Abstract
We describe genetically encoded sensors which transmit elevated cytosolic concentrations of O-acetyl serine (OAS) and O-acetyl homoserine (OAH)—intermediates of l-cysteine and l-methionine synthesis—into an optical output. The sensor pSenOAS3 elicits 7.5-fold-increased fluorescence in cultures of a Corynebacterium glutamicum strain that excrete l-cysteine. Determination of the cytosolic OAS concentration revealed an increase to 0.13 mM, whereas the concentration in the reference strain was below the detection limit, indicating that incorporation of assimilatory sulfur is limited in the strain studied. In another strain, overexpression of metX encoding homoserine acetyltransferase resulted in an 8-fold increase in culture fluorescence at a cytosolic OAH concentration of 0.76 mM. We also assayed for consequences of extracellular sulfur supply and observed a graded fluorescence increase at decreasing sulfur concentrations below 400 μM. Overall, this demonstrates the usefulness of the sensors for monitoring intracellular sulfur availability. The sensors also enable monitoring at the single-cell level, and since related and close homologs of the transcription factor used in the constructed sensors are widespread among bacteria, this technology offers a new possibility of assaying in vivo for sulfur limitation and of doing this at the single-cell level.
INTRODUCTION
Sulfur is an essential element for any microorganism. It is required for the synthesis of the sulfur-containing amino acids (l-cysteine and l-methionine) together with that of the sulfur-containing coenzymes or prosthetic groups and other compounds. In the context of applied microbiology, the assimilation of sulfur is crucial in at least two settings. One is related to pathogenic microorganisms in which there is an indication that such organisms face sulfur limitation in their natural environment. This is probably the case for the facultative intracellular pathogen Mycobacterium tuberculosis in its preferred host, the macrophage, since under conditions of sulfur limitation, the mycobacterial cysDNC operon enabling sulfur assimilation is upregulated (1). In addition to l-cysteine and l-methionine synthesis, the sulfate assimilation pathway of M. tuberculosis produces a number of sulfur-containing metabolites with important contributions to pathogenesis and survival (2). In different clinical M. tuberculosis isolates, significant differences in the abundance of sulfur (and iron) assimilatory proteins highlight the relevance of sulfur availability for the bacilli (3). In the opportunistic pathogen Pseudomonas aeruginosa E601, which causes infections of cystic fibrosis in patients, 43 proteins, among them a sulfatase with mucin as the substrate which makes the sulfur of the glycopeptide available, are upregulated more than 10-fold under conditions of sulfur limitation (4).
Sulfur availability is also of relevance for the large-scale synthesis of l-cysteine and l-methionine. The bacterium Corynebacterium glutamicum is a prime candidate for amino acid production, because processes for l-glutamate, l-lysine, and l-serine production with this bacterium are already established (5). The synthesis of both sulfur-containing amino acids has been explored in C. glutamicum (6–8), and l-cysteine can be produced with Escherichia coli (9, 10). C. glutamicum is not a pathogen but belongs with its close pathogenic relatives C. diphtheriae, C. ulcerans, and C. urealyticum to the large clade Corynebacteriales, which also includes the Mycobacterium species (11). Because of its ease of handling, C. glutamicum serves as a model organism for M. tuberculosis studies. Studies with C. glutamicum have contributed significantly to deciphering the mechanism of lipid synthesis (12–14) and the function of proteins involved in cell wall synthesis (15, 16) of M. tuberculosis.
To date, the consequences of sulfur availability have mostly been analyzed using DNA microarrays, as was done for P. aeruginosa (4), M. tuberculosis (1), and E. coli (17). Proteome studies have also contributed to understanding the consequences of sulfur limitation (3, 18). Here, we used a totally different approach based on a transcription factor involved in the control of the sulfur metabolism. This enabled us to get in vivo access to the specific metabolites O-acetyl serine (OAS) and O-acetyl homoserine (OAH). These metabolites appear at the junction of the respective carbon skeleton synthesis pathways and the sulfur assimilation pathway by which l-cysteine and l-methionine are formed. They are thus key compounds that may be suitable for optical assessment of sulfur availability in the bacterium. Here, we constructed genetically encoded optical sensors for OAS and OAH which, together with the high sensitivity of the system developed, offer a unique possibility to observe the effect of sulfur limitation at the single-cell level.
MATERIALS AND METHODS
Strains and their constructions and cultivations.
