Abstract
The glycosylation of five different flavonols, fisetin, quercetin, myricetin, kaempferol, and 3-hydroxyflavone, was achieved by applying YjiC. 3-Hydroxyflavone was selected as a probe for in vitro glycorandomization of all flavonols using diverse nucleotide diphosphate-d/l-sugars. This study unlocked the possibilities of the glycodiversification of flavonols and the generation of novel compounds as future therapeutics.
TEXT
All flavonol groups of compounds have a 3-hydroxyflavone (3-HF), i.e., 3-hydroxy-2-phenylchromen-4-one, backbone structure. 3-HF is a chemically synthesized (1) compound that has not been reported to occur in natural sources. Hydroxylation at either the A ring or B ring of 3-HF results in the generation of flavonols (Fig. 1). Flavonols are naturally available compounds with physiological as well as biological functions in plants and animals (2). In addition, 3-HFs are solvatochromic fluorescent dyes. The properties of these compounds are useful for biomolecular fluorescence intersection studies, and they can be applied as molecular fluorescence sensors (3, 4, 5, 6). The familiar and most abundantly found flavonols in vegetables include quercetin, fisetin, kaempferol, myricetin, rutin, and isorhamnetin (Fig. 1). According to the USDA Agricultural Research Service database (http://www.ars.usda.gov/Services/docs.htm?docid=6231), these flavonols are commonly present in apples, berries, onions, broccoli, grapes, flowers, bark (7), and other medicinally important botanicals, such as Ginkgo biloba and Hypericum perforatum. Most of these compounds exist in nature as glycosides; however, aglycones are also present in large quantities. Besides glycosylation, hydroxylation, malonylation, methylation, and sulfation are common modifications that diversify flavonols. Like other flavonoid compounds, all the flavonols have been reported to have similar properties, like antioxidative and radical-scavenging activities, anti-inflammatory, estrogenic, anticancer, and antidiabetic properties, detoxification, platelet aggregation, and immune effects (8, 9, 10, 11). Kelly summarized the clinical importance, drug bioavailability, and side effects along with the dosage of the flavonol quercetin in his monograph (12). Although quercetin is one of the most studied flavonols, other flavonols have potential health benefits comparable to those of quercetin.
Fig 1.
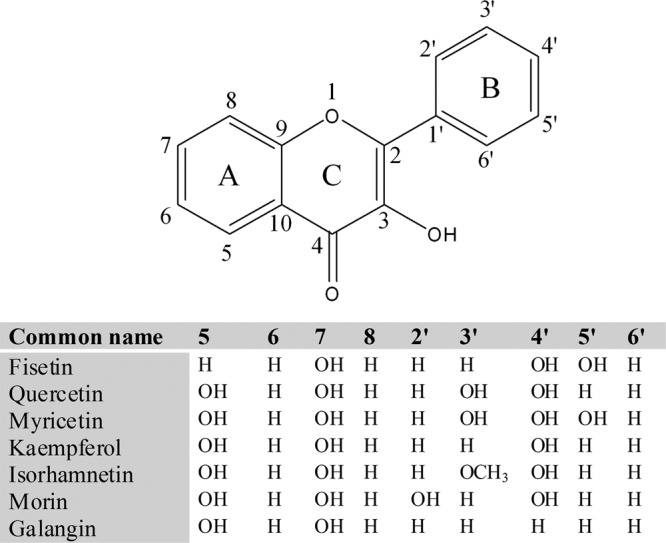
Structures of 3-hydroxyflavone (3-HF) (or 3-hydroxy-2-phenylchromen-4-one) and other common flavonols sharing a 3-HF backbone. The addition of any phenolic hydroxyl group in 3-HF results in flavonols.
