Abstract
The solventogenic clostridia have a considerable capacity to ferment carbohydrate substrates with the production of acetone and butanol, making them attractive organisms for the conversion of waste materials to valuable products. In common with other anaerobes, the clostridia show a marked dependence on the phosphoenolpyruvate (PEP)-dependent phosphotransferase system (PTS) to accumulate sugars and sugar derivatives. In this study, we demonstrate that extracts of Clostridium beijerinckii grown on N-acetylglucosamine (GlcNAc) exhibit PTS activity for the amino sugar. The PTS encoded by the divergent genes cbe4532 (encoding the IIC and IIB domains) and cbe4533 (encoding a IIA domain) was shown to transport and phosphorylate GlcNAc and also glucose. When the genes were recombined in series under the control of the lac promoter in pUC18 and transformed into a phosphotransferase mutant (nagE) of Escherichia coli lacking GlcNAc PTS activity, the ability to take up and ferment GlcNAc was restored, and extracts of the transformant showed PEP-dependent phosphorylation of GlcNAc. The gene products also complemented an E. coli mutant lacking glucose PTS activity but were unable to complement the same strain for PTS-dependent mannose utilization. Both GlcNAc and glucose induced the expression of cbe4532 and cbe4533 in C. beijerinckii, and consistent with this observation, extracts of cells grown on glucose exhibited PTS activity for GlcNAc, and glucose did not strongly repress utilization of GlcNAc by growing cells. On the basis of the phylogeny and function of the encoded PTS, we propose that the genes cbe4532 and cbe4533 should be designated nagE and nagF, respectively.
INTRODUCTION
The acetone-butanol-ethanol (ABE) fermentation of Clostridium acetobutylicum and related bacteria has a successful history of industrial-scale operation worldwide but went into decline during the latter part of the 20th century for economic reasons (1). Nevertheless, stimulated by concerns relating to the environmental effects of burning fossil fuels and the potential of butanol as a biofuel, interest in this fermentation is being revived (2). Traditionally, the industrial process used starch or molasses as the fermentable substrate, and while these may still be employed, the fermentation of the future is likely to be based on a variety of alternative feedstocks that are derived as waste products from other processes. Lignocellulose-based agricultural waste poducts have attracted considerable attention, but other materials are also being considered (3, 4). An important criterion is that the fermentable substrates should be effectively utilized to support high productivity, yield, and titer of the desired metabolic end product.
The solventogenic clostridia are capable of utilizing a wide range of carbohydrate substrates, thus displaying a metabolic capability that can be harnessed for the development of fermentation processes (5). In common with other obligately anaerobic bacteria, the principal mechanism of accumulation of fermentable monosaccharides, disaccharides, and sugar derivatives is via the phosphoenolpyruvate (PEP)-dependent phosphotransferase system (PTS), which catalyzes concomitant uptake and phosphorylation of its substrates (6, 7). The PTS comprises a phosphoryl transfer chain made up of several conserved domains that sequentially transfer phosphate from PEP to the substrate. The first two components, enzyme I (EI) and histidine-containing, phosphorylatable protein HPr, are general PTS proteins that usually contribute to all of the phosphotransferases in the cell. Substrate specificity lies in the enzyme II complex, typically made up of three domains (IIA, IIB, and IIC) but in some cases also incorporating a fourth domain (IID). The IIA and IIB domains are hydrophilic and participate in phosphate transfer, while the IIC and IID domains are within membrane-bound proteins that facilitate translocation of the substrate. In addition to its role in sugar accumulation, the bacterial PTS has been shown to play a critical role in regulation of carbohydrate metabolism, being centrally involved in the phenomenon of carbon catabolite repression (CCR) in both Gram-negative enteric bacteria and Gram-positive firmicutes (6, 8, 9). As a result of CCR, bacteria metabolize substrates selectively and sequentially when more than one option is present in the growth medium. A full appreciation of this important physiological response, which has implications for the effectiveness of a fermentation process, is therefore dependent on a thorough characterization of the PTS in individual organisms.
The genome of Clostridium beijerinckii encodes 42 complete phosphotransferases (10), suggesting a significant degree of metabolic flexibility and potential to utilize novel fermentation substrates. With the exception of genes encoding a glucitol PTS (11) and a sucrose PTS (12), none of these systems has been characterized functionally. We initially sought to examine the role of three phosphotransferases (encoded by the genes cbe4532 and cbe4533, cbe4983 and cbe4982, and cbe0751) that belong to the glucose branch of the glucose-glucoside (Glc) PTS family, since these may potentially be involved in uptake of glucose and consequent imposition of CCR in this bacterium. Furthermore, the first of these systems, which is the subject of this communication, has been annotated as an N-acetylglucosamine (GlcNAc) PTS. GlcNAc is the monomer of chitin, the second most prevalent polymer on the planet after cellulose. Chitin is found as a major component in insects, crustaceans, and fungi (13), and the aquatic food and agriculture industries can lead to production of a huge amount of chitin waste, which is considered to be a real burden on the environment (14, 15). The ABE fermentation could provide a means of dealing with this burden, employing strains with the ability to degrade chitin and take up and grow on GlcNAc. As a first step in evaluating C. beijerinckii for this purpose, the aim of this study was therefore to characterize the putative GlcNAc PTS with respect to its substrate specificity and potential physiological role.