The strains used are listed in Table 1. Molecular work is described in the supplemental material. The cultivation conditions for l-cysteine formation with C. glutamicum were identical to those described for l-serine formation (19). For sulfur limitation, C. glutamicum was pregrown overnight on 0.02 mM SO42−-MMES (8) and, after washing, was inoculated into MMES to an optical density (OD) at 600 nm of 1. A culture of C. glutamicum KK42 pregrown for 8 h on CGXII-glucose (5) and supplemented with l-leucine to a final concentration of 2 mM was used to inoculate fresh medium to which 10 to 150 ng anhydrotetracycline had been added per ml. As inocula, precultures were grown in 50 ml medium in 500-ml baffled shake flasks. The growth of the main cultures was carried out with 750 μl medium in 48-well Flowerplates (m2p-labs, Aachen, Germany) incubated in a BioLector (m2p-labs, Aachen, Germany). The Flowerplates were shaken at 990 rpm (throw of 3 mm; humidity, 80%). The BioLector system enabled the simultaneous online monitoring of the growth (backscatter at 620 nm; gain = 5) and fluorescence (λex = 510 nm and λex = 532 nm; gain = 20) of the cultures in the wells of the Flowerplates.
Table 1.
Strains and plasmids used
| Strain or plasmid | Feature(s) or usea | Source or reference |
|---|---|---|
| C. glutamicum strains | ||
| ATCC 13032 | Wild type (WT) of C. glutamicum | ATCC |
| D | WT ΔsdaA ΔpabABC | 19 |
| Ser4 | WT ΔsdaA ΔpabABC (pserA [FBR]-serB-serC); l-serine producer | 19 |
| Cys3 | Ser4ΔaecD; l-cysteine producer | This study |
| KK42 | MH20-22B lysC (FBR) ΔleuCD hom (FBR) | 20 |
| Plasmids | ||
| pJC1 | Shuttle vector, Kmr | 21 |
| pSenLys | Lysine sensor; encodes transcriptional regulator LysG and its target promoter of lysE with a transcriptional fusion to eyfP | 22 |
| pSenOAS1 | Derived from pSenLys; encodes transcriptional regulator CysR and its target promoter of NCgl1289 with a transcriptional fusion to eyfP | This study |
| pSenOAS3 | Derived from pSenLys; encodes CysR and its target promoter of cysI with a transcriptional fusion to eyfP | This study |
| pCLTON1 | Shuttle vector, Kanr | 23 |
| pCLTON2 | Shuttle vector, Spcr | This study |
| pCLTON2metX | pCLTON2 with metX of C. glutamicum | This study |
| pCR007d | pK18mobsacB carrying aecD deletion; Kmr | 8 |
Kmr, kanamycin resistance; Spcr, spectinomycin resistance; FBR, feedback-resistant enzyme.
OAS and OAH determination.
For the determination of cytosolic O-acetyl serine (OAS) and O-acetyl homoserine (OAH) concentrations, cells from cultures of the exponential-growth phase were used. They were rapidly separated from the medium and inactivated by silica oil centrifugation. This procedure and the further processing were conducted as described previously (24, 25). OAS and OAH were quantified via liquid chromatography-mass spectrometry (LC-MS) using an ultra-high-performance LC (uHPLC) 1290 Infinity system (Agilent, Santa Clara, CA) coupled to a 6130 Quadrupol LC/MS system (Agilent, Santa Clara, CA). LC separation was carried out with a Luna 5μ SCX 100 Å column (Phenomenex, Torrance, CA) (150 mm by 2.0 mm internal diameter) at 30°C. For elution of OAS and OAH, 5% acetic acid (solvent A) and 15 mM ammonium acetate (pH 6.0, adjusted with glacial acetic acid) (solvent B) were applied. The mass spectrometer was operated in the positive electrospray ionization (ESI) mode. Data acquisition was performed in selected-ion-monitoring mode.
Epifluorescence microscopic analysis.
Fluorescence imaging was performed using a fully motorized inverted microscope (Nikon Eclipse Ti; Nikon GmbH, Düsseldorf, Germany) equipped with a focus assistant (Nikon PFS; Nikon GmbH, Düsseldorf, Germany), an Apo total internal reflection fluorescence (TIRF) 100× oil differential interference contrast (DIC) N objective, Nikon DS-Vi1 color camera, Andor Luca R DL604 camera, xenon fluorescence light source, and standard filters for enhanced yellow fluorescent protein (EYFP) detection (λex = 490 to 510 nm; λem = 520 to 550 nm). Differential interference contrast (DIC) microscopy images as well as fluorescence images were captured and analyzed using the Nikon NIS Elements AR software package. Prior to analysis, cells were deposited on soft agarose-covered glass slides.
