Abstract
The maintenance of energetically costly flagella by bacteria in non-water-saturated media, such as soil, still presents an evolutionary conundrum. Potential explanations have focused on rare flooding events allowing dispersal. Such scenarios, however, overlook bacterial dispersal along mycelia as a possible transport mechanism in soils. The hypothesis tested in this study is that dispersal along fungal hyphae may lead to an increase in the fitness of flagellated bacteria and thus offer an alternative explanation for the maintenance of flagella even in unsaturated soils. Dispersal along fungal hyphae was shown for a diverse array of motile bacteria. To measure the fitness effect of dispersal, additional experiments were conducted in a model system mimicking limited dispersal, using Pseudomonas putida KT2440 and its nonflagellated (ΔfliM) isogenic mutant in the absence or presence of Morchella crassipes mycelia. In the absence of the fungus, flagellar motility was beneficial solely under conditions of water saturation allowing dispersal, while under conditions limiting dispersal, the nonflagellated mutant exhibited a higher level of fitness than the wild-type strain. In contrast, in the presence of a mycelial network under conditions limiting dispersal, the flagellated strain was able to disperse using the mycelial network and had a higher level of fitness than the mutant. On the basis of these results, we propose that the benefit of mycelium-associated dispersal helps explain the persistence of flagellar motility in non-water-saturated environments.
INTRODUCTION
Many soil bacteria have flagella (1). However, the role of these flagella for bacteria living in non-water-saturated media, such as soils, is not yet well understood (2–5). In the majority of soils, the thinness and patchiness of the liquid films restricts the dispersal of individual cells or populations on a microscopic scale (4). Indeed, flagellum-driven swimming requires bacterial cells to be fully immersed in liquid in order to avoid capillary pinning forces acting at the liquid-air interface (3, 5). Swarming, another type of bacterial motility for which flagella are required (6), is also restricted to a narrow range of wet conditions (2). Thus, flagellated bacteria would be expected to swim or to swarm only during saturation events, which are often short and sporadic (3), raising doubts as to the evolutionary advantage of maintaining the energetically costly flagella (7).
A recent study proposes that rare events of water saturation in soil yield a fitness advantage to flagellated strains through improved dispersal and hence access to novel resources (3). This advantage could theoretically suffice to explain the maintenance of flagella. However, to test this argument further, it would be necessary to investigate how often different soils are water saturated under natural conditions. Here we tested an alternative explanation for the persistence of flagella in soil bacteria that is based on the recent discovery that bacteria can disperse along fungal hyphae, which are thus used as “fungal highways” (8–12). Since hyphae from most filamentous organisms can grow through air (13, 14), the liquid film that can be present around hyphae allows bacteria to disperse over air-filled soil pores (9, 12), which are typically found in unsaturated soils.
The characterization of this dispersal mechanism carried out so far has indicated that hyphae with hydrophilic surface areas act as effective bacterial dispersal networks, i.e., allow for a liquid film for effective flagellar motility (9). In addition to flagellar motility, other features of migrating bacteria have been suggested, such as the ability to metabolize fungal exudates or the presence of molecular markers for a type III secretion system (8, 9, 12, 15–18).
An important consideration for the theory of mycelium-promoted dispersal to explain the maintenance of flagella is the ecological relevance of this mechanism. Mycelia are highly abundant in most soils, with estimates of a mycelial network that represents as much as 100 to 700 m per g of soil (9, 19). Thus, the naturally available fungal network could offer flagellated bacteria a permanent option for active migration. If so, the advantage of fungus-associated dispersal would help to explain the maintenance of flagella by soil bacteria.
Soils are physically challenging due to their opaque nature and their physical and chemical heterogeneity (20). Therefore, alternative models, such as the porous-surface model, have been used to simulate water conditions in soils (3, 21). Alternatively, culture plates with different agar concentrations simulate different hydration levels affecting bacterial dispersal and are used routinely to measure bacterial motility (15, 22) or even the evolution of cooperation in bacteria (23). Using petri dishes with variable agar concentrations, we tested whether bacterial dispersal associated with mycelial networks makes flagellar motility beneficial under conditions where swimming is otherwise impossible. We first showed flagellum-driven dispersal along mycelial networks for a wide range of motile bacteria, demonstrating the relevance of this dispersal mechanism. Then we measured the growth of Pseudomonas putida KT2440, a typical soil bacterium, and its nonflagellated mutant either alone or in direct competition under different saturation conditions. We repeated the experiments in agar plates previously inoculated with the saprophytic and mycorrhizal fungus Morchella crassipes. Because fungi can actively interact with bacteria (for example, through the secretion of exudates), in a final experiment we used glass fibers as an abiotic dispersal network instead of the fungus.
MATERIALS AND METHODS
Strains.