MATERIALS AND METHODS
Organism and growth conditions.
C. beijerinckii NCIMB 8052 was maintained as a spore suspension in water at 4°C. Aliquots of the suspension (0.8 to 1 ml) were heat shocked at 80°C for 10 min, transferred into 20 ml reinforced clostridial medium (RCM; Oxoid) and incubated overnight at 37°C in an anaerobic cabinet (MACS DG; Don Whitley Scientific) under an atmosphere of N2-H2-CO2 (80:10:10). Fresh starter cultures were used to prepare working cultures in clostridial basal medium (CBM) (16) for individual experiments. Escherichia coli TOP10 (Invitrogen) was used as a host in cloning procedures and was cultured in LB broth supplemented with 50 μg ml−1 ampicillin when required. The E. coli mutants JW0665-1 (BW25113 [nagE]) from the Keio collection (17) and ZSC113 (gpt-2 mpt-2 glk-7 strA) (18) were used in phenotype complementation studies. They were grown on LB broth and on MacConkey agar (prepared using Difco MacConkey agar base) containing the required sugar, with and without 50 μg ml−1 ampicillin as appropriate.
Preparation of cell extracts and assay of phosphotransferase activity.
As described previously, cell extracts of C. beijerinckii grown on CBM containing GlcNAc or glucose were prepared by two passages through a French press at 20,000 lb/in2, centrifuged, divided into aliquots, flash frozen in liquid nitrogen, and stored at −70°C (16). Extracts were subsequently fractionated into soluble and membrane portions by centrifugation (16), with the resulting preparations being frozen and stored in the same way. Extracts of E. coli were likewise prepared from cultures grown in LB broth. Protein concentrations of extracts were determined by a microbiuret method (19). Phosphorylation of radiolabeled GlcNAc and glucose by cell extracts was assayed as described previously (16). The substrates were d-[U-14C]glucose and N-acetyl-d-[1-14C]glucosamine (GE Healthcare), which were prepared in solution at a concentration of 9.5 mM and specific activity of 1.05 Ci mol−1 and added to the reaction mixtures at a concentration of 0.2 mM. When the effects of glucose and chitobiose as inhibitors of GlcNAc phosphorylation were tested, they were added at a concentration of 10 mM. Results are presented as nmol sugar phosphorylated per mg of extract protein and are the average of duplicate assays.
Growth and sugar utilization.
One milliliter of a C. beijerinckii starter culture was inoculated into 100 ml of CBM containing 1% (wt/vol) glucose as the only fermentable carbon source. Following overnight growth at 37°C, cells were harvested anaerobically by centrifugation, washed three times with CBM without a carbon source, and resuspended in approximately 10 ml of the wash medium (16). Washed cells were inoculated into CBM containing 5 mM glucose and 10 mM GlcNAc and incubated at 37°C, with samples being removed periodically for measurement of the optical density at 650 nm (OD650) and for sugar analysis; following centrifugation at 12,000 × g for 10 min, the sugar concentration in the supernatant was determined by high-performance liquid chromatography (HPLC). E. coli cultures were grown overnight at 37°C in 10 ml LB broth containing 50 μg ml−1 ampicillin and diluted into fresh medium containing 20 mM GlcNAc. Growth and sugar utilization were monitored as described for the C. beijerinckii cultures.
Preparation of hybridization probes.
C. beijerinckii DNA was prepared using a genomic DNA isolation kit (Gentra) by the modified method used previously for DNA extraction from C. acetobutylicum (16). Digoxigenin (DIG)-labeled probes directed against internal regions of genes of interest were generated by PCR using the primers listed in Table 1. The reaction mixtures, in a total volume of 50 μl, contained 25 μl 2× BioMix (Bioline), 1 μl (100 pmol) each primer, 1 μl (10 ng) C. beijerinckii DNA, and 0.04 mM digoxigenin-11-dUTP (Roche), under the reaction conditions described previously (16). Heat-denatured, labeled probe was used at 1.5 μl in each hybridization.
Table 1.
Oligonucleotides used in this study
| Namea | Sequence (5′→3′)b | Positions relative to start codon of gene |
|---|---|---|
| Hybridization probe primers | ||
| Cb4532-Digfwd | CATTGCTTCAGCTTTTATGC | +143–+162 |
| Cb4532-Digrev | ACAATAGCTTCACCAAATGC | +563–+544 |
| Cb4533-Digfwd | TTAGTTGCACCTATAACTGG | +52–+71 |
| Cb4533-Digrev | ATGTTTGTAACAAGGACTGG | +416–+397 |
| Cloning primers | ||
| Cb4532fwd | CATTTAGGGATATAACAATC | −67–−48 |
| Cb4532revSal | ACAGTCGACATATCATCAATCTCTTTTCC | +1523–+1504 |
| Cb4533fwdSal | ACAGTCGACAAGGAAGTGACTGTTCCTG | −39–−21 |
| Cb4533revXba | ACATCTAGACTAGCAATGCATATAGAGAG | +592–+573 |
fwd, forward primer; rev, reverse primer.
Restriction sites are underlined.