RESULTS AND DISCUSSION
Genetically encoded sensors for optical detection of cytosolic OAS and OAH.
The relevance of OAS and OAH as key compounds in the sulfur metabolism is reflected by the fact that bacteria have proteins that sense their concentrations. In E. coli and Salmonella enterica serovar Typhimurium, the transcription factor CysB serves this purpose. It activates in response to OAS or its derivative N-acetylserine in transcription of the cysteine regulon (26). The functional analogue of CysB in C. glutamicum is CysR. This regulatory protein requires OAS or OAH to activate transcription of sulfate assimilation genes (8).
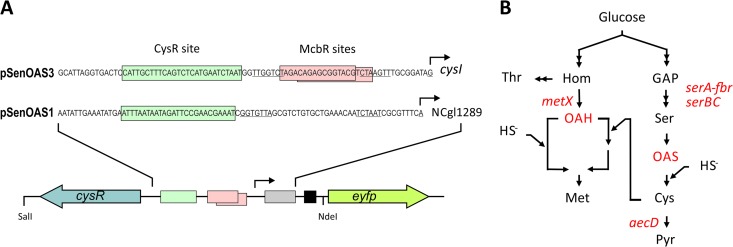
We expected that fusion of the appropriate CysR binding region with eyfP would result in the presence of fluorescent cells at elevated OAS or OAH concentrations. One target region chosen was that in front of cysI (Fig. 1A), where CysR binding dependent on either OAS or OAH was demonstrated by in vitro experiments (8). The other target region chosen was NCgl1289. The latter gene was chosen since it is one of the rare genes in C. glutamicum that is apparently controlled only by CysR and is devoid of McbR control—another regulator involved in controlling the sulfur metabolism (27). To construct pSenOAS1, a 1,359-bp fragment encoding cysR together with its promoter region was amplified. In addition, a 302-bp fragment was amplified which contained the CysR binding site in front of NCgl1289 as well as 96 bp of the 5′ end of NCgl1289, including the promoter (Fig. 1A). Using overlap extension PCR, both fragments were fused, and the resulting cassette of 1,633 bp flanked by NdeI/SalI sites was fused with the backbone of pSenLys (22), from which the l-lysine sensing cassette had been deleted previously by NdeI/SalI digestion. The sensor pSenOAS3 was built in the same manner, except that the fragment that included the CysR binding site in front of cysI was used (Fig. 1A).
Fig 1.
The metabolite pSenOAS sensors. (A) The pSenOAS sensors encode the transcription factor CysR together with parts of its target genes fused to eyfP. In front of cysI, as used for pSenOAS3, a CysR recognition site is present together with McbR sites, whereas in front of NCgl1289, as used for pSenOAS1, a CysR recognition site is predicted. To construct the pSenOAS sensors, these sequence parts, together with 126 and 96 bp of their respective genes (gray box), were fused to eyfP preceded by a ribosome binding site (black box) and to cysR, encoding the transcriptional regulator. (B) Sketch of l-cysteine and l-methionine synthesis with OAS and OAH as intermediates which serve as acceptors for sulfide. Genes relevant to this study are also given.
l-Cysteine producers exhibit increased fluorescence.
l-Cysteine is made in two steps from l-serine via the use of OAS (Fig. 1B). In prior work, we constructed the l-serine producer C. glutamicum strain Ser4 by transformation of C. glutamicum strain D with pserACB encoding the enzymes of serine biosynthesis with the serA-encoded d-3-phosphoglycerate dehydrogenase deregulated (19). In addition to l-serine, C. glutamicum strain Ser4 accumulates a small but significant concentration of 1 mM l-cysteine, whereas C. glutamicum strain D and the wild type (WT) do not (data not shown). Since the aecD-encoded cystathionine beta-lyase is known to degrade l-cysteine (10), we constructed plasmid pK19mobsacBΔaecD to delete aecD in two rounds of positive selection in C. glutamicum strain D. Transformation of the resulting strain with pserACB resulted in C. glutamicum strain Cys3, which is isogenic to C. glutamicum strain Ser4 but in which aecD is additionally deleted (see Table S1 in the supplemental material). As expected, C. glutamicum strain Cys3 accumulated an increased l-cysteine concentration of about 4 mM.
