Abstract
Biofiltration of industrial carbon disulfide (CS2)-contaminated waste air streams results in the acidification of biofilters and therefore reduced performance, high water use, and increased costs. To address these issues, we isolated 16 extremely acidophilic CS2-converting Acidithiobacillus thiooxidans strains that tolerated up to 6% (vol/vol) sulfuric acid. The ecophysiological properties of five selected strains (2Bp, Sts 4-3, S1p, G8, and BBW1) were compared. These five strains had pH optima between 1 (2Bp) and 2 (S1p). Their affinities for CS2 ranged between 80 (G8) and 130 (2Bp) μM. Strains S1p, G8, and BBW1 had more hydrophobic cell surfaces and produced less extracellular polymeric substance than did strains 2Bp and Sts 4-3. All five strains converted about 80% of the S added as CS2 to S0 when CS2 was supplied in excess. The rate of S0 consumption varied between 7 (Sts 4-3) and 63 (S1p) nmol O2 min−1 ml culture−1. Low S0 consumption rates correlated partly with low levels of cell attachment to externally produced S0 globules. During chemostat growth, the relative amount of CS2 hydrolase in the cell increased with decreasing growth rates. This resulted in more S0 accumulation during CS2 overloads at low growth rates. Intermittent interruptions of the CS2 supply affected all five strains. Strains S1p, G8, and BBW1 recovered from 24 h of starvation within 4 h, and strains 2Bp and Sts 4-3 recovered within 24 h after CS2 was resupplied. We recommend the use of mixtures of Acidithiobacillus strains in industrial biofilters.
INTRODUCTION
Carbon disulfide (CS2) is a toxic, volatile, flammable, and explosive solvent widely used in, e.g., the viscose rayon industry (1). Because of its toxicity and the increasingly stringent rules governing the emission of harmful gases, it is necessary to treat CS2-containing industrial waste gases. Biological treatment of CS2 (and hydrogen sulfide, H2S) with sulfur-oxidizing bacteria provides an attractive alternative to conventional treatment systems (e.g., active carbon, incineration, caustic scrubbing) (2, 3). Typical concentrations of CS2 in contaminated air from viscose industries are around 4 to 20 nmol ml−1 (100 to 500 ppm) (2).
The number of microorganisms known to be able to grow chemolithoautotrophically on CS2 is limited to some Thiobacillus species, Thiothrix ramosa, Paracoccus denitrificans, and a Thiomonas sp. (4–8). All of these CS2-utilizing bacteria grow at neutral pH. Thus far, only one CS2-utilizing species (Thiobacillus sp. strain TJ330, DSM8985) capable of growth under acidic conditions (as low as pH 0.5) has been described (9). The only reported screening of 10 (Acidi)Thiobacillus strains showed that CS2 conversion is not a general trait of (acidi)thiobacilli (4). In that screening, only one strain, Thiobacillus thioparus TK-m, was found to be capable of CS2 conversion. We recently discovered that CS2 conversion is not limited to the domain Bacteria; the hyperthermophilic archaea Acidianus sp. strain A1-3 and Sulfolobus solfataricus P2 can also grow on CS2 as a main carbon and energy source (10). However, these archaea are not able to grow at the extremely low pH values that acidithiobacilli can cope with.
CS2-converting sulfur oxidizers in operating biofilters are acidophilic bacteria (2, 11–13). They convert CS2 via the two hydrolysis reaction steps CS2 + H2O → COS + H2S and COS + H2O → CO2 + H2S and obtain their energy from the oxidation of H2S via S0 and SO32− to SO42− as follows: H2S + 2O2 → SO42− + 2H+ (14). Therefore, an inherent result of CS2 conversion is acidification of the biofilters, which can be limited only by flushing the trickling filters with fresh water. Operating at a pH as low as possible will considerably reduce the volume of fresh water used for neutralization. Water use would be further reduced if the H2SO4 produced could be reused in the viscose-rayon industry. This becomes economically feasible when the H2SO4 concentration in the reactor effluent is at least 10% (wt/vol) (5.6% [vol/vol]). However, the performance of biotrickling filters is compromised by severe acidification and buildup of elemental sulfur (S0) that can clog the filters. Therefore, we set out to isolate new CS2-converting bacterial strains able to tolerate extremely low pH values with variable CS2 loads without loss of CS2 conversion efficiency and without the production of large amounts of elemental sulfur.
MATERIALS AND METHODS
Media and culture conditions.
Strains were enriched and cultured in basal salt mineral medium (MM) with CS2 as the sole carbon and energy source as described previously (15). Bacteria were grown at room temperature (RT, 22°C) in 120-ml bottles containing 20 ml MM acidified with sulfuric acid. Alternatively, bacteria were grown on MM plates solidified with 1% (vol/vol) Gelrite (16) and acidified with 0.1% (vol/vol) sulfuric acid. This was the maximum [H2SO4] at which plates could still be poured without the Gelrite solution immediately solidifying when brought into contact with the H2SO4-containing MM solution. In the case of Gelrite, H2SO4 increases solidification while the opposite occurs with agar(ose). Plates were incubated in an airtight jar. Unless stated differently, sulfuric acid concentrations are reported as percentages (vol/vol) (1% [vol/vol] is equivalent to 1.8% [wt/vol] and 0.18 M sulfuric acid). Strains were also grown in minichemostat reactors as described previously (15).
The headspaces of the bottles and jars used were continuously flushed with a CS2-containing air stream from a purpose-built distribution system (see the supplemental material).
Enrichment and isolation.
Volumes of 0.5 to 1 ml of environmental or industrial samples were inoculated into 120-ml serum bottles with 20 ml acidified MM. The initial H2SO4 concentration was 0.5 to 1% (vol/vol) in the environmental samples and 2% in the samples from the biotrickling filters. CS2 was supplied as the sole energy source via the distribution system. When a visually dense culture was obtained, the enrichment was transferred to fresh MM. The maximum H2SO4 concentration at which growth occurred was determined by the subsequent transfer of enrichments to MM with higher H2SO4 concentrations. Pure cultures were obtained from enrichment cultures grown in 4 to 6% H2SO4 on Gelrite plates. Single colonies were serially transferred three times to fresh plates and checked microscopically for purity. Fungal contamination (present mainly in the enrichments from industrial samples) was eliminated by adding 150 μg ml−1 chlorothalonil either to the plates or to liquid cultures. The H2SO4 tolerance of each isolated strain was confirmed by repeated subculturing at least three times in liquid MM containing 4 to 5% H2SO4.
Screening of known Acidithiobacillus strains for CS2 conversion capacity.
Five Acidithiobacillus strains were obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ) and grown in batch cultures in the media suggested (DSMZ medium number in parentheses after the strain name) or as otherwise stated, i.e., A. ferrooxidans DSM14882 (882), A. caldus DSM8584 (150a), A. thiooxidans DSM14887 (71), A. thiooxidans DSM504 (MM plus 10 g liter−1 sterile S0), and A. albertensis DSM14366 (71). After growth for 1 week, the headspace of the cultures was supplemented with CS2 (20 to 30 nmol ml−1). The CS2 concentration and the presence of intermediates (H2S and COS) in the headspace were monitored over time by gas chromatography (17).
PCR, cloning, and sequencing.
DNA was isolated from each strain by phenol extraction (18). The 16S rRNA gene and the 16S-23S intergenic spacer region (ISR) of 16 isolated CS2-hydrolyzing bacterial strains were amplified by hot-start PCR with the GoTaq Green buffer system (Fermentas) with 2.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphates, 1 μl bovine serum albumin, 0.4 μM each primer (see Table S1 in the supplemental material), and 1 μl Taq polymerase (Fermentas). The PCR protocol consisted of 2 min at 95°C; 30 cycles of 1 min at 95°C, 1 min at 54 to 65°C, and 2 min at 72°C; and a final elongation step of 10 min at 72°C. PCR products were ligated into pGEM-T Easy (Promega) and transformed into Escherichia coli strain TOP10 (Invitrogen) according to the manufacturers' instructions. Plasmids with a correct insert were Sanger sequenced by the sequencing facility at the Department of Human Genetics of the University Medical Center St. Radboud, Nijmegen, The Netherlands. 16S ISR contigs were assembled by using Contig Express (Vector NTI; Invitrogen), and phylogenetic relationships were inferred by the neighbor-joining method (19) in MEGA 4.0 (20) with the following settings: maximum composite likelihood nucleotide substitution model, gaps and missing data eliminated, transitions and transversions included, uniform rates among sites, and homogeneous pattern among lineages. Sequences of the ribosomal operon of A. thiooxidans ATCC 19377 and A. caldus ATCC 51756 were kindly made available by Jorge Valdés and David Holmes, CBGB, Santiago, Chile (21).
Cryo-SEM.
