following parental strains are included: RO29R4, JRCT1, SC5314, and 3153A. See Table 2 for the relationships between strains. Each data point for monosomic mutants, reduplicated strains, or parentals is averaged from three independent experiments. Each data point for control derivatives of 3153A is averaged from two independent experiments.
Ch5 copy number controls the level of 1,3-β-glucan and chitin of the cell wall and the cellular level of ergosterol.
We asked whether resistance to zymolyase, which hydrolyzes 1,3-β bonds of glucan (42), could be due to a decreased number of 1,3-β bonds within the cell wall glucan. Using aniline blue, which fluoresces when bound to 1,3-β bonds, we determined fluorescence intensities from three independent experiments and calculated the results as means ± standard deviations (SD). We found that fluorescence intensity decreased for mutants Sor125(55) and Sor1210 (43), 9.2 ± 1.3 and 9.6 ± 0.2, respectively, compared to that of the parental strain 3153A, 13.9 ± 0.4. This decrease corresponds to an approximately 1.5-fold decrease of 1,3-β-glucan in the cell wall.
We also asked whether a decrease in cell wall 1,3-β-glucan is accompanied by an increase in the cell wall chitin by measuring the absorbance of glucosamine released from hydrolyzed cell walls (44). We compared 16 monosomic strains to 5 parental and 9 reduplication strains, and we found that chitin levels in monosomic strains increased significantly, ranging from 112 to 169% compared to the parentals (P < 0.05 by unpaired t tests). However, no significant difference was found in revertant strains with a reduplicated Ch5 compared to their parentals (Table 4). Similarly, control strains with various chromosomal alterations (Sor5, m2, m5, m6, m7, m8, and m16) retained the parental 3153A level of chitin (Table 4).
Table 4.
Cell wall content of chitin in strains monosomic or disomic for Ch5
| Ch5 condition | Strain | % Chitina (means ± SD) |
|---|---|---|
| Parental | RO29R4 | 6.5 ± 0.2 |
| Monosomic | Sor321, Sor322 | 9.7 ± 1.5, 9.2 ± 1.1 |
| Duplicated | Sor321-R | 6.7 ± 1.0 |
| Parental | JRCT1 | 6.8 ± 0.9 |
| Monosomic | Sor91, Sor92, Sor93 | 13.1 ± 0.6, 11.8 ± 0.9, 11.1 ± 0.6 |
| Duplicated | Sor91-R, Sor92-R, Sor93-R | 7.1 ± 0.1, 7.0 ± 0.2, 7.3 ± 1.1 |
| Parental | RO20G1 | 6.8 ± 0.3 |
| Monosomic | Sor341, Sor343 | 7.6 ± 0.2, 8.7 ± 0.3 |
| Parental | SC5314 | 5.3 ± 0.1 |
| Monosomic | Sor71, Sor72, Sor73 | 7.0 ± 0.3, 7.1 ± 0.6, 7.3 ± 0.1 |
| Duplicated | Sor71-R, Sor72-R, Sor73-R | 4.9 ± 0.3, 5.3 ± 0.2, 5.5 ± 0.2 |
| Parental | 3153A | 5.1 ± 0.1 |
| Monosomic | Sor41, Sor43, Sor1210, Sor125(55), Sor122, Sor123 | 7.0 ± 0.3, 6.8 ± 0.5, 6.8 ± 0.1, 6.8 ± 0.4, 9.3 ± 0.6, 8.1 ± 0.1 |
| Duplicated | Sor122-R, Sor123-R | 6.1 ± 0.4, 5.4 ± 0.8 |
| Control derived from 3153A | Sor5 | 5.2 ± 0.1 |
| m2, m8, m7 | 4.8 ± 0.5, 4.9 ± 0.1, 5.0 ± 0.4 | |
| m5, m6, m16 | 5.2 ± 0.6, 5.2 ± 0.1, 5.0 ± 0.3 |
The amount of chitin per dry weight of the cell wall.
We also asked whether the amount of ergosterol changed in monosomic mutants, because the level of ergosterol has been shown to correlate with resistance to fluconazole or amphotericin B (45). The amount of cellular ergosterol was determined from three independent experiments and calculated as means ± standard deviations with two parental strains RO29R4 (1.10% ± 0.01%) and 3153A (1.14% ± 0.14%), as well as with one monosomic mutant of each: Sor321 (0.43% ± 0.02%) and Sor125(55) (0.48% ± 0.02%), respectively. Our data showed an approximately 2-fold decrease in the level of ergosterol in both monosomic mutants compared to their parental strains.
