Abstract
To date, no information has been made available on the genetic traits that lead to increased carbon flow into the fatty acid biosynthetic pathway of Corynebacterium glutamicum. To develop basic technologies for engineering, we employed an approach that begins by isolating a fatty acid-secreting mutant without depending on mutagenic treatment. This was followed by genome analysis to characterize its genetic background. The selection of spontaneous mutants resistant to the palmitic acid ester surfactant Tween 40 resulted in the isolation of a desired mutant that produced oleic acid, suggesting that a single mutation would cause increased carbon flow down the pathway and subsequent excretion of the oversupplied fatty acid into the medium. Two additional rounds of selection of spontaneous cerulenin-resistant mutants led to increased production of the fatty acid in a stepwise manner. Whole-genome sequencing of the resulting best strain identified three specific mutations (fasR20, fasA63up, and fasA2623). Allele-specific PCR analysis showed that the mutations arose in that order. Reconstitution experiments with these mutations revealed that only fasR20 gave rise to oleic acid production in the wild-type strain. The other two mutations contributed to an increase in oleic acid production. Deletion of fasR from the wild-type strain led to oleic acid production as well. Reverse transcription-quantitative PCR analysis revealed that the fasR20 mutation brought about upregulation of the fasA and fasB genes encoding fatty acid synthases IA and IB, respectively, by 1.31-fold ± 0.11-fold and 1.29-fold ± 0.12-fold, respectively, and of the accD1 gene encoding the β-subunit of acetyl-CoA carboxylase by 3.56-fold ± 0.97-fold. On the other hand, the fasA63up mutation upregulated the fasA gene by 2.67-fold ± 0.16-fold. In flask cultivation with 1% glucose, the fasR20 fasA63up fasA2623 triple mutant produced approximately 280 mg of fatty acids/liter, which consisted mainly of oleic acid (208 mg/liter) and palmitic acid (47 mg/liter).
INTRODUCTION
Lipids and related compounds comprise a variety of useful materials, such as arachidonic, eicosapentaenoic, and docosahexaenoic acids that are functional lipids (1); prostaglandins and leukotrienes that are used as pharmaceuticals (2); biotin and α-lipoic acid that have pharmaceutical and cosmetic uses (3–5); and hydrocarbons and fatty acid ethyl esters that are used as fuels (6, 7). Since most of these compounds are derived via the fatty acid synthetic pathway, increasing carbon flow into this pathway is an important consideration in producing these compounds by the fermentation method.
Although there are numerous articles on lipid production by oleaginous fungi and yeasts (8, 9), attempts to use bacteria for that purpose remain limited (10–12). A pioneering study that showed the bacterial production of fatty acids with genetically engineered Escherichia coli was performed by Cho and Cronan (11). They demonstrated that cytosolic expression of the periplasmic enzyme acyl-acyl carrier protein (acyl-ACP) thioesterase I (TesA) resulted in the extracellular production of free fatty acids. This phenomenon has been reasonably explained by avoidance of the regulatory mechanism of fatty acid synthesis through the TesA-catalyzed cleavage of acyl-ACP, which acts as a feedback inhibitor of fatty acid synthetic enzymes acetyl coenzyme A (acetyl-CoA) carboxylase, FabH, and FabI (11). Most of the later studies on the bacterial production of fatty acids and their derivatives have been based on this technique (13, 14). Another representative work is the establishment of a reversal β-oxidation cycle in E. coli, which also led to the extracellular production of free fatty acids (12). The advantage of this approach is that the engineered pathway directly uses acetyl-CoA instead of malonyl-CoA for acyl-chain elongation and can thus bypass the ATP-consuming step required for malonyl-CoA formation. Despite these positive results, fatty acid productivities remain far below a practical level. In addition, the bacterial production platform has exclusively depended on E. coli, except for one example of a cyanobacterium to which the E. coli TesA technique has been applied (13).
Our objective is to develop the basic technologies to produce fatty acids by using Corynebacterium glutamicum. This bacterium has long been used for the industrial production of a variety of amino acids, including l-glutamic acid and l-lysine (15). It has also recently been developed as a production platform for various commodity chemicals (16, 17, 18), fuel alcohols (19, 20), carotenoids (21), and heterologous proteins (22). However, there are no reports of fatty acid production by this bacterium, except for undesired production of acetate, a water-soluble short-chain fatty acid, as a by-product (23). To the best of our knowledge, no attempts have been made to improve carbon flow into the fatty acid biosynthetic pathway. In this context, it seems worthwhile to verify the feasibility of this bacterium as a potential workhorse for fatty acid production.
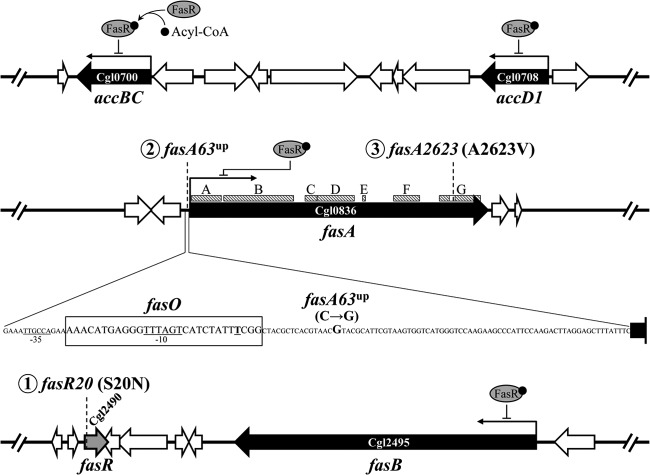
With respect to fatty acid biosynthesis in C. glutamicum, there are some genetic and functional studies on the relevant genes (24–28). Unlike the majority of bacteria, including E. coli and Bacillus subtilis, coryneform bacteria, such as members of the genera Corynebacterium and Mycobacterium, are known to possess type I fatty acid synthase (Fas) (29), a multienzyme that performs successive cycles of fatty acid synthesis, into which all activities required for fatty acid elongation are integrated (29). In addition, Corynebacterium fatty acid synthesis is thought to differ from that of common bacteria in that the donor of two-carbon units and the end product are CoA derivatives instead of ACP derivatives. This was demonstrated by using the purified Fas from Corynebacterium ammoniagenes (30), which is closely related to C. glutamicum. With regard to the regulatory mechanism of fatty acid biosynthesis, the details are not fully understood. It was only recently shown that the relevant biosynthesis genes were transcriptionally regulated by the TetR-type transcriptional regulator FasR (28). Fatty acid metabolism and its predicted regulatory mechanism in C. glutamicum are shown in Fig. 1.
Fig 1.