Most of these polyphenols are restricted to pharmacological applications because they lack water solubility and stability. Glycosylation is one way to enhance the water solubility as well as the stability of aglycones. Additionally, glycosylation alters the pharmacokinetic properties and selectivities of compounds (13, 14). Thus, the biological activities of most natural products and therapeutics are attributed to the sugar moieties attached to them. For example, doxorubicin, vancomycin, avermectin, and nystatin are biologically potent because of their sugar moieties. Hence, engineering a sugar moiety attached to these natural products is in development as one of the promising tools for engineering and developing novel compounds with potential biological applications, as is altering their physical and chemical properties. Such compounds could be developed as future drugs for treating diverse kinds of fatal diseases. Therefore, glycorandomization (the diversification of a compound by conjugating different kinds of sugar moieties) of such compounds has attracted significant interest in the scientific community. Thorson and colleagues have made noteworthy progress in the glycorandomization of a number of natural products. For example, glycorandomization of novobiocin (15), digitoxin (16), vancomycin (17), colchicine (18), and 4-methylumbelliferone (19) has been carried out. Unfortunately, the in vitro glycorandomization of such natural products for industrial production has two limits: first, the lack of economic availability of nucleotide diphosphate (NDP) sugar donors, and second, the lack of flexible glycosyltransferase enzymes for reaction catalysis. Glycorandomization of any potential compound may contribute to its further development into a novel therapeutic agent.
Based on these studies, we employed 3-HF as a probe for glycorandomization of flavonols by using a glycosyltransferase (YjiC) from Bacillus licheniformis DSM-13. YjiC can glucosylate geldanamycin analogues (20) as well as phloretin, a chalcone (21) that shows flexible glucosylation at different positions. In this study, we tested the activity of the same enzyme with different flavonols, including 3-HF, for glucosylation. Thereafter, we probed 3-HF, the backbone of all flavonols, for in vitro glycorandomization using different NDP-sugars. Moreover, we studied the donor substrate specificity of the enzyme using diverse NDP-d-glucoses as glucose moiety donors and 3-HF as an acceptor.
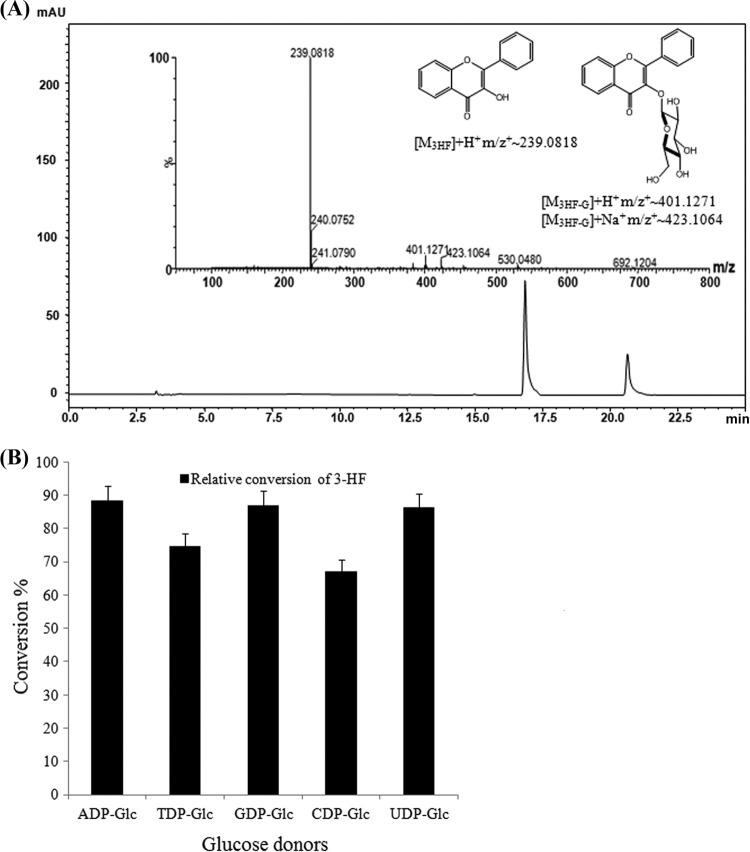