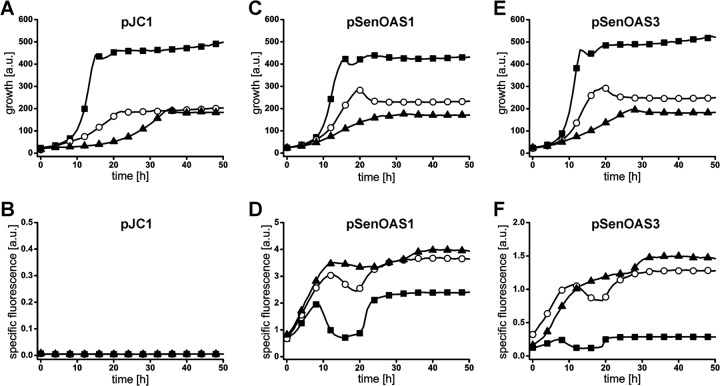
C. glutamicum strain D, strain Ser4, and strain Cys3 were transformed with empty vector pJC1, pSenOAS1, and pSenOAS3, respectively. Growth and fluorescence were recorded in microtiter plate cultivations. Compared to C. glutamicum strain D, the growth of C. glutamicum strain Ser4 and strain Cys3 was reduced apparently due to significant carbon flux redirectioning due to pserACB present in these strains (Fig. 2A, C, and E). Importantly, the presence of both pSenOAS1 and pSenOAS3 in the l-cysteine producers resulted in a strong specific fluorescence increase compared to the level seen with the nonproducer (Fig. 2D and F). Cultures of C. glutamicum strain Cys3 show a slight tendency toward increased specific fluorescence compared to C. glutamicum strain Ser4 cultures. These data, together with the intracellular accumulation of OAS (see below), verify that functional CysR recognition sites are present in front of NCgl1289 and cysI. The two sensors exhibit substantial differences regarding background fluorescence and total fluorescence, which could be due to different promoter strengths, and in the case of pSenOAS3, to an interaction of the two regulators CysR and McbR (Fig. 1A).
Fig 2.
Culture fluorescence in l-cysteine producers due to pSenOAS1 and pSenOAS3. (A) Growth of C. glutamicum strain D, strain Ser4, and strain Cys3 with control plasmid pJC1. (B) Fluorescence of the cultures shown in panel A. a.u., arbitrary units. (C) Growth of C. glutamicum strain D, strain Ser4, and strain Cys3 with pSenOAS1. (D) Fluorescence of cultures shown in panel C due to pSenOAS1. (E) Growth of C. glutamicum strain D, strain Ser4, and strain Cys3 with pSenOAS3. (F) Fluorescence of cultures shown in panel C due to pSenOAS3. C. glutamicum strain D, ■; C. glutamicum strain Ser4, ○; C. glutamicum strain Cys3, ▲.
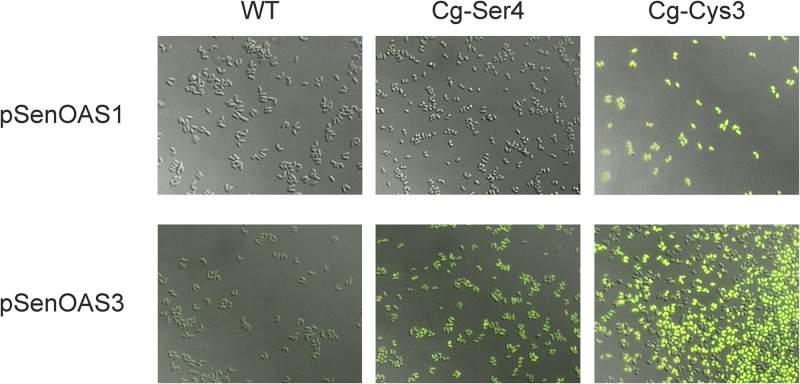
We were also interested in examining the fluorescence of populations at the single-cell level. To do this, cells grown for 30 h were inspected by epifluorescence microscopy (Fig. 3). As expected, cells of C. glutamicum strain Cys3 exhibited the strongest fluorescence (right column), whereas controls of WT cells (left column) or C. glutamicum strain D (not shown) exhibited the lowest fluorescence. The level of fluorescence of C. glutamicum strain Ser4 was in between (middle column). A significantly reduced exposure time for pSenOAS1-carrying cells of 10 ms was used compared to that used for pSenOAS3-carrying cells, reflecting the stronger fluorescence due to pSenOAS1 as already apparent from culture fluorescence (Fig. 2D).