RNA isolation and analysis of gene expression.
One milliliter of a C. beijerinckii starter culture was inoculated into 100 ml of CBM containing a 10 mM concentration of the required carbon source and incubated overnight at 37°C. A sample was then transferred to a fresh CBM culture containing the same carbon source. When the OD650 reached 0.4 to 0.6 (mid-exponential phase), 750 μl of culture was placed into a 1.5-ml Eppendorf tube with 750 μl of RNA stabilization reagent (Qiagen), and the mixture was incubated at room temperature for 10 min and then centrifuged at 13,000 × g for 3 min. The supernatant was removed, and the pellet was flash-frozen in liquid nitrogen and stored at −70°C.
Total RNA was extracted using a RNeasy minikit (Qiagen) according to the manufacturer's instructions and stored at −70°C. RNA samples (500 ng) were prepared for slot blotting as described previously (16). Prehybridization and overnight hybridization were carried out in DIG Easy Hyb buffer (Roche) at 60°C, followed by washing three times for 15 min with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and twice for 15 min with 0.2× SSC at 60°C. The membrane was then transferred to blocking solution (DIG wash and block buffer set; Roche) and incubated for 45 min with shaking at room temperature. Antidigoxigenin–alkaline phosphatase (AP) Fab fragments (Roche) at 1:10,000 were added, and incubation continued for 30 min. The membrane was washed and RNA-DNA hybrids were detected by chemiluminescence using CDP-Star according to the manufacturer's instructions.
Cloning of genes encoding the GlcNAc PTS and in vitro construction of a nag operon.
The cbe4532 and cbe4533 genes were cloned separately in pJET1.2/blunt (Fermentas) following PCR amplification from C. beijerinckii DNA using the primers listed in Table 1. Amplification was catalyzed by 1.5 U of Pfu DNA polymerase (Promega) in a total volume of 50 μl containing Pfu buffer, 2 mM MgCl2, 0.2 mM (each) dATP, dCTP, dGTP, and dTTP, 100 pmol of each primer, and 10 ng of C. beijerinckii DNA. Following heating at 95°C for 5 min, 30 reaction cycles were performed as for preparation of hybridization probes. Two microliters of the PCR product was ligated into pJET1.2/blunt according to the manufacturer's instructions, and the ligation mixture was used to transform E. coli TOP10 by the standard heat shock procedure. Transformants were selected on LB agar containing 50 μg ml−1 ampicillin and screened by PCR to confirm the presence of the insert and its orientation. The selected pJET1.2/blunt recombinants carried their respective inserts in the orientation that placed them under the control of the T7 promoter. Plasmid from a selected clone of each gene was purified and sequenced to confirm the absence of mutations.
The vector pJET-cbe4532 was digested with XhoI and SalI, and the purified 1,610-bp fragment carrying the cbe4532 gene was ligated into pJET-cbe4533 digested with the same enzymes. A recombinant carrying the two genes in the same orientation, with cbe4532 upstream of cbe4533, was selected following transformation of E. coli TOP10, and the structure of the purified plasmid was confirmed by PCR screening using the forward and reverse cloning primers for the genes. The plasmid was digested with BglII and XbaI to release a 2,263-bp fragment carrying the cloned genes, and this purified fragment was then ligated into pUC18 digested with BamHI and XbaI. The resulting plasmid recovered from transformed cells of E. coli TOP10 carried the artificially constructed nag operon under the control of the pUC18 lac promoter.
Complementation of E. coli mutants by phenotype screening.
Recombinant and control pUC18 plasmids were transformed into strains JW0665-1 and ZSC113 by the heat shock procedure. The fermentation phenotype was assessed after incubation for 48 h at 37°C on MacConkey agar containing the appropriate sugar.
Sequence analysis.
Protein sequences were obtained using the BLAST service at the National Center for Biotechnology Information (20). Multiple alignment of protein sequences was performed using ClustalW2 of the European Bioinformatics Institute (21), and phylogenetic trees were drawn using TreeView (22).
RESULTS
Phosphorylation of GlcNAc by cell extracts of C. beijerinckii.
Extracts of C. beijerinckii prepared from cells grown on GlcNAc exhibited PEP-dependent phosphorylation of the amino sugar, indicating the presence of a GlcNAc phosphotransferase system (Fig. 1A). On the other hand, phosphorylation was not supported by ATP, suggesting that the PTS is the only route of uptake and phosphorylation of GlcNAc in C. beijerinckii. Further analyses demonstrated that both soluble and membrane fractions of the extract were required for phosphorylation, as would be expected for a PTS, and also that glucose, but not chitobiose (the dimer of GlcNAc), could inhibit GlcNAc phosphorylation (data not shown). Thus, on the basis of substrate recognition, these results suggested that the GlcNAc PTS belongs to the glucose-glucoside PTS family rather than the lactose-diacetylchitobiose PTS family (23).
Fig 1.
Phosphorylation of GlcNac by extracts of C. beijerinckii grown on (A) GlcNAc or (B) glucose. ■, phosphorylation in the presence of PEP; ▲, phosphorylation in the presence of ATP; ○, control with neither PEP nor ATP. The values shown are the average of duplicate experiments.