A suite of bacteria tagged constitutively with fluorescent proteins was selected for the migration experiments. Tagging with fluorescent proteins allows a rapid, nondestructive assessment of mycelium colonization and bacterial migration by epifluorescent binocular observation. The tagged bacteria included Pseudomonas azelaica HBP1, Pseudomonas knackmussii B13, Sphingomonas wittichii RW1, and Cupriavidus necator JMP289, kindly provided by Jan van der Meer (University of Lausanne). Finally, the untagged bacteria Pseudomonas aeruginosa NEU1023, Cupriavidus oxalaticus NEU1047, Escherichia coli K-12, and Bacillus subtilis NEU1, from the culture collection of the Laboratory of Microbiology of the University of Neuchâtel, were also included. In addition, Pseudomonas putida KT2440, a flagellated bacterium isolated from the rhizosphere (24), and its ΔfliM nonflagellated mutant were used for specific experiments to assess the effect of flagellar motility on fitness. The ΔfliM nonflagellated mutant was obtained by allelic exchange with a truncated version of fliM and is an antibiotic resistance-free mutant. Both strains were tagged by inserting a constitutively expressed fluorescent-protein-encoding gene at a neutral position of the genome; the gfp gene was inserted into the wild-type strain, and the DsRed gene was inserted into the nonflagellated mutant (3). These strains were kindly provided by Arnaud Dechesne (Technical University of Denmark). The growth parameters of the two strains under different conditions are given in Table S1 in the supplemental material.
The saprophytic and mycorrhizal fungus Morchella crassipes was isolated from soil. This strain was affiliated with the species Morchella crassipes based on the internal transcribed spacer 1 (ITS 1) and ITS 2 sequences and on phenotypic observations (25). M. crassipes hyphae have a diameter of 4.5 to 7.5 μm.
Media and culture conditions.
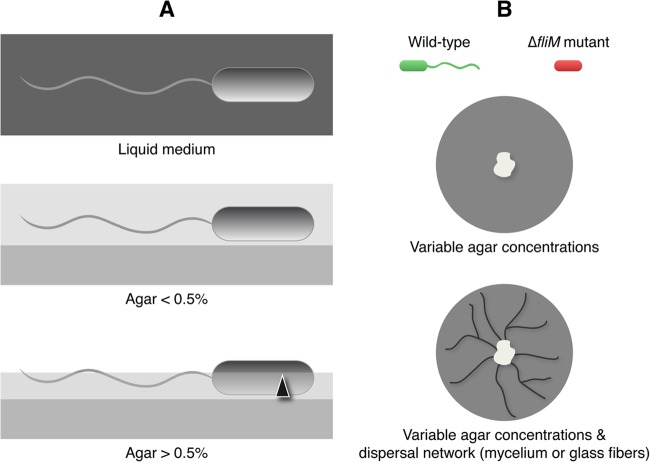
Nutrient agar (NA) medium (Biolife, Italy) was used for the regular maintenance of the bacterial cultures. Malt extract (Mycotec, Switzerland) was used for the culture of M. crassipes. The latter medium was also used as the substrate for all the experiments, because it allowed the rapid growth of M. crassipes, as well as the growth and dispersal of P. putida. The agar used was agar-agar (Merck, Germany). The medium was poured at 60°C into small petri dishes (diameter, 50 mm). The dishes contained 8 ml of the medium. Because the drying period of the agar has a strong influence on bacterial dispersal (22), after pouring, the plates were dried for 15 min under a constant laminar flow (470 m3 h−1) and were inoculated immediately thereafter. Plates were then wrapped with Parafilm and Saran Wrap to prevent dehydration (26). The experiments were conducted at 21°C. Four agar concentrations, ranging from 0.3 to 1.5%, were used. In agar concentrations up to 0.3%, swimming is possible (27). At 0.5% agar, Pseudomonas putida KT2440 exhibits a swarming-like motility (28). Above 0.5% agar, the liquid film over the agar medium is too thin for swimming or swarming (2, 3, 29, 30), so these concentrations were considered in this study to constitute a condition mimicking limited dispersal in soil (Fig. 1).
Fig 1.
Description of the experimental design. (A) Changes in the agar concentration added to the culture medium modify the liquid film formed over the solid agar, allowing (<0.5%) or restricting (>0.5%) bacterial dispersal. (B) Dispersal was estimated by using a flagellated wild-type strain labeled with GFP versus a nonflagellated mutant (ΔfliM mutant) labeled with the fluorescent protein DsRed. The strains were inoculated in media with variable concentrations of agar and in the absence or presence of a fungal dispersal network.
