Abstract
The ability of bifidobacteria to adhere to the intestine of the human host is considered to be important for efficient colonization and achieving probiotic effects. Bifidobacterium bifidum strains DSM20456 and MIMBb75 adhere well to the human intestinal cell lines Caco-2 and HT-29. The surface lipoprotein BopA was previously described to be involved in mediating adherence of B. bifidum to epithelial cells, but thioacylated, purified BopA inhibited the adhesion of B. bifidum to epithelial cells in competitive adhesion assays only at very high concentrations, indicating an unspecific effect. In this study, the role of BopA in the adhesion of B. bifidum was readdressed. The gene encoding BopA was cloned and expressed without its lipobox and hydrophobic signal peptide in Escherichia coli, and an antiserum against the recombinant BopA was produced. The antiserum was used to demonstrate the abundant localization of BopA on the cell surface of B. bifidum. However, blocking of B. bifidum BopA with specific antiserum did not reduce adhesion of bacteria to epithelial cell lines, arguing that BopA is not an adhesin. Also, adhesion of B. bifidum to human colonic mucin and fibronectin was found to be BopA independent. The recombinant BopA bound only moderately to human epithelial cells and colonic mucus, and it failed to bind to fibronectin. Thus, our results contrast the earlier findings on the major role of BopA in adhesion, indicating that the strong adhesion of B. bifidum to epithelial cell lines is BopA independent.
INTRODUCTION
The adhesion of pathogenic and commensal bacteria to host cells and tissues is considered an important step in the initiation of disease and in mediating beneficial effects for the host, respectively. In the gastrointestinal tract, adhesion is considered an essential colonization factor for both commensal and probiotic bacteria, and binding is known to be mediated by the surface proteins and structures of both bacterial and host cells. Human commensal and pathogenic bacteria may share some mechanisms of adhesion, and thus commensal bacteria may protect against pathogens by occupying adhesion sites (1, 2). The adhesion mechanisms of commensal and probiotic bacteria are currently under intensive investigation; for example, the adhesion of probiotic strain Lactobacillus rhamnosus GG was found to be mediated by a pilus, a structure known to mediate the adhesion of many pathogens (2, 3). Furthermore, adhering commensals and probiotics have close contact with the host epithelium and are proposed to function in immune stimulation and gut maturation and to enhance epithelial integrity (2, 4, 5). Bifidobacteria are commensal inhabitants of the human gastrointestinal tract, and they can constitute a considerable part of an individual's gut microbiota, especially in breast-fed infants (6, 7). Many Bifidobacterium species and strains adhere strongly to human intestinal epithelial cells in vitro (8, 9), but only a few adhesion molecules have been described thus far (9–11). The expression of moonlighting proteins on the cell surface of Bifidobacterium seems to be common, and under certain circumstances these proteins could facilitate colonization of the human gut (12–14). Recently, moonlighting transaldolase was reported to play a role in the autoaggregation and adhesion of Bifidobacterium bifidum to mucin (10).
The proteins of tight adherence (Tad) pili have been shown to be essential for the efficient gut colonization of Bifidobacterium breve in vivo in mice, and a role for these proteins favoring the adhesion of B. breve to the intestinal epithelium has been suggested (11). Gene clusters responsible for the biosynthesis of pili have been identified in the genomes of B. bifidum (15). Recently, it was demonstrated that the major subunit protein of the pil3 locus of B. bifidum PRL2010, coding for the sortase-dependent pili, is involved in the adhesion to Caco-2 cells and binding to extracellular matrix (ECM) proteins (16). Pil3 seems to be partially, but not solely, responsible for the adhesion of B. bifidum PRL2010 to Caco-2 cells via ECM proteins and autoaggregation (16). The same genetic locus has been found in all sequenced B. bifidum strain genomes and also in the genome of the probiotic strain B. bifidum MIMBb75 (S. Guglielmetti, unpublished data).
In B. bifidum, lipoprotein BopA (an outer surface lipoprotein) was the first surface protein described to be involved in the adhesion to intestinal epithelium. BopA purified from the cell wall of the probiotic strain B. bifidum MIMBb75 partly inhibited the adhesion of B. bifidum to Caco-2 cell line in a competitive adhesion assay (9). However, the inhibition was successful only when BopA was used at a remarkably high concentration, indicating that this was a result of unspecific inhibition rather than a specific competition for the adhesion sites (9). Recently, Gleinser and colleagues (17) expressed BopA in a poorly adherent strain, Bifidobacterium longum subsp. infantis E18, and detected increased adhesion of the strain (17). However, even with overexpression of BopA in the recipient strain, the adhesion was at a very low level compared with that of the parental B. bifidum strain (source of the bopA gene) (17), and therefore, the role of BopA as a true adhesin is still questionable.
In this study, we readdressed the role of BopA in the adhesion of B. bifidum. We cloned and expressed BopA without its hydrophobic signal sequence and lipobox (and therefore without the thioacyl group) in Escherichia coli and produced an antiserum against the recombinant BopA. The role of BopA in the adhesion of B. bifidum to intestinal epithelium was reevaluated by using a number of functional assays with the cell lines Caco-2 and HT-29, four ECM proteins, and human intestinal mucus.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Bifidobacterium bifidum type strain (DSM20456) and Bifidobacterium bifidum MIMBb75 (from the Industrial Microbiology Culture Collection DiSTAM, University of Milan, Milan, Italy) were cultivated in De Man, Rogosa, and Sharpe broth (MRS; Difco) supplemented with 500 μg ml−1 l-cysteine (Sigma-Aldrich) and were incubated at 37°C in an anaerobic chamber.
Cloning, overproduction, and purification of recombinant BopA.
Chromosomal DNA of B. bifidum DSM20456 was extracted by using a Wizard genomic DNA purification kit (Promega) according to the manufacturer′s instructions after mechanical lysis of bacterial cells by bead beating with zirconium beads (diameter, 0.1 mm) 3 times for 60 s each. The bopA gene, without the regions encoding the signal sequence and lipobox, was obtained by amplifying the chromosomal DNA of B. bifidum DSM20456 with a primer pair, one containing the NcoI site (5′AATTACCATGGGCAACAACGGCGC3′) and another containing the HindIII site (5′TAATTAAGCTTCTTCTCCCAGCCGAG3′) (designed on the basis of the available bopA sequence of B. bifidum MIMBb75). The gene encoding BopA was cloned into the vector pET28b+ (Novagen) for expression as a C-terminal His6 fusion protein in E. coli strain BL21(DE3)(pLysS). The BopA protein was purified from the cytoplasm of E. coli under native conditions using the Qiaexpress protein purification system (Qiagen) according to the manufacturer′s instructions.
Generation of the polyclonal anti-BopA antibodies.
The immunization protocol described by Johnston et al. (18) was used to raise polyclonal rabbit antibodies against His6-BopA of B. bifidum DSM20456 (Laboratory Animal Centre of University of Helsinki). Briefly, a 1:1 mixture of 400 ng of the purified recombinant His6-BopA and Freund′s complete adjuvant was administered by subcutaneous injection. Preimmunization serum was collected before the primary immunization. The initial injection was followed by 3 consecutive booster injections at 3-week intervals with 1:1 mixtures of 200 ng of the His6-BopA and Freund's incomplete adjuvant. Blood was collected 10 days after the last booster injection, and antiserum was prepared according to standard protocols (19).
Subcellular localization of BopA.
