Abstract
Campylobacter jejuni is a widespread pathogen responsible for most of the food-borne gastrointestinal diseases in Europe. The use of natural antimicrobial molecules is a promising alternative to antibiotic treatments for pathogen control in the food industry. Isothiocyanates are natural antimicrobial compounds, which also display anticancer activity. Several studies described the chemoprotective effect of isothiocyanates on eukaryotic cells, but the antimicrobial mechanism is still poorly understood. We investigated the early cellular response of C. jejuni to benzyl isothiocyanate by both transcriptomic and physiological approaches. The transcriptomic response of C. jejuni to benzyl isothiocyanate showed upregulation of heat shock response genes and an impact on energy metabolism. Oxygen consumption was progressively impaired by benzyl isothiocyanate treatment, as revealed by high-resolution respirometry, while the ATP content increased soon after benzyl isothiocyanate exposition, which suggests a shift in the energy metabolism balance. Finally, benzyl isothiocyanate induced intracellular protein aggregation. These results indicate that benzyl isothiocyanate affects C. jejuni by targeting proteins, resulting in the disruption of major metabolic processes and eventually leading to cell death.
INTRODUCTION
Food-borne diseases are a major public health concern. In Europe, Campylobacter jejuni remains the most detected bacterial pathogen on fresh broiler meat and the major cause of human bacterial gastrointestinal disease. C. jejuni colonizes asymptomatically the gut of several farm animals, mainly poultry. C. jejuni is therefore highly prevalent in broiler flocks and persists through different stages of food production until the consumption of contaminated meat (1).
In humans, campylobacteriosis results in severe diarrhea, abdominal pain, nausea, and fever (2). Moreover, campylobacteriosis has been associated with long-term complications such as Guillain-Barré syndrome, a neurological disorder that can result in paralysis, respiratory failure, or even death (3).
Food products can be treated with antimicrobial compounds or sanitizers to limit contamination by food-borne pathogens. However, the use of antibiotics remains controversial due to the rapid development of antibiotic resistance among many common pathogens, including C. jejuni (4, 5). As an alternative, natural antimicrobials have been used for centuries to protect food products from bacterial or fungal spoilage (6). Among them, isothiocyanates (ITCs) are promising molecules with antimicrobial activities against a large number of microorganisms (7–9). The safety of allyl isothiocyanates (AITCs) as a food additive compound was previously demonstrated (10). In fact, AITCs extracted from plant sources are already authorized as natural food preservatives in Japan (11). Interestingly, ITCs have also been reported to display anticancer activity in humans (12). Because of the antimicrobial and anticancer properties of ITCs, several studies have proposed the use of ITCs as food preservatives (11, 13–16).
ITCs are products of the degradation of plant glucosinolates by the enzyme myrosinase, found in plants and in microbes of the gut microbiota (17). Plants of the Brassicaceae family produce glucosinolates; these plants include mustard, wasabi, horseradish, and broccoli. The general structure of ITCs is R—N=C=S, with a highly electrophilic carbon atom, allowing them to react readily with cellular thiols such as cysteines in proteins and low-molecular-weight thiols (especially glutathione), resulting in dithiocarbamate derivatives. This modification leads to a loss of protein structure and function.
In eukaryotes, ITCs react with a large number of proteins containing cysteines involved in inflammation, the stress response, cell cycle arrest, or apoptosis (12). ITCs also generate oxidative stress by depleting intracellular glutathione or damaging mitochondria (18). The inactivation of proteins and the associated oxidative stress lead to apoptosis. Interestingly, ITCs appear to be more toxic to malignant cells than to healthy ones (19, 20). In addition, at low concentrations, the prooxidant effect of ITCs on eukaryotic cells stimulates the antioxidant cellular response (18). Indeed, ITCs act as stress inducers in eukaryotes, whose overall effect is beneficial to the organism at relatively low doses. Nevertheless, ITCs exhibit cytotoxic effects at higher dosages (21).
In contrast to the large number of studies on the effects of ITCs on eukaryotic cells, little is known about the mode of action of ITCs on bacteria. ITCs appear to alter intracellular structures of Listeria monocytogenes (22) and provokes membrane protrusion on Aggregatibacter actinomycetemcomitans (9). A recent study identified the sax (survival in Arabidopsis extracts) genes to be involved in the resistance of Pseudomonas syringae to the ITC sulforaphane, produced by Arabidopsis plants (23). However, the mechanism of sax-mediated resistance to ITCs remains unknown. The ITCs iberin and sulforaphane have been identified as quorum-sensing-inhibiting compounds in Pseudomonas aeruginosa (24, 25). Additionally, an in vitro study demonstrated that AITC inhibits the enzymatic activity of Escherichia coli thioredoxin and acetate kinase (26).
To date, the only study characterizing the global response of bacteria exposed to ITCs consists of a whole-transcriptome analysis of P. aeruginosa exposed to iberin (25). This work demonstrated the quorum-sensing-inhibiting effect of iberin but also the modulation of other cellular processes, including aerobic respiration. Among the upregulated genes were genes encoding oxidoreductases and the heat shock proteins GroEL and DnaK, suggesting a perturbation of the redox homeostasis and the presence of misfolded proteins. Iberin also induced the expression of the genes coding for the MexEF-OprN efflux pump and a glutathione S-transferase (GST); this suggests that efflux of ITCs and/or GST-glutathione detoxification of ITCs can be a resistance mechanism, as GSTs have been reported to be an ITC detoxification enzyme in cyanobacteria (27) and eukaryotes (28).
In our previous study, we assayed the sensitivity of C. jejuni to AITC and benzyl isothiocyanate (BITC) (29). BITC was bactericidal against C. jejuni at low concentrations (with a minimal bactericidal concentration in Mueller-Hinton [MH] broth of 1.25 mg liter−1). Since C. jejuni does not possess the pathways for glutathione biosynthesis or GST-encoding genes (30), the cellular response of C. jejuni to ITCs would likely be different from the response of P. aeruginosa.
A report by Davidson and Harrison highlighted the need for a better understanding of the mechanisms of action of natural antimicrobial compounds (31). The aim of the present work was to characterize the early response of C. jejuni to BITC, using a genome-wide transcriptomic approach. This study demonstrates that exposure to BITC elicits a heat shock response and an oxidative stress response and induced the expression of genes involved in energy production and electron transport. In addition, we investigated the effect of BITC on oxygen respiration and ATP production by high-resolution respirometry and a luciferase-based assay. Finally, we examined the aggregation of proteins as a result of BITC exposure.
MATERIALS AND METHODS
Bacterial strains, culture conditions, chemicals, and standard molecular biology procedures.
C. jejuni NCTC11168 cells were grown at 37°C under microaerophilic conditions (10% O2, 5% CO2, and 85% N2) in minimum essential medium α (MEMα) or Dulbecco's MEM (DMEM; Invitrogen) supplemented with 20 mM pyruvate, in MH broth, or on Columbia agar plates (Oxoid). BITC (Sigma) stock solutions were prepared in absolute ethanol.
Survival assay.
C. jejuni NCTC11168 cells were grown to mid-log phase (optical density at 600 nm [OD600] of ∼0.2) in DMEM with 20 mM pyruvate before the addition of BITC in ethanol solution. Final concentrations ranged from 1 mg liter−1 to 5 mg liter−1. Alternatively, ethanol was added as a negative control. Samples were removed at different time points after treatment, and cells were immediately diluted and plated onto Columbia agar plates. The viability of C. jejuni after BITC or ethanol treatment was determined by cell counting after 48 h of incubation under microaerophilic conditions.
Total RNA isolation.
C. jejuni NCTC11168 cells were cultured overnight in MH broth in biphasic flasks under microaerophilic conditions (83% N2, 4% H2, 8% O2, and 5% CO2) at 37°C. Bacterial cultures were centrifuged, washed in phosphate-buffered saline (PBS), and resuspended in MEMα (Invitrogen) supplemented with 20 mM sodium pyruvate to an OD600 of 0.05 in 25 ml. A total of 2 mg liter−1 BITC in ethanol (BITC treatment) or ethanol (control) was added to C. jejuni cells grown to mid-log phase (OD600 of 0.2). Ten or fifteen minutes following BITC or ethanol addition, a cold RNA degradation stop solution (2.5 ml of 10% buffer-saturated phenol [pH 4.3] in ethanol) was added. Bacterial cells were harvested by centrifugation and resuspended in Tris-EDTA (TE) buffer, followed by total RNA isolation using the hot phenol-chloroform method, as described previously (32). Extracted RNA samples were treated twice with DNase I (Epicenter) and purified by using the RNeasy kit (Qiagen), followed by PCR amplification to confirm the absence of contaminating genomic DNA. The Experion RNA STDsens Analysis kit (Bio-Rad Laboratories) was used to quantify RNA and confirm RNA integrity.
Microarray hybridization and data analysis.
Preparation of fluorescently labeled cDNA and microarray slide hybridization were performed as described previously (32). Briefly, each of the 16-μg extracted RNA samples was reverse transcribed by using Superscript II (Invitrogen) in the presence of random hexamers and aminoallyl-dUTP. The aminoallyl-labeled cDNA was purified by using Micron YM-30 spin columns (Millipore) prior to fluorescent labeling by coupling with Cy3 or Cy5 monoreactive fluorescent dyes (CyDye Fluor; Amersham), according to a method described previously (32). Fluorescently labeled cDNA samples from the BITC-treated and nontreated C. jejuni cells were combined and processed for hybridization. Microarray slides (32, 33) consisting of PCR products for each open reading frame of C. jejuni NCTC11168 were prepared for hybridization. The fluorescently labeled, pooled cDNA samples were hybridized to the microarray slide as described previously (32). The slides were scanned at a 10-μm resolution with a PerkinElmer ScanArray Express laser-activated confocal scanner. Next, the obtained data were normalized by using MIDAS software (available from the J. Craig Venter Institute [http://www.jcvi.org/cms/research/software/]) and locally weighted linear regression (Lowess), as described previously (32). The ratio of the fluorescence intensity values in the Cy3 channel to those in the Cy5 channel was calculated and converted into a log2 value. Statistical analysis was performed by using the Bayes empirical method (32) with CyberT software (http://molgen51.biol.rug.nl/cybert/). Genes were considered significantly differentially expressed with Bayesian P values of <10−4 and a fold change of at least 1.5 for at least one of the two time points. Genes were clustered according to their expression profiles by using Genesis software (http://genome.tugraz.at/). For the 10-min time point, two independent RNA extractions were performed, and each sample was hybridized to three microarray slides. For the 15-min time point, RNA extraction was performed in triplicates, and each sample was hybridized to three microarray slides.