Agar plates containing a dispersal network (1% and 1.5% agar) were prepared as described above with few modifications. To grow the fungus, we inoculated a plug obtained from the edge of a 5-day-old culture by using the wider end of a glass Pasteur pipette as a device with which to inoculate a regular amount of mycelium. The plug (a cylinder with a diameter of 5 mm and a height of 5 mm) was placed in the center of the plate and was removed after the fungus had colonized the entire plate (2 days). Bacteria were inoculated after the removal of the inoculation plug (Fig. 1B). In addition to the fungal mycelial network, glass fibers (Cole-Parmer, USA) with a diameter of approximately 8 μm were used as an abiotic network mimicking the fungal mycelia, as has been done in previous studies (15). For the abiotic network, ca. 40 fibers (approximately 2 cm long) were placed randomly in the plate. To maintain conditions as similar as possible, bacteria placed on an abiotic network were also inoculated 2 days after the medium was poured.
Motility assays.
One bacterial colony from an overnight culture (in NA) was inoculated into the center of a petri dish. The medium was either the swimming or the swarming medium as described previously (26). The mean diameter reached after 40 h was then measured in four plates. For direct observation of flagella, the bacterial strains were stained as described previously (31) and were observed under a light microscope.
Analysis of bacterial dispersal.
Bacterial dispersal was measured as the diameter reached by bacterial colonies after incubation in malt agar medium. To determine the diameter, bacterial colonies were observed directly after 40 h of incubation by using an epifluorescence stereomicroscope (Nikon SMZ1000) in four replicates for each agar concentration. Samples of agar from inside and outside of the observed colonized area were repicked to confirm the limits of the growing colony. Individual measurements were taken for the wild-type and ΔfliM mutant strains inoculated separately into media with agar concentrations from 0.3 to 1.5%. The same experiment was conducted with a preexisting dispersal network of either M. crassipes or glass fibers on 1 or 1.5% agar. The bacterial inoculum was prepared from an overnight culture grown on nutrient agar and suspended in a saline solution. Prior to inoculation, the solution was adjusted to obtain a density close to 7 × 104 cells μl−1. Three microliters of this suspension was used as the sole inoculum in the center of the plate. The small volume of inoculum prevented the passive spreading of bacteria with the liquid droplet.
Dispersal to the single-cell level was assessed by observing samples by confocal laser scanning microscopy (CLSM). CLSM observations were performed with a TCS SP5X system (Leica) attached to an upright microscope equipped with a 63× (numerical aperture, 1.2) water immersion objective. Excitation was performed with a white laser line at 488 nm for green fluorescent protein (GFP) (80% light intensity) and 557 nm for DsRed (50% light intensity), and emission signals were detected at 500 to 550 nm (GFP) and 575 to 625 nm (DsRed). The transmission channel was used to visualize fungal hyphae in a nonconfocal mode.
Determination of relative fitness.
For analysis of relative fitness, the two bacterial strains were coinoculated at a ratio of ≈1:1 (3 μl of a suspension with 3.5 × 104 cells of each strain μl−1). The diameter of the area colonized by each bacterial strain was measured before the faster-dispersing strain reached the edge of the petri dish (40 h in the biotic network and 20 h in the abiotic network). The diameter was measured as described above using the different fluorescence labels for each strain (GFP for the wild type and DsRed for the nonflagellated mutant). The confirmation of the colonized area for each strain was obtained after repicking, plating, and observation of the respective fluorescence of individual colonies under the epifluorescence stereomicroscope.
To measure the fitness of the wild-type strain relative to that of the nonflagellated mutant, specific plates prepared as described above were sacrificed entirely by intense washing with a saline solution (0.9% [wt/vol] NaCl). Total-cell numbers for each type of strain in the suspension obtained were determined by counting green and red CFU, with a detection threshold of ≈10 cells/sample. Counting was carried out in five independent replicates. The numbers of colonies corresponding to the wild-type and mutant strains were established by observation of the respective fluorescence (GFP for the wild type and DsRed for the nonflagellated mutant) under the epifluorescence stereomicroscope. In solid medium with or without mycelia, CFU counting was carried out after 40 h. For experiments with fibers, CFU counting was carried out after 24 h. Since the results of different treatments were not compared, the difference in incubation time was not taken into account for the analysis of the results.
The fitness of the wild-type strain (x) relative to that of the mutant strain (y) was calculated according to reference 32, as ln(xF/x0)/ln(yF/y0), where x0 and y0 are the initial densities and xF and yF are the final densities at the end of the experiment.
Statistical analysis.
One-sample t tests were used to determine whether relative fitness (W) was significantly different from 1. W data sets met the normality criterion. Two-sample t tests were used to confirm the one-sample t tests using red and green CFU data (after log transformation for compliance with the normality assumption) to perform paired t tests on CFU counting. The results (t statistics and P values) from the two tests led to the same degree of significance. Likewise, we compared the t tests to a test of proportion (i.e., testing for a proportion different from 1 green CFU/1 red CFU), which has better statistical power for the same number of samples. The results (in terms of significance) were similar. Two-sample t tests were used to compare the diameters reached by the two strains in the competition experiment.