Cryo-scanning electron microscopy (cryo-SEM) was used to study the morphology of colonies growing on 1% Gelrite plates acidified with 0.018 M H2SO4 by a method similar to that described in reference 22. Blocks containing colonies were cut out of the Gelrite, mounted on electroconductive aqueous colloidal graphite (DAG; Agar Scientific) on a mounting stub, and quickly frozen by submersion in degassed liquid N2 (−196°C). While under vacuum, the sample was transferred to the Gatan cryotransfer box. Sections through colonies were made with a razor blade at this stage when required. The temperature was subsequently increased to −100°C to sublime off water that had settled on top of the specimen by condensation for a period of 5 min. When all of the surface water had been removed, the temperature was reduced to between −100 and −150°C. The sample was then sputter coated for 45 s with a mixture of 60% gold and 40% palladium and transferred to a JEOL 6330F scanning electron microscope.
Yield determinations.
For total carbon measurements, 3- to 4-ml reactor samples were centrifuged and the pellets were resuspended in 2 ml 1 mM HCl, pH 3. The washed cells were dried overnight under vacuum at 70°C. The C/N ratio of the dried material (0.3 to 0.4 mg) was determined by elemental analysis with a Thermo Fisher Scientific EA 1110 CHN element analyzer coupled to a Finnigan DELTAplus mass spectrometer. For protein determinations, 2-ml reactor samples were centrifuged at 4°C for 30 min at 18,000 × g. The pellets were resuspended in 0.5 ml 1 M NaOH, boiled for 5 min, and neutralized with 0.5 ml 1 M HCl. Alternatively, 0.5 ml 1 M NaOH was added directly to 200-μl reactor samples, and after boiling, the mixture was neutralized with 0.3 ml 1 M HCl. Protein concentrations were determined with the Bio-Rad protein assay kit according to the manufacturer's instructions. Culture density, determined by both cell density and the presence of S0, was measured spectrophotometrically as the optical density at 600 nm (OD600).
Determination of pH optima.
pH optima were determined by a floating-filter method. Samples taken from the steady-state chemostats (1% H2SO4; measured pH, 0.72) were diluted 106-fold in MM at pH 2, and 1 ml of this dilution was filtered through a sterile, 0.2-μm, 25- or 47-mm-diameter Cyclopore polycarbonate filter (Whatman). Filters were then floated on 20 ml MM acidified with sulfuric acid to pHs of 0.5 to 6. They were placed in airtight jars and incubated at RT for 16 days with a continuous flow of 45 ml min−1 air containing 10 nmol CS2 ml−1. Growth at different pHs was determined by measuring colony diameters and counting colonies. The growth of strain BBW1 was quantified visually, as cells had spread over the filter and over the surface of the medium.
Cell surface hydrophobicity.
Cell surface hydrophobicity was determined by a modified form of the method of Rosenberg et al. (23). The pH of culture samples from the minichemostats was adjusted to 3 or 7 with PUM buffer (23). The suspensions were diluted to an OD400 of 0.45 (A0) in PUM buffer with the appropriate pH. In a test tube, a 1-ml suspension was mixed with 200 μl n-octane or n-hexane. The mixture was incubated at RT for 10 min, mixed vigorously for 1 min, and left to stand at RT for at least 25 min. OD400 was measured (At), and the percent adherence to the solvent was calculated with the equation (1 − At/Ao) × 100. The use of OD600 to measure suspension turbidity yielded similar results.
Preparation of cell extracts.
Cell extracts from steady-state reactor-grown bacterial cells were prepared as follows. Thirty to 50 ml was removed from the reactors and centrifuged at 4°C for 30 min at 12,000 × g. The cell pellets were washed with 15 ml sterile distilled H2O and resuspended in 0.5 ml 20 mM KPi at pH 7. Approximately 350 μl glass beads (80- to 110-μm size) were added, and the cells were broken by bead beating for 2 × 2 min at 30 Hz (Retsch) with intermittent cooling on ice. The broken cell mixtures were centrifuged for 5 min at 16,000 × g, and the supernatants were stored at −20°C with a final concentration of 10% glycerol.
Enzyme kinetics based on H2S measurements.
The Michaelis-Menten constants Km and Vmax were determined for the CS2 conversion rates of cell extracts of the different strains by measuring the H2S production rate with an H2S microsensor (Unisense) in 20 mM HEPES (pH 7) as described previously (15). Experiments were performed at pH 7, as the CS2 hydrolase is predicted to reside in the cytoplasm of the cell because of the absence of a signal sequence at the N terminus of the enzyme. The Km and Vmax values were calculated from Michaelis-Menten plots by nonlinear regression with the Michaelis-Menten equation V = Vmax × [CS2]/(Km + [CS2]). Experiments were repeated at least three times, and average values of three independent experiments ± the standard errors of the means were calculated. The same method was used to determine Vmax values (with 600 μM CS2 as the substrate) for steady-state cells from the minichemostats (D = 0.02) diluted 200× in reactor medium (MM containing 1% H2SO4). Rates were normalized to the reactor OD600.
Sulfur (S0) determination.
Sulfur was determined by a modified form of the method of Sorbo (24) to reduce interference from medium components and loss of sulfur during processing. Polycarbonate filters (0.1-μm Whatman Cyclopore track etched membrane 7060-2501 or Millipore Isopore VCTP filters) were rinsed with MilliQ water and placed on a vacuum filter unit. Liquid samples from batch and continuous cultures, as well as bacteria removed from Gelrite plates and resuspended in 0.5 ml sterile demineralized water, were immediately filtered to prevent the bacteria from metabolizing the S0 before processing. Up to 5 ml culture was carefully loaded directly onto the membrane to prevent S0 from sticking to the glass of the vacuum unit and filtered under vacuum. Controls consisted of 3 ml MM containing 1% H2SO4. The filters were rinsed with 1 ml MilliQ water and inserted into a 2-ml Eppendorf tube, and 1.5 ml 0.1 M KCN was added immediately to stop the cells from metabolizing the S0. The samples were incubated at 90°C for 10 min and cooled to RT, and 200 μl 0.75 M FeNO3 in 20% HNO3 was added. Samples were centrifuged for 5 min at 16,000 × g to pellet precipitate cell debris and filters, and the supernatant absorbance was measured immediately at a wavelength of 460 nm. Standard curves were prepared in the same manner, with a solution of S0 in acetone (259 mg in 100 ml) diluted in MilliQ water to a final amount of up to 75 μg S0 per filter.
S0, H2S, and SO32− accumulation in bacterial cells.
The accumulation of intermediates during CS2 respiration was measured simultaneously but in separate reaction chambers as follows. Samples from the minichemostats were diluted 10-fold with O2-saturated MM containing 1% H2SO4. The diluted cultures were used to fill three glass cuvettes containing stir bars, after which the cuvettes were closed and placed in a 22°C water bath for 35 min with vigorous stirring. CS2 (17 μM) was added to each cuvette, and simultaneous measurements were then made of (i) O2 respiration with an O2 sensor (Unisense), (ii) H2S production with an H2S sensor (Unisense), and (iii) S0 production by following the change in OD480 with an Agilent 8453 spectrophotometer. In addition and simultaneously, a 100-ml glass syringe containing a stir bar was filled with diluted culture and incubated in a 22°C water bath for 35 min with continuous stirring. CS2 was also added to the syringe, and at intervals during the respiration and H2S production curves, samples were pushed out of the syringe into an Eppendorf tube and frozen immediately for subsequent SO3− determination.
In order to study H2S and S0 formation kinetics during CS2 respiration in more detail, these parameters were also simultaneously measured in one cylindrical cuvette with three entry ports, which was positioned in a single-beam spectrophotometer. Two ports were used for O2 and H2S sensors, and the third served for the addition of CS2.
The percentage of S0 accumulated was calculated as follows. The total O2 consumption is represented by the formula H2S + 2O2 → H2SO4. In a respiration curve, the total O2 consumed (A) will depend on the total amount of H2S (produced from CS2), and so, A/2 represents the amount of sulfide oxidized to sulfate. Sulfur S0 is an intermediate and is oxidized according to the formula S0 + 1.5O2 + H2O→ H2SO4. At the sulfur peak (A480), only S0 is present and the amount of oxygen consumed (a) from then until the end represents sulfur oxidation, and so, a/1.5 represents the amount of S0 present at the peak. This gives a S0/total sulfide ratio of [a/1.5]/[A/2] = 1.33 × a/A, where a/A is the fraction of the oxygen consumption after the sulfur peak compared to the total consumption, determined from the respiration curve.
Sulfite (SO32−) determination.
Sulfite (SO32−) was measured by the method described by Trueper and Schlegel (25) but with reduction of the amount of H2SO4 in the assay to take into account the amount of H2SO4 that is present in the samples (in these experiments, 1% [vol/vol]). Dilutions of an anaerobic 1 M stock of Na2SO3 were used as standards.