ATP assay.
C. jejuni NCTC11168 cells were grown to mid-log phase in DMEM with 20 mM pyruvate. BITC (2 mg liter−1 or 5 mg liter−1) or ethanol (negative control) was added to the cultures. The ATP content was assayed at 15, 30, 60, 90, and 120 min after BITC or ethanol addition by using the BacTiterGlo kit (Promega) according to the manufacturer's instructions. Culture medium with or without added BITC or ethanol was used as a blank. The ATP content was measured three times on three distinct bacterial cultures with a LumiCount luminescence microplate reader (Packard). The average noise signal was subtracted from each value, and the results were expressed as relative lights units (RLU). A Student t test with a significance value of 0.05 was used for statistical analysis of the results.
Respirometry.
The oxygen concentration was measured by high-resolution respirometry with the Oroboros Oxygraph-2k instrument (34), in a standard configuration, with a 2-ml volume of the two chambers, a temperature of 37°C, and a 500-rpm stirrer speed. Data were recorded at 1-s intervals by using Datlab 4 Acquisition software (Oroboros, Innsbruck, Austria). Standardized calibration procedures for the oxygen signal were carried out by using DMEM supplemented with 20 mM pyruvate. Respiration was automatically corrected for contributions of the polarographic oxygen sensor and of oxygen diffusion to the total apparent respiration, as a continuous function of the oxygen concentration (35).
C. jejuni cells grown at 37°C under microaerobic conditions (Campygen; Oxoid) were harvested in the exponential growth phase (OD600 = 0.2), quickly resuspended to an OD600 of 0.1 in their own culture supernatant, and transferred into the Oxygraph chambers. In control experiments, cells were washed twice (5,000 × g for 10 min) and resuspended in fresh MEM supplemented or not with 20 mM pyruvate. Acquisition was started after a few minutes of equilibration at 37°C under oxic conditions and closing of the chambers. Identical volumes (4 μl for 2 ml) of ethanol (negative control) and a BITC-ethanol solution (2 mg liter−1 BITC in ethanol) were added to each chamber at steady states of respiration, and their effects were monitored simultaneously for about 1 h. The experiment started at the initial O2 saturation level and continued until cultures became hypoxic and then anaerobic. Measurements were performed on three independent cultures, and data shown represent characteristic results.
Isolation of protein aggregates.
C. jejuni cells were grown to mid-log phase in 500 ml DMEM–20 mM pyruvate before the addition of 1 ml BITC to reach a final concentration of 2 mg liter−1 or 5 mg liter−1 BITC or before the addition of 1 ml ethanol as a negative control. Samples of 100 ml were removed after 15 min, 1 h, or 2 h for extraction and purification of protein aggregates, according to a method described previously by Tomoyasu et al. (36). Isolated protein aggregates were then analyzed on a 12% (wt/vol)-acrylamide SDS-PAGE gel followed by Coomassie staining, as previously reported (37). Aggregated proteins and total proteins were quantified by using a bicinchoninic acid (BCA) assay (Thermo Scientific).
Microarray data accession number.
Microarray data were deposited in the Gene Expression Omnibus database under accession number GSE45823.
RESULTS
Survival of C. jejuni in the presence of BITC.
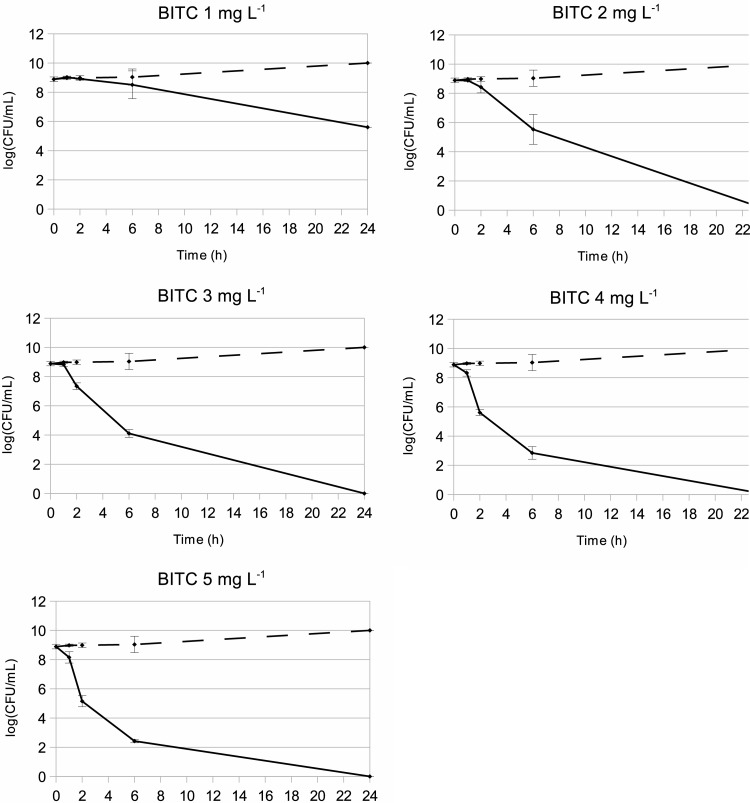
To study the early response of C. jejuni to BITC, we monitored its survival against low concentrations of BITC following short exposure times. In our previous study (29), a concentration of 1.25 mg liter−1 was found to be bactericidal to C. jejuni NCTC11168 after 18 h of exposure in MH medium. As shown in Fig. 1, a concentration of 2 mg liter−1 of BITC did not noticeably affect the viability of C. jejuni NCTC11168 with up to 2 h of exposure. However, each assayed concentration caused a loss of viability of >3 log CFU ml−1 after longer exposure times (Fig. 1). Therefore, a concentration of 2 mg liter−1 appeared appropriate to study the early response to BITC, as it affects cell viability only after prolonged exposure while still impacting importantly the cell physiology.
Fig 1.
Survival rate of C. jejuni NCTC11168 exposed to BITC. Cells were grown in DMEM–20 mM pyruvate at 37°C under microaerobic conditions, and BITC was added to final concentrations of 1 to 5 mg liter−1 (solid lines). Ethanol (dashed lines) was added as a negative control. Viable cells were enumerated at 1 h, 2 h, 6 h, or 24 h after BITC or ethanol addition. The experiment was performed twice with technical assays. Each dot represents the mean of 4 data points with standard deviations.
Transcriptomic response of C. jejuni to BITC.
A microarray approach was used to assess the early transcriptomic response of C. jejuni to BITC. The transcription of 81 genes was significantly modulated after exposure to 2 mg liter−1 BITC for 10 min or 15 min, as shown in Table 1.
Table 1.