RESULTS
Dispersal of flagellated bacteria on fungal hyphae.
On the basis of previous findings by Kohlmeier et al. (9), a broad selection of environmental bacteria were tested for their abilities to disperse along the hyphae of the model organism (M. crassipes) used in this study. The tests were carried out under limited-dispersal conditions (1.5% agar). The bacterial species selected corresponded mainly to soil or rhizospheric bacteria but also included a bacterium isolated from a wastewater treatment plant (Pseudomonas azelaica HBP1) and a human commensal (Escherichia coli K-12). Two of the 11 strains did not produce flagella and accordingly did not spread in swimming or swarming media after 40 h (Table 1). Except for Cupriavidus necator JMP289 and Cupriavidus oxalaticus NEU1047, all the flagellated bacteria dispersed in the swimming medium. Pseudomonas aeruginosa NEU1023, C. necator JMP289, and Bacillus subtilis NEU1 were able to disperse in the swarming medium. The majority of strains that had flagella and that could disperse in swimming and swarming media were able to disperse on the mycelium of M. crassipes or on glass fibers. The sole exception was E. coli K-12, which could not disperse on fungal hyphae but could disperse on glass fibers.
Table 1.
Bacterial dispersal along fungal hyphae
| Strain | Tagginga | Environmental habitat | Motilityb |
Dispersalc on: |
|||
|---|---|---|---|---|---|---|---|
| SI (mm) | SA (mm) | Fl | M. crassipes mycelium | Glass fibers | |||
| Pseudomonas putida KT2440 | GFP | Rhizosphere | 85 | 7.0 ± 0.1 | + | + | + |
| Pseudomonas putida KT2440 Δfli | DsRed | Rhizosphere | 19.3 ± 5.4 | 6.3 ± 0.6 | − | − | − |
| Pseudomonas putida UWC1 | GFP | Rhizosphere | 85 | 6.7 ± 0.6 | + | + | + |
| Pseudomonas aeruginosa NEU1023 | Soil | 54.3 ± 9.8 | 66.7 ± 4.2 | + | + | + | |
| Pseudomonas azelaica HBP1 | GFP | Wastewater | 5.8 ± 0.5 | 4.8 ± 1.3 | − | − | − |
| Pseudomonas knackmussii B13 | GFP | River | 27.5 ± 3.9 | 7.0 ± 0.1 | + | + | + |
| Sphingomonas wittichii RW1 | GFP | River | 85 | ND | + | + | + |
| Cupriavidus necator JMP289 | GFP | Soil | 8.0 ± 1.7 | 34.0 ± 3.5 | + | + | + |
| Cupriavidus oxalaticus NEU1047 | mCherry | Soil | 6.7 ± 1.2 | 8.7 ± 2.3 | + | − | − |
| Escherichia coli K-12 | Intestinal lumen | 85 | 9.0 ± 1.7 | + | − | + | |
| Bacillus subtilis NEU1 | Soil | 85 | 85 | + | +* | + | |
GFP, green fluorescent protein. DsRed and mCherry are red fluorescent proteins.
SI, swimming in NA; SA, swarming in NA; Fl, presence (+) or absence (−) of flagella. ND, not determined.
+, positive; −, negative; *, antagonism observed.
Cost of flagellar motility under limited-dispersal conditions.
In order to test the hypothesis that dispersal along fungal hyphae may lead to an increase in the fitness of flagellated bacteria, two strains of P. putida, a wild-type flagellated strain and an isogenic nonflagellated (ΔfliM) mutant, were used. Flagellar motility allowed the wild-type strain to disperse in swimming (0.3%) and swarming (0.5%) malt agar, while for the nonflagellated mutant, dispersal was reduced (Fig. 1A). Motility had an effect on the area colonized by the bacteria growing on a petri dish (Fig. 2B). When the concentration of agar was increased to restrict dispersal (1% and 1.5%), however, the area colonized by the nonflagellated mutant was larger than the area colonized by the wild-type strain (Fig. 2C). Comparison of the colonized areas and the relative abundances of the strains showed that flagellar motility was beneficial in terms of dispersal (by two-sample t tests for 0.3% and 0.5% agar; n [replicates], 8 for each test; P, <10−3) and fitness (by one-sample t tests for 0 to 0.5% agar; n, 10 for each test; W, >1; P, <0.02) only for agar concentrations lower than 0.5% (Table 2). In contrast, for agar concentrations higher than 0.5%, the absence of flagella allowed the ΔfliM mutant to spread faster than the wild type and to colonize a larger area (by two-sample t tests for 1.0% and 1.5% agar; n, 8 for each test; P, <10−3) (Table 2). Under these conditions, the nonflagellated mutant outgrew the wild type and had greater fitness (by one-sample t tests for 1.0% and 1.5% agar; n, 10 for each test; W, <1; P, <0.05).
Fig 2.