The FKS1 (GSC1) gene does not harbor point mutations and is not upregulated in Ch5 monosomic mutants.
The C. albicans FKS1 (GSC1) (orf19.2929) gene, residing on Ch1 and encoding the catalytic subunit of the 1,3-β-glucan synthase (22), harbors frequent mutations in regions HS1 and HS2. Those mutations render decreased susceptibility to caspofungin in clinical isolates (46, 47). We addressed the question of whether the decreased susceptibility to caspofungin in many Ch5 monosomic mutants is due to mutations in FKS1 (GSC1). We conducted PCR amplifications with primers for region HS1 or HS2 of FKS1 (GSC1) (Table 1) from genomic DNA that was extracted from six pairs of parent/Ch5 monosomic derivative strains: JRCT1/Sor91, RO29R4/Sor321, RO29R4/Sor322, RO20G1/Sor343, 3153A/Sor1210 (43), and SC5314/Sor71 (Table 2). No DNA mutations were detected in monosomic mutants compared to their parentals. No gene expression change of FKS1 (GSC1) was detected in three comparisons of monosomic strains to their parentals (see Table S2 in the supplemental material).
Expression level of some established genes that are implicated in chitin synthesis or drug-resistant mechanisms.
We next determined the expression levels of two genes involved in chitin synthesis, CHS7 (orf19.2444) on Ch1 and CHT2 on Ch5 (orf19.3895), in the monosomic mutant Sor321 and its duplicated derivative Sor321-R. For this purpose, we used the semiquantitative RT-PCR method (examples of gels are presented in Fig. S7 in the supplemental material). We conducted three (for CHS7) or four (for CHT2) independent experiments. Expression levels were determined as mutant Sor321/parental RO29R4 ratios or reduplication derivative Sor321-R/parental RO29R4 ratios and calculated as mean ratios ± standard deviations (Table 5). In the monosomic mutant, we found slight but statistically significant upregulation (1.4-fold) of CHS7, which encodes the protein required for wild-type chitin synthase III activity. In contrast, CHT2, encoding glycosylphosphatidylinositol-linked chitinase, was 2-fold downregulated. As expected, no expression changes occurred in the strain having two Ch5 copies. Previously generated data using a less precise expression array method showed consistent expression changes of CHS7 and CHT2 in monosomic strain Sor125(55) (Table 5).
Table 5.
Monosomic mutant/parent or duplicated derivative/parent expression ratios of two genes involved in metabolism of cell wall chitina
| Gene | Mutant/parent ratio (means ± SD) and detection method |
||
|---|---|---|---|
| Monosomic Ch5 |
Duplicated Ch5 (RT-PCR; Sor321-R/RO29R4) | ||
| RT-PCR (Sor321/RO29R4) | Arrays [Sor125(55)/3153A] | ||
| CHS7 | 1.4 ± 0.06 | 1.5 ± 0.44 | 1.1 ± 0.04 |
| CHT2 | 0.4 ± 0.04 | 0.3 ± 0.23 | 0.9 ± 0.03 |
Gene expression was determined by the monosomic mutant/parent or duplicated derivative/parent mean ratio from three (CHS7) or four (CHT2) independent experiments using semiquantitative RT-PCR or expression arrays in the indicated pairs of strains. For strain information, see Table 2.
We also determined the expression level of TAC1 (orf19.3188) on Ch5, an activator of CDR genes for fluconazole efflux pumps on Ch3, and of ERG11 (orf19.922) on Ch5, encoding the target for fluconazole. Using four pairs of strains, we found that expression levels of TAC1 and ERG11 on Ch5 are partially compensated for by the dose and mutant/parent expression ratios, ranging from 0.6 to 0.8 (see Table S2 in the supplemental material). CDRs seemed to be slightly downregulated at least in some mutants. Interestingly, the MDR1 gene (orf19.5604) encoding multidrug efflux pumps had no change in expression.
Finally, we evaluated the expression level of FUR1 (orf19.2640) on Ch5, a gene in which decreased activity due to mutations is associated with flucytosine resistance in clade I clinical isolates (48), and found that the expression of this gene was fully compensated for at the diploid level.