Recombinant poly-His-tagged soluble functional YjiC (GenBank sequence accession no. AAU40842) was expressed in Escherichia coli BL21(DE3) and purified as described in our previous report (21). The 46-kDa purified YjiC was applied to an in vitro reaction mixture with 3-HF along with UDP-d-glucose. The high-performance liquid chromatography photodiode array (HPLC-PDA) analysis of the reaction mixture revealed a new peak at a retention time (tR) of 16.8 min, which was further analyzed by HPLC-PDA coupled with quantitative time of flight–high-resolution electrospray ionization–mass spectrometry (Q-TOF-HR ESI/MS). The high-resolution exact-mass analysis confirmed the product to be 3-HF 3-O-β-d-glucoside ([M + H]+ m/z+ ∼401.1271). Because 3-HF has a single hydroxyl group at the C-3 position, we did not analyze the product by nuclear magnetic resonance analyses. After confirming the 3-HF 3-O-β-d-glucoside, we carried out donor substrate flexibility analyses of the enzyme using ADP-α-d-glucose, TDP-α-d-glucose, GDP-α-d-glucose, and CDP-α-d-glucose instead of UDP-α-d-glucose as in previous glycosylation reaction mixtures. The independent reaction mixture analysis by HPLC-PDA analysis identified the production of the 3-HF 3-O-β-d-glucoside, showing a product at the same tR (16.8 min) with the exact same mass (Fig. 2A). However, the rates of conversion of the substrate were different with the NDP-d-glucoses used for the reaction (Fig. 2B). ADP-α-d-glucose, GDP-α-d-glucose, and UDP-α-d-glucose were well accepted by YjiC, showing almost equal rates of conversion (∼90%) of the substrate (3-HF) to its glucoside. However, TDP-α-d-glucose (∼75%) and CDP-α-d-glucose (∼67%) were less accepted than other NDP-d-glucoses. This result illustrates that the enzyme can accept all five different types of NDP-d-glucoses as donors for the glycosyltransferase reaction but that it preferred ADP-α-d-glucose, GDP-α-d-glucose, and UDP-α-d-glucose over TDP-α-d-glucose and CDP-α-d-glucose. Thus, we provide a further clue that all of the NDP-d-sugars can be used for the reaction with YjiC to transfer diverse sugar moieties.
Fig 2.
(A) Typical HPLC-PDA chromatogram and QTOF-HR ESI/MS spectrum of a YjiC-catalyzed reaction of 3-HF and UDP-α-d-glucose, which is representative of the glycosylation reactions of all five NDP-α-d-glucoses and 3-HF. mAU, absorbance units (in thousands). (B) Comparison of rates conversion of 3-HF to 3-HF 3-O-β-d-glucoside using A/T/G/C/U-DP-α-d-glucoses as glucose donors. The structures of all NDP-α-d-glucoses vary only in their nucleotide bases. Error bars show standard deviations of three independent experiments, which were less than 5%.
We were interested in glycorandomizing the 3-HF backbone-containing flavonols. Hence, we initially carried out reactions of fisetin, quercetin, myricetin, and kaempferol with UDP-α-d-glucose under conditions identical to those used with 3-HF. The HPLC-PDA analysis of the independent reaction mixtures disclosed multiple glucosylations of all substrates (Fig. 3A; see also Table S1 in the supplemental material). Each product was further characterized by HPLC-PDA and QTOF-HR ESI/MS (see also