Abstract
Since the first isolation in 2002, the metallo-β-lactamase GIM-1 has not been detected outside Germany. The data presented here, for 50 clinical blaGIM-1-positive isolates, including Pseudomonas spp. and Enterobacteriaceae (Enterobacter cloacae, Klebsiella oxytoca, Serratia marcescens, Escherichia coli, and Citrobacter freundii), collected between 2007 and 2012 at the original site in an ongoing outbreak, demonstrate a diverse genetic background and dissemination of the gene conferring resistance to enteric bacteria.
TEXT
Metallo-β-lactamases (MBLs) hydrolyze all β-lactams (except monobactams), including the carbapenems, and thus are an important emerging obstacle to the treatment of Gram-negative bacterial nosocomial infections. To date, at least 11 subgroups have been described, with IMP, VIM, and NDM-1 being the most geographically widespread, while others, like GIM-1, have not become globally established (1–3). The prevalence of MBLs in Germany is low and mainly due to VIM types (4; http://ecdc.europa.eu/en/activities/surveillance/EARS-net/database/Pages/database.aspx). GIM-1 was initially discovered in a Pseudomonas aeruginosa clone isolated from the surgical intensive care unit (ICU) of the University Hospital of Düsseldorf (Germany) in 2002, in which the blaGIM-1 gene was located on a small 22-kb nontransferable plasmid embedded in the class 1 integron In77 (5). Since then, GIM-1 has been described in only a few isolates of Enterobacter cloacae, Serratia marcescens, Acinetobacter pittii, and P. aeruginosa (6–9), nearly exclusively in the greater Düsseldorf region and never outside Germany.
A total of 230 clinical isolates collected between 2007 and 2012 in one hospital, fulfilling the criteria of nonsusceptibility to piperacillin-tazobactam or ceftazidime and imipenem/meropenem, were further screened for MBL by using MBL MIC test strips (Liofilchem, Roseto degli Abruzzi, Italy) and real-time in-house PCR targeting the blaIMP-1, blaVIM-1-type, blaVIM-2-type, blaGIM-1, and blaNDM-1 genes (Table 1). The blaGIM-1 gene was detected in 50 isolates (Pseudomonas aeruginosa [33], Enterobacter cloacae [7], Pseudomonas putida [5], Serratia marcescens [2], Escherichia coli [1], Klebsiella oxytoca [1], and Citrobacter freundii [1]). Other isolates contained blaVIM-1 (2), blaVIM-2 (2), or blaNDM-1 (1). All blaGIM-1-positive isolates except the single E. coli isolate (MIC, 8 mg/liter) were highly resistant to imipenem and meropenem (MIC, >32 mg/liter), and all blaGIM-1-positive isolates were additionally resistant to quinolones and aminoglycosides. The genetic relatedness of the P. aeruginosa, P. putida, and E. cloacae isolates was investigated using DiversiLab repetitive-sequence-based PCR (repPCR) (bioMérieux, Nürtingen, Germany) according to the manufacturer's protocol. A similarity of >95% between isolates was defined as representing a genetic cluster. Pulsed-field gel electrophoresis (PFGE) was performed on all P. aeruginosa and Serratia marcescens isolates by using SpeI (Fermentas, ThermoFisher, Schwerte, Germany) as described previously (10) and interpreted according to the criteria of Tenover et al. (11). One isolate of the “original” P. aeruginosa clone, 73-5671 (5), was included. These fingerprinting data (Table 2) revealed one main PFGE cluster of 24 P. aeruginosa isolates (PSA-A), including isolates from 2002 to 2012, nearly all of which (19 of 24) were isolated from the surgical ICU, suggesting an ongoing clonal spread. The genetic relatedness of all species is shown in Table 2.
Table 1.