DISCUSSION
Earlier studies with the fungi Neurospora crassa and Aspergillus nidulans revealed that toxic properties of the sugar sorbose are due to inhibition of the 1,3-β-glucan synthase activity, which kills cells by preventing the synthesis of the 1,3-β-glucan component of the cell wall (16, 19). This is similar to the action of the major antifungal caspofungin and other drugs from the echinocandin class that inhibit fungal 1,3-β-glucan synthases, including C. albicans synthase (20–22). Sorbose, echinocandin B, and papulocandin B were shown to be noncompetitive inhibitors of the 1,3-β-glucan synthase activity in N. crassa (49). We have previously found that monosomy of C. albicans Ch5 is a major mechanism to rescue cells that are dying due to sorbose exposure (see the introduction). Thus, cells monosomic for Ch5 acquire the ability to utilize sorbose and to resist sorbose.
In this study, we continued investigating the phenotypes controlled by Ch5 copy number. We designed experiments to include the related monosomic and disomic strains, the latter serving as controls, representing five different genetic backgrounds. The purpose of such a design was to enable us to study phenotypes controlled by Ch5 copy number regardless of the particular properties of a single genetic background. The growth of cells was rigorously standardized to compare cells in various tests at the same physiological state. By growing cells in the presence of the cell wall-damaging agents caffeine, hygromycin B, sodium dodecyl sulfate, and zymolyase and by measuring levels of 1,3-β-glucan and chitin, we found that monosomy of chromosome 5 causes cell wall remodeling, a decrease of 1,3-β-glucan, and an increase of chitin. Ch5 monosomy also causes a decrease of cellular ergosterol. Similar to negative regulation of growth on sorbose, our data imply that Ch5 carries genetic factors for negative control of chitin biosynthesis. Our data also suggest that Ch5 carries factors for positive control of glucan and ergosterol.
We inquired about some mechanisms controlling the level of the cell wall chitin in our monosomic mutants. Fungal cell wall chitin metabolism involves synthesis by chitin synthases and hydrolysis by chitinases. In C. albicans, of the four C. albicans genes for chitin synthesis, CHS7 on Ch1 is a key regulator (50). Its Saccharomyces cerevisiae ortholog CHS7 (YHR142W) is the only chitin synthase transcriptionally upregulated under all conditions in which chitin synthesis is increased (51). Using the RT-PCR method that can detect small transcriptional changes, we found that CHS7 was slightly upregulated in a representative Ch5 monosomic mutant but not in its derivative with reduplicated Ch5. Furthermore, mutations or downregulation of CHT2 and CHT3 (orf19.7586) was shown to be involved in the tolerance to echinocandins (52, 53). Because our expression profiling of the Ch5 monosomic mutant Sor125(55) indicated that CHT3 could be a silent gene while CHT2 on Ch5 could be downregulated (15), we extended the analysis to CHT2 and confirmed the expected 2-fold downregulation on the monosomic Ch5 but not on the reduplicated Ch5. Previously, another laboratory found point mutations in CHT2 and CHT3 of a C. albicans laboratory mutant with decreased levels of 1,3-β-glucan and increased levels of chitin (53). The authors concluded that these mutations may affect the degradation of chitin, result in accumulation of chitin, and compensate for low 1,3-β-glucan content. We propose that the combined 2-fold downregulation of CHT2 and slight upregulation of CHS7 contribute to an increased deposition of chitin in the cell wall of Ch5 monosomic mutants.
As expected, Ch5 monosomy decreased susceptibility to caspofungin in some of the tested mutants. However, susceptibility increased in some other mutants and a minor fraction of mutants had no susceptibility change. Importantly, reduplication of Ch5 reversed susceptibility changes in all analyzed derivatives, predominantly to the parental level, confirming the overall role of Ch5 copy number in the regulation of caspofungin susceptibility. On a special note, our monosomic mutants were generated on sorbose medium or occurred spontaneously; hence, we did not select for caspofungin resistance.