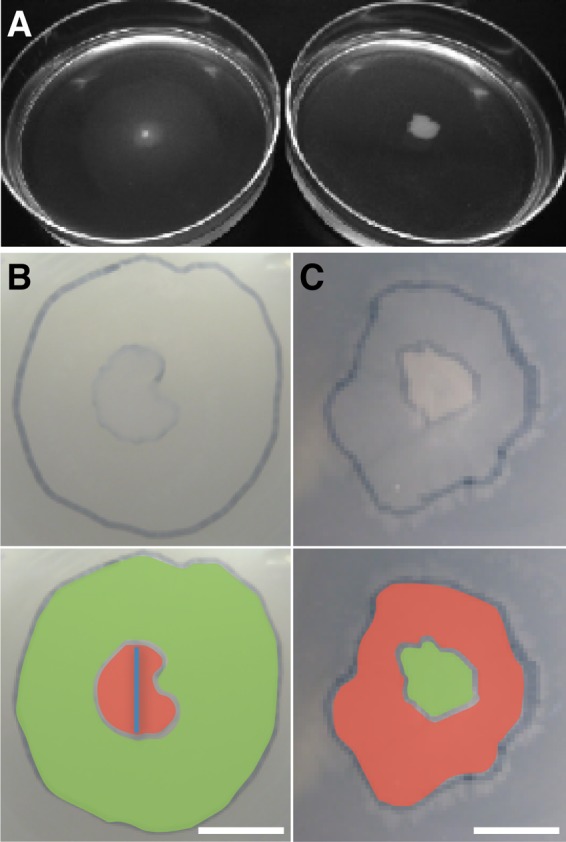
Dispersal of Pseudomonas putida on agar medium. (A) Image of the swimming test in malt agar medium. (Left) Wild-type strain; (right) ΔfliM mutant. (B) Colony diameters (image overlap) of the wild-type and mutant strains growing with 0.5% agar. The image in the lower panel is colored according to the fluorescent labels. Bar, 5 mm. (C) Colony diameters (image overlap) of the wild-type and mutant strains growing with 1.5% agar. Bar, 5 mm.
Table 2.
Dispersal and relative fitness of wild-type (flagellated) Pseudomonas putida KT2440 and its isogenic ΔfliM mutant (nonflagellated) under different agar concentrations and in the presence and absence of mycelium from Morchella crassipes (fungus) or an abiotic dispersal network (glass fibers)a
| Network | Agar concn (%) | Colony diam (mm) |
Relative abundance (CFU) (flagellated/nonflagellated strains) | Fitness | |
|---|---|---|---|---|---|
| Flagellated | Nonflagellated | ||||
| None | 0.3 | 28.7 ± 4.7 | 6.4 ± 1.3 | 6.8 ± 4.1 | 1.41 ± 0.30 |
| 0.5 | 13.1 ± 2.3 | 6.7 ± 1.0 | 6.5 ± 3.9 | 1.26 ± 0.28 | |
| 1.0 | 5.6 ± 0.4 | 9.0 ± 0.4 | 0.6 ± 0.2 | 0.96 ± 0.06 | |
| 1.5 | 5.7 ± 0.6 | 8.9 ± 0.9 | 0.4 ± 0.3 | 0.93 ± 0.15 | |
| Fungus | 1.0 | 27.3 ± 2.4 | 8.0 ± 2.7 | 2.4 ± 1.3 | 1.18 ± 0.14 |
| 1.5 | 23.9 ± 5.5 | 6.6 ± 2.1 | 2.0 ± 1.0 | 1.15 ± 0.08 | |
| Glass fibers | 1.0 | 36.1 ± 5.3 | 15.5 ± 2.7 | 1.6 ± 0.1 | 1.16 ± 0.02 |
| 1.5 | 36.7 ± 1.1 | 15.0 ± 4.0 | 1.5 ± 0.1 | 1.13 ± 0.02 | |
Dispersal was assessed using the colony diameter. Fitness was calculated based on the relative abundances, as measured by CFU counting, of the flagellated and nonflagellated strains, according to Kerr et al. (14). For the fitness calculation, the initial abundances were 30,250 and 45,150 CFU for the flagellated and nonflagellated strains, respectively.
Benefit of flagellar motility in the presence of fungal hyphae.
The effects of a mycelial network on dispersal and fitness were measured for the agar concentrations that limit dispersal (i.e., 1 and 1.5% agar). Under such conditions, the flagellated strain was able to migrate rapidly as single cells along fungal hyphae, while the nonflagellated mutant could not (Fig. 3; see also videos in the supplemental material). Consequently, for the nonflagellated mutant strain, there was no significant difference between the colony diameters in the absence and presence of fungal hyphae (by two-sample t tests for 1.0% and 1.5% agar; n, 8 for each test; P, >0.05) (Table 2). In contrast, the spread of the flagellated strain was significantly higher in the presence than in the absence of fungal hyphae (by two-sample t tests for 1.0% and 1.5% agar; n, 8 for each test; P, <0.001) (Table 2). Moreover, if the two strains were coinoculated, the wild type outgrew the nonflagellated mutant in the presence of fungal hyphae, causing a reversal in relative fitness: flagellated bacteria became more abundant and fit than the mutant (by one-sample t tests for 1.0% and 1.5% agar; n, 10 for each test; W, >1; P, <0.01) (Table 2).