Immunogold-labeled thin sections of B. bifidum DSM20456 and MIMBb75 were prepared for studying the localization of BopA on the surface of B. bifidum strains by immunoelectron microscopy (IEM) using anti-BopA antiserum or preimmune serum as described previously (20). Briefly, bacteria were cultivated, washed, and fixed prior to being embedded in Lowicryl HM20 resin. After polymerization, ultrathin sections were cut and collected on carbon-coated nickel grids. Sections were blocked (1% bovine serum albumin [BSA; Sigma-Aldrich], 0.5% fish skin gelatin [FSG; Sigma-Aldrich], and 1% fetal calf serum [FCS; Integro BV] in sodium phosphate buffer) and incubated with anti-BopA antiserum or preimmune serum (both diluted 1:100 in sodium phosphate buffer containing 2% BSA, 0.1% Tween 20, and 0.1% FSG) for 2.5 h at room temperature. Grids were then washed five times (0.2% BSA, 0.01% Tween 20, and 0.01% FSG in sodium phosphate buffer) and incubated on drops of 1:80-diluted protein A conjugated to gold particles (10 nm) for 20 min. After extensive washing in phosphate buffer and distilled water, the sections were poststained in uranyl acetate and lead citrate before examination in a JEOL EXII transmission electron microscope.
The immunofluorescence staining was used to confirm the presence of BopA on the surface of B. bifidum as detailed previously (21). Briefly, B. bifidum cells were cultivated, washed with phosphate-buffered saline (PBS), and fixed with 3.5% (wt/vol) paraformaldehyde in PBS prior labeling with anti-BopA antiserum or preimmune serum as the primary antiserum and Alexa-488 (Invitrogen)-conjugated anti-rabbit IgG (1 μg ml−1) as the secondary antibody. Bacteria were then examined with an epifluorescence microscope (Leica DM 4000B) equipped with a filter for the Alexa-488 label (excitation, 450 to 490 nm; emission, 515 nm), and images were digitally recorded using CellP̂ imaging software for life sciences microscopy (Soft Imaging System GmbH).
Isolation of cell envelope-associated proteins and the detection of BopA in the cell envelope fraction.
Cell wall-associated proteins of B. bifidum strains DSM20456 and MIMBb75 were isolated as described by Kankainen et al. (3). When the presence of BopA in the cell wall was analyzed, the isolated proteins were separated in 12% (wt/vol) SDS-PAGE and transferred onto 0.2-μm Immobilon-P polyvinylidene difluoride (PVDF) membranes (Millipore) for Western blotting with anti-BopA antiserum and horseradish peroxidase-conjugated anti-rabbit IgG (Bio-Rad). The protein was then detected by using an enhanced chemiluminescence (ECL) Advance Western blotting detection kit (Amersham) according to the manufacturer′s instructions.
Caco-2 and HT-29 cell cultures.
The human intestinal cell lines Caco-2 and HT-29 were obtained from DSMZ and were grown at 37°C in a 95% air–5% CO2 atmosphere. Caco-2 cells were grown in RPMI 1640 medium (Sigma-Aldrich) supplemented with 2 mM l-glutamine (Lonza), 20% heat-inactivated (30 min at 56°C) fetal calf serum, 100 U ml−1 penicillin-streptomycin (Lonza), and 1% (vol/vol) nonessential amino acids (Lonza), and HT-29 cells were grown in McCoy 5A medium (Lonza) supplemented with 10% fetal calf serum and 100 U ml−1 penicillin-streptomycin.
Isolation of human intestinal mucus.
Human colonic mucus was isolated from a healthy piece of tissue obtained from patients with colorectal cancer. The mucus layer was collected as previously described (22). In short, resected material was collected on ice within 20 min and processed immediately by washing gently with PBS containing 0.01% gelatin. The mucus was collected into a small amount of HEPES-Hanks buffer by gently scraping with a rubber spatula, centrifuged (13,000 × g, 10 min) and stored at −20°C until further use. In the adhesion assays, mucus preparations from several individuals were pooled in equal ratios, the pooled mucus preparation was diluted to a protein concentration of 0.5 mg ml−1 with HEPES-Hanks, and 100 μl of this solution was immobilized passively on Maxisorp microtiter plate wells (Nunc) by overnight incubation at 4°C. The use of human intestinal mucus for the adhesion studies was approved by the ethical committee of the Hospital District of Southwest Finland, and written informed consent was obtained from all patients from whom mucus was collected.
Bacterial adhesion to Caco-2 and HT-29 cells, isolated human mucus, and ECM proteins.
For the adhesion tests Caco-2 and HT-29 cells were cultivated on 96-well tissue culture plate (10,000 cells well−1; Nunc) for 3, 8, and 21 days. The cells were washed twice with culture medium before the adhesion assay. Laminin (Sigma-Aldrich), collagen I (Sigma-Aldrich), collagen IV (Sigma-Aldrich), fibronectin (Calbiochem), fetuin (Sigma-Aldrich), and BSA (Sigma-Aldrich) were immobilized on Maxisorp microtiter wells at a concentration of 2.5 pmol well−1 (23). The wells containing immobilized mucus or ECM proteins were washed twice with PBS and incubated with blocking buffer (0.5% [wt/vol] BSA in PBS) for 1 h at room temperature. The wells were washed again three times with PBS prior to the addition of bacteria. B. bifidum DSM20456 and MIMBb75 were metabolically radiolabeled by cultivating bacteria with 10 μl ml−1 [5′-3H]thymidine (17.0 Ci/mmol; PerkinElmer). The adhesion assay was performed as described previously by Vesterlund et al. (22) with the following modifications. After cultivation, bacteria were collected by centrifugation and washed with RPMI 1640 without supplements (adhesion to Caco-2 cells), McCoy 5A without supplements (adhesion to HT-29 cells), or PBS (adhesion to mucus and ECM proteins). The optical density was adjusted (A600 = 0.25) to the same culture medium or buffer used for washing. Bacteria (100 μl) were incubated on mucus or ECM proteins at 37°C or on the epithelial cells in a CO2 incubator at 37°C for 1 h, and the nonadherent bacteria were removed by washing the wells three times with PBS. Bacteria bound to cells, mucus, or ECM proteins were lysed with 1% SDS–0.1 M NaOH by incubating at 60°C for 1 h. The radioactivity of the suspension was measured by liquid scintillation. Four to five parallel wells (i.e., technical replicates) were used in each experiment, and all experiments were repeated two to seven times. The percent bacterial adhesion was determined by calculating the ratio between the radioactivity of the adherent bacteria and that of the added bacteria. In the inhibition assays, bacteria were preincubated with anti-BopA antiserum or preimmune serum diluted 1:100 in culture medium or PBS prior to addition of bacteria to the cells, mucus, or ECM proteins. In competition assays, Caco-2 and HT-29 cells were incubated with His6-BopA (100 μg well−1) in a CO2 incubator at 37°C for 1 h prior to the adhesion assay with bacteria.
Binding of 125I-BopA to epithelial cells, mucus, and ECM proteins.
Recombinant BopA was radiolabeled (125I), and the binding assays to mucus and ECM proteins were performed essentially as detailed by Kankainen et al. (3). Mucus and ECM proteins were immobilized on Maxisorp microtiter wells and blocked with BSA as described above. Caco-2 and HT-29 cells were cultivated on 96-well microtiter plates for 3, 8, and 21 days and washed twice with culture medium without supplements. The 125I-labeled BopA (50 pmol) was added to the wells and incubated for 1 h at 37°C in a CO2 incubator. The wells were then washed three times with PBS to remove any loosely bound protein, treated with 1% SDS–0.1 M NaOH, and incubated at 60°C for 1 h. The radioactivity of bound 125I-BopA was measured in a Wallac 1480 Wizard-3 automatic gamma counter. Four to six parallel wells (i.e., technical replicates) were used in each protein binding experiment, and all experiments were repeated two to four times.