Carbapenemase multiplex primers and probes
| Gene | Primer/probe | Sequence (5′→3′)a | Reference | Nucleotide positions | Size (bp) |
|---|---|---|---|---|---|
| blaGIM-1 | GIM1-F | CGACACACCTTGGTCTGAAGAA | AJ620678 | 1232–1253 | 81 |
| GIM1-R | GATGCTAGCCATAACCTGGTATCC | 1313–1290 | |||
| GIM1-P | HEX-ACACGAAGTTGTTATTATCCTGGGCGACTGAC-BHQ-1 | 1255–1286 | |||
| blaVIM-1 | VIM1-F | TGCGCTTCGGTCCAGTAGA | FJ172675 | 698–716 | 76 |
| VIM1-R | TGACGGGACGTATACAACCAGAT | 774–752 | |||
| VIM-P | FAM-CTTCTATCCTGGTGCTGCGCATTCG-BHQ-1 | 720–744 | |||
| blaVIM-2 | VIM2-F | GCGCTTCGGTCCAGTAGAAC | AF191564 | 1810–1829 | 80 |
| VIM2-R | CTCGCAGACGGGACGTACA | 1890–1872 | |||
| VIM-P | Same probe as for blaVIM-1 | 1831–1855 | |||
| blaIMP-1 | IMP1-F | GGGCGGAATAGAGTGGCTTA | AY168635 | 303–322 | 93 |
| IMP1-R | GGCTTGAACCTTACCGTCTTTTT | 396–374 | |||
| IMP1-P | HEX-CGATCTATCCCCACGTATGCATCTGAATTAACA-BHQ-1 | 328–360 | |||
| blaNDM-1 | NDM1-F | CGGCATCACCGAGATTGC | FN396876 | 2547–2564 | 73 |
| NDM1-R | CACCGACATCGCTTTTGGT | 2620–2602 | |||
| NDM1-P | FAM-GCGACTTGGCCTTGCTGTCCTTG-BHQ-1 | 2568–2590 |
HEX, hexachlorofluorescein; BHQ-1, black hole quencher 1; FAM, 6-carboxyfluorescein.
Table 2.
Characteristics of blaGIM-1-positive strains ordered by species and isolation datea
| Species and strain M no. | Date (yr or mo-yr) | First sample type | Ward | repPCR type | PFGE type | Integron type | Genetic location | Plasmid size(s) (kb) |
|---|---|---|---|---|---|---|---|---|
| Pseudomonas aeruginosa | ||||||||
| 0 | 2002 | Respiratory tract | SICU | PSA-1 | PSA-A | In77 | C/P | 22 |
| 19 | Oct-07 | Wound | SW1 | PSA-4 | PSA-B | In853 | C | |
| 1 | Dec-07 | Respiratory tract | MICU2 | PSA-5 | PSA-C | In583* | C | |
| 2 | Jan-08 | Stool | MW3 | PSA-6 | PSA-D | In851* | C | |
| 4 | Apr-08 | Urine | MW2 | PSA-1 | PSA-A | In77* | C | |
| 3 | Apr-08 | Respiratory tract | SICU | PSA-1 | PSA-A | In77 | C | |
| 5 | Jun-08 | Respiratory tract | SICU | PSA-1 | PSA-A | In77 | C | |
| 7 | Sep-08 | Respiratory tract | MICU1 | PSA-2 | PSA-E | In770 | C | |
| 8 | Sep-08 | Respiratory tract | MICU1 | PSA-2 | PSA-A | In770* | C | |
| 10 | Oct-08 | Stool | MICU1 | PSA-2 | PSA-F | In770 | C | |
| 16 | Feb-10 | Respiratory tract | SICU | PSA-1 | ND | In77 | C | |
| 25 | Feb-10 | Urine | MW2 | PSA-1 | PSA-A | In77 | C | |
| 28 | May-10 | Urine | MOP | PSA-7 | PSA-G | In770 | ND | |
| 29 | May-10 | Respiratory tract | SICU | PSA-8 | PSA-H | In851 | C | |
| 45 | Nov-10 | Respiratory tract | SICU | PSA-1 | PSA-A | In77b | C | |
| 46 | Dec-10 | Respiratory tract | SICU | PSA-1 | PSA-A | In77b | C | |
| 47 | Dec-10 | Urine | MW1 | PSA-1 | PSA-A | In77b | C | |
| 48 | Mar-11 | Wound | SICU | PSA-1 | PSA-A | In77b | C | |
| 57 | Apr-11 | Respiratory tract | SICU | PSA-1 | PSA-A | In77b | C | |
| 49 | May-11 | Respiratory tract | SICU | PSA-1 | PSA-A | In77b | C | |