Fig 3.
Fluorescence of single cells of C. glutamicum due to elevated O-acetyl-serine concentrations in the l-cysteine producers C. glutamicum strain Ser4 and strain Cys3. Fluorescence is seen in the strong producer (right) carrying pSenOAS1 or pSenOAS3 but not for cells of the nonproducer, WT strain (left). The exposure time for pSenOAS1 was 10 ms and for pSenOAS3 200 ms. Because of the short exposure time of 10 ms, the fluorescence for the intermediate producer C. glutamicum strain Ser4 pSenOAS1 is not seen in the image. The epifluorescence microscopic analysis was done at λex = 490 to 510 nm and λem = 520 to 550 nm.
Fluorescence correlates with cytosolic OAS concentration.
In the two C. glutamicum strains derived from an l-serine producer (19), elevated concentrations of the sulfur acceptor molecule OAS were apparently present. This is plausible, since the conversion of l-serine to l-cysteine involves only two steps, one catalyzed by serine acetyltransferase and the other by OAS sulfhydrylase (Fig. 1B), with the final sulfhydration regarded as a key limiting step in l-cysteine synthesis (28). To determine the concentration of OAS in the cytosol of C. glutamicum, we grew WT pSenOAS3, strain Ser4 pSenOAS3, and strain Cys3 pSenOAS3 to the exponential phase in shake flasks to perform silica oil centrifugation (24) and determined the cytosolic concentrations of OAS by LC-MS. Whereas the OAS concentration was close to the detection limit and below 0.02 mM in WT pSenOAS3, a noticeable concentration of 0.09 ± 0.02 mM was present in C. glutamicum strain Ser4 pSenOAS3, and this increased to 0.13 ± 0.02 mM in C. glutamicum strain Cys3 pSenOAS3. The increased fluorescence thus correlates with an elevated OAS concentration. OAH was not detectable in any of the strains assayed.
Expression of homoserine acetyltransferase results in increased fluorescence.
We were wondering whether it is possible to use the nanosensors for the in vivo monitoring of OAH, which is the sulfur acceptor molecule in l-methionine synthesis (Fig. 1B). OAH is derived from l-homoserine, as is l-threonine. In prior work, we generated the l-threonine producer C. glutamicum KK42 (20). In an attempt to achieve an elevated OAH concentration, we cloned metX encoding the homoserine acetyltransferase, which converts l-homoserine to OAH in pCLTON2, a vector proven to enable stringent expression control by anhydrotetracycline (atc) (23). Plasmid pCLTON2metX was introduced into strain C. glutamicum KK42, as was a control plasmid devoid of metX. The resulting strains were subsequently transformed with one of the nanosensors. As shown for the strains carrying pSenOAS3, the graded expression of metX results in graded growth reduction (Fig. 4C). This was likely due to increased homoserine acetyltransferase activity, since even at the highest concentration of 150 ng atc ml−1, no growth response was apparent with the empty vector (Fig. 4A). Interestingly, increased expression of metX also led to a graded specific fluorescence increase of the cultures (Fig. 4D). The specific fluorescence of the uninduced culture with pCLTON2metX was nearly identical to that of the control culture with pCLTON2 only (Fig. 4B). This shows the tight control of metX expression in the system used. The discernible fluorescence observed at 25 ng atc indicates the high sensitivity of the sensor for in vivo monitoring. Comparable data were obtained with pSenOAS1, and these are shown in Fig. S1 in the supplemental material.
Fig 4.
Culture fluorescence due to metX expression and pSenOAS3 in derivatives of strain KK42. (A) Growth of strain KK42 pCLTON2 ± 150 ng ml−1 anhydrotetracycline (atc). (B) Fluorescence of cultures shown in panel A. (C) Growth of strain KK42 pCLTON2 pSenOAS3 at different atc concentrations. (D) Fluorescence of cultures shown in panel C due to increasing atc concentrations. The concentrations (ng ml−1) used are indicated as follows: 0, □; 10, ○; 25, △; 50, ▽; 100, ◇; 150, ▲.