Statistical analysis.
A pairwise Student′s t test was used to determine the significant difference (P < 0.05) between the samples and controls. Results shown in Fig. 3, 4, and 5 are the means ± standard deviations for technical replicates (parallel wells) from representative experiments.
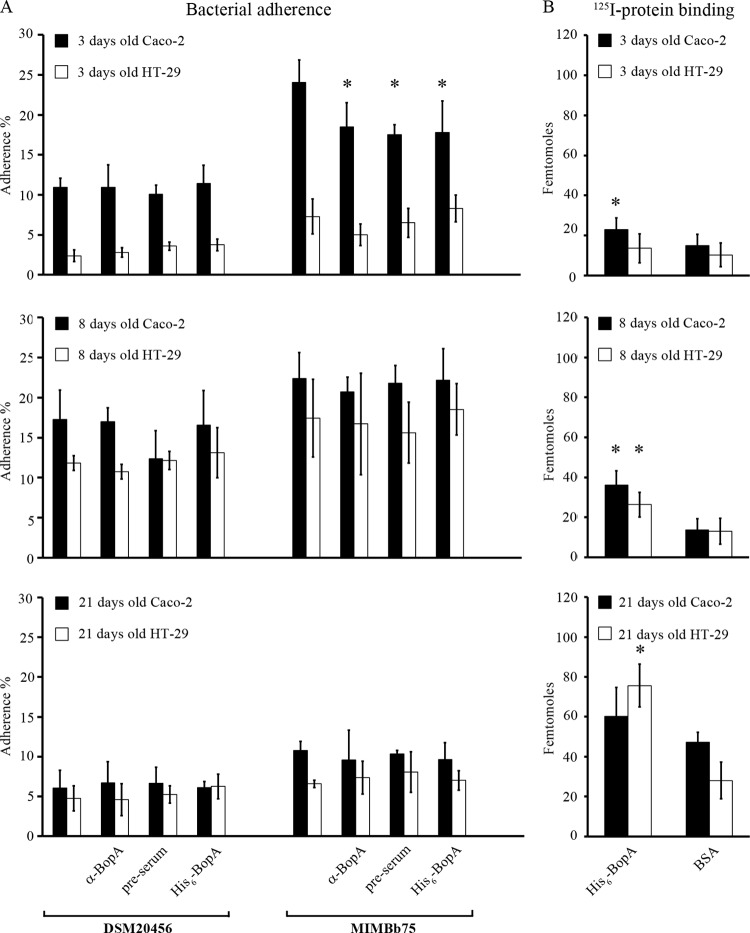
Fig 3.
Adhesion of B. bifidum (A) and binding of His6-BopA (B) to 3-, 8-, and 21-day-old epithelial cells. (A) Adhesion (%) of 3H-labeled B. bifidum DSM205456 and MIMBb75 to Caco-2 and HT-29. Anti-BopA and His6-BopA were used in the inhibition and competition assays, respectively, to assess the role of BopA in adhesion. Preimmune serum was used as a negative control. The results of 5 technical replicates (parallel wells) from the representative experiments are expressed as means ± standard deviations. Significant reductions (P < 0.05) in the adhesion in the inhibition/competition assays compared to the basic adhesion assay are indicated with asterisks. (B) Binding of 125I-labeled BopA and 125I-BSA (negative control) to Caco-2 and HT-29. Results are the means ± standard deviations for four to six technical replicates (parallel wells) of the representative experiments. Binding significantly above that of the negative control (125I-BSA) is indicated with an asterisk.
Fig 4.
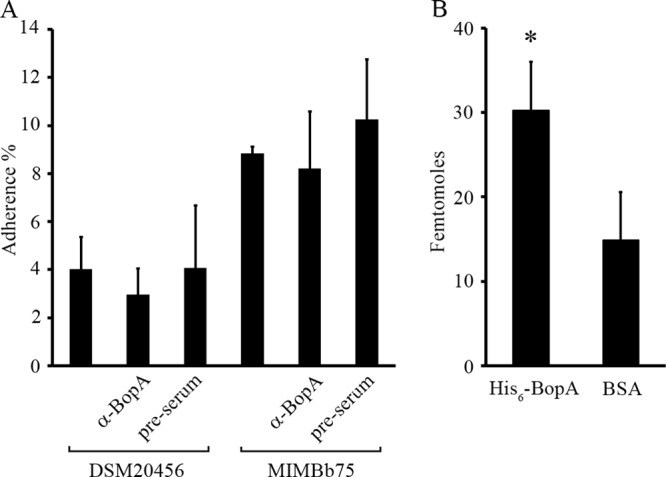
Adhesion of B. bifidum and binding of His6-BopA to colonic mucus. (A) Adhesion (%) of 3H-labeled B. bifidum DSM20456 and MIMBb75 to colonic mucus was measured. In the inhibition assays, bacteria were pretreated with anti-BopA antiserum or preimmune serum before adhesion. (B) Binding of 125I-labeled BopA and 125I-labeled BSA (negative control) to immobilized human colonic mucus. Results are the means ± standard deviations of five (A) or six (B) technical replicates (parallel wells) of the representative experiments. Binding significantly above that of the negative control (125I-BSA) is indicated with an asterisk (P < 0.05).
Fig 5.