Starvation experiments.
Starvation experiments were performed in steady-state chemostats by stopping the influent and effluent pumps and removing the CS2 supply. One-milliliter samples were removed at regular intervals from the reactors during starvation and recovery, diluted 10× in MM containing 1% (vol/vol) H2SO4, and incubated in a 7.63-ml double-port cuvette at 22.0 to 22.2°C. Ten minutes after the sample was removed from the chemostat, a 10 μM CS2 pulse from a 6 mM stock bottle (see above) was injected into the cuvette. H2S production and removal, as well as respiration, were measured simultaneously with an H2S microsensor (Unisense) and an oxygen sensor (Strathkelvin Instruments), respectively.
Nucleotide sequence accession numbers.
The 16S ISR sequences determined in this research have been deposited in the GenBank database under accession numbers KC902816 to KC902829 and KC902831.
RESULTS
Enrichment and isolation of CS2-utilizing bacteria.
To obtain extremely acidophilic CS2-converting microorganisms, samples from naturally acidic, sulfur-rich environments and from industrial biotrickling filters were incubated in acidified MM and with CS2 as the sole energy source. Significant CS2 conversion was observed within a few days, and dense cultures were obtained within 2 to 4 weeks of incubation with 1 to 4% H2SO4. Ultimately, some enrichment cultures showed growth at 6% H2SO4 (Table 1). This concentration is equal to a theoretical pH of −0.05. Enrichments containing 4 to 6% H2SO4 (Table 1) were used to isolate 16 pure cultures on Gelrite plates (0.1% H2SO4). To reconfirm their acid tolerance, all isolates were successfully transferred to 4% H2SO4 medium.
Table 1.
Comparison of isolated strains of CS2-converting microorganisms
| Sample type and origin | Sample pH | Strain | % H2SO4 (vol/vol) |
Colony morphology | |
|---|---|---|---|---|---|
| Enrichment | Max | ||||
| Environmental | |||||
| Various hot springs | 3.1 | 2Ap | 2 | 5 | Cream colored, smooth domed |
| 2.5 | 2Bp | 4 | 5 | Cream colored, smooth domed | |
| 1.9 | 1Bp | 3 | 5 | Cream colored, spread out, large | |
| Solfatara, Italy | 1.9 | S1p | 4 | 5 | White, dry, irregular domed |
| 0.3 | Sts 4-3 | 4 | 5 | Cream to brown, small | |
| Industrial | |||||
| Fabelta | ND | G8 | ND | 4 | White, spreading, flat |
| Loudon | 3.9 | BAD2 | 5 | 6 | Cream-brown, small |
| 3.9 | BAW3 | 5 | 6 | White, spread out, large | |
| 3.2 | BBF2 | 5 | 6 | Cream to brown, small | |
| 3.2 | BBW1 | 5 | 6 | White, spread out, large | |
| 3.9 | BA6-2 | 6 | 6 | Yellow or cream, large | |
| Osceola | 1 | BC5-1 | 5 | 5 | Shiny, cream colored |
| Oy Visko | 1.2 | BDW2 | 5 | 6 | White, spread out, large |
| DOW Chemical Company | 1 | BEF1 | 5 | 5 | Cream to brown, small |
| 1 | BEF3 | 5 | 5 | Cream to brown, small | |
| 1 | BED2 | 5 | 5 | Cream to brown, small | |
Descriptions of growth characteristics on plate cultures and in liquid cultures are given in the supplemental material and shown in Fig. S1 and S2 in the supplemental material. The 16 isolated strains showed distinctly different colony morphologies, ranging from large, dry, white colonies to smooth, shiny, compact colonies. Stationary-phase liquid cultures showed white or yellow aggregates or little aggregation. From the cryo-SEM analysis of surfaces and cross sections of colonies, we conclude that on solid medium, the compactly growing strains produce more extracellular polymeric substance (EPS) than the spreading strains do.
Phylogenetic analysis.
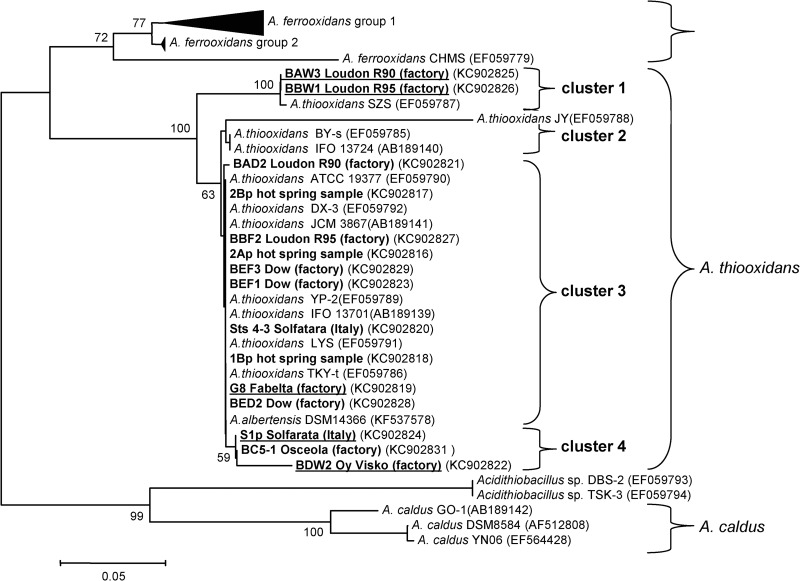
All of the new CS2-converting isolates were identified as Acidithiobacillus thiooxidans strains by conventional 16S rRNA gene analysis (data not shown). Improved discrimination was achieved by analysis of the 16S-23S ISR sequences, containing two tRNA genes and three intergenic transcribed spacers (ITS). The total length of the ISR sequences of the CS2-converting isolates varied between 456 and 460 bp, with one exception; isolate BDW2 from the Oy Visko reactor had an ISR of 439 bp (see Table S2 in the supplemental material). Within the ISR, most of the variation in nucleotide composition was observed in the third of the three ITS. Strains BAW3 and BBW1, which originated from two parallel reactors operating at the Loudon factory, were distinctly different with respect to their nucleotide composition, differing by 57 bp and harboring three insertions and three deletions compared with reference strain G8. In a neighbor-joining phylogenetic tree constructed from the 16S ISR sequences of the CS2-converting strains and other A. thiooxidans strains, strains BAW3 and BBW1 form a distinct cluster within the A. thiooxidans strains (Fig. 1). Three more clusters could be distinguished, i.e., cluster 2, which did not contain any of the CS2-converting strains; cluster 3, comprising most of the CS2-converting strains and also A. albertensis, described as a distinct species but phylogenetically indistinguishable from A. thiooxidans (26); and cluster 4, containing strain S1p from the Solfatara (Rome, Italy), strain BC6-1, and strain BDW2 from the Oy Visko plant.
Fig 1.
Phylogenetic tree based on the 16S-23S ISR of CS2-converting A. thiooxidans strains isolated from environmental samples (1Bp, 2Ap, 2Bp, Sts 4-3, and S1p) or various industrial CS2 biofilter effluents (G8, BAD2, BED2, BBF2, BBW1, BAW3, BEF1, BDW2, BC6-1, and BEF3). The optimal neighbor-joining tree with a branch length sum of 0.98269283 is shown. The tree is drawn to scale, with branch lengths in the same units as the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed by the maximum composite likelihood method and are expressed as the number of base substitutions per site. All positions containing gaps and missing data were eliminated. There were a total of 330 positions in the final data set. The percentage above 50% of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) is shown next to the branches. Four clusters of A. thiooxidans strains were identified, based on branch points that were reproduced in more than 50% of the bootstrap replicates. The type strains are A. thiooxidans ATCC 19377, A. albertensis DSM14366, and A. caldus DSM8584. In bold and underlined are strains that form white, large, dry, crusty colonies on Gelrite plates. In bold are strains that form small, shiny, smooth colonies on Gelrite plates.
Screening of Acidithiobacillus reference strains for CS2 conversion.
To check whether CS2 conversion is a general trait of acidithiobacilli under our growth conditions, we tested four publicly available Acidithiobacillus species. Of these, the three mesophilic Acidithiobacillus species (A. albertensis DSM14366, A. ferrooxidans DSM14882, and A. thiooxidans DSM504 and DSM14887) were not able to convert CS2 during the 2 to 5 days the cultures were monitored. Since a hierarchical utilization of energy sources may still be possible, CS2 utilization cannot be completely ruled out. However, the moderate thermophile A. caldus 8584 was found to be capable of CS2 conversion as soon as CS2 was added to a growing culture.
Maximum acid tolerance.