Genes differentially expressed following 10 min or 15 min of exposure to 2 μg ml−1 of BITC
| Gene and category | Gene product and/or function | Fold change (log2) |
|
|---|---|---|---|
| 10 min | 15 min | ||
| Heat shock response | |||
| clpB | ATP-dependent disaggregase | 3.13 | 7.00 |
| hrcA | Heat-inducible transcriptional regulator | 2.22 | 3.78 |
| dnaK | Chaperone | 2.40 | 3.77 |
| grpE | Cochaperone of DnaK and nucleotide exchange factor | 1.92 | 2.73 |
| groES | Chaperone | 1.06 | 2.45 |
| cbpA | DnaJ-like protein | 1.17 | 1.51 |
| Cj0760 | Hypothetical protein, metallo-beta-lactamase superfamily | 1.78 | 1.92 |
| Oxidative stress response | |||
| sodB | Superoxide dismutase | 2.66 | 2.69 |
| rrc | Rubredoxin oxidoreductase/rubrerythrin-like protein | 1.10 | 1.83 |
| Cj0379c | Putative sulfite oxidase subunit YedY | 1.39 | 1.73 |
| Iron-sulfur cluster assembly and disulfide bond formation | |||
| Cj0239c | Putative sulfite oxidase | 1.93 | 1.82 |
| trxA | Thioredoxin | 1.22 | 1.69 |
| Cj0017c | Disulfide bond formation protein DsbI | −1.14 | −1.58 |
| Electron transport/energy metabolism | |||
| napA | Nitrate reductase | 1.77 | 2.48 |
| napG | Quinol dehydrogenase | 1.84 | 1.63 |
| hydA | Ni-Fe hydrogenase, small chain | 2.68 | 2.78 |
| hydB | Ni-Fe hydrogenase, large subunit | 2.66 | 2.40 |
| hydC | Ni-Fe hydrogenase | 1.90 | 2.12 |
| frdA | Fumarate reductase, flavoprotein subunit | 2.51 | 2.17 |
| frdB | Fumarate reductase, iron-sulfur subunit | 1.51 | 1.80 |
| frdC | Fumarate reductase, cytochrome b-556 subunit | 2.43 | 2.63 |
| Cj0414 | Gluconate dehydrogenase, oxidoreductase subunit | 2.18 | 2.60 |
| Cj0415 | Gluconate dehydrogenase, GMCa oxidoreductase subunit | 1.78 | 2.19 |
| Cj1476c | Pyruvate:flavodoxin oxidoreductase PFOR | 2.40 | 2.34 |
| Cj0265c | Putative c-type cytochrome | 1.12 | 2.40 |
| Cj0037c | Putative c-type cytochrome | 2.36 | 3.13 |
| Cj1153 | Putative c-type cytochrome | 1.49 | 2.09 |
| Cj0074c | l-Lactate oxidase, iron-sulfur protein | 1.37 | 1.80 |
| Cj0075c | l-Lactate oxidase, oxidoreductase iron-sulfur subunit | 1.47 | 2.01 |
| Cj1514c | Hypothetical protein FdhM | 1.69 | 1.60 |
| Cj0264c | Molybdopterin-containing oxidoreductase TorA | −1.05 | 2.06 |
| Cj0559 | Flavodoxin:quinone reductase | 1.38 | 1.75 |
| oorA | 2-Oxoglutarate-acceptor oxidase reductase, subunit OorA | 2.03 | 1.68 |
| oorB | 2-Oxoglutarate-acceptor oxidase reductase, subunit OorB | 1.57 | 1.45 |
| acnB | Bifunctional aconitate hydratase 2/2-methylisocitrate dehydratase | 1.09 | 1.78 |
| aspA | Aspartate ammonia-lyase | 1.58 | 1.19 |
| dcuA | Anaerobic C4-dicarboxylate transporter | 2.32 | 1.64 |
| glnA | Glutamine synthase | 1.16 | 2.00 |
| IlvD | Dihydroxy acid dehydratase | 1.37 | 1.55 |
| gapA | Glyceraldehyde-3-phosphate dehydrogenase | −1.94 | −1.34 |
| fba | Fructose-bisphosphate aldolase | −1.36 | −1.62 |
| atpE | c-ring subunit of the FoF1 ATP synthase | 1.52 | 1.53 |
| atpA | FoF1 ATP synthase, subunit alpha | −1.17 | −1.55 |
| atpD | FoF1 ATP synthase, subunit beta | −1.35 | −1.80 |
| atpG | FoF1 ATP synthase, subunit gamma | −1.18 | −1.65 |
| Ribosome assembly, modification, and translation | |||
| rplB | 50S ribosomal protein L2 | −1.83 | −1.71 |
| rplE | 50S ribosomal protein L5 | −1.26 | −1.52 |
| rpsC | 30S ribosomal protein S3 | −1.58 | −1.58 |
| rpsD | 30S ribosomal protein S4 | −1.44 | −1.72 |
| rpsE | 30S ribosomal protein S5 | −1.37 | −1.51 |
| rpsG | 30S ribosomal protein S7 | −1.28 | −1.59 |
| rpsQ | 30S ribosomal protein S17 | −1.42 | −1.64 |
| ksgA | Dimethyladenosine transferase | −1.28 | −1.96 |
| Cj1710c | Putative metallo-beta-lactamase family protein | −1.46 | −2.14 |
| fusA | Elongation factor G | −1.18 | −1.70 |
| tuf | Elongation factor Tu | −1.12 | −2.09 |
| Miscellaneous | |||
| accC | Biotin carboxylase | −1.57 | −2.05 |
| cft | Ferritin | −2.13 | 1.03 |
| chuA | Hemin uptake system, outer membrane receptor | −1.50 | −1.34 |
| chuC | Putative hemin uptake system ATP binding protein | −1.30 | −1.77 |
| chuD | Putative hemin uptake system periplasmic hemin binding protein | −1.47 | −1.52 |
| Cj0391c | Hypothetical protein | −2.19 | −1.36 |
| Cj0628 | Putative lipoprotein | 1.32 | 1.85 |
| Cj0700 | Hypothetical protein | −1.53 | −1.28 |
| Cj0772c | Putative NlpA family lipoprotein | −1.59 | 1.19 |
| Cj0937 | Membrane transport protein | 1.68 | 1.11 |
| Cj1170c | Omp50 | −1.84 | −2.10 |
| Cj1324 | Hypothetical protein | −1.33 | −1.52 |
| Cj1383c | Hypothetical protein | −1.30 | −1.71 |
| Cj1429c | Hypothetical protein | −1.37 | −1.51 |
| Cj1516 | Putative periplasmic oxidoreductase | −1.70 | −1.24 |
| Cj1564 | Putative methyl-accepting chemotaxis signal transduction protein | −1.66 | −1.15 |
| Cj1725 | Putative periplasmic protein | −1.89 | −1.22 |
| fabF | 3-Oxoacyl-(acyl carrier protein) synthase II | −1.05 | −1.73 |
| flaC | Flagellin | −1.86 | 1.23 |
| nrdA | Ribonucleotide-diphosphate reductase subunit alpha | −1.35 | −1.65 |
| peb4 | Major antigenic peptide PEB4 | −1.77 | −1.78 |
| purB | Adenylosuccinate lyase | −1.07 | −1.59 |
| thiC | Thiamine biosynthesis protein | 1.37 | 1.53 |
GMC, oxidoreductase:glucose-methanol-choline oxidoreductase.
The most highly induced genes were involved in the heat shock response. The clpB gene encodes an ATP-dependent chaperone/disaggregase. In Escherichia coli, ClpB works with the DnaK-GrpE-DnaJ complex to disaggregate misfolded proteins by refolding them (38–40). In C. jejuni, the transcript level of clpB has been found to be induced following a temperature shift from 37°C to 42°C (41) and is known to be regulated by the heat shock regulator HspR (42). Six other genes encoding heat shock proteins are upregulated by BITC treatment. Among them, the genes encoding the heat shock proteins HrcA, DnaK, and GrpE have been suggested to be cotranscribed with and regulated by HspR via the HspR-associated inverted repeat (HAIR) binding site of the hrcA promoter (42). DnaK is a chaperone required for the ClpB-mediated refolding of aggregated proteins (40). GrpE is a cochaperone required for DnaK activity and a nucleotide exchange factor that allows the release of ADP after ATP hydrolysis by DnaK (43). HrcA is a heat-inducible transcriptional repressor responsible for the regulation of the expression of the GroES/EL chaperones (42).
BITC treatment increased the expression level of the groES gene, whereas the groEL gene was also upregulated by BITC treatment but did not appear in our final results due to an expression level increase just under our 1.5-fold-change cutoff value. Both genes are cotranscribed in an operonic structure in Helicobacter pylori (44) and are colocalized in C. jejuni. The GroES-GroEL complex is a chaperone involved in protein folding during the stress response and protein synthesis (45, 46).
In E. coli, cbpA encodes a DnaJ-like protein acting as a cochaperone with DnaK and was found to be upregulated in our study (47). Furthermore, cbpA was suggested to be the major DnaJ protein in C. jejuni (42). The Cj0760 gene (1.78- and 1.92-fold changes at 10 and 15 min, respectively) is located downstream of the hrcA-grpE-dnaK locus and has been shown to be a part of the HspR regulon (42).
HrcA is also a transcriptional repressor of genes encoding heat shock proteins. The hcrA gene is upregulated by BITC exposure, but the overexpression of the heat shock response can also be explained by posttranscriptional regulation by HcrA. The folding of HcrA into an active conformation needs the assistance of GroEL/GroES, chaperones which are titrated away by the increased levels of unfolded proteins (48).
Another interesting set of upregulated genes encodes several proteins involved in the oxidative stress response, including sodB, rrc, and Cj0379c. The sodB gene encodes the sole superoxide dismutase in C. jejuni, which is crucial for oxidative stress defenses and host colonization (49, 50). Rrc is a rubredoxin oxidoreductase/rubrerythrin-like protein, which is very sensitive to oxidative stress (51). Rubredoxin and rubrerythrin are involved in the detoxification of reactive oxygen species (ROS) in bacteria (52–54). The Cj0379c gene codes for a putative sulfite oxidase, which is involved in the detoxification of reactive nitrogen species (55).
In addition, genes involved in iron-sulfur cluster homeostasis were upregulated by ITC treatment. The Cj0239c gene codes for a NifU protein homologue and is potentially involved in the assembly of iron-sulfur clusters (56), which are highly sensitive to damage by oxygen radicals. The thioredoxin-encoding gene trxA, induced by BITC, is also involved in Fe-S cluster assembly (57) and in many other cellular processes, such as oxidative stress responses and the biosynthesis of macromolecules. In contrast, the dsbI gene, involved in the formation of disulfide bonds of periplasmic proteins, is downregulated after BITC exposure.
Exposure to BITC also induced the expression of a large number of genes involved in electron transport and energy metabolism. Sixteen upregulated genes are involved in the use of alternative electron donors (hydrogen, succinate, gluconate, and lactate) and acceptors (fumarate, trimethylamine N-oxide [TMAO]/dimethyl sulfoxide [DMSO], and nitrate): hydABC (Ni-Fe hydrogenase), frdABC (fumarate reductase), Cj0414-Cj0415 (gluconate dehydrogenase), napA (nitrate reductase), napG (quinol dehydrogenase), Cj0074-Cj0075c (l-lactate oxidase), Cj1514c (FdhM, required for formate dehydrogenase activity), Cj0264c (torA), and the putative cytochromes Cj0037c, Cj1153, and Cj0265c (55, 58–60).
Treatment with BITC also induced the expression of genes involved in the tricarboxylic acid (TCA) cycle and other metabolic pathways. All these gene products mediate a link between the central carbon metabolism and respiration or redox homeostasis. The upregulated acnB and ilvD genes encode metabolic enzymes, which have been reported to be inactivated by oxidation (61–63). AspA and DcuA play a crucial role in amino acid utilization and fumarate respiration during oxygen-limited growth (64).