Fig 3.
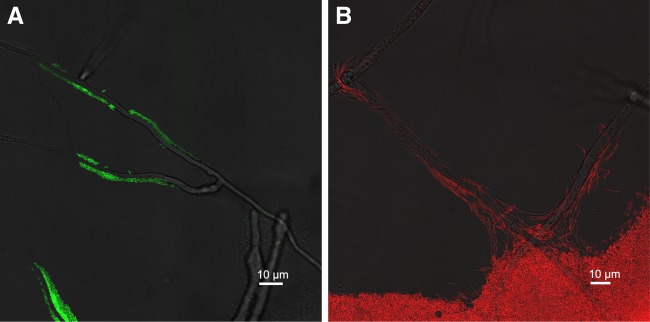
Dispersal of Pseudomonas putida on Morchella crassipes hyphae. (A) Colonization by wild type P. putida of M. crassipes hyphae. Bacteria are shown in a confocal fluorescent green channel, and hyphae are shown in a nonconfocal transmission channel. (B) Colonization by P. putida ΔfliM of M. crassipens hyphae. Bacteria are shown in a confocal fluorescent red channel, and hyphae are shown in a nonconfocal transmission channel.
Flagellated bacteria also migrated rapidly along an abiotic network composed of glass fibers, and colonies reached diameters close to 4 cm (Table 2). Like fungi, the abiotic network provided an important dispersal gain (by two-sample t tests for 1.0% and 1.5% agar; n, 8 for each test; P, <10−3) (Table 2) and made flagellar motility beneficial in terms of relative fitness (by one-sample t tests for 1.0% and 1.5% agar; n, 10 for each test; W, >1; P, <0.01) (Table 2).
DISCUSSION
Our experiments demonstrate that the presence of a dispersal network, such as fungal mycelia or glass fibers, renders flagellar motility beneficial under conditions that limit the dispersal of microbes, such as environments of low water potential or discontinuous water films. The results also show that swimming along mycelia is common to a diverse set of flagellated soil bacteria. We thereby confirmed earlier studies showing that the formation of flagella by P. putida may constitute a burden that affects its fitness relative to that of an isogenic mutant lacking flagellum formation under conditions of limited dispersal (3). Indeed, it has been observed that genes involved in the synthesis of flagella in P. putida KT2440 are upregulated under water conditions in which flagellar dispersal is impossible, probably raising the energetic cost of flagella under conditions where they are of no apparent use (33). Therefore, losing the flagellum would be beneficial for this typical soil bacterium. However, the increased dispersal and concomitant fitness of flagellated bacteria in the presence of dispersal networks may compensate for this energetic disadvantage. In the presence of fungal hyphae, the fitness (W) and dispersal (D) gains of the flagellated wild-type strain make flagellar motility beneficial under environmental conditions that impair free swimming. We also show that glass fibers mimicking the mycelial network provide the same benefits, suggesting that purely physicochemical properties can explain the migration and fitness gain of flagellated bacteria.
Much as our results offer an evolutionary explanation for the maintenance of flagellar motility in soils, they also affect the current model of bacterial dispersal in non-water-saturated media. These models predict that bacteria can disperse only under a narrow range of water-saturated conditions (2–5); however, our results suggest that bacteria can disperse under a broader range of water conditions when they can migrate on dispersal networks such as fungal mycelia. Consequently, these results indicate that bacterial dispersion in soil is probably less static than was supposed previously. Also, a recent study has shown the importance of conditional dispersal based on resource availability in bacterial dispersal models (34). Thus, a combination of conditional dispersal, chemotaxis, and the fitness advantage of flagellar motility in networks should be taken into account in order to obtain more-realistic models of bacterial dispersal in soils.
Interestingly, our results showed the dispersal of bacteria from the point of inoculation, along the preformed networks (fungal hyphae or glass fibers). Structured dispersal is known to affect local relatedness coefficients (35), which, in turn, affect the behavior of communities, but this had not yet been addressed in studies on fungus-driven bacterial dispersal. The consequences of the resulting population structure for cooperative versus competitive interactions (23, 36) need to be studied in more detail in the future. Likewise, the role of flagella in other cellular processes that might affect fitness in soils, such as biofilm formation and cell-surface (37) or cell-cell interactions, cannot be ignored for a comprehensive evaluation of the evolutionary role of flagellar persistence in soil bacteria.