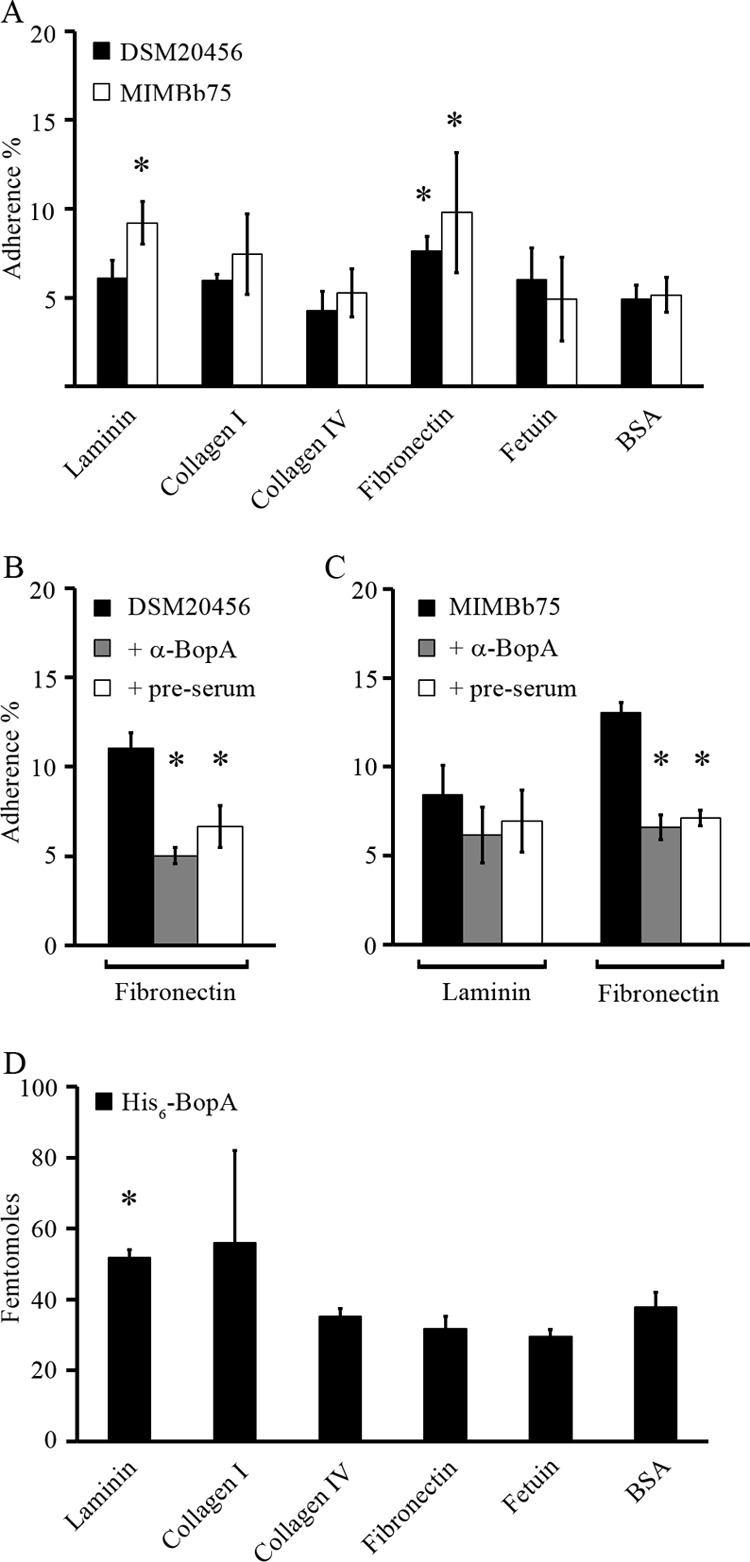
Adhesion of B. bifidum and binding of His6-BopA to extracellular matrix (ECM) proteins. (A) Adhesion (%) of radiolabeled (3H) B. bifidum DSM20456 and MIMBb75 to immobilized ECM proteins and BSA (background level of adherence). Adhesion significantly above the background level is indicated with an asterisk (P < 0.05). (B and C) Inhibition of adhesion with anti-BopA antiserum and preimmune serum treatment of ECM proteins. ECM proteins to which adhesion was above the background level were included in the inhibition assays. B. bifidum DSM20456 (B) and B. bifidum MIMBb75 (C) were pretreated with anti-BopA antiserum (gray columns) or preimmune serum (white columns) or left untreated (black columns) before adhesion. Significant reduction (P < 0.05) in the adhesion in the inhibition assays compared to the basic adhesion assay is indicated with an asterisk. (D) Binding of 125I-labeled BopA to ECM proteins and BSA (background level of binding). Binding significantly above the background level is indicated with an asterisk (P < 0.05). Results are the means ± standard deviations of four (A and D) or three (B and C) technical replicates (parallel wells) of the representative experiments.
RESULTS
Purification of BopA and production of specific antiserum.
The gene coding for B. bifidum DSM20456 lipoprotein BopA missing the signal sequence and lipobox (i.e., the recognition signal for lipid modification by thioacylation) was cloned into an expression vector. The bopA sequence of B. bifidum MIMBb75 was available from the previous studies (9), and it was utilized in designing the primers used for the amplification of bopA gene of B. bifidum DSM20456. The gene was then sequenced, and the obtained DNA sequence was identical to that of bopA of B. bifidum MIMBb75 (9). The BopA was expressed as a His6 fusion in E. coli under native conditions and purified by nickel-nitrilotriacetic acid (Ni-NTA) affinity chromatography. The purified protein was analyzed by SDS-PAGE and Western blotting (Fig. 1). The fusion protein showed an expected apparent molecular size of 61 kDa (Fig. 1A), and no other peptides were detected in the SDS-PAGE, indicating a high purity of the protein. Next, antiserum against His6-BopA was produced, and its reactivity was tested by Western blotting. The hyperimmune serum against BopA reacted well with the recombinant protein (Fig. 1B), whereas the preimmune serum failed to recognize it (Fig. 1B).
Fig 1.
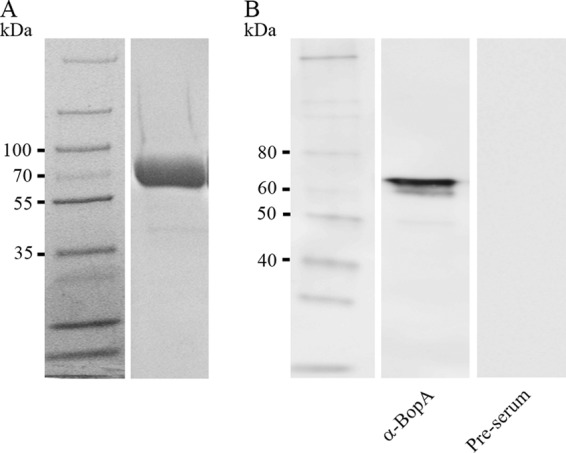
Purified recombinant BopA protein without the signal peptide and lipobox motif. SDS-PAGE (A) and Western blotting (B) analysis of purified His6-BopA were carried out. The Western blotting detection of 5 ng of purified His6-BopA was done by using anti-BopA antiserum. The reactivity of preimmune serum is shown as a control. The molecular masses (in kDa) of the standard proteins are indicated on the left.
Cellular location of BopA.
Three different methods were used to study the cellular localization of BopA in B. bifidum DSM20456 and MIMBb75. First, BopA was visualized in thin-sectioned B. bifidum cells by IEM using antiserum produced against His6-BopA in a combination with the protein A conjugated to 10-nm gold particles. IEM revealed that the majority of BopA is within or in close proximity to the cell wall, whereas preimmune serum failed to detect BopA in the thin sections (Fig. 2A). Second, the surface localization of BopA was verified in B. bifidum DSM20456 and MIMBb75 cells by indirect immunofluorescence staining (Fig. 2B). BopA was present on the cell surfaces of both strains (Fig. 2B), but the staining was not evenly distributed around the cells, suggesting a localized distribution of BopA on the cell surface. The preimmune serum did not give any signal with B. bifidum cells in immunofluorescent staining (Fig. 2B). Third, the cell envelopes were extracted from B. bifidum DSM20456 as well as MIMBb75, and BopA was detected among the cell surface-associated proteins by Western blotting using the anti-BopA antiserum. The antiserum recognized the native, cell envelope-associated BopA, and the amount of BopA in the cell surface extracts was found to be comparable in the two strains (Fig. 2C). In conclusion, BopA was shown to be abundantly present in the cell envelopes of both studied strains of B. bifidum.
Fig 2.
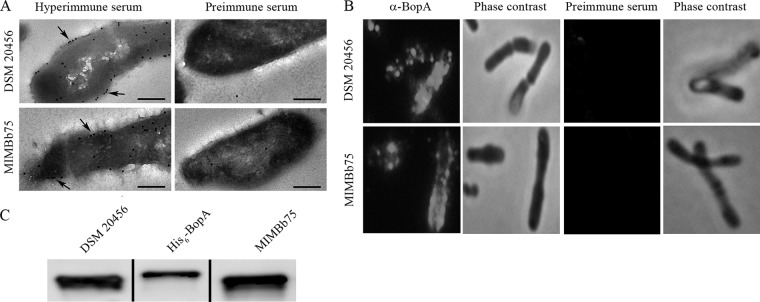
Subcellular localization of BopA in B. bifidum DSM20456 and MIMBb75. (A) Immunoelectron microscopy images of the localization of BopA in B. bifidum cells. Detection was done on thin sections by using anti-BopA (hyperimmune serum) and protein A-gold particles (pAp). The arrows indicate the pAp binding to the cell surface. Preimmune serum labeling of cells was used as a negative control. Bar, 200 nm. (B) Immunofluorescence staining of B. bifidum DSM20456 and MIMBb75 cells with anti-BopA and Alexa-488-conjugated secondary IgG. Preimmune serum was used as a negative control. Phase-contrast images of the same microscopic fields are shown on the right. (C) Western blotting of BopA in the cell wall extracts of B. bifidum DSM20456 and MIMBb75. His6-BopA (5 ng) is shown for comparison.