Cj0559 is a homologue of fqrB in H. pylori (65), which encodes a flavodoxin:quinone reductase responsible for flavodoxin oxidation coupled to NADPH production, in association with the pyruvate:flavodoxin oxidoreductase (PFOR) (encoded by Cj1476c). Interestingly, flavodoxin in C. jejuni is an electron acceptor for 2-oxoglutarate oxidoreductase (OOR), involved in the tricarboxylic acid cycle, but is also an electron donor for the complex I-mediated respiratory pathway and therefore functions as an intermediate between central carbon metabolism and the electron transport chain (66). The PFOR and OOR enzymes are oxygen labile and therefore contribute to the microaerophilic phenotype of C. jejuni (67).
Among the downregulated genes, two are involved in the gluconeogenesis pathway (gapA and fba). The downregulation of gluconeogenesis while other genes involved in energy metabolism are upregulated suggests that under these conditions, the metabolism is oriented toward energy production via the tricarboxylic acid cycle.
The atpE gene is slightly upregulated after 10 min of BITC exposure (fold change, 1.52), while the atpAGD genes are downregulated after 15 min of treatment. The atpE gene codes for the c-ring subunit of the FoF1 ATP synthase, encoded by atpAGDEFF′GH. The atpE gene is usually more expressed than the other atp genes (68, 69).
BITC treatment downregulates the expression of 11 genes involved in ribosome assembly (rpsCEGQD and rplEB), ribosome modification (ksgA and Cj1710c), and translation processes (the elongation factors fusA and tuf). Downregulation of genes involved in ribosome biosynthesis and translation may indicate a translational pause after BITC exposure, as has been observed with several stress responses (70, 71).
BITC-mediated aggregation of proteins.
The transcriptomic response of C. jejuni to BITC indicated the upregulation of genes encoding heat shock proteins, including chaperones. Therefore, we investigated the aggregation of proteins in bacterial cells following BITC treatment. The effect of BITC on protein aggregation was assayed after 15 min, 1 h, and 2 h of treatment. Aggregated proteins were separated from soluble and membrane proteins prior to electrophoresis (Fig. 2). The relative amount of aggregated protein compared to the total protein content of the same sample was calculated (reported as the percentage of total proteins in Fig. 2). The amount of aggregated proteins increased noticeably after 2 h of exposure to 2 mg liter−1 or 5 mg liter−1 BITC (Fig. 2). Levels of aggregated proteins were below our detection limit following 15 min or 1 h of treatment.
Fig 2.
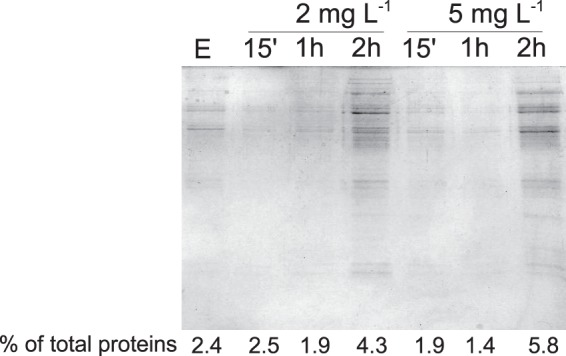
Protein aggregation in C. jejuni treated with BITC. Cells were grown to mid-log phase in DMEM–20 mM pyruvate at 37°C under microaerobic conditions before the addition of 2 mg liter−1 BITC or 5 mg liter−1 BITC. Alternatively, cells were exposed to ethanol (E) for 2 h as a negative control. Aggregated proteins were isolated by using a method described previously by Tomoyasu et al. (36) and then analyzed on a 12% (wt/vol)-acrylamide SDS-PAGE gel, followed by Coomassie staining (37). Aggregated proteins and total proteins were quantified with a BCA assay (Thermo Scientific).
Effects on ATP content.
As BITC induced the expression of several genes involved in energy metabolism, the ATP content of C. jejuni cells treated with 2 mg liter−1 of BITC, 5 mg liter−1 of BITC, or absolute ethanol (control) was determined over a 2-h incubation period. The results were expressed as a percentage of the luminescence above the luminescence of the ethanol control (Fig. 3). After 15 min of treatment, the relative ATP concentration was 8 to 19% higher in BITC-treated cultures than in control cultures, in a dose-dependent manner. The ATP concentration continued to increase in BITC-treated cultures from 15 min to 90 min of exposure. After 90 min of exposure, the ATP concentrations in BITC-treated cultures were 17% (2 mg liter−1 BITC) and 45% (5 mg liter−1) higher than those in the control cultures. Interestingly, after 120 min of BITC treatment, when noticeable cell death occurs (Fig. 1), the ATP content of BITC-treated cells dropped to the ATP contents of control cultures.
Fig 3.
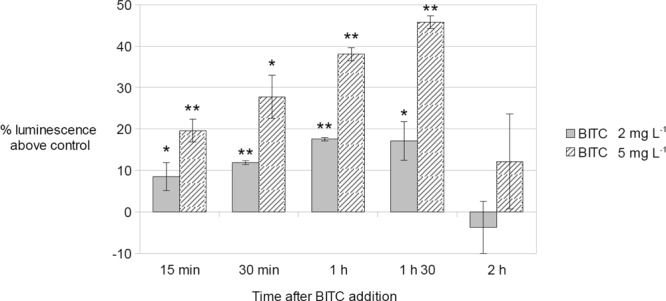
C. jejuni ATP content after addition of 2 mg liter−1 BITC or 5 mg liter−1 BITC. C. jejuni NCTC11168 cells were grown to mid-log phase in DMEM–20 mM pyruvate. BITC (2 mg liter−1 or 5 mg liter−1) or ethanol (negative control) was added to the cultures. The BacTiterGlo kit (Promega) was used to assay the ATP content at 15 min, 30 min, 1 h, 1 h 30 min, and 2 h after BITC or ethanol addition. Results are expressed as a percentage of the luminescence of the treated sample above the luminescence of the ethanol-treated control samples. The values were measured three times for each experiment, and the experiment was conducted in triplicate assays. The data consist of the means of all 9 measurements, with standard deviations. Statistical analysis was performed by using a Student t test with a significance value of 0.05. ∗, P < 0.05; ∗∗, P < 0.01.
Effects on oxygen consumption.
To further investigate the impact of BITC on electron transport pathways, high-resolution respirometry was used to examine the effect of 2 mg liter−1 of BITC on C. jejuni O2 consumption (Fig. 4). The addition of ethanol (solvent control) did not affect C. jejuni O2 consumption, which was maintained at high rates even at near-anoxic O2 concentrations. Importantly, the addition of BITC was found to progressively affect C. jejuni O2 consumption over time. Inhibition reached 30% and 50% after 15 and 25 min of treatment, respectively. Despite the strong inhibition of O2 consumption, BITC-treated cells retained a high apparent O2 affinity, as revealed by the respiration rate values at near-anoxic O2 concentrations. A similar inhibition pattern was observed with 5 mg liter−1 BITC, but the progressive impairment of O2 consumption was more rapid (50% inhibition after a 10- to 12-min exposure to BITC [data not shown]). These data also illustrate the drastic and rapid (but progressive) effects of BITC on C. jejuni cells.
Fig 4.
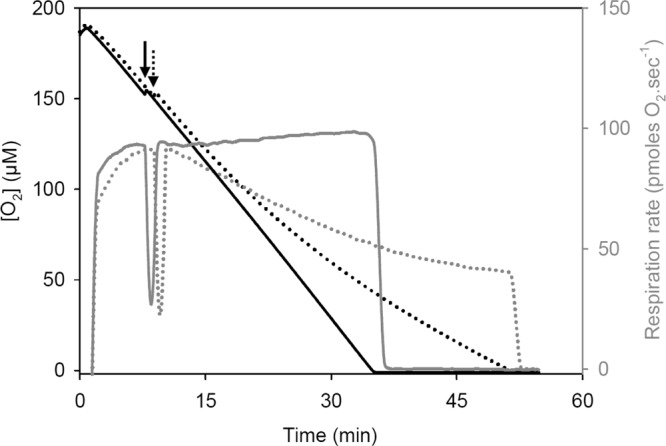
Typical high-resolution respirometry assay showing the effect of 2 mg liter−1 BITC on C. jejuni O2 respiration. Mid-log-phase cells were quickly air saturated and transferred into the two Oxygraph chambers (final OD600 = 0.1). O2 concentrations (black lines) and instantaneous respiration rates (gray lines) were monitored simultaneously. The arrows indicate the addition of ethanol (negative control [solid lines]) or a BITC-ethanol solution (dotted lines) to each chamber. Similar inhibition patterns were observed when cells were resuspended in fresh DMEM supplemented or not with 20 mM pyruvate (data not shown).
DISCUSSION
In order to gain a better understanding of the early response of C. jejuni to BITC, we analyzed the transcriptomic profile of C. jejuni 10 min and 15 min following treatment with 2 mg liter−1 BITC. While this concentration is lethal, it impacts Campylobacter viability only following at least 2 h of exposure. Therefore, RNA was extracted from viable, living cells facing a relevant challenge.
The increased expression levels of several known heat shock genes in response to BITC treatment indicate that BITC triggers a heat shock response in C. jejuni. More precisely, BITC-induced genes are involved mainly in misfolded-protein repair. The upregulation of clpB, dnaK, grpE, cbpA, and groES by C. jejuni exposed to BITC reveals the need for protein refolding. In E. coli, aggregated proteins are resolubilized in the presence of ATP by the ClpB chaperone-disaggregase, helped by the DnaK-GrpE-DnaJ system (72). In the case of C. jejuni, the complex can include CbpA, a DnaJ-like protein which has been suggested to be the major DnaJ protein in C. jejuni (42, 47). The GroES-GroEL complex can similarly contribute to the refolding of denatured proteins (46). Additionally, all the heat shock genes induced by BITC treatment are part of the HrcA and HspR regulons, as defined by Holmes et al. (42). Despite a global upregulation of every member of the HspR regulon, the hspR transcript was not differentially expressed in our study, consistent with the findings of Holmes et al. (42), who concluded that the activity of the repressor HspR is controlled posttranscriptionally. DnaK is required to stabilize HspR binding to its target DNA to repress its regulon (73). In response to heat shock or similar stresses, such as ITC exposure, unfolded proteins compete with HspR for DnaK, resulting in decreased HspR activity. Therefore, we hypothesize that BITC treatment provokes protein unfolding, leading to the derepression of HspR-regulated genes through titration of the corepressor DnaK and of HcrA via the titration of GroEL/ES.