Phylogeny of C. beijerinckii glucose-glucoside family phosphotransferase systems.
Of the 42 complete phosphotransferase systems encoded by the C. beijerinckii genome, seven are members of the glucose-glucoside family (sucrose subfamily), among which three (encoded by cbe0751, cbe4532 and cbe4533, and cbe4983 and cbe4982) are within the branch that contains characterized GlcNAc systems from other bacteria. The system encoded by the genes cbe4532 and cbe4533 has been annotated in the genome sequence database as an N-acetylglucosamine PTS, and phylogenetic analysis clearly shows that of the three C. beijerinckii systems, the Cbe4532 IIC domain is most closely related to previously characterized GlcNAc phosphotransferases (Fig. 2A). Overall, the Cbe4532 protein, which includes the IIC and IIB domains of the PTS, shows 45% identity to the NagP protein (IICB) of Bacillus subtilis and 43% identity to the corresponding region of the NagE protein (IICBA) of E. coli, while the IIA domain protein Cbe4533 has 45% identity to the IIA domain of E. coli NagE. The cbe4532 and cbe4533 genes are arranged divergently in the C. beijerinckii genome, and the adjacent gene, cbe4534, encodes a putative transcriptional antiterminator belonging to the BglG family (Fig. 2B). Since BglG-like antiterminators are characteristically involved in regulation of PTS-encoding operons (6, 8, 9), it seems likely that the function of Cbe4534 is to regulate expression of the cbe4532 and cbe4533 genes.
Fig 2.
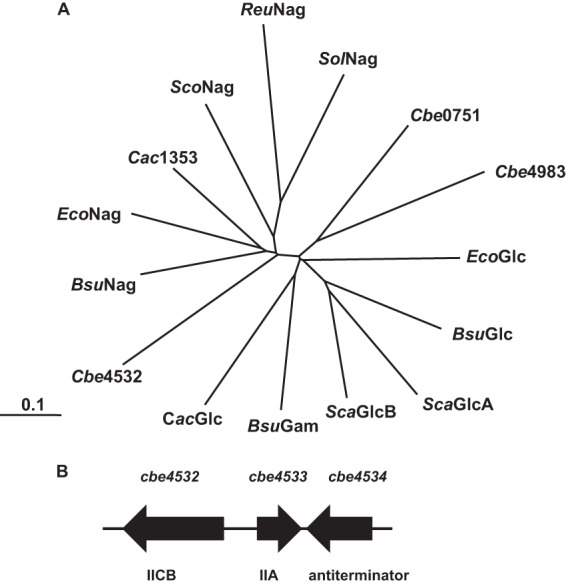
The putative N-acetylglucosamine PTS of C. beijerinckii. (A) Phylogeny of C domains of glucose-glucoside family phosphotransferase systems. The systems shown (GenBank accession numbers in parentheses) are all members of the glucose subfamily, and with the exception of numbered clostridial proteins, they have been characterized as either glucose (Glc), N-acetylglucosamine (Nag), or glucosamine (Gam) phosphotransferases: Cbe0751, Clostridium beijerinckii putative glucose PTS (YP_001307891); Cbe4983, C. beijerinckii putative glucoside PTS (YP_001312045); EcoGlc, Escherichia coli glucose (WP_000317748); BsuGlc, Bacillus subtilis glucose (NP_389272); ScaGlcA, Staphylococcus carnosus glucose (YP_002634092); ScaGlcB, S. carnosus glucose (YP_002634093); BsuGam, B. subtilis glucosamine (NP_388117); CacGlc, Clostridium acetobutylicum glucose (NP_347209); Cbe4532, C. beijerinckii putative N-acetylglucosamine PTS (YP_001311597); BsuNag, B. subtilis N-acetylglucosamine (NP_388651); EcoNag, E. coli N-acetylglucosamine (NP_415205); Cac1353, C. acetobutylicum putative N-acetylglucosamine PTS (NP_347981); ScoNag, Streptomyces coelicolor N-acetylglucosamine (NP_627133); ReuNag, Ralstonia eutrophus N-acetylglucosamine (YP_724831); SolNag, Streptomyces olivaceoviridis N-acetylglucosamine (CAD29623). (B) Chromosomal arrangement of genes cbe4532, cbe4533 and cbe4534. The genes encode the IICB domains of the PTS, IIA domain of the PTS, and BglG-like antiterminator, respectively.
Cloning and functional characterization of the cbe4532 and cbe4533 genes.
A direct approach toward characterization of pts genes from bacteria such as the clostridia is to clone them in E. coli and examine their ability to complement host activities (24, 25). Since it was possible that the unusual divergent gene arrangement of cbe4532 and cbe4533 and potential dependence of expression on Cbe4534 might compromise attempts to achieve that expression in E. coli, the strategy adopted involved construction of an artificial nag operon in which the two genes were aligned in the same orientation. Following PCR amplification and cloning of cbe4532 and cbe4533 individually, the genes were recombined as described in Materials and Methods. When transformed into the E. coli nagE mutant JW0665-1 and incubated on MacConkey agar containing 0.5% (wt/vol) GlcNAc for 48 h, a strong fermentation-positive phenotype was observed. On the other hand, a control transformant carrying the empty pUC18 vector was unable to ferment the amino sugar (Fig. 3A). In addition, the recombinant strain utilized GlcNAc at a much higher rate than the control strain when grown in LB broth (Fig. 3B). When extracts prepared from the two strains were assayed for GlcNAc PTS activity, the recombinant extract showed PEP-dependent GlcNAc phosphorylation, whereas the control extract lacked this activity (Fig. 3C). The results therefore demonstrated that Cbe4532 and Cbe4533 constitute a functional GlcNAc PTS.