Role of BopA in B. bifidum adhesion to epithelial cell lines.
The adherence of B. bifidum DSM20456 and MIMBb75 as well as the binding of recombinant BopA protein to the epithelial cells of different ages was tested. Caco-2 cells differentiate in 14 days after confluence, and the adhesion and binding properties were tested with undifferentiated cells (3 days) in addition to cells at two differentiation stages (8 and 21 days). The number of added cells (10,000 cells well−1) resulted in confluence in 3 days; i.e., at all growth times, the cell cultures were confluent. For comparison, the same growth times were used for HT-29 cell line. Both strains, B. bifidum DSM20456 and MIMBb75, were found to adhere well to Caco-2 and HT-29 cells. The adhesion was most efficient (12 to 23%) with 8-day-old epithelial cells and the weakest (5 to 11%) with 21-day-old cells (Fig. 3A). The binding of 125I-labeled His6-BopA to 3- and 8-day-old Caco-2 and 8- and 21-day-old HT-29 cells was found to be statistically significantly (P < 0.05) higher than the background level binding of 125I-BSA (Fig. 3B). However, the moderate level of His6-BopA binding (maximum, 2.5 times greater than the background binding level of BSA) and the relatively high adhesion (up to 23% adherence) of B. bifidum to the epithelial cells indicated that BopA may not have a major role in mediating bifidobacterial adhesion to Caco-2 and HT-29 cells. To readdress the role of BopA as an adhesin, we tested the inhibition of bacterial adhesion to epithelial cell lines in the presence of anti-BopA antiserum, preimmune serum, and purified His6-BopA. In the inhibition assays, where bacterial cells were pretreated with the antiserum (diluted 1:100) prior to addition of bacteria to the cells, no significant reduction in the bacterial adherence was observed (Fig. 3A). As an exception, the adherence of B. bifidum MIMBb75 to 3-day-old Caco-2 cells was significantly diminished upon pretreatment with anti-BopA antiserum. However, the preimmune serum also inhibited adhesion, indicating a BopA-independent inhibitory effect of the antiserum in the binding assay (Fig. 3A). The results were not affected when the serum was used at higher concentrations (diluted 1:50 and 1:10) (data not shown). In the competition assays, the epithelial cells were first incubated with an extremely large amount (1 mg ml−1) of recombinant BopA prior to the bacterial adherence. Similarly to the inhibition assays, no significant reduction was observed in the adherence of B. bifidum to epithelial cells of different ages in the competition assay, with the exception of the diminished adherence of MIMBb75 to 3-day-old Caco-2 cells in the competition with His6-BopA (Fig. 3A). A slight variation in the adhesion levels as well as in the inhibition of the adhesion with the addition of anti-BopA antiserum or His6-BopA was seen between the experiments. Figure 3 shows that B. bifidum DSM20456 and MIMBb75 are highly adherent to epithelial cells (5 to 25% of added bacteria adhered) and that in general the adhesion could not be inhibited with the pretreatment of bacterial cells by anti-BopA antiserum or outcompeted with the addition of recombinant BopA protein.
Role of BopA in B. bifidum adhesion to mucus.
B. bifidum strains DSM20456 and MIMBb75 were found to be adherent to mucus (Fig. 4A), and the recombinant His6-BopA was found to bind moderately to human colonic mucus (Fig. 4B). To study the role of BopA in mediating the adhesion of B. bifidum to mucus, the anti-BopA antiserum was used to inhibit the bacterial adhesion, and preimmune serum was used as a control. A minor decrease was seen in the adhesion when bacterial cells were pretreated with the anti-BopA antiserum, but the decrease was not statistically significant. These results suggest that BopA has only a minor role, if any, in the adherence of B. bifidum to human colonic mucus.
Role of BopA in B. bifidum adhesion to ECM proteins.
The adherence of B. bifidum DSM20456 and MIMBb75 cells and the binding of His6-BopA to ECM proteins laminin, collagen I, collagen IV, fibronectin, and fetuin (highly glycosylated protein) were studied. Both strains of B. bifidum showed adhesiveness to fibronectin, and B. bifidum MIMBb75 also adhered to laminin (Fig. 5A). Based on these results, we selected laminin and fibronectin for the inhibition experiments. The bacteria were incubated with anti-BopA antiserum or preimmune serum before being added to the immobilized ECM proteins. Significant inhibition in the adhesion of both strains of B. bifidum to fibronectin was detected (Fig. 5B and C), whereas antiserum did not affect the adherence to laminin (Fig. 5C). However, since the preimmune serum caused a similar reduction in bacterial adhesion, these results suggest an unspecific effect, rather than a specific BopA-dependent inhibition, brought about by the anti-BopA antiserum. This was further supported by the results of His6-BopA binding to ECM proteins, as recombinant BopA showed moderate binding to laminin, whereas it failed to bind fibronectin (Fig. 5D).
DISCUSSION
Commensal intestinal bacteria in the genus Bifidobacterium are thought to balance the intestinal microbiota and to exert health-promoting effects on the host (24–26), and some strains are also used as probiotics. Adhesion of probiotic and commensal bacteria to the intestinal epithelial cells, mucus, and ECM proteins could help these organisms to persist in the intestinal tract and enable close contact with the host. In healthy individuals, the gastrointestinal epithelium is covered with mucus, which forms a thick, continuous layer in the large intestine. However, in the small intestine, the mucus layer is thinner and discontinuous, allowing direct contact between epithelial cells and luminal bacteria. Also, under certain conditions, the mucus barrier is reduced, and bacteria can penetrate the layer and adhere to the underlying epithelial cells and ECM proteins (27).
The bifidobacterial lipoprotein BopA, which has homology with the solute-binding protein of the ABC transport system in Gram-positive bacteria, has been described as an adhesive surface protein of B. bifidum MIMBb75 (9). The previous studies of the localization of BopA on the cell surface as well as its role in the adhesion were performed with a native BopA purified from the cell envelope of B. bifidum (9). In the present study, the role of BopA as a bifidobacterial adhesion molecule was readdressed by exploiting antiserum against His6-BopA and by using a recombinant BopA without the membrane-spanning lipid moiety covalently linked to the cysteine residue in the lipobox and thus avoiding possible unspecific effects resulting from the hydrophobic nature of the N-terminal part of the native protein. The recombinant BopA was produced with a C-terminal His6 tag, and it could not be ruled out that it had the same conformation as the native protein; consequently, the recombinant protein may have had altered binding properties compared to the native one. However, the antiserum raised against the recombinant protein also recognized the native BopA, and therefore, both His6-BopA and the antiserum were considered suitable for the subsequent experiments.