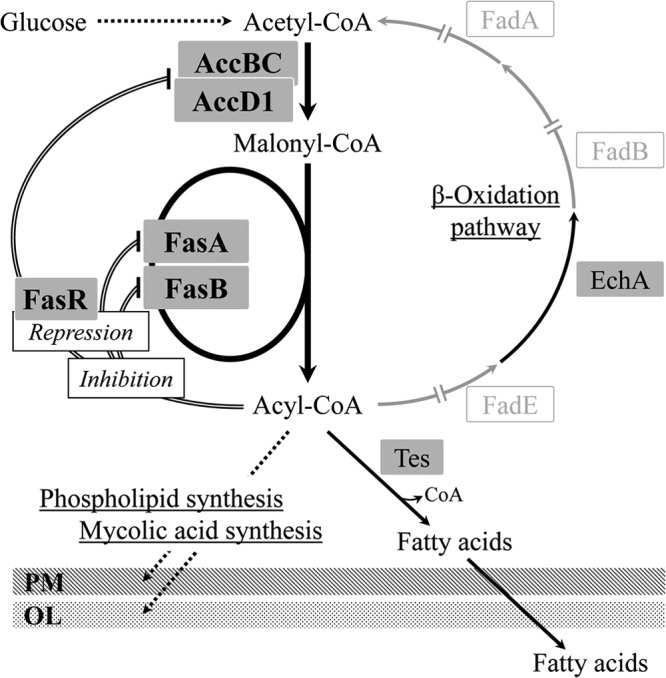
Fatty acid metabolism and its predicted regulatory mechanism in C. glutamicum. In coryneform bacteria, fatty acids are believed to be synthesized as acyl-CoAs (30), which are destined for incorporation into the membrane phospholipid and the outer layer component mycolic acid. Three genes responsible for the β-oxidation of fatty acids are missing from the C. glutamicum genome (gray arrows) (47). The Tes enzyme is assumed to be involved in the cleavage of oversupplied acyl-CoA to produce free fatty acids, considering the predicted role of the enzyme in fatty acid production in E. coli (11). The process of free fatty acid excretion remains to be elucidated. Acyl-CoA is thought to inhibit acetyl-CoA carboxylase (a complex of AccBC and AccD1), FasA, and FasB on the basis of the knowledge of related bacteria (52, 53). The repressor protein FasR, combined with the effector acyl-CoA, represses the genes for these four proteins (28). Repression and predicted inhibition are indicated by double lines. Arrows with solid and dotted lines represent single and multiple enzymatic processes, respectively. AccBC, acetyl-CoA carboxylase α subunit; AccD1, acetyl-CoA carboxylase β subunit; FasA, fatty acid synthase IA; FasB, fatty acid synthase IB; Tes, acyl-CoA thioesterase; FadE, acyl-CoA dehydrogenase; EchA, enoyl-CoA hydratase; FadB, hydroxyacyl-CoA dehydrogenase; FadA, ketoacyl-CoA reductase; PM, plasma membrane; OL, outer layer.
In this study, we initially investigated whether a desired fatty acid-producing mutant can be obtained from wild-type C. glutamicum. Our strategies were (i) to isolate a mutant that secretes oleic acid, a major fatty acid in the C. glutamicum membrane lipid (27), as an index of fatty acid production and (ii) to identify the causal mutations through genome analysis. For this purpose, we attempted to induce mutants that acquired desired phenotypes without using mutagenic treatment. Compared to the conventional mutagenic procedure, which depends on chemical mutagens or UV, the selection of a desired phenotype by spontaneous mutation is undoubtedly less efficient but seems to permit the accumulation of a minimum number of beneficial mutations even if the process is repeated. If this is true, genome analysis can be expected to directly decipher the results leading to desired phenotypes and thereby define the genetic background that is required to achieve production. Described here is the first demonstration of such strain development undertaken toward fatty acid production by C. glutamicum.
MATERIALS AND METHODS
Bacterial strains, plasmids, primers, and chemicals.
Wild-type C. glutamicum strain ATCC 13032 was used in this study. C. glutamicum OLA-15, which was used as an indicator strain for agar piece assays, is an oleic acid-auxotrophic mutant derived by a round of mutagenesis from the wild-type strain. E. coli DH5α was used as a host for DNA manipulation. Plasmid pCS299P (31), a C. glutamicum-E. coli shuttle vector, was used to clone the PCR products. Plasmid pESB30 (31), which is nonreplicative in C. glutamicum, is a vector for gene replacement in C. glutamicum. For the primer sequences used in this study, see Table S1 in the supplemental material. All of the primers were designed on the basis of the genomic sequence of C. glutamicum ATCC 13032 (BA000036), which is publicly available at http://www.genome.jp/kegg/genes.html (32). The chemical compounds Tween 40 and cerulenin were purchased from Nakalai Tesque (Kyoto, Japan) and Wako Pure Chemical Industries, Ltd. (Osaka, Japan), respectively.
Media and culture conditions.
Complete medium BY (33) and minimal medium MM (33) were used for the cultivation of wild-type ATCC 13032 and derivatives thereof. MM medium contained 1% glucose as the sole carbon source. Solid plates were made by the addition of Bacto agar (Difco) to 1.5%. For lipid production in liquid culture, a 3-ml sample of the seed culture grown in BY medium to the mid-exponential phase at 30°C was inoculated into a 300-ml baffled Erlenmeyer flask containing 30 ml of MM medium, followed by cultivation at 30°C on a rotary shaker at 200 rpm.
Agar piece assays for oleic acid production.
Microbiological assay for oleic acid was performed by an agar piece method essentially as described previously (34).
Recombinant DNA techniques.
Standard protocols (35) were used for the construction, purification, and analysis of plasmid DNA and for the transformation of E. coli. The extraction of C. glutamicum chromosomal DNA and transformation of C. glutamicum by electroporation were carried out as described previously (33).
Identification of mutations in fatty acid-producing mutants.
Mutations in strain PCC-6 were identified via a comparative genome analysis with the wild-type ATCC 13032 genome as a reference (http://www.genome.jp/kegg/genes.html). Whole-genome sequencing of strain PCC-6 was conducted by TaKaRa Bio Inc. (Shiga, Japan) with Illumina Genome Analyzer IIx (Illumina, San Diego, CA). In regard to the three specific mutations found in strain PCC-6, allele-specific PCR (36) was conducted to examine the presence or absence of each specific mutation in strains PAS-15 and PC-33.
Introduction of specific mutations into the genome.
Plasmids pCfasR20, pCfasA63up, and pCfasA2623, which were used for the introduction of specific mutations into the C. glutamicum genome, were constructed as follows. The mutated fasR gene region was PCR amplified with primers Cgl2490up700F and Cgl2490down500RFbaI with the genomic DNA from strain PCC-6 as a template, producing the 1.3-kb fragment. On the other hand, a region upstream of the fasA gene of strain PCC-6 was amplified with Cgl0836up900FFbaI and Cgl0836inn700RFbaI, producing the 1.7-kb fragment. Similarly, the mutated fasA gene region was amplified with primers Cgl0836inn700FFbaI and Cgl0836down200RFbaI with the genomic DNA of strain PCC-6, producing the 2.1-kb fragment. After verification by DNA sequencing, each PCR fragment that contained the corresponding point mutation in its middle portion was digested with BclI and then ligated to BamHI-digested pESB30 to yield the intended plasmid. The introduction of each specific mutation into the C. glutamicum genome was accomplished with the corresponding plasmid via two recombination events, as described previously (37). The presence of the mutation(s) was confirmed by allele-specific PCR and DNA sequencing.
Chromosomal deletion of the fasR gene.
Plasmid pCΔfasR containing the internally deleted fasR gene was constructed as follows. The 5′ region of the fasR gene was amplified with primers fasRup600FBglII and fasRFusR with wild-type ATCC 13032 genomic DNA as the template. Similarly, the 3′ region of the gene was amplified with primers fasRFusF and fasRdown800RBglII. The 5′ and 3′ regions were fused by PCR with primers fasRup600FBglII and fasRdown800RBglII. The resulting 1.6-kb fragment containing the deleted fasR gene, which was shortened by an in-frame deletion from 639 to 60 bp, digested with BglII, and then ligated to BamHI-digested pESB30 to yield pCΔfasR. Defined chromosomal deletion of the fasR gene was accomplished via two recombination events with the plasmid.