Misfolded proteins often form insoluble aggregates that can be isolated from soluble and membrane proteins by using a method developed previously by Tomoyasu et al. (36). As expected, a 2-h BITC treatment induced protein aggregation, which is in agreement with the upregulation of heat shock response genes. Specifically, genes encoding chaperones were upregulated within 10 min following exposure to BITC. While we were not able to isolate protein aggregates following 15 min or 1 h of exposure, the activity of these chaperones could have been sufficient to overcome the aggregation of proteins at early stages, so aggregated proteins in quantities large enough to be isolated may appear only after longer exposure times.
Interestingly, the whole-transcriptome profile of P. aeruginosa exposed to iberin conducted by Jakobsen et al. (25) showed a similar upregulation of groEL and dnaK. While no other heat shock proteins were induced in this study, the different methods and the chosen cutoff values for analysis could explain the discrepancies between the two studies. In eukaryotes, the sulforaphane analogue 6-methylsulfinyl hexyl isothiocyanate provokes transthiocarbamoylation of proteins, including the heat shock protein HSP90β, which results in the induction of the heat shock response (74) and the stimulation of proteasome activity (75). This shared heat shock induction in both prokaryotic and eukaryotic cells in response to ITCs is likely caused by the molecule's intrinsic chemistry. The electrophilic properties of ITCs enable them to react with cellular sulfhydryl groups, including cysteines in proteins, leading to a modification of the protein structure or activity. ITC can also react with amine groups but with a lesser affinity (76). Not all cysteines are capable of reacting with ITCs: hydrophobicity and steric hindrance influence the reactivity of cysteines toward electrophilic compounds (74, 77). We demonstrate in our study that BITC provoked sufficient protein modification to elicit the heat shock response and to induce noticeable protein aggregation in C. jejuni. The induction of the heat shock response was confirmed by the detection of aggregated proteins.
Enzymes harboring reactive cysteines or cysteines involved in metal coordination are putative targets of ITCs. Motifs such as CXXC, CXXS, or CXXT are commonly found in disulfide reductases or enzymes with metal coordination centers (78, 79). The oxygen requirement and oxygen sensitivity influence the number of enzymes with CXXC/CXXS/CXXT motifs; these motifs are found significantly more frequently in the proteins of microaerobes and anaerobes than in aerobic bacteria (80, 81). Indeed, approximately 40% of the proteins of C. jejuni contain at least one of these motifs (80); therefore, both the microaerophilic phenotype of C. jejuni and the large number of cysteine-based motifs encoded by its genome can explain the higher ITC sensitivity of this organism than of other prokaryotes such as P. aeruginosa (29; C. Baysse, unpublished data). Given the ability of ITCs to react with thiol groups in cysteine residues and the large number of cysteine-containing proteins in the genome of C. jejuni, the protein modifications caused by ITCs are likely aspecific and would lead to pleiotropic physiological effects.
Our microarray study indicates that the Cj0239c gene (encoding a NifU protein homologue) and the trxA gene are induced by BITC. In bacteria and eukaryotes, thioredoxins maintain the redox balance of proteins by reducing disulfide bonds, therefore playing a pleiotropic role. The upregulation of Cj0239c and trxA suggests the need for the reconstruction of Fe-S clusters and for the reduction of oxidized proteins. However, the activity of thioredoxins and other disulfide reductases relies on reactive cysteines that are potential targets for ITC inactivation. In E. coli, the inactivation of the activity of thioredoxin reductase by AITC was previously shown in vitro (26). Thus, the very same enzymes that are responsible for the maintenance of the redox status of proteins are inactivated by ITCs, which disrupt the oxidative state of thiols present within proteins.
Several major physiological processes, such as the tricarboxylic acid (TCA) cycle, respiration, oxidative stress defenses, or amino acid metabolism, involve redox reactions catalyzed by enzymes harboring reactive cysteines or iron-sulfur clusters. The BITC-mediated inactivation of proteins involved in these pathways may disrupt crucial metabolic functions and contribute to BITC-mediated killing. In mitochondria, ITCs inhibit O2 respiration, leading to reactive oxygen species (ROS) production and thus inducing an oxidative stress response (82–84). Here, we showed that BITC progressively impairs oxygen consumption in C. jejuni and induces the upregulation of the superoxide dismutase gene sodB (cluster A; 2.65- and 2.69-fold changes at 10 and 15 min after treatment), suggesting a superoxide ion-induced oxidative stress response in BITC-treated cells. Superoxide stress can also be generated during metabolic activity (85). The microaerophilic nature of C. jejuni contributes to its sensitivity to oxidative stress and ITCs. The genes encoding the rubredoxin oxidoreductase/rubrerythrin Rrc and the putative sulfide oxidase Cj0379c were also upregulated by BITC. These two proteins are potentially involved in the detoxification of reactive oxygen and nitrogen species, respectively. The induction of the expression of rrc and Cj0379c, along with sodB, corroborates the hypothesis of oxidative stress being induced by BITC.
The repair of misfolded proteins by chaperones requires a large amount of energy provided as ATP (86). According to our results, genes involved in energy production (TCA cycle and respiration) were induced by BITC, and the ATP content of cells was significantly increased within 15 min following BITC exposure. The increased metabolic activity may result from the increased energy demands of the stress responses but may also contribute to the generation of ROS. In addition, several key proteins involved in energy production are iron-sulfur cluster-containing hydratases or oxidoreductases, known to be sensitive to the oxidative damage induced by BITC exposure (61, 87). The inactivation of iron-sulfur-containing proteins may in fact hamper the metabolic effort needed to respond to BITC-triggered protein aggregation.
Further evidence of the suppression of oxygen use in response to BITC is further supported by shifts in the expression of genes involved in the use of alternative electron donors (hydrogen, succinate, gluconate, and lactate) and acceptors (fumarate, trimethylamine N-oxide [TMAO]/DMSO, and nitrate). This suggests that under these conditions, the cell drives the electron transport pathways toward non-oxygen-dependent respiration. Indeed, none of the complex I genes (nuo operon) and neither of the two oxygen-accepting terminal oxidases (the cytochrome cb′-type oxidase ccoNOQP and the cyanide-resistant oxidase cioAB) were induced by BITC, while genes related to the use of nitrate (napAG) and fumarate (frdABC) as alternative electron acceptors were induced. Similar shifts in metabolism were observed for an E. coli mutant strain deleted for the three terminal cytochrome oxidase genes and therefore unable to use oxygen for oxidative phosphorylation and ATP synthesis. In this mutant, genes for anaerobic respiration were induced even under oxic conditions, as were glycolysis-related genes, probably to sustain ATP generation through substrate-level phosphorylation (88).
Reduced oxygen consumption following BITC treatment was also observed previously in Saccharomyces cerevisiae, Candida krusei, and Candida utilis by Kojima and Ogawa (89). In C. jejuni, the upregulation of metabolic genes and those involved in alternative electron transport pathways, as well as the potential inactivation of some energy-consuming enzymatic processes by ITC conjugation, may help to account for the higher levels of ATP in BITC-treated cells than in untreated cells despite the inhibition of oxygen consumption. In contrast, in E. coli, where oxygen is the preferential electron acceptor when grown aerobically, treatment with 2-(4-hydroxyphenyl)ethyl ITC decreased the ATP cellular content (90). The switch from oxygenic respiration to alternative electron transport pathways in C. jejuni may also help to prevent further formation of ROS by components of the electron transport chain.
From this, we conclude that BITC affects the functions of a large number of thiol-containing proteins in C. jejuni, leading to pleiotropic effects on metabolism, electron transport, and stress responses.
Taken together, our results allow us to propose a model for the mechanism of the antibacterial effect of BITC on C. jejuni. The electrophilic BITC targets the thiols in proteins, leading to a loss of protein structure and function and thus displaying pleiotropic effects. A heat shock-like response allows chaperones to cope with the protein aggregates resulting from BITC conjugation with cysteine thiols or amine groups. The metabolic activity is shifted due to the inhibition of O2 consumption in an attempt to maintain the energy needed for the stress responses and survival. Finally, disruption of the activity of disulfide reductases and electron transport provokes the generation of ROS, eliciting an oxidative stress response.
In eukaryotes and some prokaryotes, glutathione maintains the redox balance but can also form conjugates with ITC, either spontaneously or by the action of the enzyme glutathione S-transferase, thus limiting the interaction of ITC with other thiols in proteins. In mammals, the enzyme glutathione S-transferase forces the conjugation of ITC with glutathione to form soluble glutathione-ITC conjugates, which are then metabolized through the mercapturic acid pathway and excreted in the urine as S-(N-acetyl)cysteine conjugates (12). In the cyanobacteria Thermosynechococcus elongatus and Synechococcus elongatus, glutathione S-transferase of the chi class can catalyze the conjugation of ITC to glutathione at high rates (27). Additionally, a gene encoding a putative glutathione S-transferase is induced by iberin in P. aeruginosa (25). However, C. jejuni does not produce glutathione or glutathione S-transferases. Additionally, the inability of C. jejuni to synthesize glutathione may increase its sensitivity, since glutathione plays a known protective role toward ITCs. To date, no detoxification pathways have been identified in C. jejuni, and the full characterization of potential pathways of ITC resistance in other bacteria remains incomplete.