Fig 3.

Complementation of E. coli JW0665-1 (nagE) by genes cbe4532 and cbe4533. A transformant containing recombinant pUC18 carrying cbe4532 and cbe4533 was compared with the control strain transformed with pUC18 alone. (A) Fermentation of GlcNAc on MacConkey agar. (B) Utilization of GlcNAc in cultures growing on LB broth. ■, growth of control; ●, growth of transformant; □, broth GlcNAc concentration of control; ○, broth GlcNAc concentration of transformant. (C) GlcNAc PTS activity in cell extracts. ■, transformant uninhibited; □, control uninhibited; ●, transformant in the presence of 10 mM glucose. The values shown in panels B and C are the average of duplicate experiments.
The recombinant vector carrying cbe4532 and cbe4533 was also transformed into E. coli ZSC113, which as a result of mutations in the glucose PTS, the mannose PTS, and glucokinase, is totally unable to phosphorylate either glucose or mannose (18). Transformants screened on MacConkey agar containing 1% (wt/vol) glucose showed a positive fermentation phenotype (Fig. 4A), but they showed no ability to ferment mannose at the same concentration (Fig. 4B). The ability to ferment glucose was reflected in the demonstration of glucose PTS activity in cell extracts (Fig. 4C). The GlcNAc PTS of C. beijerinckii therefore also has the ability to transport and phosphorylate glucose but apparently not mannose. Recognition of glucose was further demonstrated via the inhibition by glucose of GlcNAc phosphorylation catalyzed by the extract of recombinant JW0665-1 cells (Fig. 3C).
Fig 4.

Complementation of E. coli ZSC113 (Glc− Man−) by genes cbe4532 and cbe4533. The transformant containing recombinant pUC18 carrying cbe4532 and cbe4533 was compared with the control strain transformed with pUC18 alone. (A) Fermentation of glucose on MacConkey agar. (B) Fermentation of mannose on MacConkey agar. (C) Glucose PTS activity in cell extracts. ■, transformant in the presence of PEP; □, transformant in the absence of PEP; ●, control in the presence of PEP. The values shown are the average of duplicate experiments.
Expression of the C. beijerinckii nag pts genes.
Glucose has generally been observed to act as a repressor of utilization of alternative carbon sources by the solventogenic clostridia (5, 26). However, the dual substrate specificity of the PTS encoded by cbe4532 and cbe4533 may have consequences for control of expression of these genes. Their expression was therefore analyzed by hybridization of individual gene probes to RNA prepared from exponentially growing cells. Both cbe4532 and cbe4533 were expressed in cells growing on GlcNAc and apparently to a greater extent in cells grown on glucose (Fig. 5), but no hybridization was detected with RNA isolated from cells grown on glucitol. Therefore, it is clear that glucose did not repress expression of the cbe4532 and cbe4533 genes. On the contrary, since glucitol is not a repressing sugar in the clostridia (11, 27), the implication of these results is that both GlcNAc and glucose can act as inducers of gene expression.
Fig 5.

Slot blot analysis of C. beijerinckii nag gene expression. Expression of (A) cbe4532 and (B) cbe4533 was monitored following growth of C. beijerinckii in CBM containing GlcNAc, glucose, or glucitol.
Consistent with the observed expression of the putative nag genes in glucose-grown cells, an extract prepared from a glucose-grown culture of C. beijerinckii exhibited GlcNAc PTS activity at a rate slightly greater than that for GlcNAc-grown cells (Fig. 1B). Furthermore, when C. beijerinckii was grown in CBM containing glucose and GlcNAc, cbe4532 and cbe4533 were expressed (data not shown), and the two substrates were used simultaneously (Fig. 6). Therefore, although glucose has previously been shown to strongly repress utilization of disaccharides and glucitol in C. beijerinckii (11, 12, 27, 28), it has no such effect on utilization of GlcNAc. The increase in rate of GlcNAc utilization observed after 10 h did, however, suggest that glucose may have exerted a competitive inhibition on uptake of GlcNAc, which was relieved as glucose became exhausted.
Fig 6.
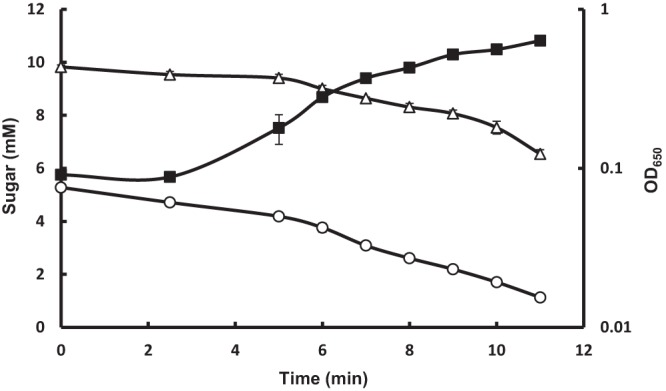
Growth and sugar utilization by C. beijerinckii in CBM containing glucose and GlcNAc. The inoculum cells were grown in CBM containing glucose and then washed and inoculated into medium containing both glucose and GlcNAc at the indicated concentrations. The values shown are means ± standard errors of the means of results from four cultures. ■, growth; ○, glucose concentration; △, GlcNAc concentration.