Fig. S1 to S4 in the supplemental material). The mass analysis identified a monoglucoside derivative of fisetin (F1) (tR = 11.8 min, [M + H]+ m/z+ ∼449.1083), two diglucoside derivatives of fisetin (F2, tR = 11.4 min, [M + H]+ m/z+ ∼611.1613, and F3, tR = 10.7 min, [M + H]+ m/z+ ∼611.1616), and a triglucoside derivative of fisetin (F4, tR = 10.0 min, [M + H]+ m/z+ ∼773.2134). Similarly, with quercetin, a monoglucoside (Q1, tR = 14.2 min, [M + H]+ m/z+ ∼465.1053), two diglucosides (Q2, tR = 13.1 min, [M + H]+ m/z+ ∼627.1549, and Q3, tR = 12.7 min, [M + H]+ m/z+ ∼627.1547), two triglucosides (Q4, tR = 12.2 min, [M + H]+ m/z+ ∼789.2085, and Q5, tR =11.3 min, [M + H]+ m/z+ ∼789.2080), and a tetraglucoside (Q6, tR = 10.7 min, [M + H]+ m/z+ ∼951.2606) were confirmed by mass analysis. With myricetin, a monoglucoside (M1, tR = 12.1 min, [M + H]+ m/z+ ∼481.0992), a diglucoside (M2, tR = 11.6 min, [M + H]+ m/z+ ∼643.1513), a triglucoside (M3, tR = 11.3 min, [M + H]+ m/z+ ∼805.2034), and a tetraglucoside (M4, tR = 10.5 min, [M + H]+ m/z+ ∼967.2551) were detected in the reaction mixture. However, in the case of kaempferol, one monoglucoside (K1, tR = 13.9 min, [M + H]+ m/z+ ∼449.1083), two diglucosides (K2, tR = 12.3 min, [M + H]+ m/z+ ∼611.1611, and K3, tR = 12 min, [M + H]+ m/z+ ∼611.1597), and a triglucoside (K4, tR = 10.3 min, [M + H]+ m/z+ ∼773.2169) were produced (Fig. 3A, and see Fig. S1 to S4 and Table S1). The 3-h reaction mixture analysis resulted in approximately 98% conversion of fisetin, 94% conversion of quercetin, 76% conversion of myricetin, and almost 100% conversion of kaempferol (Fig. 3B). Moreover, we found nonregiospecific glucosylation of all flavonols by YjiC, which resulted in multiple glucosylated products, along with mono-, di-, tri-, and even tetraglucosides. The structural elucidation of each glycorandomized product of all flavonols might be difficult because of the high cost and limited availability of all rare NDP-sugars. The chemical, enzymatic, or chemoenzymatic synthesis of all rare NDP-sugars requires long, time-consuming reaction steps. Moreover, the product yield is very low. Thus, we applied 3-HF as a probe for glycorandomization of flavonols for three reasons: (i) 3-HF along with other flavonols has been demonstrated to be glucosylated by YjiC, (ii) all flavonols have the same backbone structures, and (iii) 3-HF has only one reactive hydroxyl group, which helps to determine the conjugation of diverse sugars with 3-HF simply by mass analysis.
Fig 3.
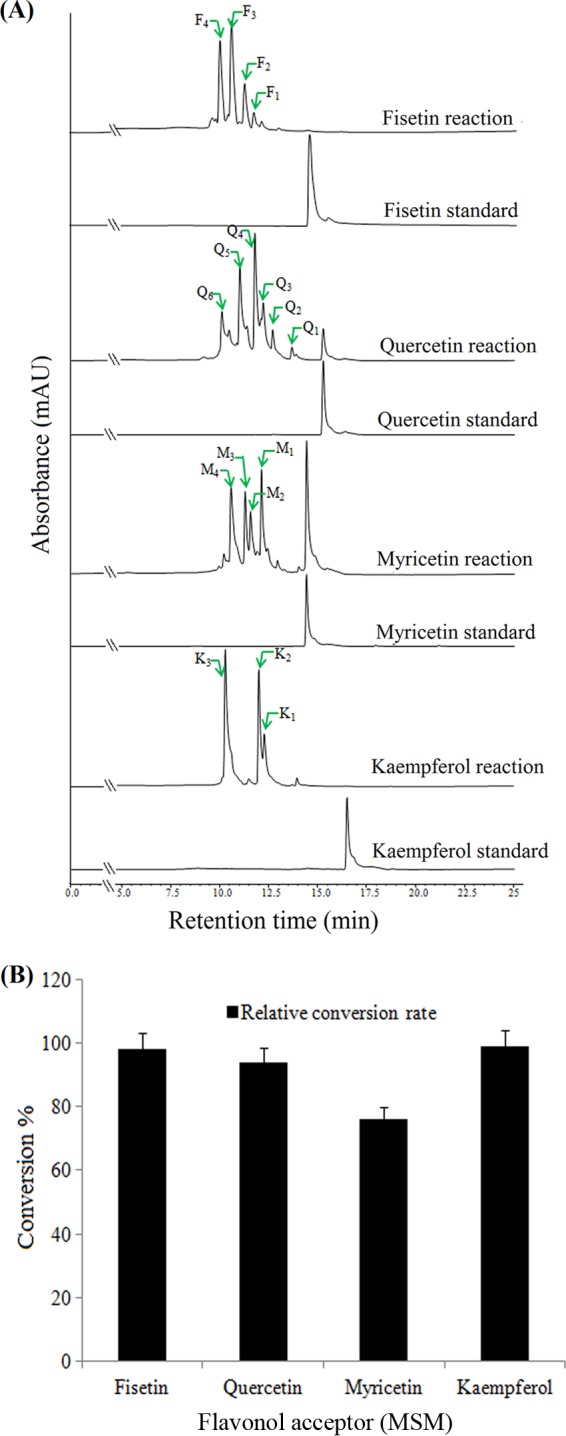
(A) HPLC-PDA analysis of flavonol reaction mixtures with UDP-α-d-glucose and YjiC; (B) comparison of the conversion rates of the substrates (fisetin, quercetin, myricetin, and kaempferol). Error bars show the standard deviations of three independent experiments, which were less than 5%.