We have previously shown that exposure of cells to toxic agents is highly mutagenic, resulting at least in increased levels of homologous recombination and chromosome instability (12, 26, 54). On the other hand, control by the monosomic Ch5 is a complex process involving, for example, a balance between a 2-fold reduction of gene copy number and a counteracting expression compensation for the dose that upregulates many Ch5 genes to disomic or close to disomic levels (see the introduction). Because we previously identified the CSU genes on Ch5 that encode negative regulators of growth on sorbose (see the introduction), we favor a model of Ch5 also carrying a CSC (for control of susceptibility to caspofungin) gene(s) to negatively regulate the susceptibility to CAS. The sense CSC transcripts could be fine-tuned by counteracting antisense transcripts, similar to ASU antisense transcripts that are imbedded in and regulate the CSU transcripts. Complex regulation is expected to have various outcomes that are more frequent in unstable genetic backgrounds. Indeed, the laboratory strain 3153A, which had a history of chromosome instability before being studied in our laboratory (55) and which could still retain a certain level of instability higher than that in other strains, showed the most variable caspofungin phenotype. Taken together, multiple known and unknown factors associated with control by Ch5 monosomy could be involved in determining levels of caspofungin susceptibility.
One generally recognized mechanism of caspofungin resistance found in clinical isolates that are associated with therapeutic failure consists of point mutations in FKS1 (GSC1) on Ch1, the gene encoding the catalytic subunit of the synthase for the cell wall 1,3-β-glucan (47). Such mutations were not found in our monosomic mutants. We did not find any change in transcription of FKS1 (GSC1) either. In some clinical isolates that have increased MICs but are not associated with therapeutic failure or in some laboratory strains, caspofungin resistance was also correlated with an increased level of cell wall chitin and cell wall thickness. In some of these strains, increased levels of chitin were coupled with decreased levels of glucan. Elevated chitin levels were proposed to contribute to cell wall rigidity and also to increased caspofungin resistance in C. albicans or increased resistance to echinocandins in other fungi (21, 44, 46, 47, 56–58). Our two representative monosomic mutants, either more resistant or more sensitive to caspofungin, each showed decreased levels of 1,3-β-glucan in the cell wall. Other monosomic mutants are also expected to have decreased levels of glucan, as they all became resistant to zymolyase, similar to the two representative monosomic mutants. Our data clearly indicated that Ch5 monosomy is associated with cell wall remodeling, resulting in decreased levels of glucan and increased levels of chitin. However, the genetic factors controlling chitin levels and caspofungin laboratory resistance seem to be different. The relationship between chitin level and susceptibility to caspofungin, as controlled by Ch5 copy number, requires further study.
To more fully understand the consequences of Ch5 monosomy, we tested our strains for susceptibility to other major antifungals, such as 5-fluorocytosine, fluconazole, and amphotericin B, each representing the major class of antifungals 5-fluorinated cytosine, azoles, and polyenes, respectively. We found that Ch5 monosomy resulted in decreased susceptibility to 5-fluorocytosine accompanied by increased susceptibility to fluconazole and amphotericin B. Interestingly, one proposed mechanism of resistance to 5-fluorocytosine is downregulation of the FUR1 gene (59), which resides on Ch5. However, FUR1 is transcriptionally compensated for to the disomic level in Ch5 monosomic mutants, which strongly suggests that a different mechanism of resistance is involved. One such mechanism could be a 2-fold decrease of the copy number of a single or multiple putative regulatory genes on Ch5, whose products need to be diminished in order for 5-fluorocytosine susceptibility to decrease. We cannot exclude differences in expression of other genes that affect transport or metabolism of 5-fluorocytosine (43). The duplication of the Ch5 left arm was previously shown to confer fluconazole resistance due to an increased copy number of two Ch5 genes, TAC1, a positive regulator of CDR genes for fluconazole efflux (6, 10) on Ch3, and ERG11, encoding the target of azoles (60, 61). In our monosomic mutants, an opposite rearrangement that produces a 2-fold decrease in the copy number of TAC1 and ERG11 produced the expected and dramatic increase in susceptibility to fluconazole. However, no obvious 2-fold downregulation of TAC1 and ERG11 on the monosomic Ch5 was observed, as could be expected, and these genes seem to partially compensate for the DNA loss. CDRs that are activated by TAC1 were not substantially downregulated. However, a slight downregulation of TAC1 and, subsequently, CDRs might be sufficient for the strong suppression of growth in the presence of fluconazole. Alternatively, another CDR-controlled efflux mechanism could be involved. This mechanism, however, does not involve MDR1 encoding an efflux pump of the major facilitator superfamily on Ch6 (62), because MDR1 transcription in the monosomic derivatives did not change, i.e., transcription remained at the diploid level. In addition, our data indicate that monosomic mutants acquired increased susceptibility to amphotericin B, which might be explained by a 2-fold decrease in ergosterol, which is the target of amphotericin B.