By using the antiserum, it was evident that BopA is an abundantly expressed surface protein of B. bifidum. It was found to be localized unevenly on the cell surface rather than being distributed uniformly around the cells. Asymmetric localization of the surface proteins, including the proteins involved in transporting of substrates across the cytoplasmic membrane, was described earlier and seems to be a common mechanism in Gram-positive bacteria (28, 29). It has been speculated that the cell wall passage of proteins is restricted to a limited number of sites to maintain the cell wall rigidity and withstand the turgor pressure (28). Alternatively, the cell surface of B. bifidum is covered with an uneven layer of exopolysaccharides, which results in an uneven staining and consequently gives an image of patchy localization of BopA on the cell surface.
The human intestinal isolate B. bifidum MIMBb75 and the type strain of B. bifidum, DSM20456, display an adhesive phenotype on the epithelial cell lines Caco-2 and HT-29 depending on the age of the epithelial cell culture. The differentiation stage of the epithelial cells affects the expression of surface molecules (30, 31), which affects the bacterial adhesion to the cells (32). In accordance, our results show various levels of adhesion to the epithelial cells of different ages or at different stages of differentiation. Only minimal effects or no effect at all were caused by the BopA-specific antiserum (inhibitory experiments) or the recombinant His6-BopA (competitive experiments) on the adhesion of B. bifidum DSM20456 and MIMBb75 to Caco-2 or HT-29 cells. Furthermore, a reduction in the adhesion of B. bifidum MIMBb75 to the 3-day-old, nondifferentiated Caco-2 cells was observed when bacteria were pretreated with the anti-BopA antiserum, but also, the preimmune serum inhibited the adhesion, indicating a non-BopA-based inhibition effect of the antisera.
In the competitive adhesion assays, only a moderate 28% reduction in the binding of B. bifidum MIMBb75, but not DSM20456, to the 3-day-old Caco-2 cells was observed when the epithelial cells were pretreated with the recombinant His6-BopA, suggesting that BopA may be a minor accessory adhesin. His6-BopA did not inhibit the adhesion of B. bifidum MIMBb75 to HT-29 or to older Caco-2 cells.
The previous reports on the role of BopA as an adhesin for Caco-2 cells were based on competitive binding experiments done with remarkably high concentrations (375 mg ml−1) of BopA containing the hydrophobic lipid moiety, which likely consists of a diglyceride molecule covalently linked to the N-terminal cysteine of BopA (9). In a more recent study, Gleinser and colleagues (17) observed a reduction in the adhesion of B. bifidum when the epithelial cells were pretreated with a recombinant His6-BopA protein, which was prepared by cloning the complete bopA gene. It includes a DNA sequence coding for an N-terminal hydrophobic signal peptide of 25 amino acids, which meets all of the requirements for a transmembrane helix, and contains the lipobox motif (17). In this study, no inhibitory effect on the bifidobacterial binding to the epithelial cells could be confirmed by using a recombinant His6-BopA devoid of the membrane-spanning lipid moiety and the 25-amino-acid hydrophobic signal sequence, or by blocking the bifidobacteria with the BopA-specific antiserum. The inhibition observed in the previous study may have resulted from an unspecific effect caused by the higher hydrophobicity of the BopA (lipo)proteins used in the study. Accordingly, the surface layer (S-layer) proteins of Lactobacillus, which are rich in hydrophobic amino acids (33), have been shown to inhibit the adhesion of S. aureus to Caco-2 cells (34). Furthermore, it has been shown that cell surface hydrophobicity indicates good adhesion potential for bacteria and that the high hydrophobicity of the cell surface of a probiotic strain correlates well with its capacity to inhibit pathogen adhesion through steric hindrance (35). In other words, it seems that hydrophobic molecules bind efficiently to the epithelial cells and thereby block the bacterial binding sites by steric hindrance. Similarly, the competitive inhibition of bifidobacterial adhesion to the epithelial cells caused by a lipid- or signal peptide-containing hydrophobic BopA could have resulted from a steric hindrance to the adhesion sites following an aspecific hydrophobic interaction of BopA protein with Caco-2 cell surface.
Recently, Gleinser and colleagues (17) also used bifidobacteria overexpressing BopA to address its role as an adhesin. The overexpression of BopA in B. longum subsp. infantis E18, a strain that does not contain the gene coding for BopA and shows only a very weak adhesion to the epithelial cells, increased the adhesion of the strain to T84, Caco-2, and HT-29 cells by 511, 180, and 209%, respectively (17). However, the adhesion of E18 to Caco-2 was below 3%, and adhesion to T84 and HT-29 was even lower. Therefore, a 180% rise in the relative binding of the recombinant B. longum subsp. infantis E18 to Caco-2 cells would mean an absolute adhesion level of <5.4% (17), which is still far from the adherence of the wild-type-BopA-bearing B. bifidum strains, which show adherence above 30%. Thus, the increase in the adhesion obtained by overexpressing bopA does not explain the high adherence of wild-type B. bifidum observed here and by others (9, 17). The slight increase in the adhesion of B. longum subsp. infantis E18 overexpressing bopA indicates, however, that BopA may serve as a minor adhesin (17).
In healthy individuals, the gastrointestinal tract is covered by two layers of mucus. The inner layer is firmly attached to the epithelium, and the outer layer is more loosely attached and colonized by bacteria (36–38). B. bifidum DSM20456 and MIMBb75 showed approximately 4 and 8% adhesion to human colonic mucus, respectively, whereas the recombinant His6-BopA bound moderately to mucus. However, the antiserum produced against His6-BopA failed to decrease the adhesion of B. bifidum to mucus, indicating that BopA is not the major adhesin mediating the bifidobacterial adherence to mucus either. Previously, both DSM20456 and MIMBb75 were found to adhere to immobilized, commercially available mucus, and the surface-exposed proteins have been suggested to be involved in the binding (10, 39). Transaldolase was reported to mediate the autoaggregation of the bifidobacterial cells at acidic pH, which was linked to the mucus-binding capacity of B. bifidum (10). However, as the autoaggregating phenotype of B. bifidum is strictly dependent on acidic pH (39), and since in this study all binding experiments were performed at neutral pH and autoaggregation was not observed, autoaggregation cannot explain the strong adhesion of B. bifidum to mucus at neutral pH. The recombinant His6-BopA bound, though moderately, to mucus, and therefore, it seems that BopA may serve as a minor moonlighting adhesin. It has been suggested that specific ABC transporter proteins are involved in host-bacterium interactions and act as moonlighting adhesins (40–43), particularly in mucus binding (40, 43). Similarly, BopA may serve as a minor moonlighting adhesin mediating bifidobacterial adhesion to mucus at neutral pH.
Next, the binding of His6-BopA and adherence of B. bifidum to ECM proteins was studied. The ECM is known to serve as a substrate for the attachment of colonizing microorganisms (44). ECM is a highly structured network of four main components, collagens, laminin, fibronectin, and elastin (45). Both B. bifidum strains studied adhered to fibronectin, and B. bifidum MIMBb75 adhered to laminin, but adherence to collagen I or IV was not observed. The adhesion to fibronectin was inhibited when bacterial cells were pretreated with anti-BopA antiserum but also when they were pretreated with preimmune serum, again suggesting an unspecific inhibitory effect of the antisera. The inhibitory effect of the antisera on the bacterial adhesion to fibronectin could have resulted from (i) the binding of antisera directly to the immobilized fibronectin or (ii) the serum itself containing fibronectin, which binds to fibronectin receptors on the bacterial surface. Similarly, the inhibitory effect of antisera on the adhesion of B. bifidum MIMBb75 to 3-day-old Caco-2 cells may have resulted from antiserum binding to fibronectin, which can be produced by Caco-2 cells (46). The recombinant BopA, however, did not bind to fibronectin and excluded the role of BopA in mediating the bifidobacterial adhesion to fibronectin. Instead of fibronectin, the recombinant BopA bound to laminin, indicating that BopA may act as a minor adhesin to ECM.