Although our experimental model is a mimic of real dispersal, considering the extent of the already existing fungal network potentially available for bacterial migration (as much as 100 to 700 m · g of dry soil−1 (9, 19), it is unlikely that this dispersal mechanism is irrelevant. As expected from previous reports in the literature (9–12), fungus-driven bacterial dispersal was confirmed for a set of bacterial species (Table 1). Thus, the facilitated dispersal of flagellated bacteria in media that limit dispersal is clearly not restricted to M. crassipes and P. putida. The importance of fungi on the evolution and selection of soil bacteria has been suggested in the past (38, 39). Here we propose a novel role for fungi as an important evolutionary force affecting bacterial fitness and dispersal that can have a strong influence on processes determining bacterial colonization, activity, and biodiversity.
Supplementary Material
ACKNOWLEDGMENTS
We thank Arnaud Dechesne and Jan van der Meer for providing bacterial strains, as well as T. R. Neu and U. Kuhlicke for assistance in using laser microscopy. We also thank Thomas Junier and three anonymous reviewers for comments on earlier versions of this article.
Footnotes
Published ahead of print 30 August 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01393-13.
REFERENCES
- 1.Czaban J, Gajda A, Wroblewska B. 2007. The motility of bacteria from rhizosphere and different zones of winter wheat roots. Pol. J. Environ. Stud. 16:301–308 [Google Scholar]
- 2.Dechesne A, Smets BF. 2012. Pseudomonad swarming motility is restricted to a narrow range of high matric water potentials. Appl. Environ. Microbiol. 78:2936–2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dechesne A, Wang G, Gulez G, Or D, Smets BF. 2010. Hydration-controlled bacterial motility and dispersal on surfaces. Proc. Natl. Acad. Sci. U. S. A. 107:14369–14372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Or D, Smets BF, Wraith JM, Dechesne A, Friedman SP. 2007. Physical constraints affecting bacterial habitats and activity in unsaturated porous media—a review. Adv. Water Resour. 30:1505–1527 [Google Scholar]
- 5.Wang G, Or D. 2010. Aqueous films limit bacterial cell motility and colony expansion on partially saturated rough surfaces. Environ. Microbiol. 12:1363–1373 [DOI] [PubMed] [Google Scholar]
- 6.Kirov SM, Tassell BC, Semmler AB, O'Donovan LA, Rabaan AA, Shaw JG. 2002. Lateral flagella and swarming motility in Aeromonas species. J. Bacteriol. 184:547–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaiser D. 2007. Bacterial swarming: a re-examination of cell-movement patterns. Curr. Biol. 17:R561–R570 [DOI] [PubMed] [Google Scholar]
- 8.Furuno S, Pazolt K, Rabe C, Neu TR, Harms H, Wick LY. 2010. Fungal mycelia allow chemotactic dispersal of polycyclic aromatic hydrocarbon-degrading bacteria in water-unsaturated systems. Environ. Microbiol. 12:1391–1398 [DOI] [PubMed] [Google Scholar]
- 9.Kohlmeier S, Smits THM, Ford RM, Keel C, Harms H, Wick LY. 2005. Taking the fungal highway: mobilization of pollutant-degrading bacteria by fungi. Environ. Sci. Technol. 39:4640–4646 [DOI] [PubMed] [Google Scholar]
- 10.Warmink JA, Nazir R, Corten B, van Elsas JD. 2011. Hitchhikers on the fungal highway: the helper effect for bacterial migration via fungal hyphae. Soil Biol. Biochem. 43:760–765 [Google Scholar]
- 11.Warmink JA, van Elsas JD. 2009. Migratory response of soil bacteria to Lyophyllum sp. strain Karsten in soil microcosms. Appl. Environ. Microbiol. 75:2820–2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wick LY, Remer R, Wurz B, Reichenbach J, Braun S, Scharfer F, Harms H. 2007. Effect of fungal hyphae on the access of bacteria to phenanthrene in soil. Environ. Sci. Technol. 41:500–505 [DOI] [PubMed] [Google Scholar]
- 13.Wösten HAB. 2001. Hydrophobins: multipurpose proteins. Annu. Rev. Microbiol. 55:625–646 [DOI] [PubMed] [Google Scholar]
- 14.Wösten HAB, van Wetter MA, Lugones LG, van der Mei HC, Busscher HJ, Wessels JGH. 1999. How a fungus escapes the water to grow into the air. Curr. Biol. 9:85–88 [DOI] [PubMed] [Google Scholar]
- 15.Banitz T, Fetzer I, Johst K, Wick LY, Harms H, Frank K. 2011. Assessing biodegradation benefits from dispersal networks. Ecol. Model. 222:2552–2560 [Google Scholar]
- 16.Harms H, Schlosser D, Wick LY. 2011. Untapped potential: exploiting fungi in bioremediation of hazardous chemicals. Nat. Rev. Microbiol. 9:177–192 [DOI] [PubMed] [Google Scholar]