In summary, we found that Ch5 copy number controls cell wall metabolism and ergosterol biosynthesis. Ch5 monosomy causes increased levels of cell wall chitin and decreased levels of cell wall 1,3-β-glucan and cellular ergosterol. Chitin increases are associated with Ch5 monosomy but not with aneuploidy of other chromosomes. A partial explanation for this association could be attributed to 2-fold downregulation of metabolic CHT2 on Ch5 that hydrolyzes chitin, as well as slight upregulation of CHS7 on Ch1 that is involved in chitin synthesis. Importantly, we demonstrate that Ch5 monosomy results in changing susceptibility to toxic agents, among them being four major antifungals, caspofungin, 5-fluorocycotine, fluconazole, and amphotericin B, each representing a different class of antifungals acting upon the cell wall, plasma membrane, or RNA synthesis. Ch5 monosomy can be associated with resistance to caspofungin, an important drug for the treatment of candidiases.
Supplementary Material
ACKNOWLEDGMENTS
We thank Damian Krysan for providing some reagents and sharing instruments with us, Constantin Haidaris for providing some antifungals, Roche NimbleGen Inc. for support, and Merck Inc. for donating caspofungin. We thank Melanie Wellington for collecting clinical isolate JRCT1 from an AIDS patient. We thank Jeffrey Hayes, Mark Dumont, and Jeffrey Becker for reading the manuscript and stimulating discussions.
These studies were supported by The University of Rochester Funds and R01 grant GM012702.
Footnotes
Published ahead of print 29 July 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00516-13.
REFERENCES
- 1.Rustchenko E, Sherman F. 2003. Genetic instability of Candida albicans, p 723–776 In Howard DH, Fungi pathogenic for humans and animals, 2nd ed. Marcel Dekker, Inc., New York, NY [Google Scholar]
- 2.Rustchenko E. 2007. Chromosome instability in Candida albicans. FEMS Yeast Res. 7:2–11 [DOI] [PubMed] [Google Scholar]
- 3.Rustchenko E. 2008. Specific chromosome alterations of Candida albicans: mechanism for adaptation to pathogenicity, p 197–212 In Nombela C, Cassel G, Baquero FGutí, errez-Fuentes J. (ed), Evolutionary biology of bacterial and fungal pathogens. ASM Press, Washington, DC [Google Scholar]
- 4.Ahmad A, Kabir MA, Kravets A, Andaluz E, Larriba G, Rustchenko E. 2008. Chromosome instability and unusual features of some widely used strains of Candida albicans. Yeast 25:433–448 [DOI] [PubMed] [Google Scholar]
- 5.Selmecki A, Forche A, Berman J. 2010. Genomic plasticity of the human fungal pathogen Candida albicans. Eukaryot. Cell 9:991–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coste AT, Karababa M, Ischer F, Bille J, Sanglard D. 2004. TAC1, transcriptional activator of CDR genes, is a new transcription factor involved in the regulation of Candida albicans ABC transporters CDR1 and CDR2. Eukaryot. Cell 3:1639–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diogo D, Bouchier C, d'Enfert C, Bougnoux ME. 2009. Loss of heterozygosity in commensal isolates of the asexual diploid yeast Candida albicans. Fungal Genet. Biol. 46:159–168 [DOI] [PubMed] [Google Scholar]
- 8.Legrand M, Lephart P, Forche A, Mueller FM, Walsh T, Magee PT, Magee BB. 2004. Homozygosity at the MTL locus in clinical strains of Candida albicans: karyotypic rearrangements and tetraploid formation. Mol. Microbiol. 52:1451–1462 [DOI] [PubMed] [Google Scholar]
- 9.Selmecki A, Forche A, Berman J. 2006. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science 313:367–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Selmecki A, Gerami-Nejad M, Paulson C, Forche A, Berman J. 2008. An isochromosome confers drug resistance in vivo by amplification of two genes, ERG11 and TAC1. Mol. Microbiol. 68:624–641 [DOI] [PubMed] [Google Scholar]