RNA extraction, cDNA synthesis, and qPCR.
Extraction of total RNAs from C. glutamicum strains and subsequent purification were performed as described previously (38). Synthesis of cDNA was performed with 300 ng of RNA as described by Kind et al. (17). Quantitative PCR (qPCR) analysis was performed by the method described by Katayama et al. (39). The gene expression levels were standardized to the constitutive level of 16S rRNA expression and calculated by the comparative cycle threshold method (40).
Quantitative determination of lipids.
Total lipids were extracted from culture supernatant by the Bligh-Dyer method (41). The culture supernatant was prepared by removing cells by centrifugation at 10,000 × g for 20 min and subsequent filtration with a Millex-MA filtration unit (0.45-μm pore size; Millipore Corporation, Billerica, MA). The extracted total lipids were dissolved in 2 ml of chloroform (here, the solution is referred to as extract A). Quantitative determination of lipids was conducted by the Toray Research Center (Kanagawa, Japan) by gas chromatography and thin-layer chromatography (TLC) as follows.
For free fatty acid analysis, 1 ml of extract A was evaporated under a nitrogen stream; suspended in a solvent containing 0.5 ml of benzene, 0.2 ml of methanol, and 1 ml of trimethylsilyldiazomethane; and then incubated at 60°C for 1 h for methyl-esterification of the free fatty acids. After the reaction, the mixture was evaporated under a nitrogen stream, dissolved in 1.0 ml of chloroform containing 0.005% methyl heneicosanoate as an internal standard, and applied to a GC-2010 gas chromatograph (Shimadzu, Kyoto, Japan) equipped with a flame ionization detector and an Omegawax 320 column (Sigma-Aldrich, St. Louis, MO). The column temperature was kept at 50°C for 1 min and then ramped to 270°C at a rate of 8°C/min. The injector and detector temperatures were held at 250°C and 270°C, respectively. Fatty acids were identified and quantified by using authentic fatty acid methyl ester standards.
For phospholipid analysis, 1 ml of extract A was evaporated under a nitrogen stream, dissolved in 0.1 ml of chloroform, and applied to HPTLC plates with Silica Gel 60 (Merck, Darmstadt, Germany). The solvent was chloroform-methanol-acetic acid-water at 125:75:6.5:5 (vol/vol/vol/vol). After separation, the plates were sprayed with 10% copper sulfate in 8% phosphoric acid solution and baked for 30 min at 150°C. The position of each lipid species was identified by comparison with the corresponding standard supplied by Doosan Serdar Research Laboratories (Toronto, Ontario, Canada). The intensities of the spots were measured with an Image Master 1D Elite ver. 3.00 (Amersham Bioscience, Tokyo, Japan). Lipid species were quantified by using the standard curves for each lipid drawn with serial dilutions of the standard substance.
Analysis.
Bacterial growth was monitored by measuring the optical density at 660 nm (OD660) of the culture broth with a Miniphoto 518R spectrophotometer (Taitec, Saitama, Japan). Glucose concentration was determined with Determinar GL-E (Kyowa Medex, Tokyo, Japan).
RESULTS
Screening of compounds to induce oleic acid-producing mutants.
A chemical substance that satisfies the following criteria is assumed to be a specific inhibitor of fatty acid biosynthesis in C. glutamicum. Mutants resistant to the compound are likely to overproduce oleic acid, a major component of C. glutamicum membrane lipid (27); (i) C. glutamicum cells are subject to growth inhibition in the presence of the compound, and (ii) the growth inhibition is restored by the copresence of oleic acid. After screening a variety of chemical substances, including known inhibitors of bacterial fatty acid biosynthesis (42), for such compounds, we found that the palmitic acid ester surfactant Tween 40, as well as the antibiotic cerulenin, satisfied the above criteria. Both of these compounds have been suggested to have targets involved in fatty acid biosynthesis in coryneform bacteria; the presence of Tween 40 in the culture caused a decreased amount of the acetyl-CoA carboxylase β subunit in C. glutamicum ATCC 13869 (24), whereas cerulenin inhibited fatty acid synthase from C. ammoniagenes in vitro (43). Both compounds have also been reported to trigger l-glutamate production by C. glutamicum, presumably by membrane destabilization (44, 45).
Selection of spontaneous mutants resistant to Tween 40.
Although both compounds met our criteria, the phenotype of growth recovery by oleic acid was more prominent when Tween 40 was used. Thus, we first attempted to isolate spontaneous Tween 40-resistant mutants from wild-type C. glutamicum ATCC 13032. For this purpose, appropriate dilutions (105 to 106 cells/ml) of the culture were spread onto MM agar plates containing the MIC of Tween 40 (approximately 1.5 g/liter), and colonies that emerged on the plates after a 5-day cultivation were isolated. These Tween 40-resistant colonies were obtained at a frequency of approximately 10−4. These resistant colonies were then examined for the ability to produce oleic acid by agar piece assay with the oleic acid auxotroph OLA-15 as an indicator strain. As a result, more than half of the mutants examined were found to produce oleic acid whereas the wild-type strain never produced the fatty acid. Among these, the strain that gave the largest halo of the indicator strain was designated strain PAS-15 (Fig. 2). It was used as the parent strain to induce a second mutation.
Fig 2.
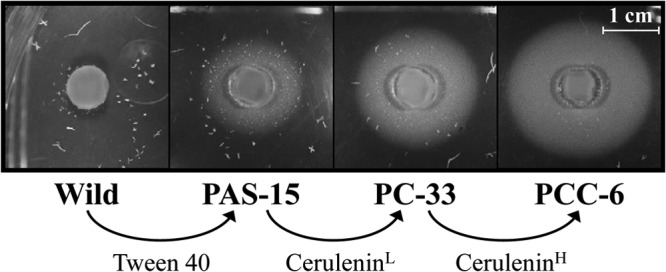
Oleic acid-producing abilities of strains PAS-15, PC-33, and PCC-6. These three strains and wild-type strain ATCC 13032 were cultivated on MM agar pieces. After cultivation for 2 days, the agar pieces were transferred onto bioassay plates containing the oleic acid auxotroph OLA-15 as an indicator strain. The plates were incubated for 1 day at 30°C. The images show one representative result from three independent experiments. Arrows represent the lineage relationships. Tween 40 and cerulenin were used as the potential specific inhibitors of fatty acid biosynthesis in C. glutamicum to induce oleic acid-producing mutants. CeruleninL, resistance to a relatively low concentration of cerulenin; CeruleninH, resistance to a relatively high concentration of cerulenin.
Repeated selection of spontaneous cerulenin-resistant mutants.