To date, another probable mechanism of resistance to ITC is encoded by the sax operon but was described only for Pseudomonas syringae by Fan et al. (23). No homologues of the sax genes were found in C. jejuni. Our previous study revealed that the antibiotic resistance profiles of 24 C. jejuni isolates did not correlate with their sensitivity to ITCs (29). However, it cannot be excluded that some antibiotic resistance mechanisms, such as efflux pumps, may be effective in C. jejuni to detoxify ITC. Indeed, in P. aeruginosa, iberin induces the expression of the efflux system MexEF-OprN (25), but none of the genes encoding putative efflux pumps in C. jejuni were induced in our transcriptomic profile. Therefore, in the absence of an identified detoxification mechanism, we may consider that the reaction of ITC with proteins in C. jejuni would be limited by the rate of ITC decomposition (91) and the efficiency of formation of thiourea conjugates.
To our knowledge, the use of BITC as a food preservative has not yet been assessed, despite the greater antibacterial effect of BITC than of AITC on C. jejuni and other bacteria (29, 92) and its anticancerous properties (93–95). The concentrations of BITC needed to inhibit C. jejuni (8.37 μmol liter−1) and human cancer cell lines (2.5 to 10 μmol liter−1) are comparable (29, 96, 97). Although the pharmacokinetics of BITC in humans is unknown to our knowledge, studies on its structural analogue phenylethyl isothiocyanate suggest that such concentrations can result from dietary intake in humans and, thus, that the level of ITC dietary intake is supposedly safe (97–100). However, genotoxic effects of BITC at doses below 5 μg ml−1 were reported in in vitro cell culture assays, while a weaker effect was observed in laboratory rodents in vivo (100). Nevertheless, it was demonstrated that BITC was detoxified by living organisms and that the doses of BITC reported to cause DNA damage in rodents largely exceeded the dietary exposure levels (100). However, experiments to determine the concentrations of BITC required for food protection and the genotoxicity of these BITC levels in animals remain to be done.
Given the sensitivity of C. jejuni toward ITCs in vitro and the absence of an identified detoxification mechanism, ITCs are promising antimicrobial compounds for the control of this pathogen in food products. ITC-based strategies to control the contamination of food products by pathogens such as Salmonella enterica serovar Typhimurium, Listeria monocytogenes, or Staphylococcus aureus have already proven their efficiency (11, 13–16, 101). The future development of an isothiocyanate-based approach to control C. jejuni contamination of food products is an exciting lead that may reduce human exposure to C. jejuni-contaminated foods.
ACKNOWLEDGMENTS
This work was supported by an ARED doctoral training grant from the Région Bretagne and by a doctoral mobility grant from the VAS graduate school (to Virginie Dufour). This work was supported by a grant from the Canadian Institutes of Health Research (MOP number 84224) to Alain Stintzi.
We thank Alain Sarniguet (INRA) for the use of the luminometer.
Footnotes
Published ahead of print 6 September 2013
REFERENCES
- 1.EFSA 2012. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2010. EFSA J. 10:2597–3039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blaser MJ. 1997. Epidemiologic and clinical features of Campylobacter jejuni infections. J. Infect. Dis. 176(Suppl 2):S103–S105 [DOI] [PubMed] [Google Scholar]
- 3.Nachamkin I, Allos BM, Ho T. 1998. Campylobacter species and Guillain-Barré syndrome. Clin. Microbiol. Rev. 11:555–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu J, Zhang Y, Hua X, Hou J, Jiang Y. 2006. Antibiotic resistance in Campylobacter. Rev. Med. Microbiol. 17:107–112 [Google Scholar]
- 5.Marshall BM, Levy SB. 2011. Food animals and antimicrobials: impacts on human health. Clin. Microbiol. Rev. 24:718–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Savoia D. 2012. Plant-derived antimicrobial compounds: alternatives to antibiotics. Future Microbiol. 7:979–990 [DOI] [PubMed] [Google Scholar]
- 7.Aires A, Mota VR, Saavedra MJ, Rosa EAS, Bennett RN. 2009. The antimicrobial effects of glucosinolates and their respective enzymatic hydrolysis products on bacteria isolated from the human intestinal tract. J. Appl. Microbiol. 106:2086–2095 [DOI] [PubMed] [Google Scholar]
- 8.Aires A, Mota VR, Saavedra MJ, Monteiro AA, Simoes M, Rosa EAS, Bennett RN. 2009. Initial in vitro evaluations of the antibacterial activities of glucosinolate enzymatic hydrolysis products against plant pathogenic bacteria. J. Appl. Microbiol. 106:2096–2105 [DOI] [PubMed] [Google Scholar]
- 9.Sofrata A, Santangelo EM, Azeem M, Borg-Karlson AK, Gustafsson A, Putsep K. 2011. Benzyl isothiocyanate, a major component from the roots of Salvadora persica is highly active against Gram-negative bacteria. PLoS One 6:e23045. 10.1371/journal.pone.0023045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aguilar F, Dusemund B, Galtier P, Gilbert J, Gott DM, Grilli S, Gürtler R, König J, Lambré C, Larsen J-C, Leblanc J-C, Mortensen A, Parent-Massin D, Pratt I, Rietjens IMCM, Stankovic I, Tobback P, Verguieva TRAW. 2010. EFSA panel on food additives and nutrient sources added to food (ANS): scientific opinion on the safety of allyl isothiocyanate for the proposed uses as a food additive. EFSA J. 8:1943–1983 [Google Scholar]
- 11.Nadarajah D, Han JH, Holley RA. 2005. Use of mustard flour to inactivate Escherichia coli O157:H7 in ground beef under nitrogen flushed packaging. Int. J. Food Microbiol. 99:257–267 [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y. 2012. The molecular basis that unifies the metabolism, cellular uptake and chemopreventive activities of dietary isothiocyanates. Carcinogenesis 33:2–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shin J, Harte B, Ryser E, Selke S. 2010. Active packaging of fresh chicken breast, with allyl isothiocyanate (AITC) in combination with modified atmosphere packaging (MAP) to control the growth of pathogens. J. Food Sci. 75:M65–M71. 10.1111/j.1750-3841.2009.01465.x [DOI] [PubMed] [Google Scholar]
- 14.Wang SY, Chen C-T, Yin J-J. 2010. Effect of allyl isothiocyanate on antioxidants and fruit decay of blueberries. Food Chem. 120:199–204 [Google Scholar]
- 15.Chen W, Jin TZ, Gurtler JB, Geveke DJ, Fan X. 2012. Inactivation of Salmonella on whole cantaloupe by application of an antimicrobial coating containing chitosan and allyl isothiocyanate. Int. J. Food Microbiol. 155:165–170 [DOI] [PubMed] [Google Scholar]
- 16.Ko JA, Kim WY, Park HJ. 2012. Effects of microencapsulated allyl isothiocyanate (AITC) on the extension of the shelf-life of kimchi. Int. J. Food Microbiol. 153:92–98 [DOI] [PubMed] [Google Scholar]
- 17.Fahey JW, Wehage SL, Holtzclaw WD, Kensler TW, Egner PA, Shapiro TA, Talalay P. 2012. Protection of humans by plant glucosinolates: efficiency of conversion of glucosinolates to isothiocyanates by the gastrointestinal microflora. Cancer Prev. Res. (Phila.) 5:603–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y, Li J, Tang L. 2005. Cancer-preventive isothiocyanates: dichotomous modulators of oxidative stress. Free Radic. Biol. Med. 38:70–77 [DOI] [PubMed] [Google Scholar]
- 19.Musk SR, Johnson IT. 1993. Allyl isothiocyanate is selectively toxic to transformed cells of the human colorectal tumour line HT29. Carcinogenesis 14:2079–2083 [DOI] [PubMed] [Google Scholar]
- 20.Gamet-Payrastre L, Lumeau S, Gasc N, Cassar G, Rollin P, Tulliez J. 1998. Selective cytostatic and cytotoxic effects of glucosinolates hydrolysis products on human colon cancer cells in vitro. Anticancer Drugs 9:141–148 [DOI] [PubMed] [Google Scholar]
- 21.Kassie F, Knasmüller S. 2000. Genotoxic effects of allyl isothiocyanate (AITC) and phenethyl isothiocyanate (PEITC). Chem. Biol. Interact. 127:163–180 [DOI] [PubMed] [Google Scholar]
- 22.Ahn E, Kim J, Shin D. 2001. Antimicrobial effects of allyl isothiocyanate on several microorganisms. Korean J. Food Sci. Technol. 31:206–211 [Google Scholar]
- 23.Fan J, Crooks C, Creissen G, Hill L, Fairhurst S, Doerner P, Lamb C. 2011. Pseudomonas sax genes overcome aliphatic isothiocyanate-mediated non-host resistance in Arabidopsis. Science 331:1185–1188 [DOI] [PubMed] [Google Scholar]
- 24.Ganin H, Rayo J, Amara N, Levy N, Krief P, Meijler MM. 2013. Sulforaphane and erucin, natural isothiocyanates from broccoli, inhibit bacterial quorum sensing. Med. Chem. Commun. 4:175–179. 10.1039/C2MD20196H [DOI] [Google Scholar]