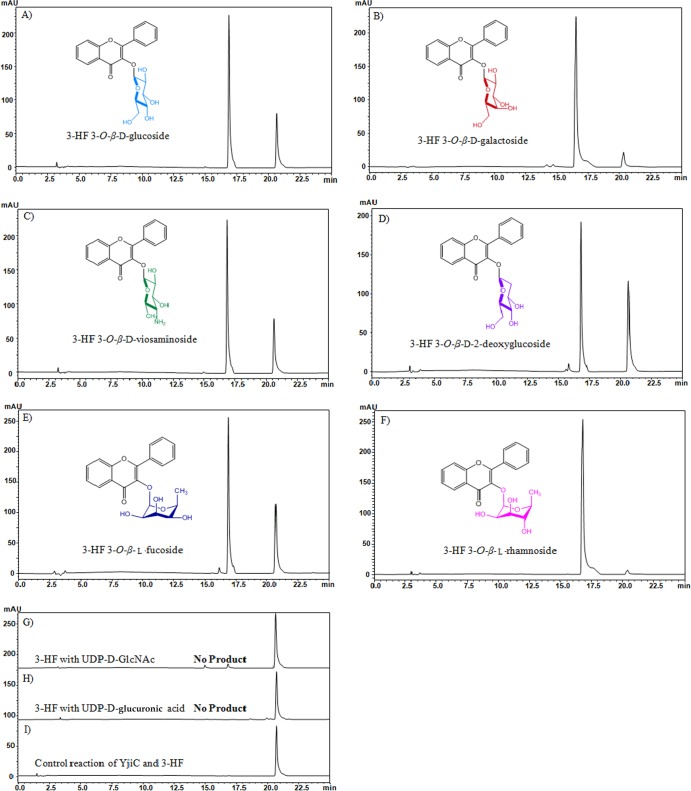
Both NDP-d-sugars and NDP-l-sugars were used for the 3-HF glycorandomization reactions under conditions identical to those described above. Because the aforementioned results showed the acceptance of diverse kinds of NDP-d-glucoses by YjiC, five different NDP-d-sugars, UDP-α-d-galactose, UDP-N-acetylglucosamine (UDP-GlcNAc), UDP-α-d-glucuronic acid, TDP-α-d-viosamine, and TDP-α-d-2-deoxyglucose, and two different NDP-l-sugars, TDP-l-rhamnose and GDP-l-fucose, were examined with 3-HF using YjiC in lab-scale reaction mixtures. The HPLC-PDA (Fig. 4) and HPLC-PDA-QTOF-HR ESI/MS analyses (see Fig. S5 and S6 in the supplemental material) confirmed the production of 3-HF 3-O-β-d-galactoside (tR = 16.8 min, [M + H]+ m/z+ ∼401.1298, or [M + Na]+ m/z+ ∼423.1057), 3-HF 3-O-β-d-viosaminoside (tR = 17.0 min, [M + H]+ m/z+ ∼384.1453), 3-HF 3-O-β-d-2-deoxyglucoside (tR = 17.0 min, [M + Na]+ m/z+ ∼407.1098), 3-HF 3-O-β-l-fucoside (tR = 16.9 min, [M + Na]+ m/z+ ∼407.1108), and 3-HF 3-O-β-l-rhamnoside (tR = 16.8 min, [M + Na]+ m/z+ ∼407.1094). However, we could not detect mass with UDP-GlcNAc or UDP-α-d-glucuronic acid by HPLC-PDA-QTOF-HR ESI/MS analysis. YjiC transferred diverse kinds of sugars, such as NDP-d-sugars modified at C-4 (galactose), C-2 (2-deoxyglucose), and C-4 and C-6 (viosamine) and NDP-l-sugars modified at C-4 (fucose and rhamnose), but the conversion rate of each reaction was variable. The rate of conversion of 3-HF to 3-HF 3-O-β-d-glucoside, 3-HF 3-O-β-d-galactoside, and 3-HF 3-O-β-l-rhamnoside by the enzyme under reaction conditions identical to those described above was 90% with the respective NDP-sugar donors, illustrating the high efficiency of the enzyme with those NDP-sugar donors. However, the conversion rate was limited to approximately 48% in a reaction using TDP-α-d-2-deoxyglucose, whereas the conversion was satisfactory in reactions using TDP-α-d-viosamine (∼70%) and GDP-l-fucose (∼60%). These results exemplified the idea that, though YjiC can accept diverse kinds of NDP-sugars, the nucleotide base as well as the sugar moiety may play a crucial role in determining the conversion rate of each reaction.