Since strain PAS-15 no longer exhibited sensitivity to Tween 40, even at 20 g/liter, we attempted to isolate spontaneous mutants resistant to the other compound, cerulenin, from the strain in the same way as when selecting Tween 40-resistant mutants. After cultivation for several days, colonies emerged on the MM agar plates containing the MIC (approximately 7.5 mg/liter) of cerulenin at a frequency of approximately 10−4. These resistant colonies were examined for the production of oleic acid by agar piece assay, which revealed that approximately 5% of the colonies showed higher production of the fatty acid than parental strain PAS-15. Among these, the strain that showed the highest production was designated strain PC-33 (Fig. 2). It was used as the parent strain to induce a third mutation. Because the strain still showed sensitivity to a higher concentration of cerulenin, we further induced higher resistance to cerulenin in the strain. When spontaneous selection was conducted at the MIC (approximately 15 mg/liter) for strain PC-33, colonies emerged at a frequency of approximately 10−4. Agar piece assay revealed that approximately 10% of the colonies showed higher production of the fatty acid than parental strain PC-33. From these, we selected the best producer, which was designated PCC-6 (Fig. 2).
Identification of mutations in strains PAS-15, PC-33, and PCC-6.
Since the strain obtained, PCC-6, had acquired the ability to produce a relatively large halo, for which we estimated the oleic acid level to be between 100 and 300 mg/liter, in our agar piece assay, we considered it worthwhile to analyze its genetic traits that were related to fatty acid production. To identify them, we conducted whole-genome sequencing of the strain, which revealed only three specific mutations (Fig. 3), a G-to-A exchange at nucleotide position 59 in the fasR gene, which led to the replacement of Ser-20 with Asn (designated mutation fasR20); a C-to-G exchange at 63 bp upstream of the fasA gene (designated mutation fasA63up); and a C-to-T exchange at nucleotide position 7868 in the fasA gene, which led to the replacement of Ala-2623 by Val (designated mutation fasA2623). Since the fasR and fasA genes are known to encode the transcriptional regulator FasR and the fatty acid synthase FasA, respectively (27, 28), the three mutations identified were all suggested to be related to fatty acid biosynthesis. Subsequent allele-specific PCR revealed that the strain initially obtained, PAS-15, carried the fasR20 mutation whereas the next strain, PC-33, carried the fasA63up mutation in addition to fasR20, indicating that the mutations arose in the order fasR20, fasA63up, and fasA2623 (Fig. 3). This also suggests that the fasR20 mutation is responsible for Tween 40 resistance, whereas the fasA63up and fasA2623 mutations are responsible for resistance to the lower and higher concentrations of cerulenin, respectively.
Fig 3.
Three specific mutations identified in the oleic acid-producing mutants. The locations of mutations fasR20, fasA63up, and fasA2623 are indicated by dotted lines. The order in which these mutations arose is shown by circled numbers 1 to 3. The fasR20 mutation is located at nucleotide position 59 in the fasR gene (gray gene). The fasA63up mutation is located 63 bp upstream of the fasA gene. The nucleotide sequence of its surrounding region is also shown. The fasA63up mutation is indicated by the letter larger than its neighbors. The FasR-biding site fasO is boxed (28). The −10 and −35 regions of a potential promoter of fasA are underlined, and the transcriptional start site is also indicated by a bold and underlined letter (28). Hatched boxes (boxes A to G) along the fasA gene represent nucleotide regions for putative catalytic domains involving in fatty acid synthesis (29, 48). The white part of box G represents a region for a motif sequence (PROSITE motif PS00606) for a 3-ketoacyl-ACP synthase active site. The fasA2623 mutation is located within the motif. Box A represents a region for acetyl-CoA transferase, box B represents a region for enoyl-ACP reductase, box C represents a region for 3-ketoacyl-ACP dehydratase, box D represents a region for malonyl/palmitoyl transferase, box E represents a region for a substrate binding site of ACP, box F represents a region for 3-ketoacyl-ACP reductase, and box G represents a region for 3-ketoacyl-ACP synthase. The genes whose expression is thought to depend on FasR (28) are black.
Reconstitution of defined mutations in a wild-type genome and their effects on oleic acid production.
To examine the relevance of the three mutations to oleic acid production, we first introduced them into the wild-type genome separately and examined their effects on the ability to produce oleic acid (Fig. 4). Agar piece assay showed that only fasR20 gave rise to oleic acid production in the wild-type strain, whereas the other two mutations showed no significant effect on production. We also examined the effect of the in-frame deletion of the fasR inner sequence (designated ΔfasR) on production in the wild-type strain, which revealed that the modification resulted in almost the same level of oleic acid production as in the case of fasR20 (Fig. 4). Next, we examined the effect of the combination of fasR20 with either fasA63up or fasA2623 on production (Fig. 4). When fasR20 was combined with fasA63up in the wild-type genome, increased oleic acid production was observed, compared with that obtained with fasR20 alone. The combination of fasR20 and fasA2623 resulted in an oleic acid production level that was comparable to that obtained with fasR20 alone. On the other hand, the combination of fasA63up and fasA2623 in the wild-type genome resulted in no oleic acid production. When all three mutations were combined in the wild-type genome, the highest oleic acid production of all of the combinations tested was observed, as expected (Fig. 4). These results indicate that loss of the function of fasR is of primary importance for fatty acid production by C. glutamicum and that the fasA63up and fasA2623 mutations positively affect carbon flow down the pathway. The fasA2623 mutation seemed to be effective, especially in the background of fasR20 and fasA63up.
Fig 4.
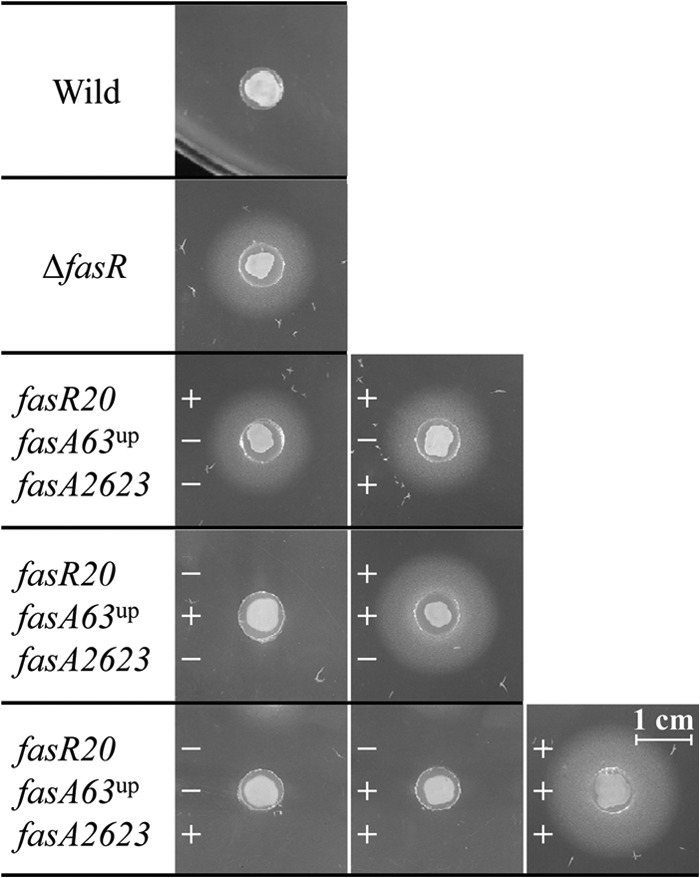
Reconstitution of defined mutations in the wild-type genome and its effect on oleic acid production. Wild-type ATCC 13032 carrying the mutations fasR20, fasA63up, fasA2623, and ΔfasR separately or in combination were examined for the ability to produce oleic acid by using the same agar piece assay as in Fig. 2. The images show one result representative of three independent experiments. Plus and minus signs represent the presence and absence of the corresponding mutation in the wild-type background, respectively. The ΔfasR mutant strain carries no other mutation, except for the deletion of the fasR gene.