| 50 | Jun-11 | Urine | SICU | PSA-1 | PSA-A | In77b | C | |
| 51 | Jun-11 | Respiratory tract | SICU | PSA-1 | PSA-A | In77b | C | |
| 53 | Jun-11 | Respiratory tract | SICU | PSA-1 | PSA-A | In77b | C | |
| 52 | Jun-11 | Respiratory tract | SICU | PSA-1 | PSA-A | In77b | C | |
| 55 | Jul-11 | Respiratory tract | SICU | PSA-1 | PSA-A | In77b | C | |
| 58 | Sep-11 | Wound | SICU | PSA-1 | PSA-A | In77b | C and P | 25 |
| 63 | Dec-11 | Respiratory tract | SICU | PSA-1 | PSA-A | In77b* | C | |
| 64 | Jan-12 | Wound | SICU | PSA-1 | PSA-A | In77b | C | |
| 67 | Mar-12 | Wound | SICU | PSA-1 | PSA-A | In77b | C | |
| 68 | Mar-12 | Urine | MW1 | PSA-3 | PSA-A | In770 | C | |
| 69 | Apr-12 | Wound | SICU | PSA-1 | PSA-A | In851 | C | |
| 70 | May-12 | Respiratory tract | SICU | PSA-1 | PSA-A | In77b | C | |
| 71 | May-12 | Respiratory tract | MW4 | PSA-3 | PSA-D | In851 | C | |
| Enterobacter cloacae | ||||||||
| 14 | Jun-09 | Urine | MW4 | ENT-1 | ND | In770 | C and two P | 25 and 220 |
| 15 | Jan-10 | Blood culture | MICU1 | ENT-1 | ND | In770* | C and two P | 25 and 220 |
| 21 | Mar-10 | Wound | MW2 | ENT-2 | ND | In770 | C and two P | 25 and 220 |
| 35 | Jul-10 | Urine | MW3 | ENT-2 | ND | In770 | C and two P | 25 and 220 |
| 44 | Aug-10 | Urine | MW3 | ENT-2 | ND | In770 | C and two P | 25 and 220 |
| 60 | Dec-11 | Wound | MW3 | ENT-1 | ND | In770 | C and two P | 25 and 220 |
| 65 | Dec-11 | Urine | MW3 | ENT-2 | ND | In770 | C and two P | 25 and 220 |
| Pseudomonas putida | ||||||||
| 18 | Sep-07 | Urine | MW3 | PUT-3 | ND | In770* | C | |
| 6 | Sep-08 | Stool | MW3 | PUT-1 | ND | In853* | C | |
| 12 | Dec-08 | Urine | MW3 | PUT-1 | ND | In853 | C | |
| 37 | Aug-10 | Urine | MW3 | PUT-2 | ND | In853 | C | |
| 38 | Aug-10 | Urine | MW3 | PUT-2 | ND | In853 | C | |
| Serratia marcescens | ||||||||
| 9 | Sep-08 | Urine | MICU2 | ND | SMA-A | In770* | P | 140 |
| 11 | Nov-08 | Urine | SW2 | ND | SMA-B | In770 | C | |
| Escherichia coli 17 | Mar-07 | Blood culture | MW5 | ND | ND | In852* | C and two P | 50 and 130 |
| Klebsiella oxytoca 43 | Oct-10 | Urine | MW3 | ND | ND | In770* | C | |
| Citrobacter freundii 56 | Jul-11 | Wound | SICU | ND | ND | In851* | C |
MBL-positive isolates were given consecutive numbers with the prefix M. DiversiLab repPCR cluster and PFGE types were given numbers and letters, respectively, with an acronym of the species. M0 (73-5671) was collected in 2002 and blaGIM-1 was described by Castanheira et al. (5) on a 22-kb plasmid, while we detected it chromosomally. *, integron sequences were published in GenBank (JX566704 to JX566715) and given INTEGRALL numbers as shown (In77b being a variant of In77). SICU, surgical intensive care unit; MICU, medical intensive care unit; SW, surgical ward; MW, medical ward (numbers indicate different wards); ND, not determined (M28 and M16 strains lost for further analysis); C, chromosome; P, plasmid.