The maximum acid tolerance of all 16 isolates in batch cultures was determined by serially transferring the strains to medium containing gradually higher concentrations of H2SO4. The maximum H2SO4 concentration in which growth could be observed within 2 months was taken as the growth limit of each organism. Table 1 shows that all of the isolated strains were able to grow at 4% H2SO4, which corresponds to a pH of 0.12. All of the strains but G8 grew at 5% H2SO4 (pH 0.03), and several of the strains isolated from industrial biofilters still showed growth at 6% H2SO4 (pH −0.05). Growth at concentrations higher than 4% H2SO4 was slow; it took up to 2 to 3 months to reach an OD600 of about 0.1 at 6% H2SO4.
Chemostat growth of CS2-converting A. thiooxidans strains 2Bp, Sts 4-3, S1p, G8, and BBW1.
To identify the strains that have the highest affinity for CS2 and produce little sulfur under highly acidic conditions, 5 of the 16 isolated CS2-converting A. thiooxidans strains were selected for further comparison. Selection was based on sample location, different genetic groups as determined by 16S ISR analysis (Fig. 1), and different colony morphologies on plates. The five strains were grown in continuous culture under identical conditions in MM with 1% H2SO4. The strains had comparable growth yields, as determined by comparing OD, total protein, and total carbon measurements (see Table S3 in the supplemental material). The OD of strain Sts 4-3 was consistently lower than that of the other four strains, probably because of the observed smaller amount of elemental sulfur in the culture.
The μmax values of the strains growing under these conditions were determined by increasing the medium flow and simultaneously increasing the [CS2] supplied to the reactors with the same factor (resulting in a constant CS2 load per milliliter of medium input), up to and beyond the point where S0 formation started becoming visible in the reactors. Strains S1p and G8 had slightly higher μmax values than the other three strains (Table 2).
Table 2.
Comparison of CS2-converting A. thiooxidans strains 2Bp, Sts 4-3, S1p, G8, and BBW1a
| Characteristic | 2Bp | Sts 4-3 | S1p | G8 | BBW1 |
|---|---|---|---|---|---|
| Colony morphology | Smooth, domed | Small, smooth, domed | White, dry, compact | White, dry, spreading | White, dry, spreading |
| A. thiooxidans 16S ISR cluster (Fig. 1) | 3 | 3 | 4 | 3 | 1 |
| Maximum acid tolerance (% [vol/vol] sulfuric acid) | 5 | 5 | 5 | 4 | 6 |
| pH optimum | 1 | 1.5 | 2 | 1.5 | 1–1.5 |
| pH tolerance | 0.5–6 | 0.5–5 | 1–6 | 0.5–6 | 0.5–3.5 |
| Cell surface hydrophobicity | Low | Low-intermediate | High | High | High |
| Reactor dilution rate at point of S0 formation (approaching μmax) | 0.09 | 0.08 | 0.1 | 0.1 | 0.09 |
| Mean cell extract Km (μM CS2) ± SEM | 130 ± 22 | 97 ± 10 | 100 ± 5 | 81 ± 7 | 116 ± 4 |
| Mean cell extract Vmax (μmol H2S min−1 mg protein−1) ± SEM | 38 ± 3 | 17 ± 0 | 48 ± 7 | 23 ± 1 | 28 ± 5 |
| kcat/Km (μM−1 s−1) | 3.5 | 2.1 | 6.5 | 3.4 | 3.3 |
| Mean whole-cell Vmax (μmol H2S min−1 ml culture−1 for OD600 of 1) ± SEM | 11 ± 0 | 10 ± 0 | 12 ± 0 | 8 ± 0 | 11 ± 0 |
| Mean calculated S0 accumulated (% of total S pulse added) ± SEM | 76 ± 4 | 83 ± 1 | 82 ± 1 | 83 ± 1 | NDb |
| Mean measured S0 accumulated (% of total S pulse added) ± SEM | 67 ± 2 | 63 ± 2 | 65 ± 1 | 61 ± 1 | ND |
| Mean respiration rate on H2S and S0 combined after a 10 μM CS2 pulse (nmol O2 min−1 ml culture−1 for OD600 of 1) ± SEM | 45 ± 9 | 43 | 87 ± 10 | 101 ± 1 | 66 ± 7 |
| Mean respiration rate on S0 only after 10 μM CS2 pulse (nmol O2 min−1 ml culture−1 for OD600 of 1) ± SEM | 41 ± 3 | 7 | 63 ± 3 | 44 ± 1 | 49 ± 1 |
| Mean % of S0 globules attached to cells after 10 17 μM CS2 pulses ± SEM | 87 ± 7 | 17 ± 4 | 9 ± 2 | 52 ± 2 | 22 ± 2 |
| Mean % of cells attached to S0 globules after 10 17 μM CS2 pulses ± SEM | 35 ± 10 | 13 ± 3 | 7 ± 2 | 30 ± 6 | 13 ± 0 |
| Resistance to CS2 stress | Low | Medium | Medium | Medium | Medium |
| Speed of recovery after CS2 starvation | Slow | Slow | Fast | Fast | Fast |
Strains were grown in continuous culture (D = 0.02) with CS2 as the sole energy source. Samples were taken from these reactors for the comparison experiments. The Km and Vmax of each strain were calculated from Michaelis-Menten plots by nonlinear regression with the Michaelis-Menten equation V = Vmax × S/(Km + S). Mean values from at least three independent experiments are shown.
ND, not determined.
Cell surface hydrophobicity.
The differences in growth characteristics on plates and in liquid culture suggested that there may be differences in the surface properties of the strains. To test this, cell surface hydrophobicity was measured by using adherence to n-octane and n-hexane (23). Strains 2Bp and Sts 4-3 had less hydrophobic cell surfaces than strains S1p, G8, and BBW1 (see Fig. S3 in the supplemental material). On plates, strains 2Bp and Sts 4-3 produced compact, smooth, and opaque colonies versus the dry and spread-out colonies of strains S1p, G8, and BBW1. Similar results were obtained when experiments were performed at pH 7 (see Fig. S3A and B) or 3 (see Fig. S3C and D); therefore, a different response to high-pH shock is not the cause of the differences in surface hydrophobicity observed.
pH optimum and pH tolerance.
The pH optima of the five selected strains were determined with a floating-filter assay on media with pHs ranging from 0.5 to 6. The pH optima varied among 1 (2Bp), 1.5 (Sts 4-3 and G8), and 2 (S1p) (Table 2). Strain BBW1 did not form colonies but showed spreading surface growth on the filter and the medium as well. On the basis of visual observations, the pH optimum of this strain was between 1 and 1.5 (Table 2).
Tolerance of different pH levels was estimated by counting the colonies that appeared on the filters after the transfer of cells growing at 1% H2SO4 (measured pH, 0.72) to media with lower or higher pHs. Survival rapidly decreased when cells were exposed to more acid (pH 0.5) and more gradually decreased when cells were exposed to higher-than-optimum pHs. Strain BBW1 was the most sensitive to high-pH stress; no growth was observed in medium with a pH higher than 3.5.
Enzyme kinetic analysis.
To assess the CS2 removal efficiencies of the five new CS2-converting A. thiooxidans isolates, the CS2 affinity constants (Km) and maximum CS2 conversion rate (Vmax) of crude protein extracts from continuous cultures were determined by adding a pulse of CS2 to diluted protein extracts and measuring the rate of H2S production. Table 2 shows that the Km values of the five strains are of the same order of magnitude, ranging between 81 and 130 μM CS2 for strains G8 and 2Bp, respectively. Comparison of the Vmax values revealed crude extracts from strain S1p to have a consistently higher Vmax under these conditions than crude extracts from the other four strains tested, resulting in a 2-fold higher kcat/Km value for S1p. In accordance, the Vmax for CS2 of steady-state, minichemostat-grown, intact S1p cells was consistently slightly higher than the Vmax of the other strains (Table 2).
Sulfur production and consumption. (i) Sulfur production.