Effects of the fasR20 and fasA63up mutations on the transcript levels of fatty acid biosynthesis genes.
Apart from the fasA2623 mutation that was thought to affect the enzymatic properties of FasA (see Discussion), the fasR20 and fasA63up mutations were both considered to affect the transcript levels of the relevant genes, because the former is a missense mutation within the transcriptional regulator FasR and the latter is located near the predicted promoter-operator regions of the fasA gene (Fig. 3). Accordingly, we used reverse transcription (RT)-qPCR to investigate the transcript levels of the fatty acid biosynthesis genes fasA, fasB, accD1, and accBC in the strains carrying the two mutations individually or in combination. As shown in Fig. 5, the fasR20 mutation increased the transcript levels of accD1 by 3.56-fold ± 0.97-fold, as well as both fasA and fasB by 1.31-fold ± 0.11-fold and 1.29-fold ± 0.12-fold, respectively, whereas the mutation had little influence on accBC gene expression. Similar changes in transcript levels were observed in the ΔfasR strain (Fig. 5). On the other hand, the fasA63up mutation led to a 2.67-fold ± 0.16-fold increase in the transcript level of fasA. The presence of both the fasR20 and fasA63up mutations resulted in an additive effect on fasA gene expression.
Fig 5.
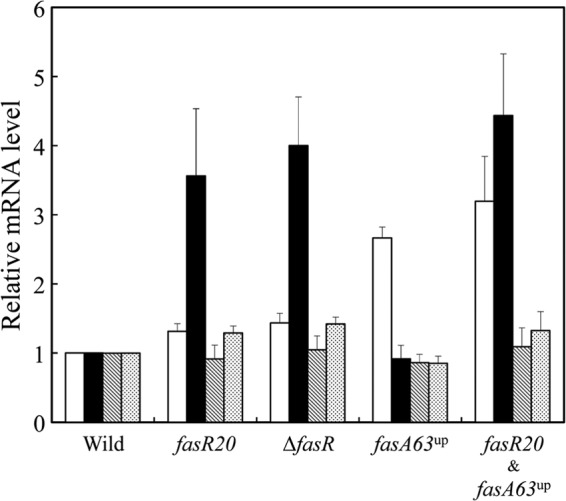
Relative mRNA levels of the fatty acid biosynthesis genes in wild-type ATCC 13032 carrying the mutations fasR20, ΔfasR, and fasA63up separately or in combination. Total RNAs were prepared from cells grown to the early exponential phase (OD660 of approximately 2.5) in MM medium. Aliquots of RNAs were reverse transcribed and subjected to qPCR. The transcript levels of fasA (white bars), accD1 (black bars), accBC (hatched bars), and fasB (dotted bars) were standardized to the constitutive expression level of 16S rRNA. The transcript levels in wild-type ATCC 13032 were set to 1.0. Data represent mean values from three independent cultures, and the standard deviation from the mean is indicated as error bars.
Lipid production by strain PCC-6.
Although strain PCC-6 produced oleic acid from glucose, we needed to determine what kinds of lipids were produced and what their yields were. To clarify this, strain PCC-6, as well as wild-type ATCC 13032, was aerobically cultivated in 30 ml of MM medium containing 1% glucose in a 300-ml baffled Erlenmeyer flask (Fig. 6). Under these conditions, strain PCC-6 showed a lower growth rate and a lower final OD660 than the wild-type strain, probably because of the production of fatty acids and their negative effects on cell physiology (46). After glucose was consumed, the cells were removed by centrifugation, followed by filtration, and the culture supernatant was subjected to lipid analysis. As shown in Table 1, wild-type ATCC 13032 produced only a trace amount of lipids. In contrast, strain PCC-6 produced 279.95 ± 8.50 mg of free fatty acids and 43.18 ± 1.84 mg of phospholipids/liter. The fatty acids consisted mainly of oleic acid (208.10 ± 5.67 mg/liter) and palmitic acid (46.93 ± 2.03 mg/liter), both accounting for 91.10% of the total free fatty acids produced in the culture supernatant. The conversion yield of the total fatty acids on glucose was 2.80% ± 0.09% (wt/wt). Since the theoretical yield of oleic acid on glucose is estimated to be 34.8% (wt/wt) on the basis of our calculation, the production level of strain PCC-6 is considered to be less than 10% of the theoretical yield.
Fig 6.

Time course of growth and glucose consumption of wild-type ATCC 13032 and strain PCC-6. The two strains were cultivated in 30 ml of MM medium with rotary shaking. Symbols: ●, growth of wild-type ATCC 13032; ■, growth of strain PCC-6; ○, residual glucose in ATCC 13032; □, residual glucose in strain PCC-6. Values are means of replicated cultures, which showed <5% difference from each other. Arrows indicate the time points at which culture supernatants were prepared for lipid analysis.
Table 1.
Lipid production by wild-type ATCC 13032 and strain PCC-6a
| Lipid | Wild type |
Strain PCC-6 |
||
|---|---|---|---|---|
| Production (mg/liter) | Wt % | Production (mg/liter) | Wt % | |
| Free fatty acids | ||||
| C15:1 | 1.61 ± 0.04 | 50.00 ± 0.16 | 2.93 ± 0.06 | 1.05 ± 0.02 |
| C16:0 | —b | — | 46.93 ± 2.03 | 16.76 ± 0.22 |
| C16:1 | 0.71 ± 0.04 | 21.95 ± 0.68 | 6.39 ± 0.21 | 2.28 ± 0.00 |
| C18:0 | — | — | 12.35 ± 0.46 | 4.41 ± 0.03 |
| C18:1 | 0.90 ± 0.01 | 28.06 ± 0.84 | 208.10 ± 5.67 | 74.34 ± 0.23 |
| C20:0 | — | — | 2.50 ± 0.06 | 0.89 ± 0.11 |
| C20:1 | — | — | 0.77 ± 0.03 | 0.28 ± 0.00 |
| Total | 3.21 ± 0.06 | 100.00 ± 0.00 | 279.95 ± 8.50 | 100.00 ± 0.00 |
| Phospholipids | ||||
| DPG | 9.76 ± 0.47 | 100.00 ± 0.00 | 43.18 ± 1.84 | 100.00 ± 0.00 |
| Total | 9.76 ± 0.47 | 100.00 ± 0.00 | 43.18 ± 1.84 | 100.00 ± 0.00 |
Culture supernatants were prepared at the points indicated by the arrows in Fig. 6 and then subjected to lipid analysis. The amounts of lipids were determined by using two independent cultures performed as described in the legend to Fig. 6. Values are means ± standard deviations. DPG is diphosphatidylglycerol. Other phospholipids (e.g., phosphatidylinositol, phosphatidylglycerol, and phosphatidic acid) were not detected in either strain.
—, not detected.