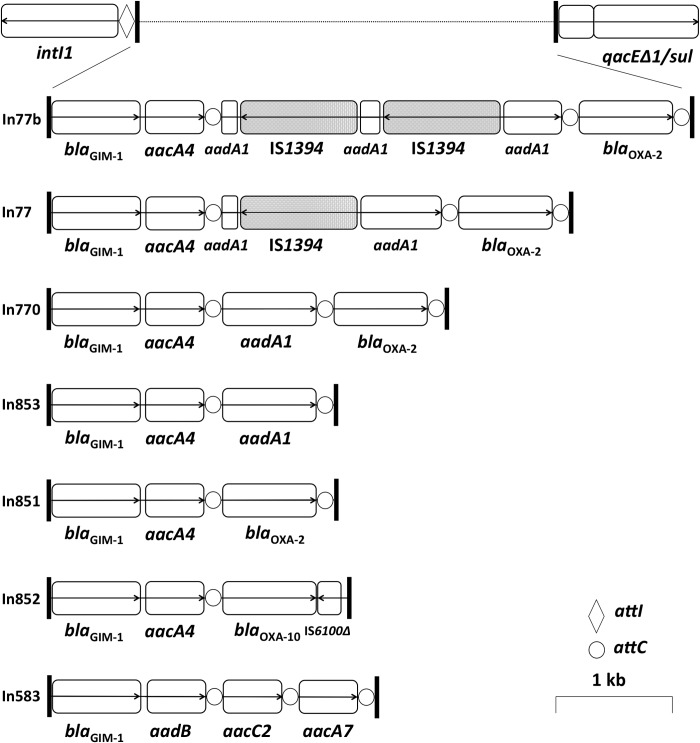
We determined the genetic environment of the blaGIM-1 gene by targeting conserved sequences of class 1 integrons by PCR and using a walking sequencing strategy. In addition to detecting the three integron types described before (5, 6, 8, 9), we detected four new arrays (In77b, In851, In852, and In853) (Fig. 1). The blaGIM-1 gene cassette was fused to aacA4 in all isolates, except in P. aeruginosa M1 (fused to aadB). Five integron arrays (In77, In77b, In770, In851, and In853) were related to each other, sharing identical elements (aacA4, aadA1, and blaOXA-2). The integron found in E. coli M17 (In852) had a unique structure with different elements (ΔIS1600 and blaOXA-10) suggesting an enhanced degree of mobility of the blaGIM-1-aacA4 gene pair. All integron types (except In852) were found in P. aeruginosa, and the appearance of the same types, e.g., In851 and In770, in other Pseudomonas spp. and Enterobacteriaceae (Table 2) clearly demonstrates horizontal gene transfer of a large block of genetic information rather than an individual gene.
Fig 1.
Class 1 integron arrays associated with blaGIM-1 found in this study. Open reading frames are represented by open boxes with arrows indicating the reading direction. IS1394 inserting into gene aadA1 is shown in gray.