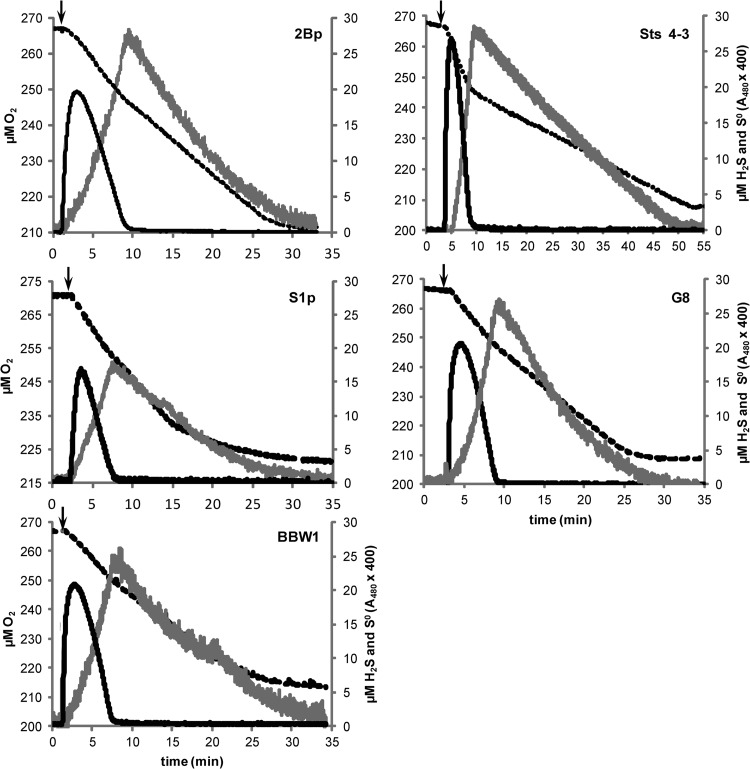
Production of S0 by CS2-converting acidithiobacilli as an intermediate in the oxidation of H2S to SO42− can block biotrickling filters and subsequently decrease performance. Therefore, we compared the H2S and S0 production and consumption of the five selected A. thiooxidans strains upon CS2 pulses. When concentrations as low as 1.8 μM CS2 (equivalent to 36 ppm CS2 in the gas phase) were added to a cuvette containing diluted samples from the minichemostats, there was an almost immediate increase in S0, just above the detection level. Peak concentrations of H2S ranged from 0.5 to 1.0 μM under these conditions, and so, S0 formation already started at H2S concentrations below this value. In subsequent experiments, H2S and S0 production was measured simultaneously in one cuvette. Addition of 17 μM CS2 resulted in an immediate and large accumulation of H2S for all strains. Also, S0 formation started almost without delay and continued until virtually all of the H2S had been consumed (Fig. 2). The respiration curves show an initial period of fast respiration when H2S is still present, followed by a sudden decrease in the respiration rate when the cells start respiring solely on S0. This decrease in the respiration rate indicates that the processing of the S intermediates in the cell, and not respiration, is the rate-limiting step. This was observed in all of the strains, but it was most prominent in strain Sts 4-3 (Fig. 2). The chemical reaction of H2S with O2 was less than 2 μM H2S h−1 and therefore did not significantly contribute to the observed oxygen consumption rates.
Fig 2.
Conversion of CS2 by five A. thiooxidans strains. The arrows indicate the times when 17 μM CS2 was injected into a cuvette containing cells from a continuous culture growing at D = 0.02, diluted 6× in MM containing 1% H2SO4. Respiration (O2 consumption, black dashed line), H2S production (black solid line), and S0 production (gray line) were monitored. The production and consumption rates in these graphs were used to determine the percentages of S0 accumulation in Table 2. Note the different scale on the x axis of the graph for strain Sts 4-3.
During the period of S0 respiration, we often observed a temporary reduction in the respiration rate. Although it is possible that an intermediate temporarily accumulated that inhibited respiration on S0, at this point, the S0 consumption rate did not decrease. Also, although detectable levels of the potentially inhibiting intermediate SO32− were present throughout the experiment, there was no increase at the reduced respiration “bump.” Therefore, the apparent temporary reduction in respiration is more likely due to the sensor being affected by an intermediate at that point in the experiment.
At the point where S0 peaks, the H2S and CS2 had been completely consumed and only 30 to 40% of the total oxygen was consumed. Therefore, the remaining O2 consumption resulted entirely from S0 oxidation, if the accumulation of any other intermediates is excluded. With this O2 consumption, and taking into account that this is an underestimate of the actual O2 consumption because of a small baseline drift upward of the O2 sensor during the experiment, we calculated that at least 80% of the S added as CS2 accumulated as S0. This was similar for all of the strains tested (Table 2), and the percentage also remained identical when CS2 concentrations as low as 1.8 μM were added. Below this value, H2S formation could not be observed any more, making such calculations impossible.
The rates of S0 formation and consumption were calculated from the slopes before and after the apex of the OD480 trace (Fig. 2). Consistently, S0 production (corrected for consumption) was between four times (strains 2Bp, S1p, G8, and BBW1) and nine times (strain Sts 4-3) faster than S0 consumption. This matches with a buildup of S0 of at least 80% of the total S added, as calculated above from the oxygen respiration. In separate experiments, chemical analysis for S0 at the moment OD480 reached its maximum value consistently showed S0 recovery between 60 and 67% (Table 2). Therefore, chemical analysis appeared to underestimate the amount of S0 produced during CS2 respiration. However, both methods of determining S0 accumulation upon the application of CS2 pulses indicated that all of the strains tested produced about equal amounts of extracellular S0 under conditions of excess CS2.
(ii) Sulfur consumption.
Although all of the strains tested behaved similarly in terms of S0 production, there were some obvious differences in subsequent S0 respiration (Fig. 2); strain Sts 4-3 respired on S0 five to eight times more slowly than the other four strains. The H2S combined with S0 respiration rates (the first part of the respiration curve) showed a maximum 2-fold difference between the strains (Fig. 2 and Table 2). Although strain Sts 4-3 respired more slowly on S0, the respiration rate did not decline until all of the S0 had been depleted. This was in contrast to the other strains, especially strain S1p, which showed a continuous decline in the rate of respiration and concurrent S0 depletion.
We hypothesized that different strains produce differently bioavailable S0; strains 2Bp, G8, and BBW1 produce S0 that can be efficiently metabolized again, whereas strains Sts 4-3 and S1p produce S0 that is difficult to remove. To test this hypothesis, we examined sulfur accumulation by different strains upon the application of repeated CS2 pulses to undiluted chemostat samples by counting S0 globules under a light microscope (Table 2). We counted 160 to 240 cells of each strain, and a representative picture of strains 2Bp, Sts 4-3, S1p, and G8 is shown in Fig. S4 in the supplemental material. All five of the strains tested showed the accumulation of S0 globules of similar sizes, either attached to the bacteria or loose in the medium (see Fig.S4). There were large differences in the observed cell attachment to S0 globules. Although a correlation between the attachment of cells to S0 globules and cell surface hydrophobicity might be expected, we did not observe this despite the fact that all of the cells had identical physiological backgrounds. Strain S1p clearly had the fewest S0 globules that were attached to cells (9% ± 2%), while in cultures of strain 2Bp, most of the S0 globules were attached to cells (87% ± 7%, Table 2). This supports our hypothesis that S1p produces S0 that appears less bioavailable; attachment of S0 globules to bacteria is required for rapid S0 consumption upon H2S depletion. Strain Sts 4-3, which consumes S0 very slowly, also had a low percentage (13%) of cells attached to S0 globules, and 17% of the S0 globules were attached to cells (Table 2). This percentage was not as low as that of strain S1p, and it therefore cannot entirely explain the very low observed S0 consumption rate of this strain. However, strain Sts 4-3 produced more H2S than the other strains when pulsed with CS2 (25 versus 20 μM, Fig. 2), which could have had an inhibitory effect on overall respiration.
CS2 stress and starvation.
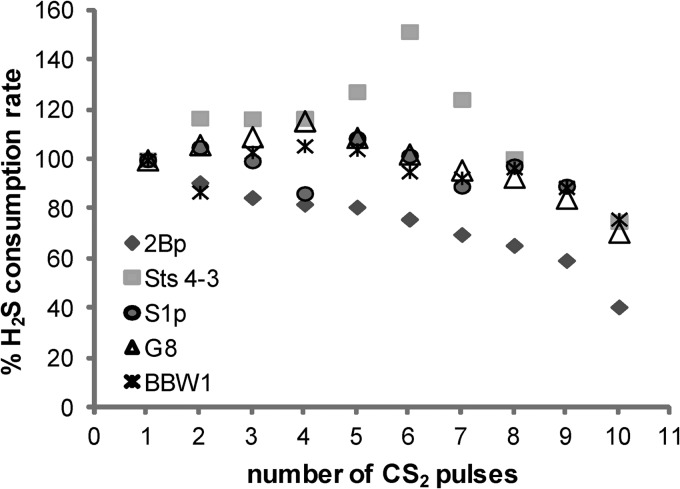
The effect of fluctuations in CS2 concentrations in biofilters can be simulated by applying CS2 pulses to undiluted culture samples from CS2-limited steady-state chemostats in MM with 1% M H2SO4. Therefore, chemostat samples of the newly isolated A. thiooxidans strains were incubated and subjected to repeated 17 μM CS2 pulses. Each new pulse was given when the H2S produced from the previous pulse had been consumed by the cells (after about 1.5 to 3 min). The H2S consumption rates after the first CS2 pulse varied between 1.9 (strain BBW1) and 3.4 μmol H2S liter−1 min−1 unit of OD600−1 (strain G8) and decreased for all of the strains after repeated CS2 pulses, with strain 2Bp appearing to be the most strongly affected (1.0 μmol H2S liter−1 min−1 unit of OD600−1, 41% of the initial rate) and strain BBW1 appearing to be the least strongly affected (1.5 μmol H2S liter−1 min−1 unit of OD600−1, 76% of the initial rate, Fig. 3). This indicates that the cells were becoming increasingly stressed when subjected to CS2 loads above the concentration the cells were adapted to in the chemostat. This is probably due to the toxic effect of the repeated buildup of H2S in the cells.