- 11.Janbon G, Sherman F, Rustchenko E. 1998. Monosomy of a specific chromosome determines l-sorbose utilization: a novel regulatory mechanism in Candida albicans. Proc. Natl. Acad. Sci. U. S. A. 95:5150–5155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janbon G, Sherman F, Rustchenko E. 1999. Appearance and properties of l-sorbose-utilizing mutants of Candida albicans obtained on a selective plate. Genetics 153:653–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greenberg JR, Price NP, Oliver RP, Sherman F, Rustchenko E. 2005. Candida albicans SOU1 encodes a sorbose reductase required for l-sorbose utilization. Yeast 22:957–969 [DOI] [PubMed] [Google Scholar]
- 14.Ahmad A, Kravets A, Rustchenko E. 2012. Transcriptional regulatory circuitries in the human pathogen Candida albicans involving sense-antisense interactions. Genetics 190:537–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kravets A, Qin H, Ahmad A, Bethlendy G, Gao Q, Rustchenko E. 2010. Widespread occurrence of dosage compensation in Candida albicans. PLoS One 5:e10856. 10.1371/journal.pone.0010856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahadevan PR, Tatum EL. 1965. Relationship of the major constituents of the Neurospora crassa cell wall to wild-type and colonial morphology. J. Bacteriol. 90:1073–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crocken B, Tatum EL. 1968. The effect of sorbose on metabolism and morphology of Neurospora. Biochim. Biophys. Acta 156:1–8 [DOI] [PubMed] [Google Scholar]
- 18.Elorza MV, Arst HN., Jr 1971. Sorbose resistant mutants of Aspergillus nidulans. Mol. Gen. Genet. 111:185–193 [DOI] [PubMed] [Google Scholar]
- 19.Mishra NC, Tatum EL. 1972. Effect of l-sorbose on polysaccharide synthetases of Neurospora crassa. Proc. Natl. Acad. Sci. U. S. A. 69:313–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurtz MB, Abruzzo G, Flattery A, Bartizal K, Marrinan JA, Li W, Milligan J, Nollstadt K, Douglas CM. 1996. Characterization of echinocandin-resistant mutants of Candida albicans: genetic, biochemical, and virulence studies. Infect. Immun. 64:3244–3251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stevens DA, Ichinomiya M, Koshi Y, Horiuchi H. 2006. Escape of Candida from caspofungin inhibition at concentrations above the MIC (paradoxical effect) accomplished by increased cell wall chitin; evidence for beta-1,6-glucan synthesis inhibition by caspofungin. Antimicrob. Agents Chemother. 50:3160–3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balashov SV, Park S, Perlin DS. 2006. Assessing resistance to the echinocandin antifungal drug caspofungin in Candida albicans by profiling mutations in FKS1. Antimicrob. Agents Chemother. 50:2058–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rustchenko-Bulgac EP, Sherman F, Hicks JB. 1990. Chromosomal rearrangements associated with morphological mutants provide a means for genetic variation of Candida albicans. J. Bacteriol. 172:1276–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rustchenko EP, Howard DH, Sherman F. 1994. Chromosomal alterations of Candida albicans are associated with the gain and loss of assimilating functions. J. Bacteriol. 176:3231–3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sherman F. 2002. Getting started with yeast. Methods Enzymol. 350:3–41 [DOI] [PubMed] [Google Scholar]
- 26.Wellington M, Rustchenko E. 2005. 5-Fluoro-orotic acid induces chromosome alterations in Candida albicans. Yeast 22:57–70 [DOI] [PubMed] [Google Scholar]
- 27.Clinical and Laboratory Standards Institute 2008. M27-A3 reference method for broth dilution antifungal susceptibility testing of yeasts: approved standard, 3rd ed. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 28.Munro CA, Selvaggini S, de Bruijn I, Walker L, Lenardon MD, Gerssen B, Milne S, Brown AJ, Gow NA. 2007. The PKC, HOG and Ca2+ signalling pathways coordinately regulate chitin synthesis in Candida albicans. Mol. Microbiol. 63:1399–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shedletzky E, Unger C, Delmer DP. 1997. A microtiter-based fluorescence assay for (1,3)-beta-glucan synthases. Anal. Biochem. 249:88–93 [DOI] [PubMed] [Google Scholar]