S1 restriction and in-gel detection, performed as previously described (10), using a 32P-radiolabeled blaGIM-1 probe (product of primers 5.1.R2 [CCAAGCAGCAAGCGCGTTAC] and GIMR [ACTCATGACTCCTCACGAGG] [5]), demonstrated the chromosomal location of the gene in all isolates other than S. marcescens M9, in which it was found only on a 140-kb plasmid. In addition to being localized on the chromosome, the gene was present on two plasmids in all E. cloacae isolates (approximately 25 kb and 220 kb) and in E. coli M17 (approximately 50 kb and 130 kb). Interestingly, we were able to detect blaGIM-1 on a plasmid (25 kb), similar in size to that originally described in 2004 (5), in only one P. aeruginosa isolate (M58, belonging to PFGE type PSA-A). S1 nuclease treatment, uncommon at the time of the first description of GIM-1, should detect plasmids of this small size, and thus we suggest that the plasmid originally described is probably unstable and has been lost during cultivation or storage. As there is a dominant chromosomal blaGIM-1-mediated resistance in our isolates and as the blaGIM-1 gene cassette has moved into plasmids of different sizes, transfer options besides plasmid-mediated transfer, such as transfer of transposons or integrative and conjugative elements (ICE) (12), must be considered. Horizontal gene transfer was demonstrated by conjugation experiments using the recipient E. coli rifampin-resistant C600 or sodium azide-resistant J53 strain on sheep blood agar at a recipient/donor ratio of 1:10 at three temperatures (18°C, 30°C, and 37°C) as previously described (13). Selective media contained 100 mg/liter ampicillin and either 100 mg/liter rifampin or 100 mg/liter sodium azide. Successful transfer was confirmed by real-time PCR targeting blaGIM-1 and by use of random amplified polymorphic DNA (RAPD) with four primers: Eric1-R, ERIC2 (14), 272 (15), and 1254 (16). We were able to prove only horizontal gene transfer in vitro for the E. coli M17 isolate; the acquired resistance had no impact on carbapenem susceptibility in the transconjugant.
The emergence of metallo-β-lactamase genes, first recovered in nonfermenters and recently spread to Enterobacteriaceae, poses a serious threat, since these bacteria are a common cause of severe nosocomial infections. This is the first description of a large number of isolates expressing the blaGIM-1 gene, including a much broader range of clinically relevant enteric bacteria than previously described. The originally described P. aeruginosa clone (5) may have acted as a genetic pool in the hospital environment over 10 years for the bacterial community, since it was the first described host of GIM-1. Although there is a considerable transfer of patients between hospitals in the greater Düsseldorf area, there does not seem to be an equivalent spread of our blaGIM-1-carrying organisms, as there are only few isolates described elsewhere (6, 8, 9). Despite the data presented here, carbapenemase production in Gram-negative bacteria remains relatively uncommon in Düsseldorf, but these data may herald a rise in GIM-1-mediated carbapenem resistance in this region, which is of great concern.
Nucleotide sequence accession numbers.
Integron sequences are published in GenBank under accession numbers JX566704 to JX566715.
ACKNOWLEDGMENTS
We thank, from our Institute of Medical Microbiology and Hospital Hygiene, Dana Belick, Raquel Guadarrama-Gonzalez, Birgit Lamik-Wolters, and Ulrike Strenz for their excellent technical assistance and Susanne Kolbe-Busch for help with DiversiLab. We thank Martin Kaase (Institute of Medical Microbiology, Ruhr University Bochum, Germany) for providing the E. coli C600 and J53 strains and aid in conducting the conjugation experiments, Michael Kresken and Barbara Körber-Irrgang (Antiinfectives Intelligence, Campus Hochschule Bonn-Rhein-Sieg, Germany) for providing many P. aeruginosa isolates for testing, Yunsop Chong and Kyungwon Lee (Department of Laboratory Medicine and Research Institute of Bacterial Resistance, Yonsei University College of Medicine, Seoul, South Korea) for providing a blaIMP-1-positive P. aeruginosa strain, and Maurine Leverstein-van Hall (Department of Medical Microbiology, University Medical Centre Utrecht, The Netherlands) for providing a blaNDM-1-positive Klebsiella pneumoniae strain (JS-37).
This work was supported by the Medical Faculty of the Heinrich-Heine-University, Düsseldorf, Germany, in collaboration with the ESCMID Study Group on Molecular Diagnostics (ESGMD), Basel, Switzerland.
We have no transparency declarations.