Fig 3.
Effect of repeated addition of CS2 on the H2S consumption capacity of minichemostat-grown cells of A. thiooxidans strains 2Bp, Sts 4-3, S1p, G8, and BBW1. Reactor samples were starved for 5 min in a cuvette, and 10 pulses of 17 μM CS2 were sequentially injected. Strain Sts 4-3 was starved for 15 min before the first injection. The experiment was repeated three times independently, and data from a representative experiment are shown as percentages of the H2S consumption rate after the first CS2 pulse. Measurements were done with an H2S sensor, and rates (in nanomoles of H2S per minute per milliliter of culture) were normalized to a culture OD600 of 1.
To test the effect of intermittent interruption of the factory CS2 supply, the five strains growing in chemostats as described above were subjected to CS2 starvation by temporarily shutting down the medium and CS2 supply for 24 h. During this period of starvation and the subsequent 24-h recovery period when the CS2 supply was reconnected again, samples were taken from the chemostats and subjected to a CS2 pulse. Subsequent H2S production and consumption rates and respiratory activity were measured. Results indicated that the strains cope reasonably well with short periods of starvation. After 4 h of CS2 starvation, there was only modest to no reduction in respiration activity when the cells were pulsed with 10 μM CS2. Of the four strains tested at this time point, G8 appeared to be the most strongly affected, with a 40% reduction of its initial respiration activity, when both H2S and S0 were present and a 22% reduction in S0 respiration activity (Fig. 4). After 24 h of CS2 starvation, all of the strains showed reduced respiratory activity. Although not tested extensively, the reduction was largest for strain 2Bp, where H2S/S0 respiration activity was reduced by 80% and S0 respiration was reduced by 85% below the steady-state rates. Strain 2Bp also seemed to recover slowly from starvation, whereas the other four strains showed a doubling of initial respiratory activity back to steady-state levels after only 4 h of resumed CS2 supply, the activity of strain 2Bp increased only 1.4-fold to 29% of the steady-state level. This slow recovery corresponds to the observation described above that strain 2Bp suffers more from CS2 stress than the other four strains do. At 24 h of recovery, the culture density of all five strains was higher than during steady state. This indicates that the cultures had started growing and/or had S0 present.
Fig 4.
Effects of CS2 starvation and recovery on respiration rates ± the standard error of the mean (n = 3) on internally produced H2S and S0 (A) and on only S0 (B) after a CS2 pulse. The experiment was performed with five CS2-converting A. thiooxidans strains (2Bp, Sts 4-3, S1p, G8, and BBW1), all growing simultaneously in five identically run chemostats. Rates are given in nanomoles of O2 per minute per milliliter of culture. Note that the respiration rates of strain 2Bp were monitored only at steady state, after 24 h of starvation, and after 4 h of recovery.
DISCUSSION
Our approach to the enrichment of extremely acidophilic CS2-converting microorganisms from volcanic regions and CS2-converting trickling filters proved successful. Conversion of CS2 was observed within days after inoculation. Furthermore, the enrichments and isolated Acidithiobacillus strains could grow at very high H2SO4 concentrations. The only acidophilic CS2-converting Acidithiobacillus strain described to date, TJ330, was isolated from an acidic (pH 6.1 to 1.4) CS2- and H2S-treating peat biofilter (9). The neutrophilic CS2-converting species Paracoccus denitrificans was isolated from oak leaves and soil beneath the leaf canopy (7, 27). Our attempt to enrich acidophilic microorganisms from soil samples taken under an oak tree and from compost was unsuccessful. Of the Acidithiobacillus strains from culture collections screened for CS2 conversion (at micromolar concentrations), only A. caldus DSM 8584 was positive. This trait was not previously reported for this species. Our screening confirmed the observation of Smith and Kelly (28), although they used a potentially toxic liquid CS2 concentration (2 mM).
The new Acidithiobacillus isolates even showed slow growth in 6% H2SO4. The constant supply of CS2 to the cultures was the key factor in the maintenance of cultures at these very high H2SO4 concentrations. Culture activity decreased dramatically upon storage without a substrate at or above 4% H2SO4. Acidophiles require a continuous and high supply of maintenance energy to be able to actively pump out protons that leak from the acidic environment into the nearly neutral pH cell cytoplasm (29). Operation of biofilters under highly acidic conditions therefore puts additional stress on the microorganisms during periods of fluctuating CS2 concentration and factory shutdown.
Although growth was very slow at 6% H2SO4, dense growth was already observed at 4% for all new Acidithiobacillus strains. Similar H2SO4 tolerance was reported for Acidithiobacillus sp. strain AZ11, which is able to respire on elemental sulfur in the presence of 4.2% H2SO4 (30). The pH optima determined for 5 of the 16 new CS2-converting strains were between 1 and 2. This is slightly lower than the optimal-pH range of 1.8 to 2.5 described for other Acidithiobacillus strains (29, 31).
Growth at around pH 0, as observed for 15 of our A. thioooxidans isolates, is among the lowest pHs reported in the literature for any organism. Ferroplasma acidarmanus (32), Picrophilus oshimae, and Picrophilus torridus could growth at pH 0 (optimum pH, 0.7) (33). Picrophilus torridus adapted to growth at pH 0.1 even showed significant growth at 1.2 M H2SO4 (pH −0.06) (34). The eukaryotic red alga Cyanidium caldarium was cultured at 0.5 M H2SO4 (35), and the green alga Dunaliella acidophila is able to survive pH 0.2 (36). Also, some fungal species were reported to grow at pH 0, i.e., Acontium cylatium (37), Cephalosporium sp., and Trichosporon cerebriae (38). We also observed fungal growth at 6% H2SO4 (pH −0.05) in our enrichment cultures but did not further investigate the species present in these cultures. With growth at pH as low as −0.05 (6% H2SO4), the new Acidithiobacillus isolates obtained in this research exceed the previously reported pH limit of 0.5 for microbial CS2 conversion for Acidithiobacillus TJ330 (9).
The newly isolated CS2-converting A. thiooxidans strains differed in colony morphology. The genus Acidithiobacillus comprises a physiologically and genetically heterogeneous group of microorganisms (39, 40), despite the often low sequence diversity in the 16S rRNA gene. For that reason, the 16S-23S ISR is used to discriminate at the intra species level (41, 42).
Two main colony types on Gelrite plates were distinguished, compact, creamy, shiny colonies and dry, white, spreading colonies (40). Reversible variation in colony morphology of several A. ferrooxidans strains has been described, resulting in the same large, white spreading colonies, as opposed to compact colonies, as observed here for some of the newly isolated A. thiooxidans strains (43, 44). A. thiooxidans strains may be motile via a polar flagellum (40). A. ferrooxidans ATCC 19859 spreading variants displayed increased motility and chemotaxis toward thiosulfate, which may be a selective advantage over biofilm growth during periods of low substrate concentrations. These variants arose through the rearrangement of insertion sequences in the genome, potentially acting as a genetic switch (44). The A. ferrooxidans group of strains has recently been reclassified into four separate species (45). Of these, only A. ferrivorans and some A. ferridurans strains were shown to be motile (45). The type strain A. ferrooxidans ATCC 23270 lacks flagellum and chemotaxis genes (46). The draft genome of A. thiooxidans ATCC 19377 (47) and the draft genomes of both A. thiooxidans strains S1p and G8 (Daan Speth, personal communication) do contain the operons for flagellum biosynthesis and chemotaxis. However, we did not observe swimming motility in liquid cultures of our strains when we examined them microscopically. Therefore, the mechanism and role of the spreading colony phenotype in these strains are not clear.