To prove that induction of metX caused an increase in the OAH concentration, we grew KK42 pCLTON2metX pSenOAS3 ± 150 ng ml−1 atc to the early exponential phase and determined the cytosolic OAH concentration for both cultures. The concentrations were 0.11 ± 0.03 mM for cells of the uninduced culture and 0.76 ± 0.12 mM for cells of the induced culture. These concentrations are in the range known from a different C. glutamicum strain engineered for increased sulfur supply (6). The significant increase due to pCLTON2metX confirms that a higher level of homoserine acetyltransferase activity is obtained upon induction, which leads to an increased OAH concentration in the cell. The fact that OAH accumulation occurs upon metX expression suggests that either the sulfide incorporation step or one of the other three steps of the branched pathway of l-methionine synthesis is limiting (Fig. 1B). It indicates the delicate balance within the l-methionine synthesis pathway (7).
Cellular fluorescence in response to sulfur limitation.
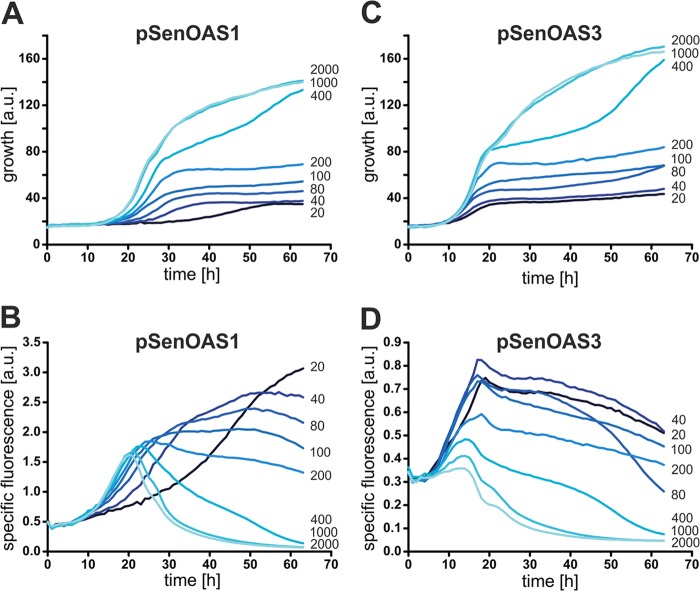
Studies with P. aeruginosa and E. coli revealed complex consequences with respect to gene expression in response to changes in sulfur supply (4, 17). Sulfur limitation also gave rise to consequences at the proteome level (3, 18). We reasoned that our sensors could also be used to visualize the consequences of sulfur limitation in vivo at the metabolite level. To this end, we grew C. glutamicum WT pSenOAS1 and WT pSenOAS3 at different ammonium sulfate concentrations. As can be seen in Fig. 5A and C, growth of the strains was not limited at a 1 mM sulfate supply. However, at 400 μM, growth became limited and a further gradual decrease in the sulfur concentration resulted in a gradual decrease in the growth rate and final density of the culture. Interestingly, a gradual increase in specific fluorescence for a decreasing sulfate supply was visible with both sensors (Fig. 5B and D). The strongest response was seen with WT pSenOAS1, and maximal fluorescence for the different cultures can be seen with WT pSenOAS3 for about the same duration of incubation. The differences in fluorescence profiles compared to that in Fig. 2 might have been due to the absence of morpholinopropane sulfonic acid in the sulfate-free medium MMES used in this experiment. The significant fluorescence response of the cultures (Fig. 5B and D) indicates that the concentration of OAS/OAH increases in the cytosol when the level of sulfur becomes limiting. The sulfur stress imposed on the cells results in increased synthesis of the carbon skeletons of the sulfur-containing amino acids rather than in reduced synthesis. Since we cannot differentiate between OAH and OAS, the observed increase could in principle also have been due to the presence of only one of these metabolites. The fluorescence increase reflects an imbalance under the stress conditions of sulfur limitation which the cell is unable to counteract by regulatory mechanisms.
Fig 5.
Growth and fluorescence in response to sulfur limitation. (A) Growth of C. glutamicum WT pSenOAS1 at decreasing sulfate concentrations. (B) Fluorescence of cultures shown in panel A. (C) Growth of C. glutamicum WT pSenOAS3 at decreasing sulfate concentrations. (D) Fluorescence of cultures shown in panel C. The numbers to the right of each of the figure panels refer to the sulfate concentrations in micromolar units.