DISCUSSION
The revival of the ABE fermentation will be dependent on the identification of inexpensive and renewable substrates and the construction of strains designed to utilize these substrates efficiently. Substrate assimilation is an important control point in metabolism, and genetic modification of the ability of strains to accumulate a range of target substrates can make a major contribution toward fermentation development. Ideally, fermentation strains will exhibit rapid substrate uptake that is not subject to the normal regulatory controls (29).
Chitin-based substrates do not appear to have been considered as feedstocks for the ABE fermentation, and there have been no studies of GlcNAc uptake by the solventogenic clostridia. The present study therefore represents the first functional identification of a GlcNAc PTS in these bacteria, thus adding GlcNAc to the list of sugars and sugar derivatives that are substrates of the PTS in C. beijerinckii and related species. The PTS encoded by cbe4532 and cbe4533 comprises two substrate-specific proteins that provide the IICB and IIA domains, respectively. Cloning and complementation of E. coli mutants demonstrated that the system could transport glucose in addition to GlcNAc. This property has been observed previously for the GlcNAc PTS of Ralstonia eutropha (30), although not for the GlcNAc PTS of other organisms, including E. coli (31), B. subtilis (32), Streptomyces olivaceoviridis (33), and Streptomyces coelicolor (34). There is therefore no direct correlation between substrate specificity and the phylogenetic relatedness of GlcNAc PTS permeases (Fig. 2A). In comparison with systems found in other bacteria, we propose that the genes cbe4532 and cbe4533 be designated nagE and nagF, respectively.
Analysis of expression of nagE and nagF in C. beijerinckii showed that both substrates of the encoded PTS, GlcNAc and glucose, could act as inducers. This finding can be rationalized with the model for antiterminator-mediated control of pts operons (6, 35). Antiterminator proteins comprise a RNA-binding domain and two PTS-regulation domains (PRDs), which provide sites for phosphorylation by the PTS by which the activity of the protein is regulated. Phosphorylation of one of the PRDs by the IIB domain of the cognate PTS acts to inhibit antiterminator function. Thus, in the absence of the PTS substrate, when the PTS domains are phosphorylated, phosphate is transferred to the antiterminator and the cognate operon is not expressed. On the other hand, when a substrate of the PTS is being accumulated, phosphate is passed from the IIB domain to the incoming substrate, resulting in dephosphorylation and activation of the antiterminator and leading to induction of the operon. The PRDs of the putative antiterminator encoded by cbe4534 contain the conserved histidine residues that act as phosphorylation sites. Thus, expression of the genes encoding the GlcNAc PTS most likely responds to the substrate, as described, and since both GlcNAc and glucose are substrates, they will both induce expression. Elucidation of the role of Cbe4534 and the molecular mechanisms underlying the induction process requires further experimental analysis.
The ability of glucose to induce expression of the cbe4532 (nagE) and cbe4533 (nagF) genes has in fact been reported in a transcriptomic study of gene expression during glucose fermentation by C. beijerinckii (36). Indeed, these genes were found to be more strongly induced by glucose than those encoding the other members of the same PTS phylogenetic branch, Cbe0751 and Cbe4983, one or both of which may also transport and phosphorylate glucose. The implication is therefore that the NagE and NagF proteins may have an important role in sensing and responding to environmental glucose and may be involved in CCR. Assessment of this role may be possible by mutational analysis, although this is likely to be complicated by the large number of phosphotransferases in C. beijerinckii, many of which may have an ability to take up and phosphorylate glucose. Nevertheless in a recent study of C. acetobutylicum, although inactivation of the gene encoding the putative glucose PTS was found to have no effect on glucose PTS activity in cell extracts, the mutant did show modest relief from CCR exerted on metabolism of pentose sugars. This relief of CCR was further enhanced by cloning and overexpression of the xylose metabolic pathway in the mutant strain (37).