SEM studies of frozen colonies indicated that the difference in colony appearance may be caused by the absence of a clear EPS layer on the dry white colonies, as has been observed for colony morphology mutants of Mycobacterium smegmatis (22). In support, A. thiooxidans strains S1p, G8, and BBW1, which produced white dry colonies, had a considerably higher cell surface hydrophobicity than strains 2Bp and Sts 4-3, which produced compact shiny colonies, suggesting the presence of relatively more hydrophilic compounds on the cell surface of the latter strains. Acidithiobacillus strains produce EPS containing both neutral sugars and fatty acids (48, 49), the proportion varying depending on the substrate the cells are grown on (50, 51). The EPS of A. thiooxidans grown on S0 consists of 40% sugars and 60% fatty acids (mainly eicosanoic acid) (50). The fatty acids are released from the outer membrane by blebbing (52) and cause the “wetting” of sulfur particles described in 1961 by Jones and Starkey (53), making S0 available as an energy source. In addition, EPS is essential for successful attachment and bioleaching of A. ferrooxidans to pyrite (50). Differences in EPS production and potential motility observed in our strains may have implications for the degree of colonization and clogging of biofilters.
Efficient biofiltration of CS2-contaminated air streams requires microorganisms with a higher affinity for CS2 than the concentration present in the air stream, to ensure its removal to concentrations complying with increasingly stringent regulations, as well as resistance to changes in operating conditions, low biomass production, low S0 accumulation, and rapid S0 removal to prevent clogging of the biofilters. The affinities of the five strains tested for CS2 were similar (around 100 μM) and correspond to that reported for the purified CS2 hydrolase of Acidianus A1-3 (10). Comparison of the Vmax values revealed strain S1p to have a consistently higher Vmax under these conditions than the other four strains tested, possibly because it has a relatively larger amount of CS2 hydrolase present in the cells than the other strains (15). The Vmax is only reached at substrate concentrations of 200 μM (nmol ml−1) or higher. Bioreactors treating CS2-contaminated air from the viscose industry are operational at much lower concentrations of around 4 to 20 nmol ml−1 (100 to 500 ppm) (2), indicating that although strain S1p converts CS2 with a higher Vmax than the other strains, it would not usually reach its full potential reaction rate in a bioreactor. Indeed, a higher Vmax may even be deleterious, as it might cause H2S to accumulate to toxic levels in the cell more rapidly upon CS2 peaks.
Comparison of the resistance to changes in operating conditions in the form of an interruption in the CS2 supply revealed that all five strains were affected by 24 h of CS2 starvation in terms of the ability the respire after a CS2 pulse during starvation, but they all recovered within 24 h after reconnection of the CS2 supply. However, during recovery, all five strains will also produce S0, as we found in all of the strains tested that CS2 pulses during starvation resulted in the transient accumulation of 80% of the total S added as S0, independently of the amount of CS2 added. This implies that it will be difficult to avoid S0 accumulation in biofiltration systems with uneven CS2 loading. However, differences in S0 removal were observed, with strain Sts 4-3 having a much lower but constant S0 removal rate than the other strains and strain S1p having a long period in which the S0 respiration rate slowly declined. Whatever the cause, the strains that remove S0 more slowly will cause more clogging problems in bioreactors because of S0 accumulation.
In summary, we have successfully isolated extremely acidophilic, CS2-converting A. thiooxidans strains from both environmental and industrial ecosystems that grow optimally around pH 1 to 2 and can grow at sulfuric acid concentrations of up to 6% (vol/vol). Currently bioreactors are operated at pH 0.5 to 1 (2). Use of the new strains would reduce water use and improve the prospect for reuse of the produced sulfuric acid in the rayon/viscose industry. The isolated strains displayed different growth and colony morphology characteristics, which may be due to differences in motility and/or the presence or absence of an EPS layer surrounding the cells. To circumvent the bottlenecks in the biofiltration of CS2, cocultures of extremely acidophilic A. thiooxidans strains to combine the best acid tolerance, affinity for CS2, S0-removing potential, and stability during periods of fluctuating CS2 loads are most promising for inoculation of industrial biofilters. This application is expected to result in reduced sulfur accumulation, increased CS2 removal rates, reduced water consumption, a more stable operation, and recycling of the sulfuric acid produced.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by STW project 6353 and ERC 232937.
Bart Kraakman is thanked for providing samples. Jorge Valdés and David Holmes are thanked for the A. thiooxidans ATCC 19377 and A. caldus ATCC 51756 sequences. Markus Schmid is thanked for advice on primers, and Wendel Broek is thanked for analyzing 16S ISR sequences. Jelle Eygensteyn is acknowledged for total carbon measurements. We thank Nardy Kip for help with the pH optimum experiment and Geert-Jan Janssen for the cryo-SEM. Sacha van Hijum is thanked for genome assembly, and Daan Speth is thanked for providing an annotated protein list from the draft genome assembly.
Footnotes
Published ahead of print 30 August 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02167-13.
REFERENCES
- 1. Gelbke HP, Goen T, Maurer M, Sulsky SI. 2009. A review of health effects of carbon disulfide in viscose industry and a proposal for an occupational exposure limit. Crit. Rev. Toxicol. 39(Suppl 2):1–126 [DOI] [PubMed] [Google Scholar]
- 2. Kraakman NJR. 2003. Robustness of a full-scale biological system treating industrial CS2 emissions. Environ. Prog. 22:79–85 [Google Scholar]
- 3. Rojo N, Gallastegi G, Barona A, Gurtubay L, Ibarra-Berastegi G, Elias A. 2010. Biotechnology as an alternative for carbon disulfide treatment in air pollution control. Environ. Rev. 18:321–332 [Google Scholar]
- 4. Smith NA, Kelly DP. 1988. Isolation and physiological characterization of autotrophic sulfur bacteria oxidizing dimethyl disulfide as sole source of energy. J. Gen. Microbiol. 134:1407–1417 [Google Scholar]
- 5. Odintsova EV, Wood AP, Kelly DP. 1993. Chemolithoautotrophic growth of Thiothrix ramosa. Arch. Microbiol. 160:152–157 [Google Scholar]
- 6. Plas C, Wimmer K, Holubar P, Mattanovich D, Danner H, Jelinek E, Harant H, Braun R. 1993. Degradation of carbon disulphide by a Thiobacillus isolate. Appl. Microbiol. Biotechnol. 38:820–823 [Google Scholar]
- 7. Jordan SL, Kracziewiczdowjat AJ, Kelly DP, Wood AP. 1995. Novel Eubacteria able to grow on carbon disulfide. Arch. Microbiol. 163:131–137 [Google Scholar]
- 8. Pol A, van der Drift C, Op den Camp HJM. 2007. Isolation of a carbon disulfide utilizing Thiomonas sp. and its application in a biotrickling filter. Appl. Microbiol. Biotechnol. 74:439–446 [DOI] [PubMed] [Google Scholar]
- 9. Hartikainen T, Ruuskanen J, Raty K, von Wright A, Martikainen PJ. 2000. Physiology and taxonomy of Thiobacillus strain TJ330, which oxidizes carbon disulphide (CS2). J. Appl. Microbiol. 89:580–586 [DOI] [PubMed] [Google Scholar]
- 10. Smeulders MJ, Barends TRM, Pol A, Scherer A, Zandvoort MH, Udvarhelyi A, Khadem AF, Menzel A, Hermans J, Shoeman RL, Wessels H, van den Heuvel LP, Russ L, Schlichting I, Jetten MSM, Op den Camp HJM. 2011. Evolution of a new enzyme for carbon disulphide conversion by an acidothermophilic archaeon. Nature 478:412–416 [DOI] [PubMed] [Google Scholar]
- 11. Lobo R, Revah S, Viveros-Garcia T. 1999. An analysis of a trickle-bed bioreactor: carbon disulfide removal. Biotechnol. Bioeng. 63:98–109 [PubMed] [Google Scholar]
- 12. Alcantara S, Estrada I, Vasquez MS, Revah S. 1999. Carbon disulfide oxidation by a microbial consortium from a trickling filter. Biotechnol. Lett. 21:815–819 [Google Scholar]
- 13. Hartikainen T, Ruuskanen J, Martikainen PJ. 2001. Carbon disulfide and hydrogen sulfide removal with a peat biofilter. J. Air Waste Manage. Assoc. 51:387–392 [DOI] [PubMed] [Google Scholar]