Fig 4.
HPLC-PDA chromatogram analysis of glycosyltransferase reactions of 3-HF with different NDP-D/L-sugar donors and YjiC under identical reaction conditions. Six different types of 3-HF glycosides, which differed only in their sugar moieties, were generated. Glycosylation reactions of 3-HF and YjiC with UDP-α-d-glucose (A), UDP-α-d-galactose (B), TDP-α-d-viosamine (C), TDP-α-d-2-deoxyglucose (D), GDP-l-fucose (E), TDP-l-rhamnose (F), UDP-N-acetylglucosamine (G), UDP-α-d-glucuronic acid (H), and the control (I). Inside each HPLC-PDA chromatogram, the structure of the reaction product is given.
Although 3-HF has been exploited as a probe in this study, all glycorandomized 3-HF derivatives were novel compounds. These compounds may have novel biological functions. Thus, this approach could be applied to randomize a large number of flavonols with wide applications to human health. For example, quercetin could be a target flavonol for glycorandomization because of its broad applications. Importantly, quercetin is one of the most studied flavonoids and is in clinical trials conducted by the U.S. National Institutes of Health (www.clinicaltrials.gov/). Preliminary clinical trial studies have found that quercetin may have beneficial activities for treating aging, hypertension, Fanconi anemia, sarcoidosis, oral mucositis, cystic fibrosis, obesity, colorectal cancer, and Alzheimer's disease. Therefore, the glycorandomized products of quercetin and other flavonols, such as myricetin, kaempferol, and fisetin, may have significant health benefits. Metabolic engineering approaches have been used by our group and other groups to produce different sugar-conjugated quercetin compounds as well as kaempferol from E. coli. For example, quercetin 3-O-glucoside, quercetin 7-O-glucoside, quercetin 4′-O-glucoside (22), quercetin 3-O-xyloside (23), quercetin 3-O-rhamnoside (24, 25), quercetin 3-O-alloside (25), quercetin-3-O-4-deoxy-4-formamido-l-arabinoside (26), quercetin 3-O-6-deoxytalose (27), quercetin 3-O-N-acetylglucosamine (28), and kaempferol 3-O-rhamnoside (25) have been produced successfully using different UDP-glycosyltransferases from plants. Besides these flavonols, other flavonols, such as galangin, morin, azaleatin, isorhamnetin, and rhamnetin, might also be potential substrates for glycorandomization by YjiC. Most of the flavonols have potent antioxidant activities because of the presence of pharmacophores on catechol ring B and C-3 hydroxyl groups (29). The substituents influencing the activities of flavonols (i.e., hydroxyl groups) at C-5 and C-7 are also present in all flavonols. Hence, modifying such a well-optimized pharmacophore unit by glycorandomization may result in novel biological as well as physical and chemical properties and generate novel drugs for future use.