DISCUSSION
Despite a broad product portfolio for C. glutamicum (15, 17, 18, 19, 21), lipids and their related compounds have not been intensively developed for production. In this study, we demonstrated for the first time that this organism has the capability of producing considerable amounts of fatty acids directly from sugar, thus expanding its product portfolio to lipids. This raises the possibility of developing C. glutamicum production processes not only for fatty acids but also for other useful compounds that are derived via the fatty acid biosynthetic pathway. To date, no information is available on what kind of modifications or selections contribute to increased carbon flow into the fatty acid biosynthetic pathway of this organism. This study is the first to report not only the selection methods used but also the genetic traits that cause fatty acid production.
The three specific mutations, fasR20, fasA63up, and fasA2623, identified as genetic traits that are useful for fatty acid production are all related to fatty acid biosynthesis, and no mutation that is related to fatty acid transport is included. This suggests that deregulation of the fatty acid biosynthetic pathway would cause carbon flow down the pathway and that the oversupplied acyl-CoAs would be excreted into the medium as free fatty acids without undergoing degradation in this organism. The latter hypothesis is supported by the C. glutamicum genome information, which shows a lack of some of the genes responsible for the β-oxidation of fatty acids (Fig. 1) (47). In fact, unlike E. coli, wild-type C. glutamicum hardly grew on MM medium containing 10 g of oleic acid/liter as the sole carbon source (data not shown). The relevance of each mutation to fatty acid production is discussed below.
The fasR20 mutation conferred oleic acid production on wild-type C. glutamicum concomitantly with the Tween 40 resistance phenotype (Fig. 2 and 4). Since this mutation more or less increased the expression levels of accD1, fasA, and fasB (Fig. 5), the effect of the mutation on production is reasonably explained by derepression of the key regulatory genes in the fatty acid biosynthetic pathway. Considering that the fasR gene product is thought to be a fatty acid biosynthesis repressor protein (28) and also that its deletion of the gene from the wild-type strain caused similar oleic acid production (Fig. 4), the fasR20 mutation would cause functional impairment of the repressor protein. In this context, it has been suggested that the FasR protein, combined with the effector acyl-CoA, binds to fasO sites upstream of the corresponding genes and thereby suppresses their expression (28). On the basis of this mechanism, the fasR20 mutation is likely to interfere with the formation of the FasR-acyl-CoA complex or binding of the complex to the fasO sites. Taken together, the findings indicate that the reason why the Tween 40 resistance phenotype resulted in oleic acid production can be explained as follows. In the wild-type strain, the palmitic acid ester surfactant Tween 40 probably triggers the FasR-mediated repression of fatty acid biosynthesis, which causes deprivation of essential lipids and results in growth inhibition. However, this Tween 40-triggered repression mechanism can be bypassed in the fasR-defective mutant, thus leading to the Tween 40 resistance phenotype, accompanied by derepression of fatty acid biosynthesis and subsequent oleic acid production. This speculation is supported by our findings that the growth-inhibitory effect of Tween 40 on wild-type C. glutamicum is restored either by the copresence of oleic acid or by the loss of the function of fasR (data not shown).
The fasA63up mutation, which is located upstream of the fasA coding region, was obtained by the selection of a relatively low concentration of cerulenin in the genetic background of fasR20. Since the mutation significantly increased the transcript level of the fasA gene (Fig. 5), the effect of the mutation on oleic production is explainable by an increased amount of the FasA enzyme that is responsible for oleic acid synthesis (27, 48). Considering that cerulenin is known to inhibit Fas from the closely related species C. ammoniagenes (43), as well as E. coli FabF and FabB (49, 50), it is reasonable to assume that the agent also inhibits C. glutamicum FasA, which causes deprivation of essential lipids and results in growth inhibition. This hypothesis is consistent with the previous observation that inactivation of FasA in C. glutamicum resulted in no growth in MM medium and that this growth impairment was recovered by oleic acid supplementation (27). Presumably, the mutants with increased transcript levels of fasA could overcome the cerulenin-caused inhibition of FasA through the dosage effect of the FasA molecules. This explains why the cerulenin resistance phenotype was caused by the mutation and resulted in increased oleic acid production. Although the fasA63up mutation is located outside the putative promoter-operator regions of the fasA gene (Fig. 3), our RT-qPCR data suggest that the mutation site is involved in fasA gene expression.
The fasA2623 mutation, which is present in the fasA coding region, was obtained by the selection of a relatively high concentration of cerulenin in the genetic background of fasR20 and fasA63up. The mutation is present within a motif sequence (PROSITE motif PS00606) for a 3-ketoacyl-ACP synthase (KS) active site in the deduced amino acid sequence of FasA. In this regard, the E. coli KS enzyme FabH, which has the same motif sequence (Institute for Biomolecular Design Project CyperCell Database CCDB FABH_ECOLI), has been reported to be feedback inhibited by long-chain (12- to 20-carbon) acyl-ACPs through a mixed type of inhibition, namely, a combination of competitive and noncompetitive inhibition with respect to acetyl-CoA (51). If C. glutamicum FasA is regulated at its KS domain in the same manner as seen for E. coli FabH, it seems reasonable to speculate that the fasA2623 mutation alleviates the feedback inhibition and thereby results in increased oleic acid production. In E. coli, cerulenin is known to inhibit KS by covalently binding to the active-center cysteine (49). This cysteine residue is assumed to correspond to Cys2619 of the deduced amino acid sequence of C. glutamicum FasA, on the basis of sequence alignment. Taking this into consideration, it is likely that the fasA2623 mutation, which is located very near the predicted active center and gives rise to a change from alanine to valine with a longer side chain, may cause steric hindrance to the binding of cerulenin, thereby resulting in cerulenin resistance. This may also be the mechanism of the possible relief of the mutated FasA enzyme from feedback inhibition.
The reconstitution experiments of three specific mutations in the wild-type background (Fig. 4) have demonstrated that the fasR mutation is of primary importance for fatty acid production by C. glutamicum. To confirm this, we sequenced the fasR genes from an additional 30 oleic acid-producing mutants selected by Tween 40 resistance and found that all of the fasR genes carried mutations, including single-base substitutions (10 cases of 30 mutants), single-base insertions (3 cases), a 165-bp deletion (1 case), and insertion of ISCg1a (15 cases) or ISCg13b (1 case) (data not shown). These results strongly suggest that loss of the function of fasR is essential for fatty acid production by C. glutamicum. To date, it has not been reported that inactivation of fasR induces fatty acid production in C. glutamicum, despite the study of the fasR gene (28).
As mentioned in the introduction, E. coli has recently been used to study fatty acid production. Since the first report on fatty acid production by E. coli overexpressing the modified acyl-ACP thioesterase gene ′tesA (11), overexpression of the enzyme has become a common strategy for fatty acid production by E. coli. A basic concept in this production is avoidance of the regulatory mechanism of fatty acid synthesis through the thioesterase-catalyzed cleavage of acyl-ACP. On the other hand, in our study with C. glutamicum, the defined genetic modifications to fatty acid biosynthesis resulted in fatty acid production without modification of the acyl-ACP thioesterase enzyme. This raises the question of how the oversupplied acyl-CoAs, end products of fatty acid biosynthesis in this organism, would be excreted into the medium as free fatty acids. In regard to this, we found that C. glutamicum originally had a high level of thioesterase activity (1.27 ± 0.018 U/mg of protein) in the soluble fraction prepared from cells grown in MM medium. This activity level is comparable to that obtained from ′tesA-overexpressing E. coli (1.29 ± 0.11 U/mg of protein) and is approximately 16-fold higher than that obtained from non-′tesA-overexpressing E. coli. Taking this into consideration, it is likely that C. glutamicum possesses a specific mechanism for maintaining lipid homeostasis even in the presence of high thioesterase activity. The C. glutamicum genome indicates the presence of three putative acyl-CoA thioesterases (Cgl0091, Cgl1664, and Cgl2451). The involvement of the genes for these putative acyl-CoA thioesterases in fatty acid production, along with the mechanism of free fatty acid secretion, needs to be clarified in a future study.