A variety of patterns of genetic organization of GlcNAc metabolism are evident in bacteria. In several cases, exemplified by E. coli (38) and R. eutropha (30), genes encoding a GlcNAc PTS are associated with genes encoding enzymes for metabolism of the intracellular PTS product N-acetylgucosamine 6-phosphate. Although this is not the case for C. beijerinckii, the fact that the bacterium grows well on GlcNAc indicates that the metabolic enzymes must be present. The closest relatives to N-acetylglucosamine 6-phosphate deacetylase (NagA) and glucosamine 6-phosphate deaminase (NagB) are the gene products of cbe4564 and cbe4562, respectively. Characterization of these genes should therefore be a priority for further understanding of GlcNAc metabolism. The organization of the nag pts genes in C. beijerinckii is not, however, unique, since a direct counterpart is found in the genome of C. acetobutylicum. The proteins encoded by cac1353 (PTS IICB), cac1354 (PTS IIA), and cac1355 (antiterminator) exhibit 41%, 53%, and 51% identity to the corresponding proteins of the GlcNAc system in C. beijerinckii. In a transcriptomic analysis of carbohydrate uptake and metabolism in C. acetobutylicum, Servinsky et al. (39) grew the bacterium on 11 different fermentable carbon sources and found that expression of cac1343 and cac1344 was not significantly affected by the growth substrate. Unfortunately, GlcNAc was not one of the substrates included in their analysis. They concluded that the genes may encode a constitutively expressed glucose PTS, although the possibility that it may transport an untested substrate was acknowledged. Similarities to the GlcNAc PTS of C. beijerinckii suggest that this system is likely to be the GlcNAc PTS of C. acetobutylicum, and it may be subject to a similar pattern of regulation as observed in this study.
In summary, a GlcNAc PTS from C. beijerinckii has been identified and characterized as a system that can transport and phosphorylate both GlcNAc and glucose. Both substrates induce expression of the system, suggesting that it may play a dual role in carbohydrate uptake, and the response to glucose is consistent with a potential regulatory function, which should be further assessed. In terms of evaluating C. beijerinckii for fermentation of chitin-containing wastes, it will be necessary to investigate its potential for chitin hydrolysis. We have observed chitin degradation by the bacterium but have yet to establish the genetic and biochemical basis of the chitinase activity.
ACKNOWLEDGMENTS
N.H.A.M. is grateful to King Abdulaziz University, Saudi Arabia, for financial support of this study.
Thanks are also extended to Michael Schweizer for provision of plasmid pUC18 and James Mackinlay for HPLC analysis of sugar concentrations.
Footnotes
Published ahead of print 30 August 2013
REFERENCES
- 1. Jones DT, Woods DR. 1986. Acetone-butanol fermentation revisited. Microbiol. Rev. 50:484–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dürre P. 2007. Butanol: an attractive biofuel. Biotechnol. J. 2:1525–1534 [DOI] [PubMed] [Google Scholar]
- 3. Dürre P. 2011. Fermentative production of butanol—the academic perspective. Curr. Opin. Biotechnol. 22:331–336 [DOI] [PubMed] [Google Scholar]
- 4. Green EM. 2011. Fermentative production of butanol—the industrial perspective. Curr. Opin. Biotechnol. 22:337–343 [DOI] [PubMed] [Google Scholar]
- 5. Mitchell WJ. 1998. Physiology of carbohydrate to solvent conversion by clostridia. Adv. Microb. Physiol. 39:31–130 [DOI] [PubMed] [Google Scholar]
- 6. Deutscher J, Francke C, Postma PW. 2006. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 70:939–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mitchell WJ, Tangney M. 2005. Carbohydrate uptake by the phosphotransferase system and other mechanisms, p 155–175 In Dürre P. (ed), Handbook on clostridia. CRC Press, Boca Raton, FL [Google Scholar]
- 8. Deutscher J. 2008. The mechanisms of carbon catabolite repression in bacteria. Curr. Opin. Microbiol. 11:87–93 [DOI] [PubMed] [Google Scholar]
- 9. Görke B, Stülke J. 2008. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol. 6:613–624 [DOI] [PubMed] [Google Scholar]
- 10. Shi Y, Li Y-X, Li Y-Y. 2010. Large number of phosphotransferase genes in the Clostridium beijerinckii NCIMB 8052 genome and the study on their evolution. BMC Bioinformatics 11(Suppl 11):S9. 10.1186/1471-2105-11-S11-S9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tangney M, Brehm JK, Minton NP, Mitchell WJ. 1998. A gene system for glucitol transport and metabolism in Clostridium beijerinckii NCIMB 8052. Appl. Environ. Microbiol. 64:1612–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Reid SJ, Rafudeen MS, Leat NG. 1999. The genes controlling sucrose utilization in Clostridium beijerinckii NCIMB 8052 constitute an operon. Microbiology 145:1461–1472 [DOI] [PubMed] [Google Scholar]
- 13. Tharanathan RN, Kittur FS. 2003. Chitin—the undisputed biomolecule of great potential. Crit. Rev. Food Sci. Nutr. 43:61–87 [DOI] [PubMed] [Google Scholar]