- 30.Arthington-Skaggs BA, Jradi H, Desai T, Morrison CJ. 1999. Quantitation of ergosterol content: novel method for determination of fluconazole susceptibility of Candida albicans. J. Clin. Microbiol. 37:3332–3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Young LY, Hull CM, Heitman J. 2003. Disruption of ergosterol biosynthesis confers resistance to amphotericin B in Candida lusitaniae. Antimicrob. Agents Chemother. 47:2717–2724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muir A, Harrison E, Wheals A. 2011. A multiplex set of species-specific primers for rapid identification of members of the genus Saccharomyces. FEMS Yeast Res. 11:552–563 [DOI] [PubMed] [Google Scholar]
- 33.Schuetzer-Muehlbauer M, Willinger B, Krapf G, Enzinger S, Presterl E, Kuchler K. 2003. The Candida albicans Cdr2p ATP-binding cassette (ABC) transporter confers resistance to caspofungin. Mol. Microbiol. 48:225–235 [DOI] [PubMed] [Google Scholar]
- 34.Rustad TR, Stevens DA, Pfaller MA, White TC. 2002. Homozygosity at the Candida albicans MTL locus associated with azole resistance. Microbiology 148:1061–1072 [DOI] [PubMed] [Google Scholar]
- 35.Pujol C, Messer SA, Pfaller M, Soll DR. 2003. Drug resistance is not directly affected by mating type locus zygosity in Candida albicans. Antimicrob. Agents Chemother. 47:1207–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kabir MA, Ahmad A, Greenberg JR, Wang YK, Rustchenko E. 2005. Loss and gain of chromosome 5 controls growth of Candida albicans on sorbose due to dispersed redundant negative regulators. Proc. Natl. Acad. Sci. U. S. A. 102:12147–12152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rustchenko-Bulgac EP. 1991. Variations of Candida albicans electrophoretic karyotypes. J. Bacteriol. 173:6586–6596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moreno I, Pedreno Y, Maicas S, Sentandreu R, Herrero E, Valentin E. 2003. Characterization of a Candida albicans gene encoding a putative transcriptional factor required for cell wall integrity. FEMS Microbiol. Lett. 12:159–167 [DOI] [PubMed] [Google Scholar]
- 39.Ritch JJ, Davidson SM, Sheehan JJ, Austriaco N. 2010. The Saccharomyces SUN gene, UTH1, is involved in cell wall biogenesis. FEMS Yeast. Res. 10:168–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Nobel JG, Klis FM, Ram A, Van Unen H, Priem J, Munnik T, Van Den Ende H. 1991. Cyclic variations in the permeability of the cell wall of Saccharomyces cerevisiae. Yeast 7:589–598 [DOI] [PubMed] [Google Scholar]
- 41.Klippel N, Cui S, Groebe L, Bilitewski U. 2010. Deletion of the Candida albicans histidine kinase gene CHK1 improves recognition by phagocytes through an increased exposure of cell wall beta-1,3-glucans. Microbiology 156:3432–3444 [DOI] [PubMed] [Google Scholar]
- 42.Kitamura K, Yamamoto Y. 1972. Purification and properties of an enzyme, zymolyase, which lyses viable yeast cells. Arch. Biochem. Biophys. 153:403–406 [DOI] [PubMed] [Google Scholar]
- 43.Loeffler J, Stevens DA. 2003. Antifungal drug resistance. Clin. Infect. Dis. 36(Suppl. 1):S31–S41 [DOI] [PubMed] [Google Scholar]
- 44.Walker LA, Munro CA, de Bruijn I, Lenardon MD, McKinnon A, Gow NA. 2008. Stimulation of chitin synthesis rescues Candida albicans from echinocandins. PLoS Pathog. 4:e1000040. 10.1371/journal.ppat.1000040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shapiro RS, Robbins N, Cowen LE. 2011. Regulatory circuitry governing fungal development, drug resistance, and disease. Microbiol. Mol. Biol. Rev. 75:213–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dannaoui E, Desnos-Ollivier M, Garcia-Hermoso D, Grenouillet F, Cassaing S, Baixench MT, Bretagne S, Dromer F, Lortholary O. 2012. Candida spp. with acquired echinocandin resistance, France, 2004–2010. Emerg. Infect. Dis. 18:86–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perlin DS. 2007. Resistance to echinocandin-class antifungal drugs. Drug Resist. Updat. 10:121–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dodgson AR, Dodgson KJ, Pujol C, Pfaller MA, Soll DR. 2004. Clade-specific flucytosine resistance is due to a single nucleotide change in the FUR1 gene of Candida albicans. Antimicrob. Agents Chemother. 48:2223–2227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quigley D, Selitrennikoff C. 1984. β(1-3)Glucan synthase of Neurosporn crassa: kinetic analysis of negative regulators. Exp. Mycol. 8:320–333 [Google Scholar]