Footnotes
Published ahead of print 22 July 2013
REFERENCES
- 1.Cornaglia G, Giamarellou H, Rossolini GM. 2011. Metallo-beta-lactamases: a last frontier for beta-lactams? Lancet Infect. Dis. 11:381–393 [DOI] [PubMed] [Google Scholar]
- 2.Wachino J, Yoshida H, Yamane K, Suzuki S, Matsui M, Yamagishi T, Tsutsui A, Konda T, Shibayama K, Arakawa Y. 2011. SMB-1, a novel subclass B3 metallo-beta-lactamase, associated with ISCR1 and a class 1 integron, from a carbapenem-resistant Serratia marcescens clinical isolate. Antimicrob. Agents Chemother. 55:5143–5149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walsh TR. 2010. Emerging carbapenemases: a global perspective. Int. J. Antimicrob. Agents 36(Suppl 3):S8–S14 [DOI] [PubMed] [Google Scholar]
- 4.Canton R, Akova M, Carmeli Y, Giske CG, Glupczynski Y, Gniadkowski M, Livermore DM, Miriagou V, Naas T, Rossolini GM, Samuelsen O, Seifert H, Woodford N, Nordmann P. 2012. Rapid evolution and spread of carbapenemases among Enterobacteriaceae in Europe. Clin. Microbiol. Infect. 18:413–431 [DOI] [PubMed] [Google Scholar]
- 5.Castanheira M, Toleman MA, Jones RN, Schmidt FJ, Walsh TR. 2004. Molecular characterization of a beta-lactamase gene, blaGIM-1, encoding a new subclass of metallo-beta-lactamase. Antimicrob. Agents Chemother. 48:4654–4661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamprecht A, Poirel L, Gottig S, Seifert H, Kaase M, Nordmann P. 2013. Detection of the carbapenemase GIM-1 in Enterobacter cloacae in Germany. J. Antimicrob. Chemother. 68:558–561 [DOI] [PubMed] [Google Scholar]
- 7.Kaase M, Pfennigwerth N, Szabados F, Gatermann S. 2012. First description of the metallobetalactamase GIM-1 in Acinetobacter pittii (formerly Acinetobacter genomospecies 3), abstr P-1680. Abstr. 22nd Eur. Congr. Clin. Microbiol. Infect. Dis. (ECCMID), London, United Kingdom [Google Scholar]
- 8.Rieber H, Frontzek A, Pfeifer Y. 2012. Emergence of metallo-beta-lactamase GIM-1 in a clinical isolate of Serratia marcescens. Antimicrob. Agents Chemother. 56:4945–4947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rieber H, Frontzek A, von Baum H, Pfeifer Y. 2012. Emergence of metallo-beta-lactamases GIM-1 and VIM in multidrug-resistant Pseudomonas aeruginosa in North Rhine-Westphalia, Germany. J. Antimicrob. Chemother. 67:1043–1045 [DOI] [PubMed] [Google Scholar]
- 10.Patzer JA, Walsh TR, Weeks J, Dzierzanowska D, Toleman MA. 2009. Emergence and persistence of integron structures harbouring VIM genes in the Children's Memorial Health Institute, Warsaw, Poland, 1998–2006. J. Antimicrob. Chemother. 63:269–273 [DOI] [PubMed] [Google Scholar]
- 11.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toleman MA, Walsh TR. 2011. Combinatorial events of insertion sequences and ICE in Gram-negative bacteria. FEMS Microbiol. Rev. 35:912–935 [DOI] [PubMed] [Google Scholar]
- 13.Walsh TR, Weeks J, Livermore DM, Toleman MA. 2011. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect. Dis. 11:355–362 [DOI] [PubMed] [Google Scholar]
- 14.Versalovic J, Koeuth T, Lupski JR. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19:6823–6831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahenthiralingam E, Campbell ME, Foster J, Lam JS, Speert DP. 1996. Random amplified polymorphic DNA typing of Pseudomonas aeruginosa isolates recovered from patients with cystic fibrosis. J. Clin. Microbiol. 34:1129–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Madico G, Akopyants NS, Berg DE. 1995. Arbitrarily primed PCR DNA fingerprinting of Escherichia coli O157:H7 strains by using templates from boiled cultures. J. Clin. Microbiol. 33:1534–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]