In conclusion, this approach of probing 3-HF for glycorandomization of flavonols has unlocked the area for production of specific flavonol compounds regioselectively conjugated with target sugar moieties by engineered YjiC in the near future. Additionally, YjiC could be applied to produce diverse sugars conjugated with flavonols by constructing microbial cell factories via a metabolic engineering approach.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the Intelligent Synthetic Biology Center of the Global Frontier Project, funded by the Ministry of Education, Science and Technology (grant 2011-0031960), and by a grant from the Next-Generation BioGreen 21 Program (grant PJ0094832 to J.K.S. and grant PJ0094834 to J.W.P.), the Rural Development Administration, and the Republic of Korea.
Footnotes
Published ahead of print 23 August 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02057-13.
REFERENCES
- 1. Gunduz S, Goren AC, Ozturk T. 2012. Facile syntheses of 3-hydroxyflavones. Org. Lett. 16:1576–1579 [DOI] [PubMed] [Google Scholar]
- 2. Pollastri S, Tattini M. 2011. Flavonols: old compounds for old roles. Ann. Bot. 108:1225–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zamotaiev OM, Postupalenko VY, Shvadchak VV, Pivovarenko VG, Klymchenko AS, Mely Y. 2011. Improved hydration-sensitive dual-fluorescence labels for monitoring peptide-nucleic acid interactions. Bioconjug. Chem. 22:101–107 [DOI] [PubMed] [Google Scholar]
- 4. Hope-Roberts M, Horobin RW, Wainwright M. 2011. Identifying apoptotic cells with the 3-hydroxyflavone derivative F2N12S, a ratiometric fluorescent small molecule probe selective for plasma membranes: a possible general mechanism for selective uptake into apoptotic cells. Biotech. Histochem. 86:255–261 [DOI] [PubMed] [Google Scholar]
- 5. Xu S, Shao Y, Ma K, Cui Q, Liu G, Wu F, Li M. 2011. Fluorescence light-up recognition of DNA nucleotide based on selective abasic site binding of an excited-state intramolecular proton transfer probe. Analyst 136:4480–4485 [DOI] [PubMed] [Google Scholar]
- 6. Shynkar VV, Klymchenko AS, Kunzelmann C, Duportail G, Muller CD, Demchenko AP, Freyssinet JM, Mely Y. 2007. Fluorescent biomembrane probe for ratiometric detection of apoptosis. J. Am. Chem. Soc. 129:2187–2193 [DOI] [PubMed] [Google Scholar]
- 7. Häkkinen SH, Kärenlampi SO, Heinonen IM, Mykkanen HM, Torronen AR. 1999. Content of the flavonols quercetin, myricetin, and kaempferol in 25 edible berries. J. Agric. Food Chem. 47:2274–2279 [DOI] [PubMed] [Google Scholar]
- 8. Amado NG, Fonseca BF, Cerqueira DM, Neto VM, Abreu JG. 2011. Flavonoids: potential Wnt/beta-catenin signaling modulators in cancer. Life Sci. 89:545–554 [DOI] [PubMed] [Google Scholar]
- 9. Huxley RR, Neil HA. 2003. The relation between dietary flavonol intake and coronary heart disease mortality: a meta-analysis of prospective cohort studies. Eur. J. Clin. Nutr. 57:904–908 [DOI] [PubMed] [Google Scholar]
- 10. Boots AW, Haenen GR, Bast A. 2008. Health effects of quercetin: from antioxidant to nutraceutical. Eur. J. Pharmacol. 585:325–337 [DOI] [PubMed] [Google Scholar]
- 11. Calderon-Montano JM, Burgos-Moron E, Perez-Guerrero C, Lopez-Lazaro M. 2011. A review on the dietary flavonoid kaempferol. Mini Rev. Med. Chem. 11:298–344 [DOI] [PubMed] [Google Scholar]
- 12. Kelly GS. 2011. Quercetin. Altern. Med. Rev. 16:172–194 [PubMed] [Google Scholar]
- 13. Kren V, Martinkova L. 2001. Glycosides in medicine: the role of glycosidic residue in biological activity. Curr. Med. Chem. 8:1303–1328 [DOI] [PubMed] [Google Scholar]