In conclusion, our results show, in contrast to previous studies, that BopA has a very limited role in adhesion and that the adherence of B. bifidum to epithelial cells, mucus, or ECM proteins is BopA independent. However, BopA binds moderately to human colonic mucus and laminin and may act as a minor coadhesin of B. bifidum. Very recently, pil3 sortase-dependent pili were shown to be involved in the adhesion of B. bifidum (16). Further studies are needed to unambiguously demonstrate whether these pili or other bacterial structures are the main adhesion molecules mediating the adhesion of B. bifidum to epithelial cells.
ACKNOWLEDGMENT
This study was supported by The Academy of Finland (grant 138902).
Footnotes
Published ahead of print 6 September 2013
REFERENCES
- 1.Ashida H, Ogawa M, Kim M, Mimuro H, Sasakawa C. 2012. Bacteria and host interactions in the gut epithelial barrier. Nat. Chem. Biol. 8:36–45 [DOI] [PubMed] [Google Scholar]
- 2.Lebeer S, Vanderleyden J, De Keersmaecker SC. 2010. Host interactions of probiotic bacterial surface molecules: comparison with commensals and pathogens. Nat. Rev. Microbiol. 8:171–184 [DOI] [PubMed] [Google Scholar]
- 3.Kankainen M, Paulin L, Tynkkynen S, von Ossowski I, Reunanen J, Partanen P, Satokari R, Vesterlund S, Hendrickx AP, Lebeer S, De Keersmaecker SC, Vanderleyden J, Hamalainen T, Laukkanen S, Salovuori N, Ritari J, Alatalo E, Korpela R, Mattila-Sandholm T, Lassig A, Hatakka K, Kinnunen KT, Karjalainen H, Saxelin M, Laakso K, Surakka A, Palva A, Salusjarvi T, Auvinen P, de Vos WM. 2009. Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human-mucus binding protein. Proc. Natl. Acad. Sci. U. S. A. 106:17193–17198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ewaschuk JB, Diaz H, Meddings L, Diederichs B, Dmytrash A, Backer J, Looijer-van Langen M, Madsen KL. 2008. Secreted bioactive factors from Bifidobacterium infantis enhance epithelial cell barrier function. Am. J. Physiol. Gastrointest. Liver Physiol. 295:G1025–G1034 [DOI] [PubMed] [Google Scholar]
- 5.Maynard CL, Elson CO, Hatton RD, Weaver CT. 2012. Reciprocal interactions of the intestinal microbiota and immune system. Nature 489:231–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harmsen HJ, Wildeboer-Veloo AC, Raangs GC, Wagendorp AA, Klijn N, Bindels JG, Welling GW. 2000. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J. Pediatr. Gastroenterol. Nutr. 30:61–67 [DOI] [PubMed] [Google Scholar]
- 7.Nylund L, Satokari R, Nikkila J, Rajilic-Stojanovic M, Kalliomaki M, Isolauri E, Salminen S, de Vos WM. 2013. Microarray analysis reveals marked intestinal microbiota aberrancy in infants having eczema compared to healthy children in at-risk for atopic disease. BMC Microbiol. 13:12. 10.1186/1471-2180-13-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Li S, Jian M, Zhou Y, Li Y, Zhang X, Li S, Qin N, Yang H, Wang J, Brunak S, Dore J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, MetaHIT Consortium, Bork P, Ehrlich SD, Wang J. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guglielmetti S, Tamagnini I, Mora D, Minuzzo M, Scarafoni A, Arioli S, Hellman J, Karp M, Parini C. 2008. Implication of an outer surface lipoprotein in adhesion of Bifidobacterium bifidum to Caco-2 cells. Appl. Environ. Microbiol. 74:4695–4702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonzalez-Rodriguez I, Sanchez B, Ruiz L, Turroni F, Ventura M, Ruas-Madiedo P, Gueimonde M, Margolles A. 2012. Role of extracellular transaldolase from Bifidobacterium bifidum in mucin adhesion and aggregation. Appl. Environ. Microbiol. 78:3992–3998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Connell Motherway M, Zomer A, Leahy SC, Reunanen J, Bottacini F, Claesson MJ, O'Brien F, Flynn K, Casey PG, Munoz JA, Kearney B, Houston AM, O'Mahony C, Higgins DG, Shanahan F, Palva A, de Vos WM, Fitzgerald GF, Ventura M, O'Toole PW, van Sinderen D. 2011. Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved host-colonization factor. Proc. Natl. Acad. Sci. U. S. A. 108:11217–11222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Candela M, Bergmann S, Vici M, Vitali B, Turroni S, Eikmanns BJ, Hammerschmidt S, Brigidi P. 2007. Binding of human plasminogen to Bifidobacterium. J. Bacteriol. 189:5929–5936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Candela M, Biagi E, Centanni M, Turroni S, Vici M, Musiani F, Vitali B, Bergmann S, Hammerschmidt S, Brigidi P. 2009. Bifidobacterial enolase, a cell surface receptor for human plasminogen involved in the interaction with the host. Microbiology 155:3294–3303 [DOI] [PubMed] [Google Scholar]
- 14.Candela M, Centanni M, Fiori J, Biagi E, Turroni S, Orrico C, Bergmann S, Hammerschmidt S, Brigidi P. 2010. DnaK from Bifidobacterium animalis subsp. lactis is a surface-exposed human plasminogen receptor upregulated in response to bile salts. Microbiology 156:1609–1618 [DOI] [PubMed] [Google Scholar]
- 15.Foroni E, Serafini F, Amidani D, Turroni F, He F, Bottacini F, O'Connell Motherway M, Viappiani A, Zhang Z, Rivetti C, van Sinderen D, Ventura M. 2011. Genetic analysis and morphological identification of pilus-like structures in members of the genus Bifidobacterium. Microb. Cell Fact. 10(Suppl 1):S16. 10.1186/1475-2859-10-S1-S16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turroni F, Serafini F, Foroni E, Duranti S, O′Connell Motherway M, Taverniti V, Mangifesta M, Milani C, Viappini A, Roversi T, Sánchez B, Santoni A, Gioiosa L, Ferrarini A, Delledonne M, Margolles A, Paizza L, Planza P, Bolchi A, Guglielmetti S, van Sinderen D, Ventura M. 2013. Role of sortase-dependent pili of Bifidobacterium bifidum PRL2010 in modulating bacterium-host interactions. Proc. Natl. Acad. Sci. U. S. A. 110:11151–11156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gleinser M, Grimm V, Zhurina D, Yuan J, Riedel CU. 2012. Improved adhesive properties of recombinant bifidobacteria expressing the Bifidobacterium bifidum-specific lipoprotein BopA. Microb. Cell. Fact. 11:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnston BA, Eisen H, Fry D. 1991. An evaluation of several adjuvant emulsion regimens for the production of polyclonal antisera in rabbits. Lab. Anim. Sci. 41:15–21 [PubMed] [Google Scholar]