- 50.Sanz M, Carrano L, Jimenez C, Candiani G, Trilla JA, Duran A, Roncero C. 2005. Candida albicans strains deficient in CHS7, a key regulator of chitin synthase III, exhibit morphogenetic alterations and attenuated virulence. Microbiology 151:2623–2636 [DOI] [PubMed] [Google Scholar]
- 51.Trilla JA, Duran A, Roncero C. 1999. Chs7p, a new protein involved in the control of protein export from the endoplasmic reticulum that is specifically engaged in the regulation of chitin synthesis in Saccharomyces cerevisiae. J. Cell Biol. 145:1153–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaneko Y, Ohno H, Kohno S, Miyazaki Y. 2010. Micafungin alters the expression of genes related to cell wall integrity in Candida albicans biofilms. Jpn. J. Infect. Dis. 63:355–357 [PubMed] [Google Scholar]
- 53.Drakulovski P, Dunyach C, Bertout S, Reynes J, Mallie M. 2011. A Candida albicans strain with high MIC for caspofungin and no FKS1 mutations exhibits a high chitin content and mutations in two chitinase genes. Med. Mycol. 49:467–474 [DOI] [PubMed] [Google Scholar]
- 54.Wang YK, Das B, Huber DH, Wellington M, Kabir MA, Sherman F, Rustchenko E. 2004. Role of the 14-3-3 protein in carbon metabolism of the pathogenic yeast Candida albicans. Yeast 21:685–702 [DOI] [PubMed] [Google Scholar]
- 55.Rustchenko EP, Curran TM, Sherman F. 1993. Variations in the number of ribosomal DNA units in morphological mutants and normal strains of Candida albicans and in normal strains of Saccharomyces cerevisiae. J. Bacteriol. 175:7189–7199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee KK, Maccallum DM, Jacobsen MD, Walker LA, Odds FC, Gow NA, Munro CA. 2012. Elevated cell wall chitin in Candida albicans confers echinocandin resistance in vivo. Antimicrob. Agents Chemother. 56:208–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Plaine A, Walker L, Da Costa G, Mora-Montes HM, McKinnon A, Gow NA, Gaillardin C, Munro CA, Richard ML. 2008. Functional analysis of Candida albicans GPI-anchored proteins: roles in cell wall integrity and caspofungin sensitivity. Fungal Genet. Biol. 45:1404–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jimenez-Ortigosa C, Aimanianda V, Muszkieta L, Mouyna I, Alsteens D, Pire S, Beau R, Krappmann S, Beauvais A, Dufrene YF, Roncero C, Latge JP. 2012. Chitin synthases with a myosin motor-like domain control the resistance of Aspergillus fumigatus to echinocandins. Antimicrob. Agents Chemother. 56:6121–6131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Balkis MM, Leidich SD, Mukherjee PK, Ghannoum MA. 2002. Mechanisms of fungal resistance: an overview. Drugs 62:1025–1040 [DOI] [PubMed] [Google Scholar]
- 60.Hitchcock CA, Dickinson K, Brown SB, Evans EG, Adams DJ. 1990. Interaction of azole antifungal antibiotics with cytochrome P-450-dependent 14 alpha-sterol demethylase purified from Candida albicans. Biochem. J. 266:475–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ji H, Zhang W, Zhou Y, Zhang M, Zhu J, Song Y, Lu J. 2000. A three-dimensional model of lanosterol 14alpha-demethylase of Candida albicans and its interaction with azole antifungals. J. Med. Chem. 43:2493–2505 [DOI] [PubMed] [Google Scholar]
- 62.Wirsching S, Michel S, Morschhauser J. 2000. Targeted gene disruption in Candida albicans wild-type strains: the role of the MDR1 gene in fluconazole resistance of clinical Candida albicans isolates. Mol. Microbiol. 36:856–865 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.