- 14. Weymouth-Wilson AC. 1997. The role of carbohydrates in biologically active natural products. Nat. Prod. Rep. 14:99–110 [DOI] [PubMed] [Google Scholar]
- 15. Albermann C, Soriano A, Jiang J, Vollmer H, Biggins JB, Barton WA, Lesniak J, Nikolov DB, Thorson JS. 2003. Substrate specificity of NovM: implications for novobiocin biosynthesis and glycorandomization. Org. Lett. 20:933–936 [DOI] [PubMed] [Google Scholar]
- 16. Langenhan JM, Peters NR, Guzei IA, Hoffmann FM, Thorson JS. 2005. Enhancing the anticancer properties of cardiac glycosides by neoglycorandomization. Proc. Natl. Acad. Sci. U. S. A. 102:12305–12310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fu X, Albermann C, Zhang C, Thorson JS. 2005. Diversifying vancomycin via chemoenzymatic strategies. Org. Lett. 7:1513–1515 [DOI] [PubMed] [Google Scholar]
- 18. Ahmed A, Peters NR, Fitzgerald MK, Watson JA, Jr, Hoffmann FM, Thorson JS. 2006. Colchicines glycorandomization influences cytotoxicity and mechanism of action. J. Am. Chem. Soc. 128:14224–14225 [DOI] [PubMed] [Google Scholar]
- 19. Williams GJ, Yang J, Zhang C, Thorson JS. 2011. Recombinant E. coli prototype strains for in vivo glycorandomization. ACS Chem. Biol. 6:95–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wu CZ, Jang JH, Woo M, Ahn JS, Kim JS, Hong YS. 2012. Enzymatic glycosylation of non-benzoquinone geldanamycin analogs via Bacillus UDP-glycosyltransferase. Appl. Environ. Microbiol. 78:7680–7686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pandey RP, Li TF, Kim EH, Yamaguchi T, Park YI, Kim JS, Sohng JK. 2013. Enzymatic synthesis of novel phloretin glucosides. Appl. Environ. Microbiol. 79:3516–3521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lim EK, Ashford DA, Hou B, Jackson RG, Bowles DJ. 2004. Arabidopsis glycosyltransferases as biocatalysts in fermentation for regioselective synthesis of diverse quercetin glucosides. Biotechnol. Bioeng. 87:623–631 [DOI] [PubMed] [Google Scholar]
- 23. Pandey RP, Malla S, Simkhada D, Kim BG, Sohng JK. 2013. Production of 3-O-xylosyl quercetin in Escherichia coli. Appl. Microbiol. Biotechnol. 97:1889–1901 [DOI] [PubMed] [Google Scholar]
- 24. Lim EK, Ashford DA, Bowles JD. 2006. The synthesis of small-molecule rhamnosides through the rational design of a whole-cell biocatalysis system. Chembiochem 7:1181–1185 [DOI] [PubMed] [Google Scholar]
- 25. Simkhada D, Lee HC, Sohng JK. 2010. Genetic engineering approach for the production of rhamnosyl and allosyl flavonoids from Escherichia coli. Biotechnol. Bioeng. 107:154–162 [DOI] [PubMed] [Google Scholar]
- 26. Kim BG, Jung NR, Joe EJ, Hur HG, Lim Y, Chong Y, Ahn JH. 2010. Bacterial synthesis of a flavonoid deoxyaminosugar conjugate in Escherichia coli expressing a glycosyltransferase of Arabidopsis thaliana. Chembiochem 11:2389–2392 [DOI] [PubMed] [Google Scholar]
- 27. Yoon JA, Kim BG, Lee WJ, Lim Y, Chong Y, Ahn JH. 2012. Production of novel quercetin glycoside through metabolic engineering of Escherichia coli. Appl. Environ. Microbiol. 78:4256–4262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim BG, Sung SH, Ahn JH. 2012. Biological synthesis of quercetin 3-O-N-acetylglucosamine conjugate using engineered Escherichia coli expressing UGT78D2. Appl. Microbiol. Biotechnol. 93:2447–2453 [DOI] [PubMed] [Google Scholar]
- 29. Heijnen CG, Haenen GR, van Acker FA, van der Vijgh WJ, Bast A. 2001. Flavonoids as peroxynitrite scavengers: the role of the hydroxyl groups. Toxicol. In Vitro 15:3–6 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.