In the present work, we demonstrated that the use of the natural transcription factor CysR of C. glutamicum allows a highly sensitive optical device to be constructed. The optical device monitors cytosolic OAS and OAH concentrations, which increase as a result of imbalances in l-methionine and l-cysteine synthesis. As such, its use is not a direct method of detecting limiting intracellular sulfur or sulfide. This may be difficult or even impossible to achieve in living single bacterial cells. However, the assay in which extracellular sulfur was limiting indicates that increases in the OAS and OAH concentrations are likely to be directly related to limiting intracellular sulfur levels. This view is supported by the fact that the sulfur assimilation pathway in bacteria like E. coli is energetically very costly. It consists of six steps, whereas the conversion of OAS to l-cysteine consists of only a single step (28). CysR homologs are widespread in bacteria, where they serve to orchestrate sulfur metabolism. Sensors comparable to those developed for C. glutamicum might allow the study of consequences of sulfur limitation in other single cells such as in pathogens under the conditions that may exist in their hosts.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a grant from the CLIB-Graduate Cluster Industrial Biotechnology awarded to K.H.
We declare that we have no conflicts of interest.
Footnotes
Published ahead of print 30 August 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01804-13.
REFERENCES
- 1. Pinto R, Tang QX, Britton WJ, Leyh TS, Triccas JA. 2004. The Mycobacterium tuberculosis cysD and cysNC genes form a stress-induced operon that encodes a tri-functional sulfate-activating complex. Microbiology 150:1681–1686 [DOI] [PubMed] [Google Scholar]
- 2. Hatzios SK, Bertozzi CR. 2011. The regulation of sulfur metabolism in Mycobacterium tuberculosis. PLoS Pathog. 7:e1002036. 10.1371/journal.ppat.1002036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mehaffy C, Hess A, Prenni JE, Mathema B, Kreiswirth B, Dobos KM. 2010. Descriptive proteomic analysis shows protein variability between closely related clinical isolates of Mycobacterium tuberculosis. Proteomics 10:1966–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tralau T, Vuilleumier S, Thibault C, Campbell BJ, Hart CA, Kertesz MA. 2007. Transcriptomic analysis of the sulfate starvation response of Pseudomonas aeruginosa. J. Bacteriol. 189:6743–6750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Eggeling L, Bott M. 2005. Handbook of Corynebacterium glutamicum. CRC Press, Boca Raton, FL [Google Scholar]
- 6. Bolten CJ, Schroder H, Dickschat J, Wittmann C. 2010. Towards methionine overproduction in Corynebacterium glutamicum—methanethiol and dimethyldisulfide as reduced sulfur sources. J. Microbiol. Biotechnol. 20:1196–1203 [DOI] [PubMed] [Google Scholar]
- 7. Park SD, Lee JY, Sim SY, Kim Y, Lee HS. 2007. Characteristics of methionine production by an engineered Corynebacterium glutamicum strain. Metab. Eng. 9:327–336 [DOI] [PubMed] [Google Scholar]
- 8. Rückert C, Milse J, Albersmeier A, Koch DJ, Pühler A, Kalinowski J. 2008. The dual transcriptional regulator CysR in Corynebacterium glutamicum ATCC 13032 controls a subset of genes of the McbR regulon in response to the availability of sulphide acceptor molecules. BMC Genomics 9:483. 10.1186/1471-2164-9-483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dassler T, Maier T, Winterhalter C, Böck A. 2000. Identification of a major facilitator protein from Escherichia coli involved in efflux of metabolites of the cysteine pathway. Mol. Microbiol. 36:1101–1112 [DOI] [PubMed] [Google Scholar]
- 10. Wada M, Awano N, Haisa K, Takagi H, Nakamori S. 2002. Purification, characterization and identification of cysteine desulfhydrase of Corynebacterium glutamicum, and its relationship to cysteine production. FEMS Microbiol. Lett. 217:103–107 [DOI] [PubMed] [Google Scholar]
- 11. Gao BL, Gupta RS. 2012. Phylogenetic framework and molecular signatures for the main clades of the phylum Actinobacteria. Microbiol. Mol. Biol. Rev. 76:66–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gande R, Dover LG, Krumbach K, Besra GS, Sahm H, Oikawa T, Eggeling L. 2007. The two carboxylases of Corynebacterium glutamicum essential for fatty acid and mycolic acid synthesis. J. Bacteriol. 189:5257–5264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gande R, Gibson KJC, Brown AK, Krumbach K, Dover LG, Sahm H, Shioyama S, Oikawa T, Besra GS, Eggeling L. 2004. Acyl-CoA carboxylases (accD2 and accD3), together with a unique polyketide synthase (Cg-pks), are key to mycolic acid biosynthesis in corynebacterianeae such as Corynebacterium glutamicum and Mycobacterium tuberculosis. J. Biol. Chem. 279:44847–44857 [DOI] [PubMed] [Google Scholar]