- 14. Barrie Johnson D, Hallberg KB. 2009. Carbon, iron and sulfur metabolism in acidophilic micro-organisms. Adv. Microb. Physiol. 54:201–255 [DOI] [PubMed] [Google Scholar]
- 15. Smeulders MJ, Pol A, Venselaar H, Barends TRM, Hermans J, Jetten MSM, Op den Camp HJM. 2013. Bacterial CS2 hydrolases from Acidithiobacillus thiooxidans strains are homologous to the archaeal catenane CS2 hydrolase. J. Bacteriol. 195:4046–4056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Johnson DB. 1995. Selective solid media for isolating and enumerating acidophilic bacteria. J. Microbiol. Methods 23:205–218 [Google Scholar]
- 17. Derikx PJL, Op den Camp HJM, van der Drift C, Van Griensven LJLD, Vogels GD. 1990. Odorous sulfur compounds emitted during production of compost used as a substrate in mushroom cultivation. Appl. Environ. Microbiol. 56:176–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhou JZ, Bruns MA, Tiedje JM. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:316–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Saitou N, Nei M. 1987. The neigbor-joining method—a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 20. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 21. Valdes J, Pedroso I, Quatrini R, Holmes DS. 2008. Comparative genome analysis of Acidithiobacillus ferrooxidans, A. thiooxidans and A. caldus: insights into their metabolism and ecophysiology. Hydrometallurgy 94:180–184 [Google Scholar]
- 22. Smeulders MJ, Keer J, Speight RA, Williams HD. 1999. Adaptation of Mycobacterium smegmatis to stationary phase. J. Bacteriol. 181:270–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rosenberg M, Gutnick D, Rosenberg E. 1980. Adherence of bacteria to hydrocarbons—a simple method for measuring cell-surface hydrophobicity. FEMS Microbiol. Lett. 9:29–33 [Google Scholar]
- 24. Sorbo B. 1957. A colorimetric method for the determination of thiosulfate. Biochim. Biophys. Acta 23:412–416 [DOI] [PubMed] [Google Scholar]
- 25. Trueper HG, Schlegel HG. 1964. Sulphur metabolism in Thiorhodaceae. 1. quantitative measurements on growing cells of Chromatium okenii. Antonie Van Leeuwenhoek 30:225–238 [DOI] [PubMed] [Google Scholar]
- 26. Bryant RD, McGroarty KM, Costerton JW, Laishley EJ. 1983. Isolation and characterization of a new acidophilic Thiobacillus species (Thiobacillus albertis). Can. J. Microbiol. 29:1159–1170 [Google Scholar]
- 27. Jordan SL, McDonald IR, Kraczkiewicz-Dowjat AJ, Kelly DP, Rainey FA, Murrell JC, Wood AP. 1997. Autotrophic growth on carbon disulfide is a property of novel strains of Paracoccus denitrificans. Arch. Microbiol. 168:225–236 [DOI] [PubMed] [Google Scholar]
- 28. Smith NA, Kelly DP. 1988. Oxidation of carbon disulfide as the sole source of energy for the autotrophic growth of Thiobacillus thioparus strain Tk-m. J. Gen. Microbiol. 134:3041–3048 [Google Scholar]
- 29. Baker-Austin C, Dopson M. 2007. Life in acid: pH homeostasis in acidophiles. Trends Microbiol. 15:165–171 [DOI] [PubMed] [Google Scholar]
- 30. Lee EY, Lee NY, Cho KS, Ryu HW. 2006. Removal of hydrogen sulfide by sulfate-resistant Acidithiobacillus thiooxidans AZ11. J. Biosci. Bioeng. 101:309–314 [DOI] [PubMed] [Google Scholar]
- 31. Leduc LG, Ferroni GD. 1994. The chemolithotrophic bacterium Thiobacillus ferrooxidans. FEMS Microbiol. Rev. 14:103–119 [Google Scholar]
- 32. Edwards KJ, Bond PL, Gihring TM, Banfield JF. 2000. An archaeal iron-oxidizing extreme acidophile important in acid mine drainage. Science 287:1796–1799 [DOI] [PubMed] [Google Scholar]
- 33. Schleper C, Piihler G, Kuhlmorgen B, Zillig W. 1995. Life at extremely low pH. Nature 375(6534):741–742 [DOI] [PubMed] [Google Scholar]
- 34. Schleper C, Puehler G, Holz I, Gambacorta A, Janekovic D, Santarius U, Klenk HP, Zillig W. 1995. Picrophilus gen. nov., fam. nov.: a novel aerobic, heterotrophic, thermoacidophilic genus and family comprising archaea capable of growth around pH 0. J. Bacteriol. 177:7050–7059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Allen MB. 1959. Studies with Cyanidium caldarium, an anomalously pigmented chlorophyte. Arch. Microbiol. 32:270–277 [DOI] [PubMed] [Google Scholar]
- 36. Fuggi A, Pinto G, Pollio A, Taddei R. 1988. Effects of NaCl, Na2SO4, H2SO4, and glucose on growth, photosynthesis, and respiration in the acidophilic alga Dunaliella acidophila (Volvocales, chlorophyta). Phycologia 27:334–339 [Google Scholar]
- 37. Starkey RL, Waksman SA. 1943. Fungi tolerant to extreme acidity and high concentrations of copper sulfate. J. Bacteriol. 45:509–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sletten O, Skinner CE. 1948. Fungi capable of growing in strongly acid media and in concentrated copper sulfate solutions. J. Bacteriol. 56:679–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Harrison AP. 1982. Genomic and phsyiological diversity amongst strains of Thiobacillus ferrooxidans, and genomic comparison with Thiobacillus thiooxidans. Arch. Microbiol. 131:68–76 [Google Scholar]
- 40. Kelly DP, Wood AP. 2000. Reclassification of some species of Thiobacillus to the newly designated genera Acidithiobacillus gen. nov., Halothiobacillus gen. nov. and Thermithiobacillus gen. nov. Int. J. Syst. Evol. Microbiol. 50:511–516 [DOI] [PubMed] [Google Scholar]
- 41. Ni YQ, Yang Y, Bao JT, He KY, Li HY. 2007. Inter- and intraspecific genomic variability of the 16S-23S intergenic spacer regions (ISR) in representatives of Acidithiobacillus thiooxidans and Acidithiobacillus ferrooxidans. FEMS Microbiol. Lett. 270:58–66 [DOI] [PubMed] [Google Scholar]
- 42. Ni YQ, He KY, Bao JT, Yang Y, Wan DS, Li HY. 2008. Genomic and phenotypic heterogeneity of Acidithiobacillus spp. strains isolated from diverse habitats in China. FEMS Microbiol. Ecol. 64:248–259 [DOI] [PubMed] [Google Scholar]
- 43. Schrader JA, Holmes DS. 1988. Phenotypic switching of Thiobacillus ferrooxidans. J. Bacteriol. 170:3915–3923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chakraborty R, Singh A, Lahiri C, Deb C, Roy P. 2002. Colony morphology mutants of chemolithotrophic Acidithiobacillus ferrooxidans are associated with altered genomic distribution of family 1 repetitive DNA sequence. Curr. Sci. 82:1009–1014 [Google Scholar]
- 45. Amouric A, Brochier-Armanet C, Johnson DB, Bonnefoy V, Hallberg KB. 2011. Phylogenetic and genetic variation among Fe(II)-oxidizing acidithiobacilli supports the view that these comprise multiple species with different ferrous iron oxidation pathways. Microbiology 157:111–122 [DOI] [PubMed] [Google Scholar]
- 46. Valdes J, Pedroso I, Quatrini R, Dodson RJ, Tettelin H, Blake R, Eisen JA, Holmes DS. 2008. Acidithiobacillus ferrooxidans metabolism: from genome sequence to industrial applications. BMC Genomics 9:597. 10.1186/1471-2164-9-597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Valdes J, Ossandon F, Quatrini R, Dopson M, Holmes DS. 2011. Draft genome sequence of the extremely acidophilic biomining bacterium Acidithiobacillus thiooxidans ATCC 19377 provides insights into the evolution of the Acidithiobacillus genus. J. Bacteriol. 193:7003–7004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schaeffer WI, Umbreit WW. 1963. Phosphatidylinositol as a wetting agent in sulfur oxidation by Thiobacillus thiooxidans. J. Bacteriol. 85:492–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sand W, Gehrke T. 2006. Extracellular polymeric substances mediate bioleaching/biocorrosion via interfacial processes involving iron(III) ions and acidophilic bacteria. Res. Microbiol. 157:49–56 [DOI] [PubMed] [Google Scholar]
- 50. Harneit K, Goksel A, Kock D, Klock JH, Gehrke T, Sand W. 2006. Adhesion to metal sulfide surfaces by cells of Acidithiobacillus ferrooxidans, Acidithiobacillus thiooxidans and Leptospirillum ferrooxidans. Hydrometallurgy 83:245–254 [Google Scholar]
- 51. Rohwerder T, Gehrke T, Kinzler K, Sand W. 2003. Bioleaching review part A: progress in bioleaching: fundamentals and mechanisms of bacterial metal sulfide oxidation. Appl. Microbiol. Biotechnol. 63:239–248 [DOI] [PubMed] [Google Scholar]
- 52. Knickerbocker C, Nordstrom DK, Southam G. 2000. The role of “blebbing” in overcoming the hydrophobic barrier during biooxidation of elemental sulfur by Thiobacillus thiooxidans. Chem. Geol. 169:425–433 [Google Scholar]
- 53. Jones GE, Starkey RL. 1961. Surface-active substances produced by Thiobacillus thiooxidans. J. Bacteriol. 82:788–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.