Supplementary Material
ACKNOWLEDGMENTS
We thank Yasuo Ueda, Shin-ichi Hashimoto, Satoshi Koizumi, Tatsuya Ogawa, and Akinori Yasuhara for their encouraging support of our research. We are also grateful to John E. Cronan (University of Illinois) for the kind gift of ′tesA-overexpressing E. coli strain HC125.
Footnotes
Published ahead of print 30 August 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02003-13.
REFERENCES
- 1. Horrobin DF. 1992. Nutritional and medical importance of gamma-linolenic acid. Prog. Lipid Res. 31:163–194 [DOI] [PubMed] [Google Scholar]
- 2. Smith WL, Borgeat P. 1985. The eicosanoids: prostaglandins, thromboxanes, leukotrienes, and hydroxy-eicosaenoic acids, p 325–360 In Vance DE, Vance JE. (ed), Biochemistry of lipids and membranes. Benjamin/Cummings, Menlo Park, CA [Google Scholar]
- 3. Bilska A, Włodek L. 2005. Lipoic acid—the drug of the future? Pharmacol. Rep. 57:570–577 [PubMed] [Google Scholar]
- 4. Packer L, Witt EH, Tritschler HJ. 1995. Alpha-lipoic acid as a biological antioxidant. Free Radic. Biol. Med. 19:227–250 [DOI] [PubMed] [Google Scholar]
- 5. Streit WR, Entcheva P. 2003. Biotin in microbes, the genes involved in its biosynthesis, its biochemical role and perspectives for biotechnological production. Appl. Microbiol. Biotechnol. 61:21–31 [DOI] [PubMed] [Google Scholar]
- 6. Kalscheuer R, Stölting T, Steinbüchel A. 2006. Microdiesel: Escherichia coli engineered for fuel production. Microbiology 152:2529–2536 [DOI] [PubMed] [Google Scholar]
- 7. Metzger P, Largeau C. 2005. Botryococcus braunii: a rich source for hydrocarbons and related ether lipids. Appl. Microbiol. Biotechnol. 66:486–496 [DOI] [PubMed] [Google Scholar]
- 8. Sakuradani E, Ando A, Shimizu S, Ogawa J. 3 May 2013, posting date Metabolic engineering for the production of polyunsaturated fatty acids by oleaginous fungus Mortierella alpina 1S-4. J. Biosci. Bioeng. (Epub ahead of print.) 10.1016/j.jbiosc.2013.04.008 [DOI] [PubMed] [Google Scholar]
- 9. Beopoulos A, Nicaud JM, Gaillardin C. 2011. An overview of lipid metabolism in yeasts and its impact on biological processes. Appl. Microbiol. Biotechnol. 90:1193–1206 [DOI] [PubMed] [Google Scholar]
- 10. Lennen RM, Pfleger BF. 2012. Engineering Escherichia coli to synthesize free fatty acids. Trends Biotechnol. 30:659–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cho H, Cronan JE., Jr 1995. Defective export of a periplasmic enzyme disrupts regulation of fatty acid synthesis. J. Biol. Chem. 270:4216–4219 [DOI] [PubMed] [Google Scholar]
- 12. Dellomonaco C, Clomburg JM, Miller EN, Gonzalez R. 2011. Engineered reversal of the β-oxidation cycle for the synthesis of fuels and chemicals. Nature 476:355–359 [DOI] [PubMed] [Google Scholar]
- 13. Liu X, Brune D, Vermaas W, Curtiss R., III 2011. Production and secretion of fatty acids in genetically engineered cyanobacteria. Proc. Natl. Acad. Sci. U. S. A. 108:6899–6904 [DOI] [PubMed] [Google Scholar]
- 14. Steen EJ, Kang Y, Bokinsky G, Hu Z, Schirmer A, McClure A, del Cardayre SB, Keasling JD. 2010. Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature 463:559–562 [DOI] [PubMed] [Google Scholar]
- 15. Ikeda M, Takeno S. 2013. Amino acid production by Corynebacterium glutamicum, p 107–147 In Yukawa H, Inui M. (ed), Corynebacterium glutamicum. Microbiology monographs 23. Springer, Berlin, Germany [Google Scholar]
- 16. Okino S, Noburyu R, Suda M, Jojima T, Inui M, Yukawa H. 2008. An efficient succinic acid production process in a metabolically engineered Corynebacterium glutamicum strain. Appl. Microbiol. Biotechnol. 81:459–464 [DOI] [PubMed] [Google Scholar]
- 17. Kind S, Kreye S, Wittmann C. 2011. Metabolic engineering of cellular transport for overproduction of the platform chemical 1,5-diaminopentane in Corynebacterium glutamicum. Metab. Eng. 13:617–627 [DOI] [PubMed] [Google Scholar]
- 18. Song Y, Matsumoto K, Tanaka T, Kondo A, Taguchi S. 2013. Single-step production of polyhydroxybutyrate from starch by using α-amylase cell-surface displaying system of Corynebacterium glutamicum. J. Biosci. Bioeng. 115:12–14 [DOI] [PubMed] [Google Scholar]
- 19. Inui M, Kawaguchi H, Murakami S, Vertès AA, Yukawa H. 2004. Metabolic engineering of Corynebacterium glutamicum for fuel ethanol production under oxygen-deprivation conditions. J. Mol. Microbiol. Biotechnol. 8:243–254 [DOI] [PubMed] [Google Scholar]
- 20. Blombach B, Riester T, Wieschalka S, Ziert C, Youn JW, Wendisch VF, Eikmanns BJ. 2011. Corynebacterium glutamicum tailored for efficient isobutanol production. Appl. Environ. Microbiol. 77:3300–3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Heider SA, Peters-Wendisch P, Wendisch VF. 2012. Carotenoid biosynthesis and overproduction in Corynebacterium glutamicum. BMC Microbiol. 12:198. 10.1186/1471-2180-12-198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kikuchi Y, Itaya H, Date M, Matsui K, Wu LF. 2009. TatABC overexpression improves Corynebacterium glutamicum Tat-dependent protein secretion. Appl. Environ. Microbiol. 75:603–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Inui M, Murakami S, Okino S, Kawaguchi H, Vertès AA, Yukawa H. 2004. Metabolic analysis of Corynebacterium glutamicum during lactate and succinate productions under oxygen deprivation conditions. J. Mol. Microbiol. Biotechnol. 7:182–196 [DOI] [PubMed] [Google Scholar]
- 24. Kimura E, Yagoshi C, Kawahara Y, Ohsumi T, Nakamatsu T, Tokuda H. 1999. Glutamate overproduction in Corynebacterium glutamicum triggered by a decrease in the level of a complex comprising DtsR and a biotin-containing subunit. Biosci. Biotechnol. Biochem. 63:1274–1278 [DOI] [PubMed] [Google Scholar]
- 25. Gande R, Gibson KJ, Brown AK, Krumbach K, Dover LG, Sahm H, Shioyama S, Oikawa T, Besra GS, Eggeling L. 2004. Acyl-CoA carboxylases (accD2 and accD3), together with a unique polyketide synthase (Cg-pks), are key to mycolic acid biosynthesis in Corynebacterianeae such as Corynebacterium glutamicum and Mycobacterium tuberculosis. J. Biol. Chem. 279:44847–44857 [DOI] [PubMed] [Google Scholar]