- 14. Rath P, Demange P, Saurel O, Tropis M, Daffe M, Dotsch V, Ghazi A, Bernhard F, Milon A. 2011. Functional expression of the PorAH channel from Corynebacterium glutamicum in cell-free expression systems: implications for the role of the naturally occurring mycolic acid modification. J. Biol. Chem. 286:32525–32532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Alderwick LJ, Radmacher E, Seidel M, Gande R, Hitchen PG, Morris HR, Dell A, Sahm H, Eggeling L, Besra GS. 2005. Deletion of Cg-emb in corynebacterianeae leads to a novel truncated cell wall arabinogalactan, whereas inactivation of Cg-ubiA results in an Arabinan-deficient mutant with a cell wall galactan core. J. Biol. Chem. 280:32362–32371 [DOI] [PubMed] [Google Scholar]
- 16. Portevin D, Sousa-D'Auria CC, Houssin C, Grimaldi C, Chami M, Daffe M, Guilhot C. 2004. A polyketide synthase catalyzes the last condensation step of mycolic acid biosynthesis in mycobacteria and related organisms. Proc. Natl. Acad. Sci. U. S. A. 101:314–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gyaneshwar P, Paliy O, McAuliffe J, Popham DL, Jordan MI, Kustu S. 2005. Sulfur and nitrogen limitation in Escherichia coli K-12: specific homeostatic responses. J. Bacteriol. 187:1074–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Quadroni M, James P, Dainese-Hatt P, Kertesz MA. 1999. Proteome mapping, mass spectrometric sequencing and reverse transcription-PCR for characterization of the sulfate starvation-induced response in Pseudomonas aeruginosa PAO1. Eur. J. Biochem. 266:986–996 [DOI] [PubMed] [Google Scholar]
- 19. Stolz M, Peters-Wendisch P, Etterich H, Gerharz T, Faurie R, Sahm H, Fersterra H, Eggeling L. 2007. Reduced folate supply as a key to enhanced L-serine production by Corynebacterium glutamicum. Appl. Environ. Microbiol. 73:750–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Morbach S, Sahm H, Eggeling L. 1996. L-Isoleucine production with Corynebacterium glutamicum: further flux increase and limitation of export. Appl. Environ. Microbiol. 62:4345–4351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cremer J, Eggeling L, Sahm H. 1990Cloning the Dapa Dapb cluster of the lysine-secreting bacterium Corynebacterium glutamicum. Mol. Gen. Genet. 220:478–480 [Google Scholar]
- 22. Binder S, Schendzielorz G, Stäbler N, Krumbach K, Hoffmann K, Bott M, Eggeling L. 2012. A high-throughput approach to identify genomic variants of bacterial metabolite producers at the single-cell level. Genome Biol. 13:R40. 10.1186/gb-2012-13-5-r40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lausberg F, Chattopadhyay AR, Heyer A, Eggeling L, Freudl R. 2012. A tetracycline inducible expression vector for Corynebacterium glutamicum allowing tightly regulable gene expression. Plasmid 68:142–147 [DOI] [PubMed] [Google Scholar]
- 24. Klingenberg M, Pfaff E. 1967. Means of terminating reactions. Methods Enzymol. 10:680–684 [Google Scholar]
- 25. Stäbler N, Oikawa T, Bott M, Eggeling L. 2011. Corynebacterium glutamicum as a host for synthesis and export of D-amino acids. J. Bacteriol. 193:1702–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Colyer TE, Kredich NM. 1996. In vitro characterization of constitutive CysB proteins from Salmonella typhimurium. Mol. Microbiol. 21:247–256 [DOI] [PubMed] [Google Scholar]
- 27. Rey DA, Nentwich SS, Koch DJ, Rückert C, Pühler A, Tauch A, Kalinowski J. 2005. The McbR repressor modulated by the effector substance S-adenosylhomocysteine controls directly the transcription of a regulon involved in sulphur metabolism of Corynebacterium glutamicum ATCC 13032. Mol. Microbiol. 56:871–887 [DOI] [PubMed] [Google Scholar]
- 28. Kredich N. 1996. Biosynthesis of cysteine, p 514–527 In Neidhardt FC, Curtiss R, III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE. (ed), Escherichia coli and Salmonella: cellular and molecular biology. American Society for Microbiology, Washington, DC [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.