- 17.Warmink JA. 2010. Bacterial response to ecological opportunities offered by soil fungi. Ph.D. dissertation University of Groningen, Groningen, Germany [Google Scholar]
- 18.Warmink JA, Nazir R, van Elsas JD. 2009. Universal and species-specific bacterial ‘fungiphiles' in the mycospheres of different basidiomycetous fungi. Environ. Microbiol. 11:300–312 [DOI] [PubMed] [Google Scholar]
- 19.Jeffery S, Gardi C, Jones A, Montanarella L, Marmo L, Miko L, Ritz K, Peres J, Roembke J, van der Putten WH. 2010. European atlas of soil biodiversity. Publications Office of the European Union, Luxembourg [Google Scholar]
- 20.Young IM, Crawford JW. 2004. Interactions and self-organization in the soil-microbe complex. Science 304:1634–1637 [DOI] [PubMed] [Google Scholar]
- 21.Dechesne A, Or D, Gulez G, Smets BF. 2008. The porous surface model, a novel experimental system for online quantitative observation of microbial processes under unsaturated conditions. Appl. Environ. Microbiol. 74:5195–5200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tremblay J, Deziel E. 2008. Improving the reproducibility of Pseudomonas aeruginosa swarming motility assays. J. Basic Microbiol. 48:509–515 [DOI] [PubMed] [Google Scholar]
- 23.Kümmerli R, Gardner A, West SA, Griffin AS. 2009. Limited dispersal, budding dispersal, and cooperation: an experimental study. Evolution 63:939–949 [DOI] [PubMed] [Google Scholar]
- 24.Nakazawa T. 2002. Travels of a Pseudomonas, from Japan around the world. Environ. Microbiol. 4:782–786 [DOI] [PubMed] [Google Scholar]
- 25.Volk TJ, Leonard TJ. 1989. Physiological and environmental studies of sclerotium formation and maturation in isolates of Morchella crassipes. Appl. Environ. Microbiol. 55:3095–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rashid MH, Kornberg A. 2000. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 97:4885–4890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harshey RM. 2003. Bacterial motility on a surface: many ways to a common goal. Annu. Rev. Microbiol. 57:249–273 [DOI] [PubMed] [Google Scholar]
- 28.Matilla MA, Ramos JL, Duque E, de Dios Alche J, Espinosa-Urgel M, Ramos-Gonzalez MI. 2007. Temperature and pyoverdine-mediated iron acquisition control surface motility of Pseudomonas putida. Environ. Microbiol. 9:1842–1850 [DOI] [PubMed] [Google Scholar]
- 29.Wu Y, Berg HC. 2012. Water reservoir maintained by cell growth fuels the spreading of a bacterial swarm. Proc. Natl. Acad. Sci. U. S. A. 109:4128–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang R, Turner L, Berg HC. 2010. The upper surface of an Escherichia coli swarm is stationary. Proc. Natl. Acad. Sci. U. S. A. 107:288–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kodaka H, Armfield AY, Lombard GL, Dowell VR. 1982. practical procedure for demonstrating bacterial flagella. J. Clin. Microbiol. 16:948–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kerr B, Riley MA, Feldman MW, Bohannan BJM. 2002. Local dispersal promotes biodiversity in a real-life game of rock-paper-scissors. Nature 418:171–174 [DOI] [PubMed] [Google Scholar]
- 33.Gülez G, Dechesne A, Workman CT, Smets BF. 2012. Transcriptome dynamics of Pseudomonas putida KT2440 under water stress. Appl. Environ. Microbiol. 78:676–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Banitz T, Johst K, Wick LY, Fetzer I, Harms H, Frank K. 2012. The relevance of conditional dispersal for bacterial colony growth and biodegradation. Microb. Ecol. 63:339–347 [DOI] [PubMed] [Google Scholar]
- 35.Lehmann L, Rousset F. 2010. How life history and demography promote or inhibit the evolution of helping behaviours. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365:2599–2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Queller DC, Strassmann JE. 2009. Beyond society: the evolution of organismality. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364:3143–3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sauer K, Camper AK. 2001. Characterization of phenotypic changes in Pseudomonas putida in response to surface-associated growth. J. Bacteriol. 183:6579–6589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Boer W, Folman LB, Summerbell RC, Boddy L. 2005. Living in a fungal world: impact of fungi on soil bacterial niche development. FEMS Microbiol. Rev. 29:795–811 [DOI] [PubMed] [Google Scholar]
- 39.Nazir R, Warmink JA, Boersma H, van Elsas JD. 2010. Mechanisms that promote bacterial fitness in fungal-affected soil microhabitats. FEMS Microbiol. Ecol. 71:169–185 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.