- 26. Jäger W, Peters-Wendisch PG, Kalinowski J, Pühler A. 1996. A Corynebacterium glutamicum gene encoding a two-domain protein similar to biotin carboxylases and biotin-carboxyl-carrier protein. Arch. Microbiol. 166:76–82 [DOI] [PubMed] [Google Scholar]
- 27. Radmacher E, Alderwick LJ, Besra GS, Brown AK, Gibson KJ, Sahm H, Eggeling L. 2005. Two functional FAS-I type fatty acid synthases in Corynebacterium glutamicum. Microbiology 151:2421–2427 [DOI] [PubMed] [Google Scholar]
- 28. Nickel J, Irzik K, van Ooyen J, Eggeling L. 2010. The TetR-type transcriptional regulator FasR of Corynebacterium glutamicum controls genes of lipid synthesis during growth on acetate. Mol. Microbiol. 78:253–265 [DOI] [PubMed] [Google Scholar]
- 29. Schweizer E, Hofmann J. 2004. Microbial type I fatty acid synthases (FAS): major players in a network of cellular FAS systems. Microbiol. Mol. Biol. Rev. 68:501–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kawaguchi A, Okuda S. 1977. Fatty acid synthetase from Brevibacterium ammoniagenes: formation of monounsaturated fatty acids by a multienzyme complex. Proc. Natl. Acad. Sci. U. S. A. 74:3180–3183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mitsuhashi S, Ohnishi J, Hayashi M, Ikeda M. 2004. A gene homologous to β-type carbonic anhydrase is essential for the growth of Corynebacterium glutamicum under atmospheric conditions. Appl. Microbiol. Biotechnol. 63:592–601 [DOI] [PubMed] [Google Scholar]
- 32. Ikeda M, Nakagawa S. 2003. The Corynebacterium glutamicum genome: features and impacts on biotechnological processes. Appl. Microbiol. Biotechnol. 62:99–109 [DOI] [PubMed] [Google Scholar]
- 33. Takeno S, Ohnishi J, Komatsu T, Masaki T, Sen K, Ikeda M. 2007. Anaerobic growth and potential for amino acid production by nitrate respiration in Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 75:1173–1182 [DOI] [PubMed] [Google Scholar]
- 34. Ikeda M, Miyamoto A, Mutoh S, Kitano Y, Tajima M, Shirakura D, Takasaki M, Mitsuhashi S, Takeno S. 2013. Development of biotin-prototrophic and -hyperauxotrophic Corynebacterium glutamicum strains. Appl. Environ. Microbiol. 79:4586–4594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 36. Newton CR, Graham A, Heptinstall LE, Powell SJ, Summers C, Kalsheker N, Smith JC, Markham AF. 1989. Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS). Nucleic Acids Res. 17:2503–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ohnishi J, Mitsuhashi S, Hayashi M, Ando S, Yokoi H, Ochiai K, Ikeda M. 2002. A novel methodology employing Corynebacterium glutamicum genome information to generate a new l-lysine-producing mutant. Appl. Microbiol. Biotechnol. 58:217–223 [DOI] [PubMed] [Google Scholar]
- 38. Hayashi M, Mizoguchi H, Shiraishi N, Obayashi M, Nakagawa S, Imai J, Watanabe S, Ota T, Ikeda M. 2002. Transcriptome analysis of acetate metabolism in Corynebacterium glutamicum using a newly developed metabolic array. Biosci. Biotechnol. Biochem. 66:1337–1344 [DOI] [PubMed] [Google Scholar]
- 39. Katayama S, Kukita T, Ishikawa E, Nakashima S, Masuda S, Kanda T, Akiyama H, Teshima R, Nakamura S. 2013. Apple polyphenols suppress antigen presentation of ovalbumin by THP-1-derived dendritic cells. Food Chem. 138:757–761 [DOI] [PubMed] [Google Scholar]
- 40. Schmittgen TD, Livak K. 2008. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3:1101–1108 [DOI] [PubMed] [Google Scholar]
- 41. Bligh EG, Dyer WJ. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911–917 [DOI] [PubMed] [Google Scholar]
- 42. Heath RJ, White SW, Rock CO. 2002. Inhibitors of fatty acid synthesis as antimicrobial chemotherapeutics. Appl. Microbiol. Biotechnol. 58:695–703 [DOI] [PubMed] [Google Scholar]
- 43. Kawaguchi A, Tomada H, Okuda S, Awaya J, Omura S. 1979. Cerulenin resistance in a cerulenin-producing fungus: isolation of cerulenin insensitive fatty acid synthetase. Arch. Biochem. Biophys. 197:30–35 [DOI] [PubMed] [Google Scholar]
- 44. Asakura Y, Kimura E, Usuda Y, Kawahara Y, Matsui K, Osumi T, Nakamatsu T. 2007. Altered metabolic flux due to deletion of odhA causes l-glutamate overproduction in Corynebacterium glutamicum. Appl. Environ. Microbiol. 73:1308–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hoischen C, Krämer R. 1990. Membrane alteration is necessary but not sufficient for effective glutamate secretion in Corynebacterium glutamicum. J. Bacteriol. 172:3409–3416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Desbois AP, Smith VJ. 2010. Antibacterial free fatty acids: activities, mechanisms of action and biotechnological potential. Appl. Microbiol. Biotechnol. 85:1629–1642 [DOI] [PubMed] [Google Scholar]
- 47. Barzantny H, Brune I, Tauch A. 2012. Molecular basis of human body odour formation: insights deduced from corynebacterial genome sequences. Int. J. Cosmet. Sci. 34:2–11 [DOI] [PubMed] [Google Scholar]
- 48. Stuible HP, Wagner C, Andreou I, Huter G, Haselmann J, Schweizer E. 1996. Identification and functional differentiation of two type I fatty acid synthases in Brevibacterium ammoniagenes. J. Bacteriol. 178:4787–4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Magnuson K, Jackowski S, Rock CO, Cronan JE., Jr 1993. Regulation of fatty acid biosynthesis in Escherichia coli. Microbiol. Rev. 57:522–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Val D, Banu G, Seshadri K, Lindqvist Y, Dehesh K. 2000. Re-engineering ketoacyl synthase specificity. Structure 8:565–566 [DOI] [PubMed] [Google Scholar]
- 51. Heath RJ, Rock CO. 1996. Inhibition of β-ketoacyl-acyl carrier protein synthase III (FabH) by acyl-acyl carrier protein in Escherichia coli. J. Biol. Chem. 271:10996–11000 [DOI] [PubMed] [Google Scholar]
- 52. Erfle JD. 1973. Acetyl-CoA and propionyl-CoA carboxylation by Mycobacterium phlei: partial purification and some properties of the enzyme. Biochim. Biophys. Acta 316:143–155 [DOI] [PubMed] [Google Scholar]
- 53. Morishima N, Ikai A. 1987. Active site organization of bacterial type I fatty acid synthetase. J. Biochem. 102:1451–1457 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.