- 25.Jakobsen TH, Bragason SK, Phipps RK, Christensen LD, Van Gennip M, Alhede M, Skindersoe M, Larsen TO, Hoiby N, Bjarnsholt T, Givskov M. 2012. Food as a source for quorum sensing inhibitors: iberin from horseradish revealed as a quorum sensing inhibitor of Pseudomonas aeruginosa. Appl. Environ. Microbiol. 78:2410–2421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luciano FB, Holley RA. 2009. Enzymatic inhibition by allyl isothiocyanate and factors affecting its antimicrobial action against Escherichia coli O157:H7. Int. J. Food Microbiol. 131:240–245 [DOI] [PubMed] [Google Scholar]
- 27.Wiktelius E, Stenberg G. 2007. Novel class of glutathione transferases from cyanobacteria exhibit high catalytic activities towards naturally occurring isothiocyanates. Biochem. J. 406:115–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kolm RH, Danielson UH, Zhang Y, Talalay P, Mannervik B. 1995. Isothiocyanates as substrates for human glutathione transferases: structure-activity studies. Biochem. J. 311(Part 2):453–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dufour V, Alazzam B, Ermel G, Thepaut M, Rossero A, Tresse O, Baysse C. 2012. Antimicrobial activities of isothiocyanates against Campylobacter jejuni isolates. Front. Cell. Infect. Microbiol. 2:53. 10.3389/fcimb.2012.00053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parkhill J, Wren BW, Mungall K, Ketley JM, Churcher C, Basham D, Chillingworth T, Davies RM, Feltwell T, Holroyd S, Jagels K, Karlyshev AV, Moule S, Pallen MJ, Penn CW, Quail MA, Rajandream MA, Rutherford KM, Van Vliet AH, Whitehead S, Barrell BG. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665–668 [DOI] [PubMed] [Google Scholar]
- 31.Davidson PM, Harrison MA. 2002. Resistance and adaptation to food antimicrobials, sanitizers, and other process controls. Food Technol. 56:69–78 [Google Scholar]
- 32.Palyada K, Threadgill D, Stintzi A. 2004. Iron acquisition and regulation in Campylobacter jejuni. J. Bacteriol. 186:4714–4729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stintzi A, Whitworth L. 2003. Investigation of the Campylobacter jejuni cold-shock response by global transcript profiling. Genome Lett. 2:18–27 [Google Scholar]
- 34.Hutter E, Unterluggauer H, Garedew A, Jansen-Dürr P, Gnaiger E. 2006. High-resolution respirometry—a modern tool in aging research. Exp. Gerontol. 41:103–109 [DOI] [PubMed] [Google Scholar]
- 35.Gnaiger E. 2001. Bioenergetics at low oxygen: dependence of respiration and phosphorylation on oxygen and adenosine diphosphate supply. Respir. Physiol. 128:277–297 [DOI] [PubMed] [Google Scholar]
- 36.Tomoyasu T, Mogk A, Langen H, Goloubinoff P, Bukau B. 2001. Genetic dissection of the roles of chaperones and proteases in protein folding and degradation in the Escherichia coli cytosol. Mol. Microbiol. 40:397–413 [DOI] [PubMed] [Google Scholar]
- 37.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 38.Parsell DA, Kowal AS, Singer MA, Lindquist S. 1994. Protein disaggregation mediated by heat-shock protein Hsp104. Nature 372:475–478 [DOI] [PubMed] [Google Scholar]
- 39.Weibezahn J, Tessarz P, Schlieker C, Zahn R, Maglica Z, Lee S, Zentgraf H, Weber-Ban EU, Dougan DA, Tsai FTF, Mogk A, Bukau B. 2004. Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell 119:653–665 [DOI] [PubMed] [Google Scholar]
- 40.Mogk A, Tomoyasu T, Goloubinoff P, Rüdiger S, Röder D, Langen H, Bukau B. 1999. Identification of thermolabile Escherichia coli proteins: prevention and reversion of aggregation by DnaK and ClpB. EMBO J. 18:6934–6949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stintzi A. 2003. Gene expression profile of Campylobacter jejuni in response to growth temperature variation. J. Bacteriol. 185:2009–2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holmes CW, Penn CW, Lund PA. 2010. The hrcA and hspR regulons of Campylobacter jejuni. Microbiology 156:158–166 [DOI] [PubMed] [Google Scholar]
- 43.Harrison C. 2003. GrpE, a nucleotide exchange factor for DnaK. Cell Stress Chaperones 8:218–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Homuth G, Domm S, Kleiner D, Schumann W. 2000. Transcriptional analysis of major heat shock genes of Helicobacter pylori. J. Bacteriol. 182:4257–4263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fayet O, Ziegelhoffer T, Georgopoulos C. 1989. The groES and groEL heat shock gene products of Escherichia coli are essential for bacterial growth at all temperatures. J. Bacteriol. 171:1379–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grantcharova V, Alm EJ, Baker D, Horwich AL. 2001. Mechanisms of protein folding. Curr. Opin. Struct. Biol. 11:70–82 [DOI] [PubMed] [Google Scholar]
- 47.Chae C, Sharma S, Hoskins JR, Wickner S. 2004. CbpA, a DnaJ homolog, is a DnaK co-chaperone, and its activity is modulated by CbpM. J. Biol. Chem. 279:33147–33153 [DOI] [PubMed] [Google Scholar]
- 48.Lund PA. 2001. Microbial molecular chaperones. Adv. Microb. Physiol. 44:93–140 [DOI] [PubMed] [Google Scholar]
- 49.Garénaux A, Ritz M, Jugiau F, Rama F, Federighi M, De Jonge R. 2009. Role of oxidative stress in C. jejuni inactivation during freeze-thaw treatment. Curr. Microbiol. 58:134–138 [DOI] [PubMed] [Google Scholar]
- 50.Palyada K, Sun Y-Q, Flint A, Butcher J, Naikare H, Stintzi A. 2009. Characterization of the oxidative stress stimulon and PerR regulon of Campylobacter jejuni. BMC Genomics 10:481. 10.1186/1471-2164-10-481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamasaki M, Igimi S, Katayama Y, Yamamoto S, Amano F. 2004. Identification of an oxidative stress-sensitive protein from Campylobacter jejuni, homologous to rubredoxin oxidoreductase/rubrerythrin. FEMS Microbiol. Lett. 235:57–63 [DOI] [PubMed] [Google Scholar]
- 52.Coulter ED, Kurtz DMJ. 2001. A role for rubredoxin in oxidative stress protection in Desulfovibrio vulgaris: catalytic electron transfer to rubrerythrin and two-iron superoxide reductase. Arch. Biochem. Biophys. 394:76–86 [DOI] [PubMed] [Google Scholar]
- 53.Kawasaki S, Sakai Y, Takahashi T, Suzuki I, Niimura Y. 2009. O2 and reactive oxygen species detoxification complex, composed of O2-responsive NADH:rubredoxin oxidoreductase-flavoprotein A2-desulfoferrodoxin operon enzymes, rubperoxin, and rubredoxin, in Clostridium acetobutylicum. Appl. Environ. Microbiol. 75:1021–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Riebe O, Fischer R-J, Wampler DA, Kurtz DMJ, Bahl H. 2009. Pathway for H2O2 and O2 detoxification in Clostridium acetobutylicum. Microbiology 155:16–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hitchcock A, Hall SJ, Myers JD, Mulholland F, Jones MA, Kelly DJ. 2010. Roles of the twin-arginine translocase and associated chaperones in the biogenesis of the electron transport chains of the human pathogen Campylobacter jejuni. Microbiology 156:2994–3010 [DOI] [PubMed] [Google Scholar]
- 56.Yuvaniyama P, Agar JN, Cash VL, Johnson MK, Dean DR. 2000. NifS-directed assembly of a transient [2Fe-2S] cluster within the NifU protein. Proc. Natl. Acad. Sci. U. S. A. 97:599–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ding H. 2005. Thioredoxin reductase system mediates iron binding in IscA and iron delivery for the iron-sulfur cluster assembly in IscU. J. Biol. Chem. 280:30432–30437 [DOI] [PubMed] [Google Scholar]
- 58.Pajaniappan M, Hall JE, Cawthraw SA, Newell DG, Gaynor EC, Fields JA, Rathbun KM, Agee WA, Burns CM, Hall SJ, Kelly DJ, Thompson SA. 2008. A temperature-regulated Campylobacter jejuni gluconate dehydrogenase is involved in respiration-dependent energy conservation and chicken colonization. Mol. Microbiol. 68:474–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thomas MT, Shepherd M, Poole RK, Van Vliet AHM, Kelly DJ, Pearson BM. 2011. Two respiratory enzyme systems in Campylobacter jejuni NCTC 11168 contribute to growth on L-lactate. Environ. Microbiol. 13:48–61 [DOI] [PubMed] [Google Scholar]
- 60.Sellars MJ, Hall SJ, Kelly DJ. 2002. Growth of Campylobacter jejuni supported by respiration of fumarate, nitrate, nitrite, trimethylamine-N-oxide, or dimethyl sulfoxide requires oxygen. J. Bacteriol. 184:4187–4196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Flint DH, Tuminello JF, Emptage MH. 1993. The inactivation of Fe-S cluster containing hydro-lyases by superoxide. J. Biol. Chem. 268:22369–22376 [PubMed] [Google Scholar]
- 62.Hyduke DR, Jarboe LR, Tran LM, Chou KJY, Liao JC. 2007. Integrated network analysis identifies nitric oxide response networks and dihydroxyacid dehydratase as a crucial target in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 104:8484–8489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kuo CF, Mashino T, Fridovich I. 1987. Alpha,beta-dihydroxyisovalerate dehydratase. A superoxide-sensitive enzyme. J. Biol. Chem. 262:4724–4727 [PubMed] [Google Scholar]
- 64.Guccione E, del Rocio Leon-Kempis M, Pearson RBM, Hitchin E, Mulholland F, Van Diemen PM, Stevens MP, Kelly DJ. 2008. Amino acid-dependent growth of Campylobacter jejuni: key roles for aspartase (AspA) under microaerobic and oxygen-limited conditions and identification of AspB (Cj0762), essential for growth on glutamate. Mol. Microbiol. 69:77–93 [DOI] [PubMed] [Google Scholar]