- 14. Horn SJ, Sikorski P, Cederkvist JB, Vaaje-Kolstad G, Sorlie M, Synstad B, Vriend G, Varum KM, Eijsink VG. 2006. Costs and benefits of processivity in enzymatic degradation of recalcitrant polysaccharides. Proc. Natl. Acad. Sci. U. S. A. 103:18089–18094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hayes M, Carney B, Slater J, Bruck W. 2008. Mining marine shellfish wastes for bioactive molecules: chitin and chitosan. Part A. Extraction methods. Biotechnol. J. 3:871–877 [DOI] [PubMed] [Google Scholar]
- 16. Yu Y, Aass HC, Tangney M, Mitchell WJ. 2007. Analysis of the mechanism and regulation of lactose transport and metabolism in Clostridium acetobutylicum ATCC 824. Appl. Environ. Microbiol. 73:1842–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. 10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Curtis SJ, Epstein W. 1975. Phosphorylation of D-glucose in Escherichia coli mutants defective in glucosephosphotransferase, mannosephosphotransferase, and glucokinase. J. Bacteriol. 122:1189–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zamenhof S. 1957. Preparation and assay of deoxyribonucleic acid from animal tissue. Methods Enzymol. 3:696–704 [Google Scholar]
- 20. Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Page RDM. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357–358 [DOI] [PubMed] [Google Scholar]
- 23. Barabote RD, Saier MH., Jr 2005. Comparative genomic analyses of the bacterial phosphotransferase system. Microbiol. Mol. Biol. Rev. 69:608–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brown GD, Thomson JA. 1998. Isolation and characterization of an aryl-β-d-glucoside uptake and utilization system (abg) from the Gram-positive ruminal Clostridium species C. longisporum. Mol. Gen. Genet. 257:213–218 [DOI] [PubMed] [Google Scholar]
- 25. Lai XK, Davis FC, Hespell RB, Ingram LO. 1997. Cloning of cellobiose phosphoenolpyruvate-dependent phosphotransferase genes: functional expression in recombinant Escherichia coli and identification of a putative binding region for disaccharides. Appl. Environ. Microbiol. 63:355–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tangney M, Mitchell WJ. 2005. Regulation of catabolic gene systems, p 583–605 In Dürre P. (ed), Handbook on clostridia. CRC Press, Boca Raton, FL [Google Scholar]
- 27. Mitchell WJ. 1996. Carbohydrate uptake and utilization by Clostridium beijerinckii NCIMB 8052. Anaerobe 2:379–384 [Google Scholar]
- 28. Mitchell WJ, Albasheri KA, Yazdanian M. 1995. Factors affecting utilization of carbohydrates by clostridia. FEMS Microbiol. Rev. 17:317–329 [Google Scholar]
- 29. Alper H, Stephanopoulos G. 2009. Engineering for biofuels: exploiting innate microbial capacity or importing biosynthetic potential? Nat. Rev. Microbiol. 7:715–723 [DOI] [PubMed] [Google Scholar]
- 30. Raberg M, Kaddor C, Kusian B, Stahlhut G, Budinova R, Kolev N, Bowien B, Steinbüchel A. 2012. Impact of each individual component of the mutated PTSNag on glucose uptake and phosphorylation in Ralstonia eutropha. Appl. Microbiol. Biotechnol. 95:735–744 [DOI] [PubMed] [Google Scholar]
- 31. Vogler AP, Broekhuizen CP, Schuitema A, Lengeler JW, Postma PW. 1988. Suppression of IIIGlc-defects by enzymes IINag and IIBgl of the PEP:carbohydrate phosphotransferase system. Mol. Microbiol. 2:719–726 [DOI] [PubMed] [Google Scholar]
- 32. Mobley HLT, Doyle RJ, Streips UN, Lengemeier SO. 1982. Transport and incorporation of N-acetyl-d-glucosamine in Bacillus subtilis. J. Bacteriol. 150:8–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang F, Xiao X, Saito A, Schrempf H. 2002. Streptomyces olivaceoviridis possesses a phosphotransferase system that mediates specific, phosphoenolpyruvate-dependent uptake of N-acetylglucosamine. Mol. Genet. Genomics 268:344–351 [DOI] [PubMed] [Google Scholar]
- 34. Nothaft H, Rigali S, Boomsma B, Swiatek M, McDowall KJ, van Wezel GP, Titgemeyer F. 2010. The permease gene nagE2 is the key to N-acetylglucosamine sensing and utilization in Streptomyces coelicolor and is subject to multi-level control. Mol. Microbiol. 75:1133–1144 [DOI] [PubMed] [Google Scholar]
- 35. Stülke J, Arnaud M, Rapoport G, Martin-Verstraete I. 1998. PRD—a protein domain involved in PTS-dependent induction and carbon catabolite repression of catabolic operons in bacteria. Mol. Microbiol. 28:865–874 [DOI] [PubMed] [Google Scholar]
- 36. Wang Y, Li X, Mao Y, Blaschek HP. 2012. Genome-wide dynamic transcriptional profiling in Clostridium beijerinckii NCIMB 8052 using single-nucleotide resolution RNA-Seq. BMC Genomics 13:102. 10.1186/1471-2164-13-102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xiao H, Gu Y, Ning Y, Yang Y, Mitchell WJ, Jiang W, Yang S. 2011. Confirmation and elimination of xylose metabolism bottlenecks in glucose phosphoenolpyruvate-dependent phosphotransferase system-deficient Clostridium acetobutylicum for simultaneous utilization of glucose, xylose, and arabinose. Appl. Environ. Microbiol. 77:7886–7895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Peri KG, Goldie H, Waygood EB. 1990. Cloning and characterization of the N-acetylglucosamine operon of Escherichia coli. Biochem. Cell Biol. 68:123–137 [DOI] [PubMed] [Google Scholar]
- 39. Servinsky MD, Kiel JT, Dupuy NF, Sund CJ. 2010. Transcriptional analysis of differential carbohydrate utilization by Clostridium acetobutylicum. Microbiology 156:3478–3491 [DOI] [PubMed] [Google Scholar]