- 19.Harlow E, Lane D. (ed). 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 20.Reunanen J, von Ossowski I, Hendrickx AP, Palva A, de Vos WM. 2012. Characterization of the SpaCBA pilus fibers in the probiotic Lactobacillus rhamnosus GG. Appl. Environ. Microbiol. 78:2337–2344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kainulainen V, Loimaranta V, Pekkala A, Edelman S, Antikainen J, Kylvaja R, Laaksonen M, Laakkonen L, Finne J, Korhonen TK. 2012. Glutamine synthetase and glucose-6-phosphate isomerase are adhesive moonlighting proteins of Lactobacillus crispatus released by epithelial cathelicidin LL-37. J. Bacteriol. 194:2509–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vesterlund S, Paltta J, Karp M, Ouwehand AC. 2005. Measurement of bacterial adhesion-in vitro evaluation of different methods. J. Microbiol. Methods 60:225–233 [DOI] [PubMed] [Google Scholar]
- 23.Antikainen J, Kuparinen V, Lahteenmaki K, Korhonen TK. 2007. Enolases from Gram-positive bacterial pathogens and commensal lactobacilli share functional similarity in virulence-associated traits. FEMS Immunol. Med. Microbiol. 51:526–534 [DOI] [PubMed] [Google Scholar]
- 24.Isolauri E, Rautava S, Salminen S. 2012. Probiotics in the development and treatment of allergic disease. Gastroenterol. Clin. North Am. 41:747–762 [DOI] [PubMed] [Google Scholar]
- 25.Rautava S, Luoto R, Salminen S, Isolauri E. 2012. Microbial contact during pregnancy, intestinal colonization and human disease. Nat. Rev. Gastroenterol. Hepatol. 9:565–576 [DOI] [PubMed] [Google Scholar]
- 26.Jalanka-Tuovinen J, Salonen A, Nikkila J, Immonen O, Kekkonen R, Lahti L, Palva A, de Vos WM. 2011. Intestinal microbiota in healthy adults: temporal analysis reveals individual and common core and relation to intestinal symptoms. PLoS One 6:e23035. 10.1371/journal.pone.0023035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johansson ME, Sjovall H, Hansson GC. 2013. The gastrointestinal mucus system in health and disease. Nat. Rev. Gastroenterol. Hepatol. 10:352–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buist G, Ridder AN, Kok J, Kuipers OP. 2006. Different subcellular locations of secretome components of Gram-positive bacteria. Microbiology 152:2867–2874 [DOI] [PubMed] [Google Scholar]
- 29.Lybarger SR, Maddock JR. 2001. Polarity in action: asymmetric protein localization in bacteria. J. Bacteriol. 183:3261–3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garcia-Lorenzo A, Rodriguez-Pineiro AM, Rodriguez-Berrocal FJ, Cadena MP, Martinez-Zorzano VS. 2012. Changes on the Caco-2 secretome through differentiation analyzed by 2-D differential in-gel electrophoresis (DIGE). Int. J. Mol. Sci. 13:14401–14420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambuy Y, De Angelis I, Ranaldi G, Scarino ML, Stammati A, Zucco F. 2005. The Caco-2 cell line as a model of the intestinal barrier: influence of cell and culture-related factors on Caco-2 cell functional characteristics. Cell Biol. Toxicol. 21:1–26 [DOI] [PubMed] [Google Scholar]
- 32.Coconnier MH, Bernet MF, Kerneis S, Chauviere G, Fourniat J, Servin AL. 1993. Inhibition of adhesion of enteroinvasive pathogens to human intestinal Caco-2 cells by Lactobacillus acidophilus strain LB decreases bacterial invasion. FEMS Microbiol. Lett. 110:299–305 [DOI] [PubMed] [Google Scholar]
- 33.Hynonen U, Palva A. 2013. Lactobacillus surface layer proteins: structure, function and applications. Appl. Microbiol. Biotechnol. 97:5225–5243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ren D, Li C, Qin Y, Yin R, Li X, Tian M, Du S, Guo H, Liu C, Zhu N, Sun D, Li Y, Jin N. 2012. Inhibition of Staphylococcus aureus adherence to Caco-2 cells by lactobacilli and cell surface properties that influence attachment. Anaerobe 18:508–515 [DOI] [PubMed] [Google Scholar]
- 35.Xu H, Jeong HS, Lee HY, Ahn J. 2009. Assessment of cell surface properties and adhesion potential of selected probiotic strains. Lett. Appl. Microbiol. 49:434–442 [DOI] [PubMed] [Google Scholar]
- 36.Juge N. 2012. Microbial adhesins to gastrointestinal mucus. Trends Microbiol. 20:30–39 [DOI] [PubMed] [Google Scholar]
- 37.Kim YS, Ho SB. 2010. Intestinal goblet cells and mucins in health and disease: recent insights and progress. Curr. Gastroenterol. Rep. 12:319–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Tassell ML, Miller MJ. 2011. Lactobacillus adhesion to mucus. Nutrients 3:613–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guglielmetti S, Tamagnini I, Minuzzo M, Arioli S, Parini C, Comelli E, Mora D. 2009. Study of the adhesion of Bifidobacterium bifidum MIMBb75 to human intestinal cell lines. Curr. Microbiol. 59:167–172 [DOI] [PubMed] [Google Scholar]
- 40.Macias-Rodriguez ME, Zagorec M, Ascencio F, Vazquez-Juarez R, Rojas M. 2009. Lactobacillus fermentum BCS87 expresses mucus- and mucin-binding proteins on the cell surface. J. Appl. Microbiol. 107:1866–1874 [DOI] [PubMed] [Google Scholar]
- 41.Roos S, Aleljung P, Robert N, Lee B, Wadström T, Lindberg M, Jonsson H. 1996. A collagen binding protein from Lactobacillus reuteri is part of an ABC transporter system? FEMS Microbiol. Lett. 144:33–38 [DOI] [PubMed] [Google Scholar]
- 42.Watanabe M, Kinoshita H, Nitta M, Yukishita R, Kawai Y, Kimura K, Taketomo N, Yamazaki Y, Tateno Y, Miura K, Horii A, Kitazawa H, Saito T. 2010. Identification of a new adhesin-like protein from Lactobacillus mucosae ME-340 with specific affinity to the human blood group A and B antigens. J. Appl. Microbiol. 109:927–935 [DOI] [PubMed] [Google Scholar]
- 43.Watanabe M, Kinoshita H, Huang IN, Eguchi K, Tsurumi T, Kawai Y, Kitazawa H, Kimura K, Taketomo N, Kikuchi D, Sase T, Miura K, Ogawa H, Shibata C, Horii A, Saito T. 2012. An adhesin-like protein, Lam29, from Lactobacillus mucosae ME-340 binds to histone H3 and blood group antigens in human colonic mucus. Biosci. Biotechnol. Biochem. 76:1655–1660 [DOI] [PubMed] [Google Scholar]
- 44.Chagnot C, Listrat A, Astruc T, Desvaux M. 2012. Bacterial adhesion to animal tissues: protein determinants for recognition of extracellular matrix components. Cell. Microbiol. 14:1687–1696 [DOI] [PubMed] [Google Scholar]
- 45.Patti JM, Hook M. 1994. Microbial adhesins recognizing extracellular matrix macromolecules. Curr. Opin. Cell Biol. 6:752–758 [DOI] [PubMed] [Google Scholar]
- 46.Kolachala VL, Bajaj R, Wang L, Yan Y, Ritzenthaler JD, Gewirtz AT, Roman J, Merlin D, Sitaraman SV. 2007. Epithelial-derived fibronectin expression, signaling, and function in intestinal inflammation. J. Biol. Chem. 282:32965–32973 [DOI] [PubMed] [Google Scholar]