- 65.St Maurice M, Cremades N, Croxen MA, Sisson G, Sancho J, Hoffman PS. 2007. Flavodoxin:quinone reductase (FqrB): a redox partner of pyruvate:ferredoxin oxidoreductase that reversibly couples pyruvate oxidation to NADPH production in Helicobacter pylori and Campylobacter jejuni. J. Bacteriol. 189:4764–4773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weerakoon DR, Olson JW. 2008. The Campylobacter jejuni NADH:ubiquinone oxidoreductase (complex I) utilizes flavodoxin rather than NADH. J. Bacteriol. 190:915–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kelly DJ. 2008. Complexity and versatility in the physiology and metabolism of Campylobacter, p 41–61 In Nachamkin I, Szymanski CM, Blaser MJ. (ed),Campylobacter, 3rd ed. ASM Press, Washington, DC [Google Scholar]
- 68.McCarthy JE, Schairer HU, Sebald W. 1985. Translational initiation frequency of atp genes from Escherichia coli: identification of an intercistronic sequence that enhances translation. EMBO J. 4:519–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lang V, Gualerzi C, McCarthy JE. 1989. Ribosomal affinity and translational initiation in Escherichia coli. In vitro investigations using translational initiation regions of differing efficiencies from the atp operon. J. Mol. Biol. 210:659–663 [DOI] [PubMed] [Google Scholar]
- 70.Cameron A, Frirdich E, Huynh S, Parker CT, Gaynor EC. 2012. Hyperosmotic stress response of Campylobacter jejuni. J. Bacteriol. 194:6116–6130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Reid AN, Pandey R, Palyada K, Whitworth L, Doukhanine E, Stintzi A. 2008. Identification of Campylobacter jejuni genes contributing to acid adaptation by transcriptional profiling and genome-wide mutagenesis. Appl. Environ. Microbiol. 74:1598–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Doyle SM, Hoskins JR, Wickner S. 2007. Collaboration between the ClpB AAA+ remodeling protein and the DnaK chaperone system. Proc. Natl. Acad. Sci. U. S. A. 104:11138–11144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bucca G, Brassington AM, Schönfeld HJ, Smith CP. 2000. The HspR regulon of Streptomyces coelicolor: a role for the DnaK chaperone as a transcriptional co-repressor. Mol. Microbiol. 38:1093–1103 [DOI] [PubMed] [Google Scholar]
- 74.Shibata T, Kimura Y, Mukai A, Mori H, Ito S, Asaka Y, Oe S, Tanaka H, Takahashi T, Uchida K. 2011. Transthiocarbamoylation of proteins by thiolated isothiocyanates. J. Biol. Chem. 286:42150–42161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gan N, Wu Y-C, Brunet M, Garrido C, Chung F-L, Dai C, Mi L. 2010. Sulforaphane activates heat shock response and enhances proteasome activity through up-regulation of Hsp27. J. Biol. Chem. 285:35528–35536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Podhradsky D, Drobnica L, Kristian P. 1979. Reactions of cysteine, its derivatives, glutathione coenzyme A, and dihydrolipoic acid with isothiocyanates. Experientia 35:154–155 [DOI] [PubMed] [Google Scholar]
- 77.Nagahara N, Matsumura T, Okamoto R, Kajihara Y. 2009. Protein cysteine modifications: (2) reactivity specificity and topics of medicinal chemistry and protein engineering. Curr. Med. Chem. 16:4490–4501 [DOI] [PubMed] [Google Scholar]
- 78.Rosato V, Pucello N, Giuliano G. 2002. Evidence for cysteine clustering in thermophilic proteomes. Trends Genet. 18:278–281 [DOI] [PubMed] [Google Scholar]
- 79.Fomenko DE, Gladyshev VN. 2003. Identity and functions of CxxC-derived motifs. Biochemistry 42:11214–11225 [DOI] [PubMed] [Google Scholar]
- 80.Kaakoush NO, Sterzenbach T, Miller WG, Suerbaum S, Mendz GL. 2007. Identification of disulfide reductases in Campylobacterales: a bioinformatics investigation. Antonie Van Leeuwenhoek 92:429–441 [DOI] [PubMed] [Google Scholar]
- 81.Major TA, Burd H, Whitman WB. 2004. Abundance of 4Fe-4S motifs in the genomes of methanogens and other prokaryotes. FEMS Microbiol. Lett. 239:117–123 [DOI] [PubMed] [Google Scholar]
- 82.Nakamura Y, Kawakami M, Yoshihiro A, Miyoshi N, Ohigashi H, Kawai K, Osawa T, Uchida K. 2002. Involvement of the mitochondrial death pathway in chemopreventive benzyl isothiocyanate-induced apoptosis. J. Biol. Chem. 277:8492–8499 [DOI] [PubMed] [Google Scholar]
- 83.Tang L, Zhang Y. 2005. Mitochondria are the primary target in isothiocyanate-induced apoptosis in human bladder cancer cells. Mol. Cancer Ther. 4:1250–1259 [DOI] [PubMed] [Google Scholar]
- 84.Xiao D, Powolny AA, Moura MB, Kelley EE, Bommareddy A, Kim S-H, Hahm E-R, Normolle D, Van Houten B, Singh SV. 2010. Phenethyl isothiocyanate inhibits oxidative phosphorylation to trigger reactive oxygen species-mediated death of human prostate cancer cells. J. Biol. Chem. 285:26558–26569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kiley PJ, Storz G. 2004. Exploiting thiol modifications. PLoS Biol. 2:e400. 10.1371/journal.pbio.0020400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Szabo A, Langer T, Schröder H, Flanagan J, Bukau B, Hartl FU. 1994. The ATP hydrolysis-dependent reaction cycle of the Escherichia coli Hsp70 system DnaK, DnaJ, and GrpE. Proc. Natl. Acad. Sci. U. S. A. 91:10345–10349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hughes NJ, Clayton CL, Chalk PA, Kelly DJ. 1998. Helicobacter pylori porCDAB and oorDABC genes encode distinct pyruvate:flavodoxin and 2-oxoglutarate:acceptor oxidoreductases which mediate electron transport to NADP. J. Bacteriol. 180:1119–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Portnoy VA, Scott DA, Lewis NE, Tarasova Y, Osterman AL, Palsson BØ 2010. Deletion of genes encoding cytochrome oxidases and quinol monooxygenase blocks the aerobic-anaerobic shift in Escherichia coli K-12 MG1655. Appl. Environ. Microbiol. 76:6529–6540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kojima M, Ogawa K. 1971. Studies on the effects of isothiocyanates and their analogues on microorganisms. J. Ferment. Technol. 49:740–746 [Google Scholar]
- 90.Tajima H, Kimoto H, Taketo Y, Taketo A. 1998. Effects of synthetic hydroxy isothiocyanates on microbial systems. Biosci. Biotechnol. Biochem. 62:491–495 [DOI] [PubMed] [Google Scholar]
- 91.Conaway CC, Krzeminski J, Amin S, Chung FL. 2001. Decomposition rates of isothiocyanate conjugates determine their activity as inhibitors of cytochrome p450 enzymes. Chem. Res. Toxicol. 14:1170–1176 [DOI] [PubMed] [Google Scholar]
- 92.Jang M, Hong E, Kim GH. 2010. Evaluation of antibacterial activity of 3-butenyl, 4-pentenyl, 2-phenylethyl, and benzyl isothiocyanate in Brassica vegetables. J. Food Sci. 75:M412–M416. 10.1111/j.1750-3841.2010.01725.x [DOI] [PubMed] [Google Scholar]
- 93.Chung FL, Morse MA, Eklind KI. 1992. New potential chemopreventive agents for lung carcinogenesis of tobacco-specific nitrosamine. Cancer Res. 52:2719s–2722s [PubMed] [Google Scholar]
- 94.Zhang Y, Yao S, Li J. 2006. Vegetable-derived isothiocyanates: anti-proliferative activity and mechanism of action. Proc. Nutr. Soc. 65:68–75 [DOI] [PubMed] [Google Scholar]
- 95.Ho C-C, Lai K-C, Hsu S-C, Kuo C-L, Ma C-Y, Lin M-L, Yang J-S, Chung J-G. 2011. Benzyl isothiocyanate (BITC) inhibits migration and invasion of human gastric cancer AGS cells via suppressing ERK signal pathways. Hum. Exp. Toxicol. 30:296–306 [DOI] [PubMed] [Google Scholar]
- 96.Verkerk R, Schreiner M, Krumbein A, Ciska E, Holst B, Rowland I, De Schrijver R, Hansen M, Gerhäuser C, Mithen R, Dekker M. 2009. Glucosinolates in Brassica vegetables: the influence of the food supply chain on intake, bioavailability and human health. Mol. Nutr. Food Res. 53(Suppl 2):S219. 10.1002/mnfr.200800065 [DOI] [PubMed] [Google Scholar]
- 97.Xiao D, Vogel V, Singh SV. 2006. Benzyl isothiocyanate-induced apoptosis in human breast cancer cells is initiated by reactive oxygen species and regulated by Bax and Bak. Mol. Cancer Ther. 5:2931–2945 [DOI] [PubMed] [Google Scholar]
- 98.Liebes L, Conaway CC, Hochster H, Mendoza S, Hecht SS, Crowell J, Chung FL. 2001. High-performance liquid chromatography-based determination of total isothiocyanate levels in human plasma: application to studies with 2-phenethyl isothiocyanate. Anal. Biochem. 291:279–289 [DOI] [PubMed] [Google Scholar]
- 99.Ji Y, Morris ME. 2003. Determination of phenethyl isothiocyanate in human plasma and urine by ammonia derivatization and liquid chromatography-tandem mass spectrometry. Anal. Biochem. 323:39–47 [DOI] [PubMed] [Google Scholar]
- 100.Kassie F, Pool-Zobel B, Parzefall W, Knasmüller S. 1999. Genotoxic effects of benzyl isothiocyanate, a natural chemopreventive agent. Mutagenesis 14:595–604 [DOI] [PubMed] [Google Scholar]
- 101.Dos Santos Pires AC, Soares NFF, Andrade NJ, Mendes da Silva LH, Camilloto GP, Bernades PC. 2009. Increased preservation of sliced mozzarella cheese by antimicrobial sachet incorporated with allyl isothiocyanate. Braz. J. Microbiol. 40:1002–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]